Abstract
Purpose
Biomarkers can add substantial value to current medical practice by providing an integrated approach to prediction using the genetic makeup of the tumor and the genotype of the patient to guide patient-specific treatment selection. We discuss and evaluate various clinical trial designs for the validation of biomarker-guided therapy.
Methods
Designs for predictive marker validation are broadly classified as retrospective (ie, using data from previously well-conducted randomized controlled trials [RCTs]) versus prospective (enrichment, unselected, hybrid, or adaptive analysis). We discuss the salient features of each design in the context of real trials.
Results
Well-designed retrospective analysis from well-conducted prospective RCTs can bring forward effective treatments to marker-defined subgroups of patients in a timely manner (eg, KRAS and colorectal cancer). Enrichment designs are appropriate when preliminary evidence suggest that patients with or without that marker profile do not benefit from the treatments in question; however, this may sometimes leave questions unanswered (eg, trastuzumab and breast cancer). An unselected design is optimal where preliminary evidence regarding treatment benefit and assay reproducibility is uncertain (eg, epidermal growth factor receptor and lung cancer). Hybrid designs are appropriate when preliminary evidence demonstrate the efficacy of certain treatments for a marker-defined subgroup, making it unethical to randomly assign patients with that marker status to other treatments (eg, multigene assay and breast cancer). Adaptive analysis designs allow for prespecified marker-defined subgroup analyses of data from an RCT.
Conclusion
The implementation of these design strategies will lead to a more rapid clinical validation of biomarker-guided therapy.
INTRODUCTION
With the advent of so-called targeted therapies, biomarkers have the potential to provide substantial added value to the current practice of medical oncology. Biomarkers provide the possibility to integrate an accurate predictor of efficacy with a specific mechanism-based therapy using the genetic makeup of the tumor and the genotype of the patient to guide the selection of treatment for each individual patient. However, the validation of biomarkers through clinical research, leading to successful use of the biomarker in clinical practice, remains a considerable challenge, in large part due to the multitude of marker assessment methods (eg, immunohistochemistry, circulating tumor cells, fluorescent in situ hybridization [FISH], high-dimensional microarray, proteomics-based classifiers, and so on); feasibility of obtaining the specimens (tissue v serum based); the reliability and reproducibility of the assay (including issues of central v local testing); and additional costs involved with assessing the marker status on every patient.
Biomarkers can be broadly classified as prognostic markers (associated with disease outcome) or predictive markers (associated with drug response). A prognostic marker is a single trait or signature of traits that separates a population with respect to the outcome of interest in the absence of treatment, or despite nontargeted standard treatment. Prognostic marker validation is therefore relatively straightforward, as it is associated with the disease or the patient, and can be established (at least in theory) using data from a series of patients treated with placebo or with standard treatment. A predictive marker, on the other hand, is a single trait or signature of traits that separates a population with respect to the outcome of interest in response to a particular targeted therapy. A validated predictive marker can prospectively identify individuals who are likely to have a favorable clinical outcome, such as improved survival or decreased toxicity, from a specific treatment. Prognostic and predictive signatures are beginning to be established in various tumor types to estimate disease-related patient trajectories and to predict the patient-specific outcome to different treatments.1–6
In this article, we focus on clinical trial designs for predictive marker validation, under the assumption that the methods for assessment of the biomarker are established and initial results show promise with regard to the predictive ability of the marker(s). The systematic evaluation of these designs represents an essential step toward the goal of personalized medicine; the successful and efficient implementation of these strategies will speed the rapid clinical validation of biomarker-guided therapy.
RETROSPECTIVE VALIDATION: CHALLENGES AND OPPORTUNITIES
Prospectively designed clinical trials are the gold standard approach to validating a predictive marker. In most cases, however, due to the time and expense required for prospective trials, the possibility to test the predictive ability of a marker using data from previously well-conducted randomized controlled trials (RCTs) comparing therapies for which a marker is proposed to be predictive is a more feasible and timely option. The use of an RCT, as opposed to a cohort or single-arm study, is fundamentally essential for retrospective validation, as it assures that the patients who were treated with the agent for whom the marker is purported to be predictive are comparable to those who were not. In a nonrandomized design, it is impossible to isolate any causal effect of the marker on therapeutic efficacy from the multitude of other factors that may influence the decision to treat or not to treat a patient. For instance, in a study that attempted to evaluate the predictive utility of tumor microsatellite instability for the efficacy of fluorouracil-based chemotherapy in colon cancer using a cohort of non–randomly assigned patients, the median age of the treated patients was 13 years younger than those of the nontreated patients, rendering any meaningful statements about the predictive value of the marker impossibly confounded.7,8 The essential elements that are critical for retrospective validation studies are outlined in Table 1. Retrospective validation, when conducted appropriately, can aid in bringing forward effective treatments to marker-defined patient subgroups in a timely manner that might otherwise be impossible due to ethical and logistical (ie, large trial and long time to complete) considerations.9,10 In particular, if such a retrospective validation can be demonstrated in data from two independent RCTs, this provides in our opinion strong evidence for a robust predictive effect.
Table 1.
Requirements for a Valid Retrospective Assessment of a Predictive Biomarker
| Requirements |
|---|
|
An example of a marker that has been successfully validated using data collected from previous RCTs is KRAS as a predictor of efficacy of panitumumab and cetuximab in advanced colorectal cancer. In a prospectively specified analysis of data from a previously conducted randomized phase III trial of panitumumab versus best supportive care, KRAS status was assessed on 92% (427 of 463) of the patients enrolled, with 43% having the KRAS mutation.11 The hazard ratio for treatment effect comparing panitumumab versus best supportive care on progression free survival in the wild-type and mutant subgroups was 0.45 and 0.99, respectively, with a significant treatment by KRAS status interaction (P < .0001). In addition, multiple phase II trials have demonstrated similar results.12 Similarly, retrospective data on KRAS status and cetuximab from phase III and phase II trials have demonstrated a statistically significant advantage in the overall survival for patients with wild-type KRAS, with no survival benefit in patients with KRAS-mutant status.13–17 In summary, these well-designed retrospective validation studies have consistently demonstrated that the benefit from panitumumab and cetuximab is restricted to patients with wild-type KRAS status, with no clinical benefit for patients with mutant KRAS. Based on this strong evidence, all ongoing clinical trials sponsored by the National Cancer Institute with these agents in colorectal cancer have been or are being modified to only include patients with wild-type KRAS, and the label for panitumumab monotherapy has been restricted to patients with wild-type KRAS in Europe.
PROSPECTIVE VALIDATION: CHALLENGES AND OPPORTUNITIES
While retrospective validation may be acceptable as a marker validation strategy in selected circumstances, the gold standard for predictive marker validation continues (appropriately) to be a prospective RCT. Several designs have been proposed and utilized in the field of cancer biomarkers for validation of predictive markers. We classify these designs broadly into three categories: targeted or enrichment designs; unselected or all-comers designs, which can further be categorized into sequential testing strategy designs and marker-based designs; and hybrid designs. We discuss the salient features of these designs, along with pertinent examples.
Targeted or Enrichment Designs
This design is based on the paradigm (when there is compelling preliminary evidence) that not all patients will benefit from the study treatment under consideration, but rather that the benefit will be restricted to a subgroup of patients who express (or not express) a specific molecular feature. Consequently, all patients are screened for the presence or absence of a marker or a panel of markers, and only those with (or without) certain molecular features are included in the trial. This design therefore results in a stratification of the study population, with a goal of understanding the safety, tolerability and clinical benefit of a treatment in the subgroup of the patient population defined by a specific marker status.
An enrichment design strategy of enrolling only human epidermal growth factor receptor 2 (HER2) –positive patients demonstrated that trastuzumab (Herceptin; Genentech, South San Francisco, CA) combined with paclitaxel after doxorubicin and cyclophosphamide significantly improved disease-free survival (DFS) among women with surgically removed HER2-positive breast cancer.18 The combined analysis using data from both phase III trials with over 1,600 patients in each of the control and treatment arms provided 90% power to detect a 25% reduction in the hazard rate for DFS. In this case, the enrichment strategy clearly succeeded in identifying a subgroup of patients who received a significant benefit from this therapy. However, subsequent analyses have raised the possibility of a beneficial effect of trastuzumab in a more broadly defined patient population than that defined in the two phase III trials.19,20 While multiple possible explanations exist, two questions remain: whether trastuzumab therapy may benefit a potentially larger group than the approximately 20% of patients defined as HER2 positive in these two trials, and questions of assay reproducibility arising from local versus central testing for HER2 status were left unanswered due to the inclusion of only biomarker-defined subgroups in the two phase III clinical trials.21
Two key lessons may be learned from the HER2/trastuzumab example regarding the appropriateness of targeted or enrichment designs. First, before the launching of any trial, particularly one with an enrichment design strategy, assay reproducibility and accuracy must be well established. Second, there should be compelling preliminary evidence to suggest that patients with or without that marker profile do not benefit from the treatments in question. As a general guideline, targeted designs are appropriate when therapies have modest absolute benefit in the unselected population, but cause significant toxicity; when in the absence of selection, therapeutic results are similar whereby a selection design (even if incorrect) would not hurt; and when an unselected design is ethically impossible based on previous studies.
Unselected or All-Comers Designs
In this design, all patients meeting the eligibility criteria (which does not include the status of a biomarker characteristic) are entered into the trial. We note that the ability to provide adequate tissue may be an eligibility criterion for these designs, but not the specific biomarker result. These designs can be broadly classified into sequential testing strategy designs, marker-based designs, or hybrid designs, which are differentiated from each other by the protocol specified approach to the prespecified type I and type II error rates (influencing sample size), analysis plans (including a single hypothesis test, multiple tests, or sequential tests), and randomization schema. The key features of these designs along with examples of clinical trials that have utilized these designs are discussed.
Sequential testing strategy designs.22,23
Sequential testing designs are similar in principle to a standard RCT design with a single primary hypothesis, that is either tested in the overall population first and then in a prospectively planned subset, or in the marker-defined subgroup first, and then tested in the entire population if the subgroup analysis is significant. The first is recommended in cases where the experimental treatment is hypothesized to be broadly effective, and the subset analysis is ancillary. The latter (also known as the closed testing procedure) is recommended when there is strong preliminary data to support that the treatment effect is strongest in the marker-defined subgroup, and that the marker has sufficient prevalence that the power for testing the treatment effect in the subgroup is adequate. Both these approaches appropriately control for the type I error rates associated with multiple testing. A modification to this approach, taking into account potential correlation arising from testing the overall treatment effect and the treatment effect within the marker-defined subgroup has also been proposed.24
The approach of first testing in the subgroup defined by marker status has been implemented in the ongoing US-based phase III trial testing cetuximab in addition to infusional fluorouracil, leucovorin, and oxaliplatin as adjuvant therapy in stage III colon cancer (N0147). While the trial has now been amended to accrue only patients with KRAS–wild-type tumors, approximately 800 patients with KRAS-mutant tumors have already been enrolled. In this trial, the primary analysis will be conducted at the 0.05 level in the patients with wild-type KRAS. A sample size of 1,035 patients with wild-type KRAS per arm will result in 515 total events, providing 90% power to detect a hazard ratio of 1.33 for this comparison using a two-sided log-rank test at a significance level of 0.05. If this subset analysis is statistically significant at P = .05, then the efficacy of the regimen in the entire population will also be tested at level 0.05, as this is a closed testing procedure. This comparison using all 2,910 patients will have 90% power to detect a hazard ratio of 1.27 comparing the two treatment arms, based on a total of 735 events.
Marker-based designs.25,26
Designs that fall under this classification are the marker-by-treatment-interaction design and the marker-based strategy design. A formal comparison of these two designs in the setting of a binary marker is discussed.
The marker-by-treatment-interaction design uses the marker status as a stratification factor (ie, assumes that the overall population can be split into marker-defined subgroups) and randomly assigns patients to treatments within each marker subgroup. This is similar to conducting two independent RCTs, one in each marker-based subgroup, except that both are conducted under one large RCT umbrella. However, this design differs from a single large RCT in four essential characteristics: only patients with a valid marker result are allowed to be randomized, the sample size is prospectively specified separately within each marker-based subgroup, the randomization is stratified by marker status, and this design is clearly a prospective (and a definitive) marker validation trial.
The marker-based strategy design, on the other hand, randomly assigns patients to have their treatment either based on or independent of the marker status. A downside of this design is that it fundamentally includes patients treated with the same regimen on both the marker-based and the non–marker-based arms, resulting in a significant overlap (driven by the prevalence of the marker) in the number of patients receiving the same treatment regimen in both arms. As a consequence, the overall detectable difference in outcomes between the two arms is reduced (depending on the marker prevalence), thus resulting in a comparatively larger trial.
An example of the marker-by-treatment-interaction design is the recently activated phase III biomarker validation study, also known as MARVEL (Marker Validation of Erlotinib in Lung Cancer), of second-line therapy in patients with advanced non–small-cell lung cancer (NSCLC) randomly assigned to pemetrexed or erlotinib (Fig 1). This trial is motivated by the need to obtain prospective evidence to address the conflicting results from several retrospective analyses regarding the predictive role of epidermal growth factor receptor (EGFR) amplification by FISH in the setting of treatment with chemotherapy and EGFR tyrosine kinase inhibitors, and the fact that EGFR FISH represents a poor prognostic factor in untreated NSCLC patients, MARVEL is designed to prospectively evaluate the clinical predictive utility of EGFR copy number as measured by FISH in advanced NSCLC.4,27–33 The FISH status of the patient will be assessed before randomization (to ensure adequate number of patients with FISH+ and FISH− status) in a central location (to address issues regarding standardization of assay techniques, reproducibility and interpretability of assay results). The primary comparisons will be progression-free survival of patients treated on the erlotinib arm compared to the pemetrexed arm within the FISH+ and FISH− subgroups (286 [30%] FISH+; 670 [70%] FISH−). An overview of the statistical hypothesis testing framework for the MARVEL trial is included in the online-only Appendix.
Fig 1.
MARVEL (Marker Validation for Erlotinib in Lung Cancer) trial design. Each cycle of treatment is 21 days. Stratification factors: ECOG performance status, gender, smoking status, histology, best response to prior chemotherapy. EGFR, epidermal growth factor receptor; FISH, fluorescent in situ hybridization.
Hybrid Designs
In this design, only a certain marker-defined subgroup of patients are randomly assigned to have their treatment based on their marker status, whereas patients in the other marker-defined subgroups are assigned the standard-of-care treatment(s). This design is powered to detect differences in outcomes only in the marker-defined subgroup that is randomized to treatment choices based on the marker status, similar to an enrichment design strategy. However, unlike the enrichment design, the hybrid design provides additional value: since all patients are screened for marker status to determine whether they are randomly assigned or assigned the standard-of-care treatment(s), it seems prudent to include and collect specimens and follow-up from “all” patients in the trial to allow for future testing for other potential prognostic markers in this population. This design is an appropriate choice when there is compelling prior evidence demonstrating the efficacy of a certain treatment(s) for a marker-defined subgroup, thereby making it unethical to randomly assign patients with that particular marker status to other treatment options.
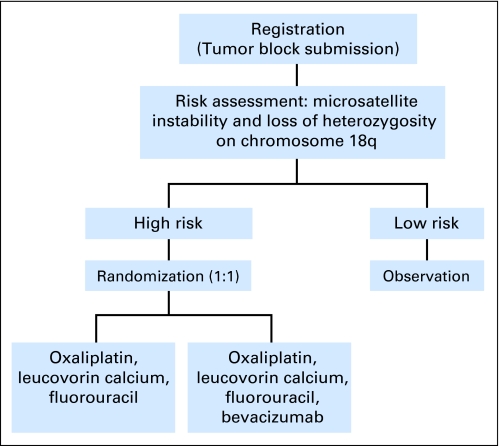
At least three large phase III marker validation trials have been recently launched with a hybrid trial design: the phase III randomized study of oxaliplatin, leucovorin calcium, and fluororacil with versus without bevacizumab in patients with resected stage II colon cancer and at high risk for recurrence based on molecular markers (ECOG [Eastern Cooperative Oncology Group] 5202; Fig 2); the TAILORx (Trial Assigning Individualized Options for Treatment; Fig 3) trial designed to evaluate the Oncotype Dx (Genomic Health, Redwood City, CA), a 21-gene recurrence score in tamoxifen-treated patients with breast cancer;34 and the MINDACT (Microarray in Node-Negative Disease May Avoid Chemotherapy; Fig 4) trial for patients with node-negative breast cancer designed to evaluate MammaPrint (Agendia, Amsterdam, the Netherlands), the 70-gene expression profile discovered at the Netherlands Cancer Institute.35,36
Fig 2.
ECOG (Eastern Cooperative Oncology Group) 5202 trial design. Cycle of treatment, 2 days every 2 weeks, repeat for 12 cycles.
Fig 3.
TAILORx (Trial Assigning Individualized Options for Treatment) trial design. RS, risk score.
Fig 4.
MINDACT (Microarray in Node-Negative Disease May Avoid Chemotherapy) trial design.
In ECOG 5202, patients with stage II colon cancer, deemed to be at a high risk for recurrence after surgery (estimated 5-year survival rate of 60%) based on two molecular markers are randomly assigned to one of two treatment arms, whereas patients deemed to be at a low risk for recurrence after surgery (5-year survival rate estimate of 90%) will not receive any adjuvant therapy (Fig 2). The trial is expected to enroll approximately 3,500 patients. With 250 eligible patients per year accrued for 5.5 years (1,375 eligible high-risk patients randomly assigned; 3,438 eligible patients total) and 3 years of follow-up, there is at least 88% power to detect a 37% difference in median DFS (absolute difference of 5%, from 80% to 85%, at 3 years) using a one-sided stratified log-rank test at 0.025 level, with stratification on stage and microsatellite instability status. Unfortunately, this design will not allow for a determination of the benefit of bevacizumab in the low-risk strata, however if the outcomes in the absence of treatment are as favorable as predicted in that group, no postsurgical therapy would generally be recommended.
The TAILORx and MINDACT trials aim to validate two new prognostic and possibly predictive tools for breast cancer, and are the first to test the feasibility of a prognostic tool in clinical application. The TAILORx trial was activated in 2006 and will accrue approximately 10,000 women with hormone receptor–positive, lymph node–negative breast cancer (Fig 3).37–40 This study uses a noninferiority design (null hypothesis of no difference) to determine whether patients with a recurrence score between 11 and 25 derive benefit from adjuvant chemotherapy with a larger type I error (one-sided, 10%) and smaller type II error (5%) than usual. A decrease in the 5-year DFS rate from 90% with chemotherapy to 87.0% or lower on hormone therapy alone would be considered unacceptable. All patients in the TAILORx trial will provide blood samples for banking and future research. The MINDACT trial is expected to enroll 6,000 patients with node-negative breast cancer (Fig 4).36 The primary test will be for group A in Figure 4, where a null hypothesis rate of 92% for the 5-year distant metastases–free survival will be tested. With 6,000 patients overall, this group is expected to enroll 672 patients, thus providing 80% power to reject the null hypothesis if the true distant metastases–free survival is 95% using a one-sided test at the 0.025 significance level. Several other tests to compare the overall efficacy between the two prognosis methods within the randomized cohort as well as comparisons between treatment alternatives (based on a subsequent randomization) within specific subgroups will be performed. The 70-gene profile used in MINDACT previously demonstrated a 97% negative predictive value across all disease types, and a 38% positive predictive value, thus decreasing the likelihood of undertreatment of patients, but having a higher chance of overtreatment.41 The interlaboratory reproducibility of this gene profile and its discriminative ability was also independently validated in retrospective analyses.42,43 Complete genome arrays will be performed on all patient samples collected in this trial.
In summary, these three trials are examples of prospective validation trials utilizing a hybrid trial design that has the potential to substantially change the management of patients in the future, allowing for a better risk assessment and improved individualized treatment.
COMPARISON OF PROSPECTIVE DESIGNS
Targeted (or enrichment) Versus Unselected (or all-comers) Designs
A recent article compared the performance of the targeted versus the randomize-all designs in terms of the sample size (both the number screened and randomized) and statistical power in the setting of a binary outcome.44 Based on the simulation studies, Hoering et al concluded that a targeted design is most efficient where there is an underlying true predictive marker and the cut point for determining the marker status is well established; and that the randomize-all design (with sequential testing strategy to test for the overall and within-marker subgroups treatment effect) is recommended in cases where the cut point for marker status determination is not well established, the marker prevalence is high, and the new treatment has the potential to benefit both marker subgroups. The findings from this article can be put in perspective using the HER2/trastuzumab example. In the HER2/trastuzumab phase III trials, an enrichment design strategy was used to bring forward an effective treatment to a subgroup of patients in an efficient and timely manner. However, if indeed trastuzumab had a beneficial effect in a more broadly defined patient population, an unselected or sequential testing design strategy including both HER2-positive and -negative patients may have provided a more definitive answer regarding the predictive utility of HER2.
A formal comparison and discussion of the statistical properties of the targeted versus the unselected designs can also be found in Simon and Maitouram45 and Maitouram and Simon.46 Simon and Maitouram evaluated the relative efficiency of the two designs for an RCT under certain assumptions, and their simulations showed that the targeted design required fewer randomly assigned (and screened) patients compared with the untargeted design. The reduction in the sample size was dependent on the accuracy of the assay, and the prevalence of the markers in patients, in addition to other limitations.45,46
Marker-Based Designs
Preliminary work suggested that the marker-by-treatment-interaction design may be superior to the marker-based strategy design in terms of the number of events (and hence the total sample size) required (while keeping all the parameters the same for both designs) under specific clinical settings, while the opposite may be true in other settings.26 In a recent article by Mandrekar and Sargent,9 a head-to-head comparison of the two designs in the setting of a single or multimarker signature that can be distilled to a binary measure over a wide spectrum of clinically relevant scenarios came to the same conclusion. Mandrekar and Sargent calculated the sample size required for all possible clinical trials for these prespecified design parameters, and demonstrated that in the setting of a binary marker designed to decide between the treatments, the marker-by-treatment-interaction design has greater efficiency in terms of overall sample size and events than the marker-based strategy design.9 While the impact of the error in measurement of the biomarker on the efficiency of these designs needs to be explored further, it is likely that it will have a similar effect on both designs by inflating the required sample size due to patient misclassification. A formal investigation of these designs in a multimarker situation or where a marker is designed to make a choice between multiple treatments remains open.
ADAPTIVE ANALYSIS DESIGNS
A number of innovative statistical designs have been recently proposed that use either an adaptive strategy for analysis, or an outcome-based adaptive randomization.47–49 We discuss briefly the key elements of three such designs.
The biomarker-adaptive threshold design48 is similar to the sequential testing strategy designs discussed earlier and can be implemented one of two ways: the new treatment is compared with the control in all patients at a prespecified significance level, and if not significant, a second stage analysis involving finding an optimal cut point for the predictive marker is performed using the remaining alpha; or under the assumption that the treatment is effective only for a marker-driven subset, no overall treatment to control comparisons are made, instead, the analysis focuses on the identification of optimal cut points. Both these approaches were concluded to be superior (in terms of the power and number of events required to detect an effect at a prespecified overall type I error rate) to the classic nonadaptive design approaches in the simulation studies. Two issues need further consideration: the added cost of a somewhat larger sample size and/or redundant power dictated by the strategy of partitioning the overall type I error rate and use of data from the same trial to both define and validate a marker cut point.
The adaptive accrual design outlines a strategy to adaptively modify accrual to two predefined marker-defined subgroups based on an interim futility analysis.47 Specifically, the trial follows the following scheme: (1) begin with accrual to both marker-defined subgroups; (2) if the treatment effect in one of the subgroups fails to satisfy a futility boundary at the interim analysis, terminate accrual to that subgroup, and (3) continue accrual to the other subgroup until the planned total sample size is reached, including accruing subjects that had planned to be included from the terminated subgroup. This design demonstrated greater power than a nonadaptive trial in simulation settings; however, this strategy might lead to a substantial increase in the accrual duration depending on the prevalence of the marker. In addition, the futility boundary is somewhat conservative and less than optimal as it is set to be in the region where the observed efficacy is greater for the control arm than the experimental regimen.
The outcome-based adaptive randomization design uses a Bayesian hierarchical framework to adaptively (based on outcome from the accumulated data in the trial) randomly assign patients to treatments based on the biomarker status.49 The design is extensively described in the context of the phase II BATTLE (Biomarker-Integrated Approaches of Targeted Therapy of Lung Cancer Elimination) trial in advanced NSCLC. Patients are classified into five biomarker subgroups based on their biomarker profile, and subsequently adaptively randomly assigned. The trial is expected to enroll 200 patients to test the null hypothesis of disease control rate at 8 weeks of 30% versus the target rate of 50%, with a false-positive rate between 15% and 19% based on simulation studies The adaptive accrual and adaptive randomization designs require a rapid and reliable end point, which is somewhat challenging as most oncology trials use time to event end points as the gold standard for validation trials.
CONCLUSION
Advancing new discoveries from bench to bedside is the ultimate goal of clinical and translational research. In this article, we have attempted to provide a comprehensive overview of the designs for predictive biomarker validation along with pertinent examples. While there is no one-size-fits-all solution, it is clear that the choice of a clinical trial design is driven by a combination of scientific, clinical, statistical and ethical considerations. The current era of novel agents and targeted therapies such as small molecules, antibodies, and vaccines are mandating intelligent clinical trial designs. Well-designed retrospective analyses of RCTs, supplemented by prospective trials whenever possible, will hasten this important progress.
Appendix
Appendix I: Details on the Design of the MARVEL (Marker Validation of Erlotinib in Lung Cancer) Trial
Erlotinib is approved for patients with previously treated non–small-call lung cancer (NSCLC) with a median progression-free survival (PFS) of 2.2 months and median survival time (MST) of 6.7 months (Shepherd FA, Pereira JR, Ciuleanu T, et al: N Engl J Med 353:123-132, 2005). Pemetrexed is also approved for patients with previously treated NSCLC with a median PFS of 2.9 months and MST of 8.3 months (Hanna N, Shepherd FA, Fossella FV, et al: J Clin Onc 22:1589-1597, 2004). Emerging data suggest that patients with fluorescent in situ hybridization (FISH) + status and/or increased epidermal growth factor receptor (EGFR) copy number as measured by FISH may benefit from this agent, while those without these characteristics may not (Bonomi PD, Buckingham L, Coon J: Clin Cancer Res 13:s4606-s4612, 2007 [suppl 2]; Jänne PA, Johnson BE: Clin Cancer Res 12:4416s-4420s, 2006). These data are however based on retrospective analyses.
The primary comparisons will be PFS for patients on erlotinib compared with pemetrexed within the FISH+ and FISH− subgroups. A total of 1,196 patients with tumor tissue samples (956 with assay results, assuming an 80% assay success rate) will be screened to identify 286 FISH+ patients (30%) and 670 FISH− patients (70%) to test the two coprimary null hypotheses of interest, defined as follows:
Null Hypothesis I: FISH+ subgroup—Erlotinib is not superior to pemetrexed.
The target sample size of 286 total patients randomly assigned 1:1 to erlotinib or pemetrexed for the FISH+ subgroup is set so that there will be at least 90% power to reject Null Hypothesis I if the truth is that erlotinib is superior to pemetrexed with a hazard ratio of 1.5 in favor of erlotinib (50% improvement, or 3.75 v 2.5 months in median PFS).
Null Hypothesis II: FISH− subgroup—Pemetrexed is equally effective as erlotinib.
The planned sample size for the FISH− subgroup (670 total patients randomly assigned 1:1 to erlotinib or premetrexed) will allow at least 90% power to reject Null Hypothesis II if the truth is that pemetrexed is superior to erlotinib with a hazard ratio of 0.77 in favor of pemetrexed (30% improvement, or 2.5 v 1.92 months median PFS). This sample size will also allow approximately 90% power to reject Null Hypothesis II if the truth is that erlotinib is superior to pemetrexed with a hazard ratio of 1.3 in favor of erlotinib (30% improvement, or 3.25 v 2.5 months median PFS).
Hypotheses I and II will jointly evaluate the clinical predictive utility of EGFR copy number as measured by FISH. 1 If Null Hypothesis I is rejected indicating superiority of erlotinib and Null Hypothesis II is rejected in the direction of superiority of pemetrexed, then EGFR-FISH will have been established as fully predictive for erlotinib benefit for FISH+ patients and pemetrexed benefit for FISH− patients. 2 If Null Hypothesis I is rejected, indicating superiorty of erlotinib, and Null Hypothesis II is not rejected, EGFR FISH may still have clinical use because a clear benefit of erlotinib will have been established for FISH+ patients, and it will have been established that there are not clinically important differences in PFS due to treatment with erlotinib versus pemetrexed for FISH− patients. (The hypothesis test in the FISH− subgroup is well-powered to detect a relatively small benefit.) In this latter situation, which will be referred to as partially predictive, erlotinib would be preferred for FISH+ patients, but decisions regarding how to treat FISH− patients could be based on other considerations such as drug toxicities, patient comorbidities, cost, and convenience. 3 If Null Hypothesis I is rejected, indicating superiority of erlotinib, and Null Hypothesis II is rejected in the direction of superiority of erlotinib, then EGFR-FISH testing will have been shown not to be clinically useful because all patients would benefit from receiving erlotinib, regardless of EGFR-FISH status.
Footnotes
Presented in part at the 2008 American Society of Clinical Oncology/National Cancer Institute/European Organisation for Research and Treatment of Cancer Annual Meeting on Molecular Markers in Cancer, October 30-November 1, 2008, Hollywood, FL.
Supported in part by National Cancer Institute Grants No. CA-15083 (Mayo Clinic Cancer Center) and CA-25224 (North Central Cancer Treatment Group).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The author(s) indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
Conception and design: Sumithra J. Mandrekar, Daniel J. Sargent
Administrative support: Sumithra J. Mandrekar
Collection and assembly of data: Sumithra J. Mandrekar, Daniel J. Sargent
Data analysis and interpretation: Sumithra J. Mandrekar, Daniel J. Sargent
Manuscript writing: Sumithra J. Mandrekar, Daniel J. Sargent
Final approval of manuscript: Sumithra J. Mandrekar, Daniel J. Sargent
REFERENCES
- 1.Conley BA, Taube SE. Prognostic and predictive markers in cancer. Dis Markers. 2004;20:35–43. doi: 10.1155/2004/202031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taube SE, Jacobson JW, Lively TG. Cancer diagnostics: Decision criteria for marker utilization in the clinic. Am J Pharmacogenomics. 2005;5:357–364. doi: 10.2165/00129785-200505060-00003. [DOI] [PubMed] [Google Scholar]
- 3.Sequist LV, Bell DW, Lynch TJ, et al. Molecular predictors of response to epidermal growth factor receptor antagonists in non–small-cell lung cancer. J Clin Oncol. 2007;25:587–595. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 4.Bonomi PD, Buckingham L, Coon J. Selecting patients for treatment with epidermal growth factor tyrosine kinase inhibitors. Clin Cancer Res. 2007;13(suppl 2):s4606–s4612. doi: 10.1158/1078-0432.CCR-07-0332. [DOI] [PubMed] [Google Scholar]
- 5.Slamon D. Herceptin: Increasing survival in metastatic breast cancer. Eur J Oncol Nurs. 2000;4:24–29. doi: 10.1054/ejon.2000.0070. [DOI] [PubMed] [Google Scholar]
- 6.Paik S. Clinical trial methods to discover and validate predictive markers for treatment response in cancer. Biotechnol Annu Rev. 2003;9:259–267. doi: 10.1016/s1387-2656(03)09005-7. [DOI] [PubMed] [Google Scholar]
- 7.Elsaleh H, Joseph D, Grieu F, et al. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet. 2000;355:1745–1750. doi: 10.1016/S0140-6736(00)02261-3. [DOI] [PubMed] [Google Scholar]
- 8.Elsaleh H, Powell B, McCaul K, Grieu F, et al. p53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin Cancer Res. 2001;7:1343–1349. [PubMed] [Google Scholar]
- 9.Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: One size does not fit all. J Biopharm Stat. 2009;19:530–542. doi: 10.1080/10543400902802458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandrekar SJ, Sargent DJ. Clinical validation of biomarkers in cancer. In: Winther H, Jørgensen JT, editors. Developing Molecular Diagnostics for Cancer. Singapore: Pan Stanford Publishing; in press. [Google Scholar]
- 11.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 12.Freeman D, Juan T, Meropol NJ, et al. Association of somatic KRAS gene mutations and clinical outcome in patients with metastatic colorectal cancer receiving panitumumab monotherapy. 14th European Cancer Conference; September 23-27, 2007; Barcelona, Spain. abstr 3014. [Google Scholar]
- 13.Jonker DJ, O'Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 14.Karapetis CS, Khambata-Ford S, Jonker DJ, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 15.Van Cutsem E, Lang I, D'haens G, et al. KRAS status and efficacy in the first-line treatment of patients with metastatic colorectal cancer (mCRC) treated with FOLFIRI with or without cetuximab: The CRYSTAL experience. J Clin Oncol. 2008;26(suppl):5s. abstr 2. [Google Scholar]
- 16.Bokemeyer C, Bondarenko I, Hartmann JT, et al. KRAS status and efficacy of first-line treatment of patients with metastatic colorectal cancer (mCRC) with FOLFOX with or without cetuximab: The OPUS experience. J Clin Oncol. 2008;26(suppl):178s. abstr 4000. [Google Scholar]
- 17.Van Cutsem E, Lang I, D'haens G, et al. The CRYSTAL study: Assessment of the predictive value of KRAS status on clinical outcome in patients with mCRC receiving first-line treatment with cetuximab or cetuximab plus FOLFIRI. 10th World Congress on Gastrointestinal Cancer; June 25-28, 2008; Barcelona, Spain. abstr O-031. [Google Scholar]
- 18.Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–1684. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 19.Paik S, Kim C, Jeong J, Geyer CE, et al. Benefit from adjuvant trastuzumab may not be confined to patients with IHC 3+ and/or FISH-positive tumors: Central testing results from NSABP B-31. J Clin Oncol. 2007;25:18s. abstr 511. [Google Scholar]
- 20.Perez EA, Romond EH, Suman VJ, et al. Updated results of the combined analysis of NCCTG N9831 and NSABP B-31 adjuvant chemotherapy with/without trastuzumab in patients with HER2-positive breast cancer. J Clin Oncol. 2007;25:18s. abstr 512. [Google Scholar]
- 21.Perez EA, Suman VJ, Davidson NE, et al. HER2 testing by local, central, and reference laboratories in specimens from the North Central Cancer Treatment Group N9831 intergroup adjuvant trial. J Clin Oncol. 2006;24:3032–3038. doi: 10.1200/JCO.2005.03.4744. [DOI] [PubMed] [Google Scholar]
- 22.Simon R, Wang SJ. Use of genomic signatures in therapeutics development. Pharmacoenomics J. 2006;6:1667–1673. doi: 10.1038/sj.tpj.6500349. [DOI] [PubMed] [Google Scholar]
- 23.Bauer P. Multiple testing in clinical trials. Stat Med. 1991;10:871–890. doi: 10.1002/sim.4780100609. [DOI] [PubMed] [Google Scholar]
- 24.Song Y, Chi GYH. A method for testing a prespecified subgroup in clinical trials. Stat Med. 2007;26:3535–3549. doi: 10.1002/sim.2825. [DOI] [PubMed] [Google Scholar]
- 25.Mandrekar SJ, Grothey A, Goetz MP, et al. Clinical trial designs for prospective validation of biomarkers. Am J Pharmacogenomics. 2005;5:317–325. doi: 10.2165/00129785-200505050-00004. [DOI] [PubMed] [Google Scholar]
- 26.Sargent DJ, Conley BA, Allegra C, et al. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol. 2005;23:2020–2027. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- 27.Cappuzzo F, Ligorio C, Toschi L, et al. EGFR and HER2 gene copy number and response to first-line chemotherapy in patients with advanced non–small-cell lung cancer (NSCLC) J Thorac Oncol. 2007;2:423–429. doi: 10.1097/01.JTO.0000268676.79872.9b. [DOI] [PubMed] [Google Scholar]
- 28.Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non–small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- 29.Hirsch FR, Scagliotti GV, Langer CJ, et al. Epidermal growth factor family of receptors in preneoplasia and lung cancer: Perspectives for targeted therapies. Lung Cancer. 2003;41(suppl 1):S29–S42. doi: 10.1016/s0169-5002(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch FR, Varella-Garcia M, Bunn PA, Jr, et al. Epidermal growth factor receptor in non–small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 31.Douillard J, Kim E, Hirsh V, et al. Gefitinib (IRESSA) versus docetaxel in patients with locally advanced or metastatic non–small-cell lung cancer pretreated with platinum-based chemotherapy: A randomized, open-label phase III study (INTEREST) J Thorac Oncol. 2007;2(suppl 4):S305–S306. [Google Scholar]
- 32.Crinò L, Zatloukal P, Reck M, et al. Gefitinib (IRESSA) versus vinorelbine in chemonaïve elderly patients with advanced non–small-cell lung cancer (INVITE): A randomized phase II study. J Thorac Oncol. 2007;2(suppl 4):S341. doi: 10.1200/JCO.2007.15.0672. [DOI] [PubMed] [Google Scholar]
- 33.Jänne PA, Johnson BE. Effect of epidermal growth factor receptor tyrosine kinase domain mutations on the outcome of patients with non–small-cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitors. Clin Cancer Res. 2006;12:4416s–4420s. doi: 10.1158/1078-0432.CCR-06-0555. [DOI] [PubMed] [Google Scholar]
- 34.Sparano JA, Paik S. Development of the 21-gene assay and its application in clinical practice and clinical trials. J Clin Oncol. 2008;26:721–728. doi: 10.1200/JCO.2007.15.1068. [DOI] [PubMed] [Google Scholar]
- 35.Cardoso F, van't Veer L, Rutgers E, et al. Clinical application of the 70-gene profile: The MINDACT trial. J Clin Oncol. 2008;26:729–735. doi: 10.1200/JCO.2007.14.3222. [DOI] [PubMed] [Google Scholar]
- 36.Bogaerts J, Cardoso F, Buyse M, et al. TRANSBIG consortium: Gene signature evaluation as a prognostic tool—Challenges in the design of the MINDACT trial. Nat Clin Pract Oncol. 2006;3:540–551. doi: 10.1038/ncponc0591. [DOI] [PubMed] [Google Scholar]
- 37.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 38.Cronin M, Sangli C, Liu ML, et al. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor–positive breast cancer. Clin Chem. 2007;53:1084–1091. doi: 10.1373/clinchem.2006.076497. [DOI] [PubMed] [Google Scholar]
- 39.Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor–positive breast cancer. J Clin Oncol. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 40.Mamounas E, Tang G, Bryant J, et al. Association between the 21-gene recurrence score assay (RS) and risk of loco-regional failure in node-negative, ER-positive breast cancer: Results from NSABP B-14 and NSABP B-20. Breast Cancer Res. 2005;94(suppl):S16. abstr 29. [Google Scholar]
- 41.van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 42.Buyse M, Loi S, van't Veer L, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 43.Mook S, van't Veer LJ, Rutgers E, et al. Individualization of therapy using MammaPrint: From development to the MINDACT trial. Cancer Genomics Proteomics. 2007;4:147–155. [PubMed] [Google Scholar]
- 44.Hoering A, Leblanc M, Crowley JJ. Randomized phase III clinical trial designs for targeted agents. Clin Cancer Res. 2008;14:4358–4367. doi: 10.1158/1078-0432.CCR-08-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon R, Maitournam A. Evaluating the efficiency of targeted designs for randomized clinical trials. Clin Cancer Res. 2004;10:6759–6763. doi: 10.1158/1078-0432.CCR-04-0496. [DOI] [PubMed] [Google Scholar]
- 46.Maitournam A, Simon R. On the efficiency of targeted clinical trials. Stat Med. 2005;24:329–339. doi: 10.1002/sim.1975. [DOI] [PubMed] [Google Scholar]
- 47.Wang SJ, O'Neill RT, Hung HMJ. Approaches to evaluation of treatment effect in randomized clinical trials with genomic subset. Pharmaceutical Statistics. 2007;6:227–244. doi: 10.1002/pst.300. [DOI] [PubMed] [Google Scholar]
- 48.Jiang W, Freidlin B, Simon R. Biomarker-adaptive threshold design: A procedure for evaluating treatment with possible biomarker-defined subset effect. J Natl Cancer Inst. 2007;99:1036–1043. doi: 10.1093/jnci/djm022. [DOI] [PubMed] [Google Scholar]
- 49.Zhou X, Liu S, Kim ES, et al. Bayesian adaptive design for targeted therapy development in lung cancer: A step towards personalized medicine. Clinical Trials. 2008;5:181–193. doi: 10.1177/1740774508091815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shepherd FA, Pereira JR, Ciuleanu T, et al. Erlotinib in previously treated non–small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 51.Hanna N, Shepherd FA, Fossella FV, et al. Randomized phase III trial of pemetrexed versus docetaxel in patients with non–small-cell lung cancer previously treated with chemotherapy. J Clin Onc. 2004;22:1589–1597. doi: 10.1200/JCO.2004.08.163. [DOI] [PubMed] [Google Scholar]