Abstract
Summary: FancyGene is a fast and user-friendly web-based tool for producing images of one or more genes directly on the corresponding genomic locus. Starting from a variety of input formats, FancyGene rebuilds the basic components of a gene (UTRs, intron, exons). Once the initial representation is obtained, the user can superimpose additional features—such as protein domains and/or a variety of biological markers—in specific positions. FancyGene is extremely flexible allowing the user to change the resulting image dynamically, modifying colors and shapes and adding and/or removing objects. The output images are generated either in portable network graphics (PNG) or portable document format (PDF) formats and can be used for scientific presentations as well as for publications. The PDF format preserves editing capabilities, allowing picture modification using any vector graphics editor.
Availability: http://bio.ifom-ieo-campus.it/fancygene
Contact: francesca.ciccarelli@ifom-ieo-campus.it
Supplementary Information: Details, examples and tutorials can be found at http://bio.ifom-ieo-campus.it/fancygene/tutorial.html and http://bio.ifom-ieo-campus.it/fancygene/help.html
1 INTRODUCTION
The mass of genomic data currently available has prompted the development of several web tools such as the UCSC genome browser (Karolchik et al., 2008), the NCBI map viewer (Sayers et al., 2009) and the Ensembl suite (Hubbard et al., 2006). The purpose of these web services is to integrate various types of data by mapping them directly onto the genome sequence and returning a visual representation of the annotation. In addition, a number of tools specialized in the representation of high-quality images have been developed for certain types of biological data. For example, idiographica (Kin and Ono, 2007) can be used to draw entire chromosomes, while gff2ps (Abril and Guigo, 2000) allows the conversion of General Feature Format (GFF) files into post-script graphical representations. None of these resources, however, are intended for the generation of customized images of single genes, nor they do allow the dynamic addition of specific features onto the gene structure.
Here, we introduce FancyGene, a web-based resource that, starting from the genomic coordinates of one ore more genes, produces a graphical representation of the corresponding gene structure. Once the basic representation of the gene locus is obtained, the user can dynamically add several features, such as the architecture of protein domains, as well as change the appearance of each object using a simple web interface. The resulting portable network graphics (PNG) images with high resolution can be freely used in screen and poster presentations. The vector graphics portable document format (PDF) files that preserve editing capability allow the user to generate high-quality images suitable for publications.
2 FEATURES
2.1 Input and output data formats
FancyGene accepts initial data in a variety of input formats. It recognizes both GFF and GTF (Gene Transfer Format) files, which are commonly used to organize genomic data. GTF files can be directly retrieved from the UCSC web site using the Wizard mode. Any track of the UCSC database for which the GTF output format is available can be converted in an input for FancyGene.
In addition, FancyGene also accepts a simpler format, which is intended for sequences not yet added into the databases. In this format, each line contains the start and end points of the gene exons, while introns are computed automatically. If known, it is possible to add a label to define the direction of transcription, as well as information on UTRs. Once uploaded, any type of input is converted into the FancyGene format. In this format, each line contains four mandatory fields: the label, corresponding to the gene name; the tag, which describes the object (intron, exon, utr, marker, domain); the start and stop of the object. Additional fields can be specified, such as the strand, the object name, the label of a marker, etc. The user can save the FancyGene input file and the corresponding configuration file and re-use them to directly generate images of the locus of interest with the same layout.
2.2 Gene structure representation
Depending on the annotations present in the input data, FancyGene is able to automatically recognize elements of the gene structure, such as coding exons, introns, UTRs and various types of labels. Default conventions are used to render exons (thick boxes), UTRs (thin boxes) and introns (straight or hat lines), but the user can modify shapes, dimensions and colors of each of the elements of the gene. In addition, several other options (gene order, gene labels; background; color mode; etc.) can be dynamically added, deleted or modified.
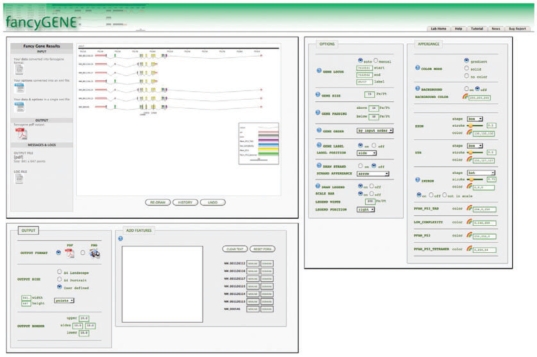
Fig. 1.
Screenshot of a Gene Representation Obtained with FancyGene. The locus on human chromosome 17 corresponding to the p53 gene (entrez id: 7157) is reported. The seven splicing variants are shown with their corresponding domain architecture. Nucleotide positions frequently mutated in cancer, as reported in the COSMIC database (Bamford et al., 2004), are also reported as examples of additional features that can be mapped into the gene structure.
2.3 Projection of domain architecture
An important feature of FancyGene is the capability to project information on protein domains directly onto the gene structure, allowing to clearly associate exon(s) to the encoded domain(s). Information on domain architecture (domain name, start and end within the protein sequence) can be uploaded either manually by the user, or automatically by using the summary provided by public domain databases, such as SMART (Letunic et al., 2009) (http://smart.embl-heidelberg.de/) and PFAM (Finn et al., 2008) (http://pfam.sanger.ac.uk/). FancyGene automatically converts the amino acidic coordinates of the protein domain into nucleotide coordinates and superimposes the domain architecture starting from the first coding exon.
2.4 Dynamic addition of features
After the initial image generation, it is possible to make substantial changes to the locus representation. The user may dynamically add features and new genes, as well as modify the appearance of any object, their order, refine the coordinates of the locus, change the background and move the positions of gene labels. Each time the image is modified, FancyGene generates a new version of the configuration and input files, which can be saved locally and used for future sessions. In addition, at each step the user can delete the last modifications as well as browse and restore all of the previous versions of the image.
3 IMPLEMENTATION
FancyGene is a web-based tool written in Perl and JavaScript. It generates images dynamically using the cairographics library (http://cairographics.org/); the rainbow palettes used to modify colors are generated using MooTools (http://mootools.net/) and MooRainbow (http://moorainbow.woolly-sheep.net/).
ACKNOWLEDGEMENTS
We thank Alessandro Riccombeni for testing FancyGene.
Funding: AIRC and Cariplo Grants to FDC.
Conflict of Interest: none declared.
REFERENCES
- Abril JF, Guigo R. gff2ps: visualizing genomic annotations. Bioinformatics. 2000;16:743–744. doi: 10.1093/bioinformatics/16.8.743. [DOI] [PubMed] [Google Scholar]
- Bamford S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJP, et al. Ensembl 2007. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC Genome Browser Database: 2008 update. Nucl. Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kin T, Ono Y. Idiographica: a general-purpose web application to build idiograms on-demand for human, mouse and rat. Bioinformatics. 2007;23:2945–2946. doi: 10.1093/bioinformatics/btm455. [DOI] [PubMed] [Google Scholar]
- Letunic I, et al. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]