Abstract
The daily sleep cycle in humans and other mammals is driven by a complex circuit within which GABAergic sleep-promoting neurons oppose arousal systems. The latter includes the circadian system, aminergic/cholinergic systems as well as neurons secreting the peptide orexin/hypocretin, which contribute to sharp behavioral transitions (Lu and Greco, 2006). Drosophila sleep has recently been shown also to be controlled by GABAergic inputs, which act on unknown cells expressing the Rdl GABAA receptor (Agosto et al., 2008). We identify here the relevant Rdl-containing cells as a subset of the well-studied Drosophila circadian clock neurons, the PDF-expressing small and large ventral lateral neurons (LNvs). LNv activity regulates the total amount of sleep as well as the rate of sleep onset, and both large and small LNvs are part of the sleep circuit. Flies mutant for either the pdf gene or its receptor are hypersomnolent, and PDF acts on the LNvs themselves to control sleep. These features of the Drosophila sleep circuit, GABAergic control of sleep onset and maintenance as well as peptidergic control of arousal, support the idea that features of sleep circuit architecture as well as the mechanisms governing the behavioral transitions between sleep and wake are conserved between mammals and insects.
Introduction
The regulation of sleep is of vast clinical importance. Insomnia and circadian disorders are costly in both economic and human health terms. Sleep is believed to be controlled by both circadian and homeostatic systems, which ensure that sleep needs are met. The heart of the mammalian sleep circuit is a switch consisting of reciprocally connected sleep and arousal centers (Fuller et al., 2006; Sakurai, 2007). The ventrolateral preoptic area of the hypothalamus contains inhibitory GABAergic sleep-promoting neurons, whereas arousal centers are more distributed and consist of both aminergic and cholinergic neurons; these cells additionally feed back to inhibit the hypothalamic sleep center. Hypothalamic neurons that release the peptide orexin/hypocretin also modulate the switch and stabilize the waking state. Loss of this peptide results in narcolepsy, an inability to maintain the waking state. In summary, the organization of the human circuit is complex and not completely understood.
Drosophila has become a well accepted behavioral model for sleep research, and we have recently shown that GABAergic transmission influences total sleep as well as sleep latency. In humans, both the initiation and maintenance of sleep are also controlled by GABAergic inputs to arousal centers, which explains why drugs that enhance transmission via GABAA receptors are among the most widely used sleep promoting agents (Roth, 2007). The conserved role of GABA even extends to this pharmacological level: the GABAA receptor subunit encoded by the Rdl gene (defined by resistance to the insecticide and GABAA antagonist dieldrin) has a role in the onset of fly sleep (Agosto et al., 2008). These findings indicate that part of the core circuitry controlling sleep in flies will consist of GABAA-regulated, wake-promoting cells like in mammals.
In this study we identify a population of circadian clock cells, the ventrolateral neurons or LNvs, that meet these criteria. Manipulation of Rdl levels within the LNvs indicates that they are a major target of sleep-promoting GABAergic neurons. Up- and down-regulation of PDF-positive LNv activity demonstrates that they control both total sleep and sleep onset. Indeed, acute activation of the large LNvs alone is sufficient to block sleep in the early evening, indicating that this subset of the LNvs responds to arousal signals and is a target of homeostatic regulation. Moreover, we show that the peptide PDF and its receptor are required to mediate the wake-promoting effects of these cells and that PDF modulation of LNvs themselves (presumably small LNvs) can regulate sleep. These results indicate that in flies, as in mammals, the sleep circuit is intimately linked to the circadian clock and that the strategies used to govern sleep in the brain are evolutionarily ancient.
Results
To identity the Rdl-expressing cells controlling sleep, anti-Rdl antisera were used to examine the pattern of Rdl protein expression in the adult brain. A strongly immunoreactive group of lateral brain neurons was identified by double staining as the PDF peptide-containing lateral clock neurons (LNvs). Figure 1a (top panel) and Supplemental Figure 1 show staining of whole adult brains from pdf-GAL4;UAS-mCD8-GFP animals, demonstrating that the pdf-GAL4-positive LNvs, i.e., both small (s-LNvs) and large LNvs (l-LNvs), express the Rdl GABAA receptor. Strong Rdl expression was also detected in a nearby pdf-GAL4-negative cell, whose position and morphology suggests it may represent the 5th small LNv (See Supplemental Figure 1). Preliminary experiments examining Rdl levels at ZT3 and ZT15 suggest that protein levels do not undergo circadian oscillations (data not shown).
Figure 1.
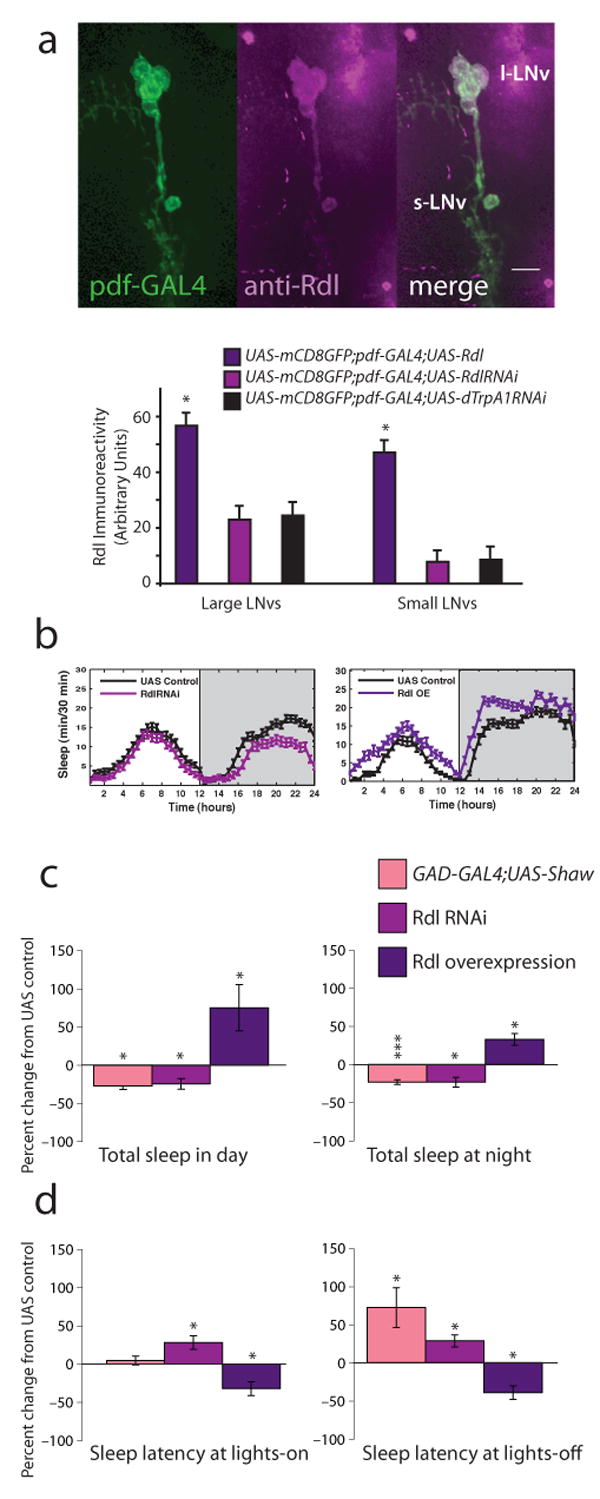
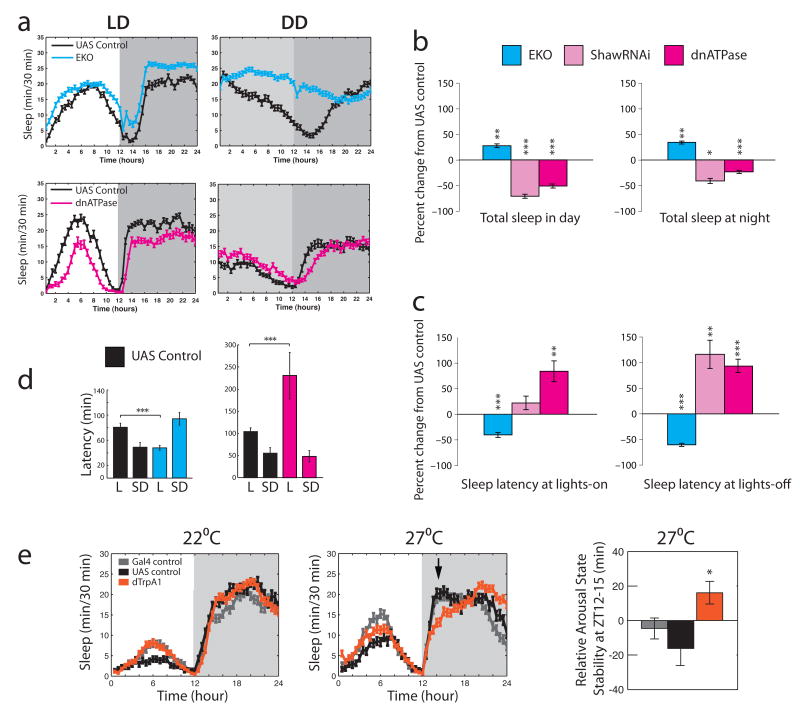
LNvs express the Rdl GABAA receptor and mediate GABAergic effects on sleep. a, Top shows images of Rdl expression in wild type LNvs. Adult brains from pdf-GAL4;UAS-mCD8-GFP animals were stained with anti-Rdl (1:100) and visualized with confocal microscopy. Rdl is shown in magenta, GFP in green, overlap in white. Scale bar = 10 μm. Bottom shows quantification of somatic Rdl levels in LNvs expressing excess Rdl, RdlRNAi or control dTrpA1RNAi. b, Standard sleep plots of control and experimental flies in 12 hour: 12 hour light:dark (LD). Left panel shows the effects of reducing Rdl levels in LNvs: pdf-GAL4;UAS-RdlRNAi, right panel shows the effects of overexpressing Rdl in LNvs: pdf-GAL4;UAS-Rdl. c, GABA regulates total sleep. 12 h sleep from the light (left) or dark (right) period in LD was assessed for animals with decreased overall GABAergic transmission (GAD-GAL4;UAS-Shaw; n=62), decreased LNv Rdl levels (pdf-GAL4;UAS-RdlRNAi; n = 93), or increased LNv Rdl levels (pdf-GAL4;UAS-Rdl; n = 21). Data are expressed as the percent change from the genetic control. d, GABA regulates sleep onset. The latency to first sleep bout during the light (left) or dark (right) period in LD was assessed for the same genotypes. Data are expressed as the percent change from the genetic control. * indicates P < 0.05, ** P < 0.005 and *** P < 0.0005 for comparisons of experimental and control using ANOVA with Tukey posthoc test.
As the circadian system and the suprachiasmatic nucleus (SCN) regulate mammalian sleep, the LNvs were attractive candidates for affecting Drosophila sleep. To determine whether Rdl activity within LNvs was relevant, we both overexpressed and downregulated the Rdl GABAA receptor exclusively in LNvs (Figure 1a, bottom panel). Expression of RNAi for Rdl using pdf-GAL4 slightly decreased somatic Rdl levels in both l- and s-LNvs, although the decrease was not statistically significant when compared to Rdl in LNvs expressing a control RNAi against dTrpA1, a channel protein expressed in a small number of non-clock adult neurons (Hamada et al., 2008). Expression of this control RNAi did not significantly affect sleep (Supplemental Figure 2). Overexpression of Rdl cDNA significantly increased somatic Rdl immunoreactivity in both l- and s-LNvs. Rdl in LNv processes was also dramatically increased going from undetectable in controls to bright in overexpressers (data not shown).
To assess the behavioral effects of altering Rdl levels in LNvs, we measured daytime and nighttime sleep in 12 hour light:12 hour dark cycles (LD). Remarkably, the nature and severity of the effects caused by LNv-specific knockdown of Rdl expression were virtually identical to the effects of reducing the excitability of GABA-producing neurons throughout the nervous system via overexpression of the hyperpolarizing Shaw potassium channel (Figure 1c and d; Agosto et al., 2008); a small but significant decrease in sleep. Only sleep latency after lights-off was more profoundly affected by the down-regulation of GABAergic tone with overexpression of Shaw (Figure 1d), perhaps reflecting the small change in Rdl levels or possibly a role for other GABA receptors; we note that cultured LNvs have been shown to have GABAB receptors (Hamasaka et al., 2005). Not only was Rdl activity within the LNvs required for sleep, the level of Rdl activity appeared important in determining the quantity of sleep, as overexpression of Rdl within the LNvs caused flies to fall asleep faster and to remain asleep longer in both the day and at night (Figure 1b, left). These results indicate that the circadian LNvs are a major target for GABAergic control of sleep and suggest that the LNvs may act as wake-promoting cells.
To more directly test this hypothesis, we used transgenes that modulate the excitability of LNvs to increase or decrease neuronal activity. A similar approach has been previously used to demonstrate a role for LNv neuronal activity in coordinating the circadian clock circuit under constant (DD) conditions (Nitabach et al., 2002; Nitabach et al., 2006). Expression of the EKO potassium channel (White et al., 2001) is expected to hyperpolarize cells, reducing their ability to be stimulated by endogenous inputs. To chronically increase activity, we developed two new molecular tools. The response to inputs was increased by either expressing an RNAi construct (Figure 2a) against the ubiquitous leak channel Shaw (Hodge et al., 2005), or by expressing a dominant negative Na+/K+-ATPase α subunit (Sun et al., 2001). Whole cell current clamp recordings from larval motor neurons expressing these transgenes indicate that they both increase resting membrane potential and the firing rate response (Figure 2b and c). These manipulations amplify the effects of endogenous inputs and allowed us to interrogate the normal function of LNvs. This is unlike the widely used bacterial sodium channel NaChBac (Nitabach et al., 2006), which imposes a novel constitutive activity pattern on neurons (Sheeba et al., 2008b). The only other putatively activity-enhancing transgene, truncated Eag (Broughton et al., 2004), has not been characterized electrophysiologically.
Figure 2.
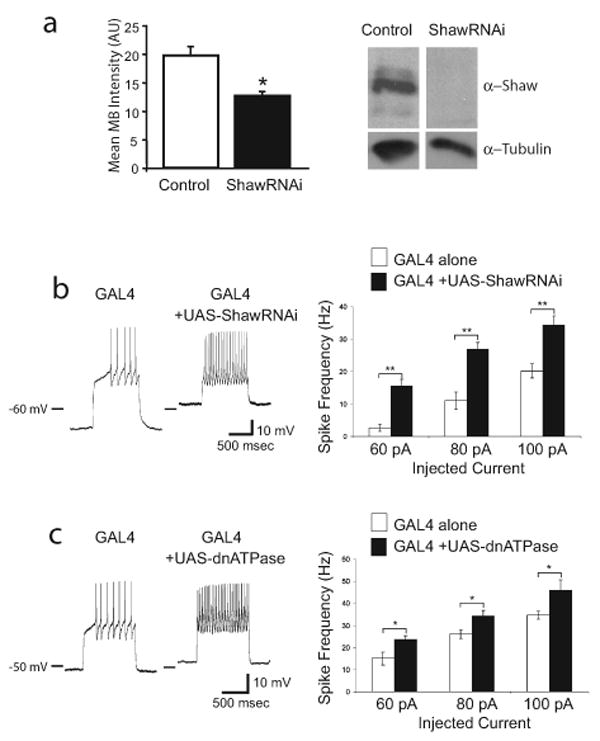
Shaw RNAi and dominant negative Na+/K+-ATPase increase neuronal excitability. a, Shaw RNAi reduces endogenous Shaw expression. (left) Expression of Shaw double-stranded RNA in adult central and motor neurons (GAL4-C380/UAS-ShawRNAi) causes reduction in Shaw levels. Endogenous Shaw levels were detected with the anti-C terminus Shaw antibody and quantified in the mushroom body calyx region with Leica confocal software. A significant reduction (P < 0.05) in intensity (arbitrary units) is seen in central neurons expressing ShawRNAi. (right) Expression of interfering Shaw double-stranded RNA (GAL4-24B/UAS-ShawRNAi) decreases endogenous Shaw compared to controls (+/UAS-ShawRNAi) when compared by immunoblot. Whole head lysates were separated by SDS-PAGE, transferred to nitrocellulose membranes and detected with a antibody (1:1000) to the C-terminus of Shaw which detects full-length Shaw protein (Hodge et al., 2005). Anti-Tubulin (1:200,000) was used to assess protein loading. b, Expression of Shaw RNAi in larval motor neurons with C380-GAL4 increases excitability. (left) Traces of whole cell current clamp recording from MNISN-Is of control (C380-GAL4 only) and experimental (C380-GAL4;UAS-ShawRNAi) animals injected with 60 pA current. (right) Quantified data for the firing rate response to various current injections. n = 7 for C380-GAL4 alone and n = 10 for C380-GAL4;UAS-ShawRNAi. c, Expression of dominant negative Na+/K+-ATPase in larval motor neurons with C380-GAL4 increases excitability. (left) Traces of whole cell current clamp recording from MNISN-Is of control (C380-GAL4 only) and experimental (C380-GAL4;UAS-dnATPase) animals injected with 60 pA current. (right) Quantified data for the firing rate response to various current injections. n = 8 for C380-GAL4 alone and n = 7 for C380-GAL4;UAS-dnATPase.
To restrict the action of these activity modulators to LNv neurons, we expressed them under the control of pdf-GAL4 and assayed sleep under standard LD conditions. Suppression of LNv activity increased both daytime and nighttime sleep (Figure 3a and b). In contrast, increasing LNv excitability using either the Shaw RNAi or the dominant negative Na+/K+-ATPase transgene decreased daytime sleep significantly (Figure 3a and b). This enhancement of LNv activity also had a suppressing effect on total nighttime sleep, further supporting a role for LNvs in sleep regulation. Importantly, sleep latency was also bidirectionally modulated by alterations in LNv excitability: decreased LNv activity caused flies to fall asleep faster in both the day and night, whereas increasing excitability suppressed initiation of the first sleep bout (Figure 3c). We found no coherent effect of manipulation of LNv excitability on locomotor activity during active periods (Supplemental Figure 3), demonstrating that the regulation of sleep is independent of basal locomotor activity, as has been previously demonstrated. We also found no effect of these manipulations on the circadian pattern of locomotor activity (Supplemental Figure 4), indicating that their effects on sleep are not secondary to disruption of the clock.
Figure 3.
Excitability of LNvs controls sleep. a, Standard sleep plots of control and experimental flies in 12 hour: 12 hour light:dark (LD) or constant darkness (DD). Top panel show the effects of reducing neuronal activity levels in LNvs: pdf-GAL4;UAS-EKO. Bottom panels show the effects of enhancing normally patterned activity in LNvs: pdf-GAL4;UAS-dnATPase. b, LNv activity controls total sleep. 12 h sleep from the light (left) or dark (right) period in LD was assessed for animals with suppressed responsiveness to inputs (pdf-GAL4;UAS-EKO; n = 55), or increased responsiveness to inputs (pdf-GAL4;UAS-ShawRNAi and pdf-GAL4;UAS-dnATPase; n = 32 and 80). Data are expressed as the percent change from the genetic control. c, LNv activity controls sleep onset. The latency to first sleep bout during the light (left) or dark (right) period in LD was assessed for the same genotypes. Data are expressed as the percent change from the genetic control. d, LNvs mediate the wake-promoting effects of light. Latency to first sleep bout during the light period in LD (L) or subjective day in DD (SD) is shown for animals with reduced responsiveness to inputs (left, pdf-GAL4;UAS-EKO) or with increased responsiveness to inputs (right, pdf-GAL4;UAS-dnATPase). e, Acute activation of LNvs disrupts nighttime sleep. pdf-GAL4;UAS-dTrpA1 and control animals (n = 32 for each genotype) were raised at the non-permissive temperature of 22°C and entrained in LD at that temperature. Data were collected for 3 days then temperature was increased to 27°C to activate dTrpA1. Left panel shows the 3 days immediately preceding the temperature increase. Middle panel shows 3 days after temperature increase. Left panel shows arousal state stability at 27°C for all genotypes in the early evening (ZT12-15; time marked by arrow in middle panel). * indicates P < 0.05, ** P < 0.005 and *** P < 0.0005 for t-test comparisons of experimental and control in panels b and c and for Tukey post-hoc test after ANOVA for panels d and e.
Control flies of all of the genotypes shown have a long sleep latency during the daytime, maintaining wakefulness for about an hour after lights on. This daytime sleep latency is strongly dependent on light, as flies fall asleep sooner after the start of the subjective day in DD (P < 0.0001 for a comparison of latency during the daytime in LD vs. during subjective day in DD, Figures 3c and 4e). Suppressing LNv activity significantly blocks the wake-promoting effect of light (Figure 3c, left panel). Interestingly, enhancing LNv activity amplifies the effect of light on sleep latency (Figure 3c, right panel). The data are consistent with light modulating LNv activity to control sleep onset and are in agreement with reports that LNvs are directly activated by light (Sheeba et al., 2007).
Figure 4.
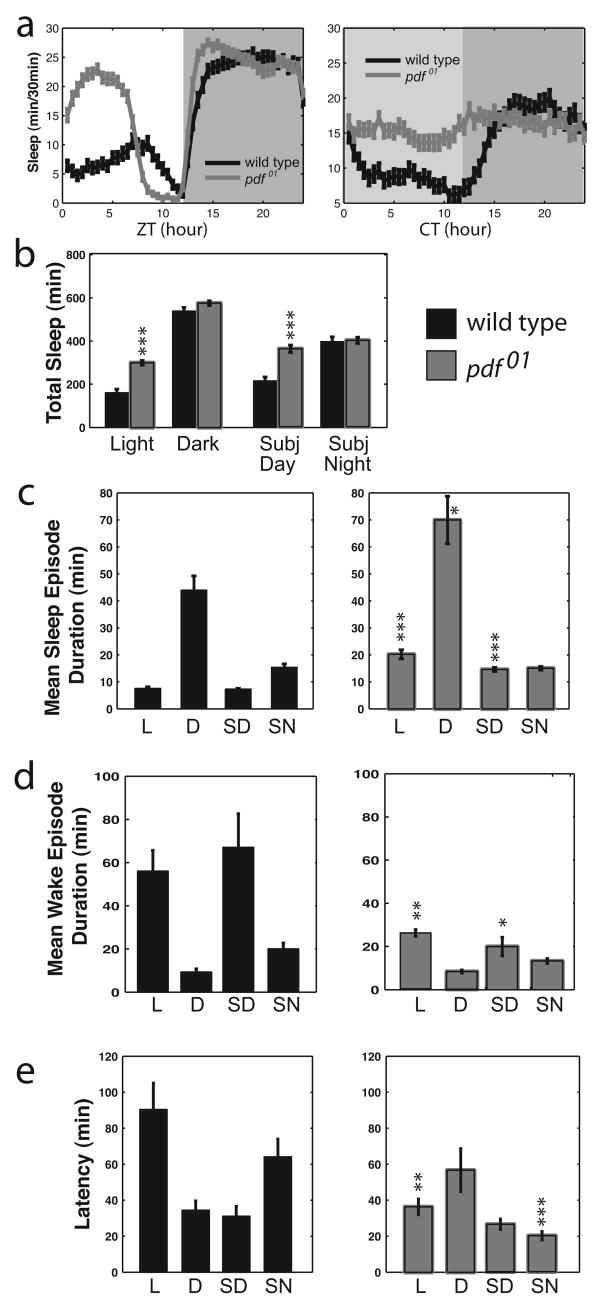
pdf01 mutants have increased total sleep and decreased sleep latency. a, Standard sleep plots of control and mutant flies in 12 hour: 12 hour light:dark (LD, left) or in constant darkness (DD, right). b, Total sleep for controls (black bars) and pdf01 mutants (gray bars) for the light period and dark period in LD and subjective day and subjective night in DD. c, Mean sleep episode duration, d, Mean wake episode duration, and e, Latency to first sleep bout, for control (left) and pdf01 mutants (right). Data are shown for light (L) and dark (D) periods in LD and for subjective day (SD) and subjective night (SN) in DD. Data are presented as means ± SEM. * indicates P < 0.05, ** P < 0.005, *** P < 0.0005 for the comparison to control by ANOVA with Tukey posthoc test. n = 106.
All of the manipulations documented above are chronic: the activity manipulation occurs throughout the lifetime of the cell. To rule out the possibility that developmental effects or circuit rewiring were responsible for the sleep phenotypes we observed, we expressed the temperature-gated non-specific cation channel, dTrpA1 in LNvs. This channel is activated at temperatures above 25°C in Drosophila larval neurons (Hamada et al., 2008). In the adult brain endogenous dTrpA1 is detectably expressed in only about a dozen cells. These cells are not known circadian cells, mushroom body cells or other circuits believed to be involved in sleep. In contrast to the chronic manipulations used above that amplify or suppress the effects of native inputs to LNvs, activated dTrpA1 imposes a fast firing pattern on the cell. Animals raised at 22°C, a temperature at which the channel is not open, show normal sleep patterns in LD compared to both GAL4 alone and UAS alone control animals (Figure 3e). Elevating the temperature to 27°C results in an immediate increase in overall sleep for all genotypes, especially in the day. Specific to flies with dTrpA1 expressed in LNvs is an increase in wakefulness in the early night. This is reflected in a significant increase in arousal state stability between ZT12 and 15, a measure of the relative length of wake and sleep bouts. Daytime sleep was largely unaffected by dTrpA1 expression in LNvs. However, the LNvs are normally activated by light (Sheeba et al., 2008b), potentially masking any additional effects of dTrpA1 on firing and wakefulness, particularly in females which generally have lower levels of daytime sleep than males (Hendricks et al., 2003).
Previous studies of LNv function have uncovered roles for the LNv-specific circadian neuropeptide PDF and perhaps other transmitters released by LNvs in the regulation of locomotor behavior (Sheeba et al., 2008b). To determine if PDF is involved in the LNv regulation of sleep, we examined the sleep behavior of pdf01 mutant flies, which lack this neuropeptide transmitter. Compared to controls, mutant flies had significantly more daytime sleep (Figure 4a-c) in LD and even under constant dark conditions (DD). Nighttime sleep was less affected, but this may be due to a ceiling effect. (The genetic controls for the pdf01 mutant had a slightly higher basal level of nighttime sleep.) This increase in daytime sleep was due primarily to a decrease in wake duration/consolidation during the day, similar to mammalian narcolepsy. EKO flies also had a similarly decreased mean wake episode duration, whereas Shaw RNAi, Rdl RNAi and dominant negative Na+/K+-ATPase animals had the opposite effect: their mean wake episode duration increased (data not shown). Loss of PDF also had effects on sleep latency, i.e. how fast the fly fell asleep after a light transition (Figure 4e). The effects of pdf01 on latency and on total sleep were similar in magnitude to the changes seen with expression of EKO or Rdl in LNvs (Figures 1 and 3).
As noted above, control flies have a longer sleep latency after lights on in LD than in DD. This light-dependent latency is totally abolished in pdf01 flies (P > 0.05 for comparison of latency during day vs. subjective day, Figure 4e, right panel). This suggests that the comparable effect in animals with decreased LNv excitability (Figure 3d, left) is due to a decrease in PDF release, as opposed to some other LNv transmitter. There is also an increase in latency in subjective night compared with subjective day, which is also eliminated in the pdf01 mutant (Figure 4e). The basis of this second alteration is unclear and could even be an indirect effect, i.e., it may mirror features of homeostatic sleep regulation and increased daytime sleep apparent in this mutant background. These factors may also contribute to the increase in latency seen during subjective day in EKO flies (Figure 3d).
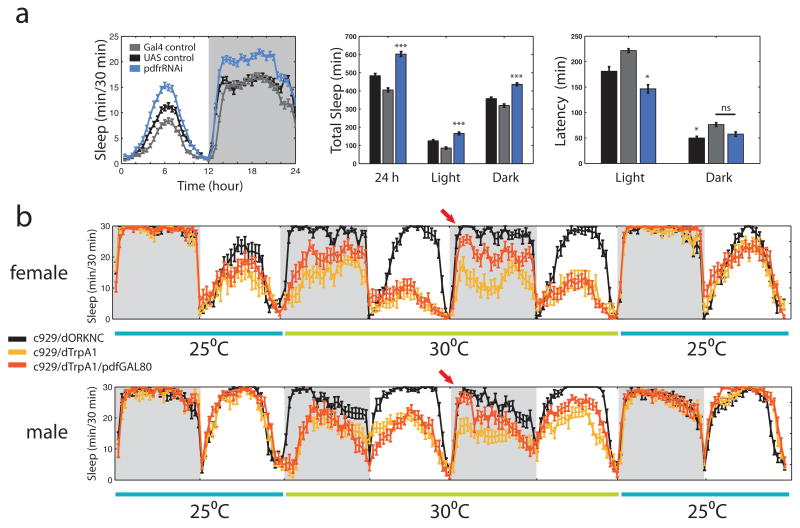
The pdf-GAL4 driver expresses in both l-LNv and s-LNv cells. s-LNvs have been postulated to be “morning cells”, which time the onset of morning behavior (Grima et al., 2004), as well as the key pacemaker cells in constant darkness (Helfrich-Forster, 1998; Stoleru et al., 2005). The function of l-LNvs has been obscure, but it has recently been shown that they respond directly to light (Sheeba et al., 2008b) and promote activity during the day, i.e., they act as dawn photoreceptors for arousal (Shang et al., 2008; Sheeba et al., 2008a). To determine the relative roles of s- and l-LNvs in sleep, we asked if PDF signaling between l- and s-LNvs was important for the wake promoting role of these cells by downregulating PDFR. Since the majority of l-LNvs do not respond to PDF (Shafer et al., 2008), this manipulation should primarily test the function of s-LNvs. We find that loss of PDFR in these cells increases total sleep in both the daytime and the nighttime (Figure 5a). Interestingly, sleep latency is only decreased compared to both GAL4 and UAS controls during the light period, suggesting that there may be other targets of PDF relevant to sleep. This is supported by the finding that a P element-generated partial deletion of the pdfr gene (Mertens et al., 2005) manifests a decrease in both daytime and nighttime sleep latency as well as increased total sleep (Supplemental Figure 5).
Figure 5.
Both large and small LNvs are involved in sleep control. a, Down regulation of the PDFR with UAS-pdfrRNAi driven by pdf-GAL4 in LNvs increases both daytime and nighttime sleep, but only significantly affects daytime latency. Standard sleep plots of female flies in 12 hour: 12 hour light:dark are shown. * indicates P < 0.05, *** indicates P < 0.0005 and ns indicates “not significant” for the comparison to other genotypes by ANOVA with Tukey posthoc test. n = 70, 71 and 75 for UAS alone (UAS-pdfrRNAi), GAL4 alone (pdfGAL4) and experimental (pdf-GAL4;UAS-pdfrRNAi) respectively. b, Continuous sleep data from flies expressing the temperature-gated cation channel dTrpA1 in peptidergic neurons ± l-LNvs. Flies were entrained in LD for 5 days at 25°C (last day is shown) and shifted to 30°C for two days, then back to 25°C. Females (n = 16 for control c929-GAL4;UAS-dORKNC), 14 for c929-GAL4;UAS-dTrpA1 and 21 for c929-GAL4;pdf-GAL80;UAS-dTrpA1) are shown at top, males (n = 23 for control, 20 for c929 and 19 for c929+pdf-GAL80) at bottom. Arrow indicates rescue of early evening sleep by suppression of dTrpA1 expression in l-LNvs by pdf-GAL80 on day 2 of elevated temperature.
To examine the specific role of l-LNvs, we first altered the temporal firing pattern of a broad set of peptidergic (PHM+) neurons, by overexpressing dTrpA1 with the c929-GAL4 driver (Park et al., 2008). The expression pattern of this driver includes l-LNvs but not s-LNvs. Flies were entrained for 5 days in LD at 25°C, switched to 30°C for two days and then back to 25°C to determine if effects were reversible. Figure 5b shows a continuous trace of sleep behavior starting during the last day of entrainment.
As in the pdf-GAL4 experiment (Figure 3e), temperature elevation increased daytime sleep even for the control genotype. In contrast, however, activation of dTrpA1 in PHM+ neurons caused a dramatic decrease in both daytime and nighttime sleep (P < 0.01 compared to control for females, P < 0.001 compared to controls for males, ANOVA with posthoc T-test). This indicates that some of these peptidergic neurons are part of the fly arousal system. Temperature elevation to 27°C also produced a decrease in sleep in the c929-GAL4;UAS-dTrpA1 animals (data not shown) but effects at 30°C were more robust.
Because PHM+ neurons include the l-LNvs and because pdf-GAL80 completely suppresses GAL4 transcription activity in all LNvs (Stoleru et al., 2004), we also assayed the sleep phenotype of c929-GAL4;UAS-dTrpA1;pdf-GAL80 flies (Figure 5a) to dissect out the specific role of l-LNvs. Immediately after temperature elevation, c929-GAL4 female flies expressing pdf-GAL80 slept significantly more than dTrpA1-expressing flies without GAL80 (P < 0.0001). On day 2 of elevated temperature, however, the both male and female flies containing pdf-GAL80 showed a markedly increased amount of sleep during the early night compared to c929-GAL4;UAS-dTrpA1 flies (arrows on Figure 5a; P < 0.0001 for females, P < 0.01 for males). Late night female sleep was also restored. This effect was harder to discern in males, perhaps because the M-peak of predawn activity causes control flies to sleep less in the late night. Because sleep homeostasis promotes rebound sleep after sleep deprivation, it seems likely that the enhanced sleep in the early night on day 2 reflects this process and that high levels of l-LNv rebound sleep stimulated by the build up of sleep pressure on the second day. We were only able to visualize the effects of sleep deprivation with the dTrpA1 animals because only the chronic situation is visible with the dnATPase and ShawRNAi tools. In aggregate, the data suggest that persistent l-LNv firing keeps flies awake at night, but that the effects are larger at the beginning of the night than at the end of the night. Therefore, activity and/or sleep circuits downstream of the l-LNvs and unknown wake-promoting non-clock peptidergic neurons may be gated differentially over the course of the night.
Although the effects on early evening sleep are almost identical, we see significant effects on daytime sleep in both c929-GAL4;UAS-dTrpA1 and c929-GAL4;UAS-dTrpA1;pdf-GAL80 flies that we did not see with pdf-GAL4;UAS-TrpA1. One possible explanation is that the innately higher levels of male and female daytime sleep in the c929-GAL4 genetic background allows a bigger dynamic range for inhibition. A more interesting possibility is that this reflects different roles for l-LNvs vs. s-LNvs in daytime sleep since the daytime loss is partially rescued on day 2 of 30°C in males (P < 0.05 for comparison of c929-GAL4;UAS-dTrpA1 and c929-GAL4;UAS-dTrpA1;pdf-GAL80 males).
Our data also imply that PDF receptors must be present on output cells downstream of the circadian/sleep circuit. The anatomical distribution of PDFR has been difficult to ascertain due to the lack of specific antibodies. Using a functional assay of cAMP accumulation, the only cells that have been definitively identified as PDF targets to date are other clock cells (Shafer et al., 2008). To determine if the PDFR might be expressed on cells more directly involved in control of motor activity, we fused 10 kilobases of genomic DNA upstream of the pdfr gene to GAL4. Figure 6a (left) shows that this promoter region drives expression in a limited number of cells that parallel the distribution of PDF in the adult brain. GFP expression is seen in optic lobes and the dorsal and lateral brain. Lateral brain staining is partly due to expression of this GAL4 line in a subset of both l-LNvs and s-LNvs (example shown in Figure 6a, right). 13 brains were imaged and GFP was typically see in at least one neuron per side in both the s-LNv and l-LNv groups. Dorsal brain staining is seen in the majority of LNds but not significantly in DNs, although GFP-positive processes from LNds project dorsally. This is consistent with our GAL4 line capturing a subset of the endogenous PDFR clock distribution that has been described. Interestingly, there is also very significant expression in neurons that innervate the ellipsoid bodies, a structure that is part of the central complex and has been implicated in motor control (Strauss, 2002). Ellipsoid body expression was consistently strong in all three of the lines examined.
Figure 6.
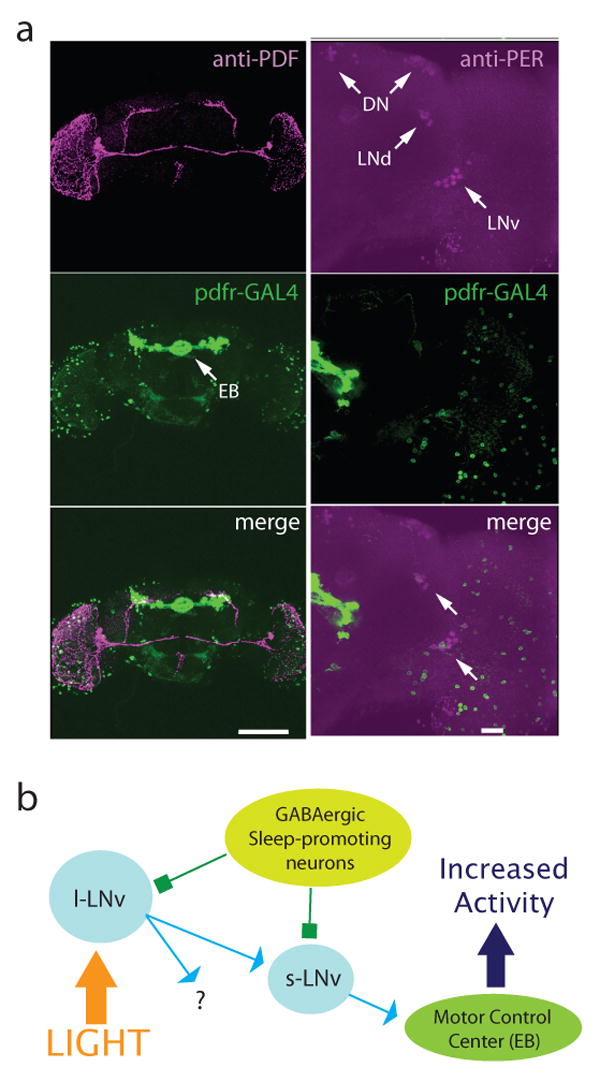
Output of the sleep circuit. a, pdfr-GAL4 marks output cells of the LNv circuit. Left panels show reconstructed pdfr-GAL4;UAS-mCD8-GFP adult brain stained with anti-PDF (1:1000) and visualized with confocal microscopy. PDF is shown in magenta, GFP in green, overlap in white. Scale bar = 150 μm. Right panels show a 37 μ section of a pdfr-GAL4;UAS-mCD8-GFP adult brain stained with anti-Per (1:100) and visualized with confocal microscopy. Per is shown in magenta, GFP in green, overlap in white. Arrows indicate clock cells. Scale bar = 20 μm. b, Model of the Drosophila sleep circuit. Light, and perhaps other arousal cues, activate l-LNvs which release PDF onto s-LNvs that project to other clock cells and also send dorsal projections that pass by pdfr-GAL4 positive cells groups such as the ellipsoid bodies that are involved in control of activity. Both l- and s-LNvs express GABAA receptors, allowing sleep promoting GABAergic neurons to suppress wakefulness.
Discussion
Using a variety of mutants and novel genetic strategies to manipulate chronic and acute circuit activity, we have shown that a small set of circadian clock cells in Drosophila has a critical role in the GABAergic initiation and maintenance of sleep. We have developed new genetic tools (dnATPase, ShawRNAi), which allow an increase in the chronic response of neurons to their endogenous inputs. This adds greatly to the arsenal of activity-manipulating tools, most of which suppress firing or neurotransmitter release. Bidirectional manipulation of activity provides much more information about circuit function and dynamics (c.f. Broughton et al., 2004). We have also shown the utility of a new tool for acute activity manipulation (dTrpA1), which can be used on small numbers of neurons deep within the fly brain. Our data suggest a model (Figure 6b) in which l-LNvs translate light inputs (and perhaps other arousal signals) into wakefulness. The release of PDF from these cells is required, and l-LNv PDF signals to s-LNvs. Our data demonstrating somnolence after downregulation of PDFR in LNvs indicates that s-LNvs participate in sleep control, although experiments in which they have been ablated suggest that they are not be the only sleep-relevant l-LNv targets (Sheeba et al., 2008a). PDF signaling to PDFR-expressing neurons outside the clock that directly control activity is likely to be important (see below). GABA may modulate the ability of LNvs to suppress sleep by acting on either or both s- and l-LNvs.
In mammals, the role of the circadian clock in sleep is not completely understood. It is nonetheless clear that there are genetic (e.g. familial advance sleep phase syndrome) and environmental (e.g. jet-lag, shift work) conditions that disrupt sleep despite primarily affecting the circadian rhythms (Gottlieb et al., 2007). The clock has been shown to regulate both when an animal sleeps and how much sleep occurs. The current consensus view is that the mammalian clock is primarily wake-promoting (Edgar et al., 1993; Laposky et al., 2005; Naylor et al., 2000), acting along with the homeostatic sleep drive to shape sleep over the day and night (Dijk and Franken, 2005).
Our data indicate that in flies PDF and the circadian LNvs more generally regulate both the maintenance of sleep as well as the ability of flies to respond to the wake-promoting effects of light. Although these effects recall the role of the mammalian SCN in sleep regulation, there are few prior links between the Drosophila circadian clock and the regulation of fly sleep (Shaw et al., 2002). The almost complete elimination of the difference in total sleep between subjective day and subjective night in the pdf01 background (Figure 4c) adds substantially to this connection, i.e., light regulation of sleep appears to be substantially circadian clock-mediated Therefore, the contribution of the circadian machinery and fly brain clock circuitry to the control of sleep will probably parallel the important role of the mammalian circadian clock and the SCN in sleep regulation (Borbely and Achermann, 1999; Edgar et al., 1993).
PDF neurons have been recently shown to be light-responsive (Sheeba et al., 2007), like some neurons of the mammalian SCN (Meijer et al., 1998). The l-LNvs also act as the dawn photoreceptor for the clock, sending a reset signal each morning to the rest of the clock (Shang et al., 2008; Sheeba et al., 2008a). There is also good evidence that fly cryptochrome responds directly to light in addition to influencing circadian timekeeping (e.g., Allada et al., 1998; Emery et al., 2000a; Emery et al., 2000b), and a cry mutant substantially decreases the PDF neuron acute light response (Sheeba et al., 2007). Therefore, some of the waking effects described here probably reflect a role of PDF cells on acute processes involving light stimulation. Indeed, the phenotypes of flies without PDF or with decreased LNv neuronal excitability resemble some of the acute effects of the loss of orexin/hypocretin in narcoleptic mice (Mochizuki et al., 2004). PDF neurons are also regulated by GABAergic inputs, analogous to those from the basal forebrain that regulate orexin/hypocretin neurons (Henny and Jones, 2006).
Despite these similarities, there are also important organizational differences between systems. Most notable is the wide distribution of sleep circuitry in mammals. There are for example many targets of sleep-promoting GABAergic neurons, and the role of the circadian clock may be largely modulatory (Mistlberger, 2005). The sleep circuitry of flies is almost certainly more circumscribed and simpler. Indeed, the surprisingly large effects of manipulating Rdl in the 16 LNvs argue that they are a principal target of sleep promoting GABAergic neurons and constitute part of the “core” sleep circuitry. The fact that activation of a subset of these cells, the l-LNvs, has an effect on sleep homeostasis, further suggests that these cells sit at the heart of the sleep circuit. The fly sleep circuitry may therefore have condensed mammalian stimulatory systems (e.g. histaminergic, cholinergic and adrenergic, as well as orexin) into a simpler and more compact region, which may even largely coincide with the sixteen PDF cells of the circadian circuit.
A limited number of other fly brain regions have been proposed to contribute to fly sleep. Our manipulations of PHM+ cells indicate that peptidergic neurons other than PDF neurons are wake promoting. An attractive hypothesis is that some these other peptidergic cells reside in the pars intercerebralis, a group of neurohumoral cells shown to an important sleep output center (Foltenyi et al., 2007). The targets of these cells may even overlap with the targets of LNvs, e.g. the ellipsoid bodies. The PDFR is a class II G-protein coupled receptor and is fairly promiscuous: PDF is the highest affinity ligand, but this receptor is also activated by DH31 and PACAP-38 (Mertens et al., 2005). Since peptidergic modulation may occur by “volume” transmission instead of by direct synaptic contact (Zoli et al., 1999), both LNv peptides and peptides from the pars could together affect this motor center to regulate sleep and activity. The role of the pars may be to inform the sleep generation machinery about nutritional and metabolic state, i.e., animals undergoing starvation exhibit hyperlocomotor activity that is believed to be evolutionarily useful as a method for finding food (Lee and Park, 2004), and alteration of this pars-generated locomotor program affects sleep (Mattaliano et al., 2007). The role of l-LNvs is clearly different from that of other PHM+ neurons, and their unique involvement in homeostatic sleep suggests they are central to sleep control.
The only other brain region that has been implicated in Drosophila sleep regulation is the mushroom bodies (Joiner et al., 2006; Pitman et al., 2006). These studies showed that GAL4-driven manipulation of signaling or of neurotransmitter release in this neuropil had complex effects on sleep, not inconsistent with a modulatory role for this sensory integration center. The exact mechanism of these effects is not clear, however, especially since all of the mushroom body GAL4 lines we have examined also express in multiple subsets of clock cells (data not shown).
The small circuit we describe presents a tractable model system for understanding the circuit-level control of sleep, the relationship between homeostatic and circadian control as well as the dynamics of sleep-wake transitions; the latter are critical to an understanding of episodic and age-related insomnia.
Methods
Animals
Flies were raised under a 12 h light: 12 h dark (LD) schedule at 24-25°C on cornmeal dextrose yeast food. Transgenic lines and mutants are as described: pdf-GAL4 (Renn et al., 1999), GAD-GAL4 (Mehren and Griffith, 2006), C380-GAL4 (Packard et al., 2002), c929-GAL4 (Park et al., 2008), UAS-mCD8-GFP (Lee and Luo, 1999), UAS-dnATPase (Sun et al., 2001), UAS-EKO (White et al., 2001), UAS-ShawWT (Hodge et al., 2005), UAS-dTrpA1 (Hamada et al., 2008), UAS-Rdl and UAS-RdlRNAi (Liu et al., 2007), UAS-NaChBacGFP and UAS-dORKNCGFP (Nitabach et al., 2006), pdf-GAL80 (Stoleru et al., 2004), pdf01 (Renn et al., 1999), UAS-pdfrRNAi (Dietzl et al., 2007), UAS-dTrpA1RNAi (Hamada et al., 2008), and pdfrP2-36 and its revertant control (Mertens et al., 2005). The UAS-ShawRNAi transgene was generated by inserting a 720 bp fragment of the 3′ end of the Shaw cDNA starting at nucleotide 881 in exon 8 through to the end of the coding region including approximately 110 bp of 3′ untranslated sequence into Sym-pUAST-w (Giordano et al., 2002). A transgenic line containing inserts on both chromosomes 2 and 3 was generated by standard methods (Robertson et al., 1988). The pdfr-GAL4 line was constructed by amplifying 10 kilobases upstream of the ATG of the pdfr gene (CG13758) by PCR, subcloning the fragment into the pPTGAL4 vector and generating transgenic flies by standard methods. Expression patterns from three independent insertion lines were analysed and found to be essentially identical. Primers used were: pdfR forward: 5′CCGGCTTTTGTTTTGTGTTTTG3′ and pdfR rev: 5′GCCATCGACCGCATAGTAAATG3′. All other lines were obtained from Bloomington Stock Center.
For each LNv manipulation, experimental animals were compared to a control line that was generated by crossing the UAS line to Df(1)w, the background strain used to make transgenic lines. This is indicated in the figures as “UAS Control”. We find that this type of control is very important to do since the genetic background of strains can have a big influence on basal sleep parameters- c.f. the controls in Figure 3a. Since pdf-GAL4 is used as a common driver for all LNv manipulations, and therefore cannot contribute to differential phenotypes. Baseline data for this GAL4 line is shown in Figure 3e. GAD-GAL4 has previously been shown to sleep normally (Agosto et al., 2008). For pdf01, the mutant was extensively outcrossed to Canton S wildtype and mutant and sibling control lines established using PCR genotyping. For pdfr the precise excision strain (Mertens et al., 2005) was used as a genetic background control. For c929-GAL4 experiments, a control line expressing a dead channel protein (dORKNC) was used as a control line since the c929-GAL4 genetic background has relatively high basal daytime sleep compared to other genotypes.
Sleep and activity assays
All behavior was done on female flies unless explicitly indicated. 5 day old flies were placed in 65 mm × 5 mm glass tubes (Trikinetics, Waltham, MA) containing 5% agarose with 2% sucrose. Flies were acclimated in behavior tubes for at least 24 h at 25°C (or 22°C where indicated) in 12 h light/12 h dark (LD) conditions before data collection. Flies were entrained at least 4 days in LD before switching to constant darkness (DD). Locomotor activity was collected with DAM System monitors (Trikinetics) in 1 min bins as previously described (Agosto et al., 2008). Sleep was measured as bouts of uninterrupted 5 minutes of inactivity. Sleep parameters were analyzed using MATLAB software (Natick, MA) of averages over four days of LD. Total sleep duration, mean sleep and wake bout duration, and latency were analyzed for each 12 h period of LD and DD and averaged over three days for each condition. Arousal state stability was calculated by subtracting the maximum sleep bout duration from the maximum wake bout duration. Values greater than one indicate a more wakeful state while values less than one are indicative of a stable sleep state.
Immunohistochemistry, imaging and quantification
For determination of Shaw RNAi efficacy, adult brains from C380-GAL4;mCD8-GFP animals were processed and stained with anti-Shaw antibody (preabsorbed and used at 1:1000) and Cy5 secondary antibody (1:180; Jackson Labs) as described (Hodge et al., 2005). All preparations were processed in parallel and images acquired with identical settings using the 50× (zoomed 1-4×) objectives of a Leica TCS SP2 confocal microscope. Care was taken to keep all intensity readings within the linear range below saturation. Quantification was performed on 1 μm sections with pixel intensity readings taken in a given region of interest (in this case the mushroom bodies) for GFP and Cy5 using the Leica TCS SP2 quantification software. Quantification was performed blind to genotype. Statistical analysis was performed in Excel (Microsoft) and JMP (SAS). Significance levels in figures were determined by one-way ANOVA unless otherwise specified and * indicates P < 0.05.
For Rdl localization adult brains from pdf-GAL4;mCD8-GFP animals that had been entrained in a 12h L: 12 h D cycle for at least 3 days were dissected, fixed and stained basically as described (Van Vactor et al., 1991) with anti-Rdl (1:100) (Liu et al., 2007) and anti-GFP (1:200, Roche Applied Biosciences), and Alexa 635 and 488 secondary antibodies (1:200, Invitrogen). For costaining with pdfr-GAL4, pdfr-GAL4;UAS-mCD8-GFP animals were entrained in a 12h L: 12 h D cycle for at least 3 days were dissected, fixed and stained as described (Van Vactor et al., 1991) with anti-GFP (1:300, Roche Applied Biosciences) + Alexa 635 secondary (1:200, Invitrogen) and either anti-PAP (PDF precursor) (1:1000) + Texas Red secondary (1:200, Jackson Labs) or anti-Per (1:1000) + Alexa 488 secondary antibody (1:200, Invitrogen). Images were acquired on a Leica TCS SP2 confocal system with Leica Confocal Software at 63×. Separate images were taken using 488 nm and 633 nm lasers and overlapped to avoid bleed through. Leica Confocal Software was used to quantify the images. Quantification was performed using the first scan taken at 633 nm excitation. After signal digitization RMS values of background were subtracted to get final values.
Immunoblotting
Age matched flies were frozen in liquid nitrogen, and decapitated by vortexing. Extracts were prepared as described (Hodge et al., 2006), separated by SDS-PAGE, and analyzed by immunoblot using rabbit anti-C terminal Shaw antibody (1:1000), monoclonal anti-tubulin (1:200,000; Sigma).
Electrophysiology
Whole cell recordings were performed on third instar larvae using methods described previously (Choi et al., 2004). Larvae were cut open dorsally and pinned down onto a sylgard-lined dish in calcium-free solution consisting (in mM) of, 128 NaCl, 2 NaOH, 2 KCl, 15 sucrose, 5 Trehalose, 4 MgCl2, and 5 HEPES, with pH 7.1–7.2. Sheath tissue surrounding dorsal motorneuron clusters was digested with 0.01% protease (type XIV, Sigma). Motor neuron MNISN-Is was targeted exclusively for all experiments. Pipette resistance was 5-10 MΩ, and solution contained (in mM) 130 potassium gluconate, 10 HEPES, 1 EGTA, 2 MgCl2, 0.1 CaCl2, 2 NaCl, 10 KOH, with pH adjusted to 7.2. An Axopatch 200B amplifier (Axon Instruments, Union City CA) was used to perform whole cell recordings and acquisition and analysis performed with IgorPro (Wavemetrics, Oswego, OR). Two tailed, unpaired t-tests or ANOVA repeated measures test was used to analyse significance of values when comparing two genotypes (Statview software package, Abacus Concepts, Cary NC). Multiple comparisons were done using ANOVA with the Tukey-Kramer posthoc test for pair-wise comparisons.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 MH067284 to LCG and US Army grant W81XWH-04-1-0158 to LCG and MR. KP was supported by NIH training grant T32 NS07292 and JA by T32 GM07122. Work at the Baylor College of Medicine was supported by NIH grant NS19904 (Ronald L. Davis, PI). We thank Ed Dougherty for imaging assistance and Todd Holmes and Ravi Allada for communication of results prior to publication. The Brandeis Biology confocal facility was supported by NIH grants P30 NS045713 and S10 RR16780.
Response to reviewers: We thank the reviewers for their support and helpful suggestions. We have revised the manuscript accordingly.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008;11:354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- Borbely AA, Achermann P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms. 1999;14:557–568. doi: 10.1177/074873099129000894. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Kitamoto T, Greenspan RJ. Excitatory and inhibitory switches for courtship in the brain of Drosophila melanogaster. Curr Biol. 2004;14:538–547. doi: 10.1016/j.cub.2004.03.037. [DOI] [PubMed] [Google Scholar]
- Choi JC, Park D, Griffith LC. Electrophysiological and morphological characterization of identified motor neurons in the Drosophila third instar larva central nervous system. J Neurophysiol. 2004;91:2353–2365. doi: 10.1152/jn.01115.2003. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dijk D, Franken P. Principles and Practice of Sleep Medicine. Saunders WB; 2005. [Google Scholar]
- Edgar DM, Dement WC, Fuller CA. Effect of SCN lesions on sleep in squirrel monkeys: evidence for opponent processes in sleep-wake regulation. J Neurosci. 1993;13:1065–1079. doi: 10.1523/JNEUROSCI.13-03-01065.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Hall JC, Rosbash M. A unique circadian-rhythm photoreceptor. Nature. 2000a;404:456–457. doi: 10.1038/35006558. [DOI] [PubMed] [Google Scholar]
- Emery P, Stanewsky R, Helfrich-Forster C, Emery-Le M, Hall JC, Rosbash M. Drosophila CRY is a deep brain circadian photoreceptor. Neuron. 2000b;26:493–504. doi: 10.1016/s0896-6273(00)81181-2. [DOI] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci. 2007;10:1160–1167. doi: 10.1038/nn1957. [DOI] [PubMed] [Google Scholar]
- Fuller PM, Gooley JJ, Saper CB. Neurobiology of the sleep-wake cycle: sleep architecture, circadian regulation, and regulatory feedback. J Biol Rhythms. 2006;21:482–493. doi: 10.1177/0748730406294627. [DOI] [PubMed] [Google Scholar]
- Giordano E, Rendina R, Peluso I, Furia M. RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics. 2002;160:637–648. doi: 10.1093/genetics/160.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb DJ, O'Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC medical genetics. 2007;8 1:S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaka Y, Wegener C, Nassel DR. GABA modulates Drosophila circadian clock neurons via GABAB receptors and decreases in calcium. J Neurobiol. 2005;65:225–240. doi: 10.1002/neu.20184. [DOI] [PubMed] [Google Scholar]
- Helfrich-Forster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol [A] 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Lu S, Kume K, Yin JC, Yang Z, Sehgal A. Gender dimorphism in the role of cycle (BMAL1) in rest, rest regulation, and longevity in Drosophila melanogaster. J Biol Rhythms. 2003;18:12–25. doi: 10.1177/0748730402239673. [DOI] [PubMed] [Google Scholar]
- Henny P, Jones BE. Innervation of orexin/hypocretin neurons by GABAergic, glutamatergic or cholinergic basal forebrain terminals evidenced by immunostaining for presynaptic vesicular transporter and postsynaptic scaffolding proteins. J Comp Neurol. 2006;499:645–661. doi: 10.1002/cne.21131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge JJ, Choi JC, O'Kane CJ, Griffith LC. Shaw potassium channel genes in Drosophila. J Neurobiol. 2005;63:235–254. doi: 10.1002/neu.20126. [DOI] [PubMed] [Google Scholar]
- Hodge JJ, Mullasseril P, Griffith LC. Activity-Dependent Gating of CaMKII Autonomous Activity by Drosophila CASK. Neuron. 2006;51:327–337. doi: 10.1016/j.neuron.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- Laposky A, Easton A, Dugovic C, Walisser J, Bradfield C, Turek F. Deletion of the mammalian circadian clock gene BMAL1/Mop3 alters baseline sleep architecture and the response to sleep deprivation. Sleep. 2005;28:395–409. doi: 10.1093/sleep/28.4.395. [DOI] [PubMed] [Google Scholar]
- Lee G, Park JH. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics. 2004;167:311–323. doi: 10.1534/genetics.167.1.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Liu X, Krause WC, Davis RL. GABA(A) Receptor RDL Inhibits Drosophila Olfactory Associative Learning. Neuron. 2007;56:1090–1102. doi: 10.1016/j.neuron.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Greco MA. Sleep circuitry and the hypnotic mechanism of GABAA drugs. J Clin Sleep Med. 2006;2:S19–26. [PubMed] [Google Scholar]
- Mattaliano MD, Montana ES, Parisky KM, Littleton JT, Griffith LC. The Drosophila ARC homolog regulates behavioral responses to starvation. Molecular and cellular neurosciences. 2007;36:211–221. doi: 10.1016/j.mcn.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehren JE, Griffith LC. Cholinergic neurons mediate CaMKII-dependent enhancement of courtship suppression. Learn Mem. 2006;13:686–689. doi: 10.1101/lm.317806. [DOI] [PubMed] [Google Scholar]
- Meijer JH, Watanabe K, Schaap J, Albus H, Detari L. Light responsiveness of the suprachiasmatic nucleus: long-term multiunit and single-unit recordings in freely moving rats. J Neurosci. 1998;18:9078–9087. doi: 10.1523/JNEUROSCI.18-21-09078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Mistlberger RE. Circadian regulation of sleep in mammals: role of the suprachiasmatic nucleus. Brain Res Brain Res Rev. 2005;49:429–454. doi: 10.1016/j.brainresrev.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Crocker A, McCormack S, Yanagisawa M, Sakurai T, Scammell TE. Behavioral state instability in orexin knock-out mice. J Neurosci. 2004;24:6291–6300. doi: 10.1523/JNEUROSCI.0586-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor E, Bergmann BM, Krauski K, Zee PC, Takahashi JS, Vitaterna MH, Turek FW. The circadian clock mutation alters sleep homeostasis in the mouse. J Neurosci. 2000;20:8138–8143. doi: 10.1523/JNEUROSCI.20-21-08138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–495. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26:479–489. doi: 10.1523/JNEUROSCI.3915-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard M, Koo ES, Gorczyca M, Sharpe J, Cumberledge S, Budnik V. The Drosophila Wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation. Cell. 2002;111:319–330. doi: 10.1016/s0092-8674(02)01047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Veenstra JA, Park JH, Taghert PH. Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS ONE. 2008;3:e1896. doi: 10.1371/journal.pone.0001896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillips RW, Johnson-Schlitz D, Benz WK, Engels WR. A stable genomic source of P-element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth T. A physiologic basis for the evolution of pharmacotherapy for insomnia. The Journal of clinical psychiatry. 2007;68 5:13–18. [PubMed] [Google Scholar]
- Sakurai T. The neural circuit of orexin (hypocretin): maintaining sleep and wakefulness. Nat Rev Neurosci. 2007;8:171–181. doi: 10.1038/nrn2092. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Griffith LC, Rosbash M. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. 2008 doi: 10.1073/pnas.0809577105. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou YT, Sharma VK, Holmes TC. Large Ventral Lateral Neurons Modulate Arousal and Sleep in Drosophila. Curr Biol. 2008a doi: 10.1016/j.cub.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Gu H, Sharma VK, O'Dowd DK, Holmes TC. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol. 2007 doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Sharma VK, Gu H, Chou YT, O'Dowd DK, Holmes TC. Pigment dispersing factor-dependent and -independent circadian locomotor behavioral rhythms. J Neurosci. 2008b;28:217–227. doi: 10.1523/JNEUROSCI.4087-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Strauss R. The central complex and the genetic dissection of locomotor behaviour. Curr Opin Neurobiol. 2002;12:633–638. doi: 10.1016/s0959-4388(02)00385-9. [DOI] [PubMed] [Google Scholar]
- Sun B, Xu P, Wang W, Salvaterra PM. In vivo modification of Na(+),K(+)-ATPase activity in Drosophila. Comparative biochemistry and physiology. 2001;130:521–536. doi: 10.1016/s1096-4959(01)00470-5. [DOI] [PubMed] [Google Scholar]
- Van Vactor DL, Jr, Cagan RL, Kramer H, Zipursky SL. Induction in the developing compound eye of Drosophila: multiple mechanisms restrict R7 induction to a single retinal precursor cell. Cell. 1991;67:1145–1155. doi: 10.1016/0092-8674(91)90291-6. [DOI] [PubMed] [Google Scholar]
- White BH, Osterwalder TP, Yoon KS, Joiner WJ, Whim MD, Kaczmarek LK, Keshishian H. Targeted attenuation of electrical activity in Drosophila using a genetically modified K(+) channel. Neuron. 2001;31:699–711. doi: 10.1016/s0896-6273(01)00415-9. [DOI] [PubMed] [Google Scholar]
- Zoli M, Jansson A, Sykova E, Agnati LF, Fuxe K. Volume transmission in the CNS and its relevance for neuropsychopharmacology. Trends Pharmacol Sci. 1999;20:142–150. doi: 10.1016/s0165-6147(99)01343-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.