Abstract
Radially encoded MR imaging (MRI) has gained increasing attention in applications such as hyperpolarized gas imaging, contrast-enhanced MR angiography, and dynamic imaging, due to its motion insensitivity and improved artifact properties. However, since the technique collects k-space samples nonuniformly, multidimensional (especially 3D) radially sampled MRI image reconstruction is challenging. The balance between reconstruction accuracy and speed becomes critical when a large data set is processed. Kaiser-Bessel gridding reconstruction has been widely used for non-Cartesian reconstruction. The objective of this work is to provide an alternative reconstruction option in high dimensions with on-the-fly kernels calculation. The work develops general multi-dimensional least square nonuniform fast Fourier transform (LS-NUFFT) algorithms and incorporates them into a k-space simulation and image reconstruction framework. The method is then applied to reconstruct the radially encoded k-space, although the method addresses general nonuniformity and is applicable to any non-Cartesian patterns. Performance assessments are made by comparing the LS-NUFFT based method with the conventional Kaiser-Bessel gridding method for 2D and 3D radially encoded computer simulated phantoms and physically scanned phantoms. The results show that the LS-NUFFT reconstruction method has better accuracy-speed efficiency than the Kaiser-Bessel gridding method when the kernel weights are calculated on the fly. The accuracy of the LS-NUFFT method depends on the choice of scaling factor, and it is found that for a particular conventional kernel function, using its corresponding deapodization function as scaling factor and utilizing it into the LS-NUFFT framework has the potential to improve accuracy. When a cosine scaling factor is used, in particular, the LS-NUFFT method is faster than Kaiser-Bessel gridding method because of a quasi closed-form solution. The method is successfully applied to 2D and 3D in vivo studies on small animals.
Index Terms: 2D and 3D radially encoded magnetic resonance imaging (MRI), projection reconstruction, least square nonuniform fast Fourier transform (LS-NUFFT), non-Cartesian reconstruction, k-space, Kaiser-Bessel gridding method, 3D isotropic radial sampling
I. INTRODUCTION
Historically, early MR images were obtained with projection reconstruction (PR) [1][2][3]. The PR technique was later generalized to radial sampling by collecting k-space data from center to periphery along a radius. The technique has recently gained more popularity [4][5][6][7] due to its advantageous motion insensitivity and relatively high signal to noise ratio (SNR) compared to the n-dimensional Fourier transform (nDFT). In a radial scan, however, the samples do not fall on a Cartesian grid, so image reconstruction requires more sophisticated processing techniques.
Recently, the older back-projection reconstruction has been supplanted by gridding method [8][9][10][11][12]. The gridding method generally consists of three steps: pre-weighting, convolving the original data with shift-invariant interpolation kernels, and inverse Fourier transform. In 2D, Jackson et al. [11] compared several convolution kernels including Gaussian, Kaiser-Bessel, and Prolate Spheroidal Wave Function (PSWF), and concluded that the Kaiser-Bessel kernel is the best in terms of minimum averaged aliasing energy. The Kaiser-Bessel gridding method was later extended to 3D and applied to radial data [7][13][12]. Beatty et al. [12] proposed a practical parameter formula for Kaiser-Bessel method that minimizes the maximum aliasing energy. In this paper, the Kaiser-Bessel gridding method is referred to as the conventional method.
The accuracy of non-Cartesian MRI image reconstruction is usually limited by the pre-weighting and interpolation. Besides gridding accuracy, the computational complexity becomes a significant issue especially in 3D when a large number of rays are acquired. Unlike nDFT methods that sample k-space uniformly on a Cartesian grid, radially encoded MRI samples the center of k-space much more densely than the periphery. In this case, the Nyquist limit is loosely defined in the radial framework as the maximum separation between neighboring data points in k-space that can eliminate aliasing. Because of the divergence of the rays, this means more samples need to be collected using a radial scan than using nDFT scans. In gridding methods, the tradeoff between accuracy and processing speed, as well as memory requirements, can be managed by changing the window size of the kernel and the oversampling rate. Some work has been done to find optimal parameters to balance accuracy and computational cost. For example, Beatty et al. [12] used a minimal oversampling rate, from 1.125 to 1.375, to reduce the high computational memory demand in 3D.
The recent developments in fast numerical algorithms [14][15][16][17][18] provide more efficient tools to evaluate the nonuniform discrete Fourier transform (NUDFT). In particular, Liu and Nguyen [16] named their inverse transform algorithm using least square error kernels the nonuniform fast Fourier transform (NUFFT). NUFFT calculates complex kernels based on the given sampling pattern to minimize interpolation error. Some of these NUFFTs have been implemented in 2D and applied to spiral MRI reconstruction [19] and sensitivity encoding (SENSE) image reconstruction [20]. In the present work, we develop multi-dimensional LS-NUFFTs of first type (NUFFT-1) and second type (NUFFT-2) by adopting the least square NUFFT framework in 1D [16][17], and apply these methods to 2D and 3D k-space data acquired using projection reconstruction trajectories. We discuss the choice of scaling factor and its impact on approximation accuracy and processing speed. Reconstruction results of both computer-simulated and physically scanned objects are compared with those obtained by the Kaiser-Bessel gridding method. Accuracy-speed efficiency and SNR comparisons are used to show performance improvements.
The paper is organized as follows. Section II formulates the multi-dimensional NUFFT-1 and NUFFT-2. A generalized multi-dimensional non-Cartesian MRI image reconstruction and k-space data simulation package is developed in Section III based on the NUFFTs. In Section IV, results are shown for both 2D and 3D imaging using a computer-simulated phantom, a physically scanned phantom, and small live animals. The choice of scaling factor is discussed. Comparisons are made with the conventional Kaiser-Bessel gridding method.
II. d-DIMENSIONAL NUFFTS
We define Equation (2) and Equation (1) as the d-dimensional nonuniform discrete Fourier transform of the first-type and second-type, respectively,
| (1) |
| (2) |
where denotes the mth non-Cartesian sample locations in the frequency domain and n denotes the Cartesian pixel index in the image domain. Note (2) and (1) become regular DFT and inverse DFT if v becomes uniform in a Cartesian grid.
A. d-dimensional least square kernel
The direct evaluations of (1) and (2) both cost operations, where Np is the number of image pixels in the pth dimension and M is the total number of frequency samples. To reduce the computational complexity, we use an approach similar to Liu and Nguyen’s 1D least square NUFFT to find the d-dimensional kernel Φm that satisfy
| (3) |
where sn is a d-dimensional accuracy factor, the choice of which drives the approximation accuracy, μ is the oversampling factor and (q + 1) is the kernel size in each dimension. [x] denotes the nearest integer of x and {x} = x − [x]. The exact solution of Φm generally does not exist, but instead a least square solution. LS-NUFFT does not define kernel function before hand, but calculates it optimally in a least square sense. The approximation error is ultimately determined by the choice of sn.
The least square solution of (3) gives Φm as a tensor product of 1D least square kernels
| (4) |
where in the pth dimension ϕm can be solved by ϕm = F−1 α(υm) where F is a (q + 1) × (q + 1) Toeplitz matrix independent of υm
| (5) |
Vector α in general has to be evaluated by the direct summation with complexity O (N (q + 1))
| (6) |
If a cosine function is chosen for sj [16][17], α has a closed-form solution that only has O (q + 1) complexity
| (7) |
Φm varies from point to point, but they only depend on the sample distributions.
In comparison, conventional gridding method uses defined kernel functions Φ, for example, [14] uses a Gaussian function and successfully bounds the approximation error. Other feasible shift-invariant kernel functions include the cosine function, Kaiser-Bessel function, and PSWF, which are well summarized and compared in [11]. In particular, the Kaiser-Bessel kernel is most favorable by the MR community due to its minimized aliasing performance. The function is based on the zero-order modified Bessel function of the first kind and its inverse transform has a closed-form solution
| (8) |
Recently, a practical formula of the shape parameter β was provided by [12] that generates a near optimal Kaiser-Bessel window. The β value corresponds to optimized maximum aliasing energy. The central lobe of the inverse Fourier transform of the window extends slightly to the edge of the first stopband.
| (9) |
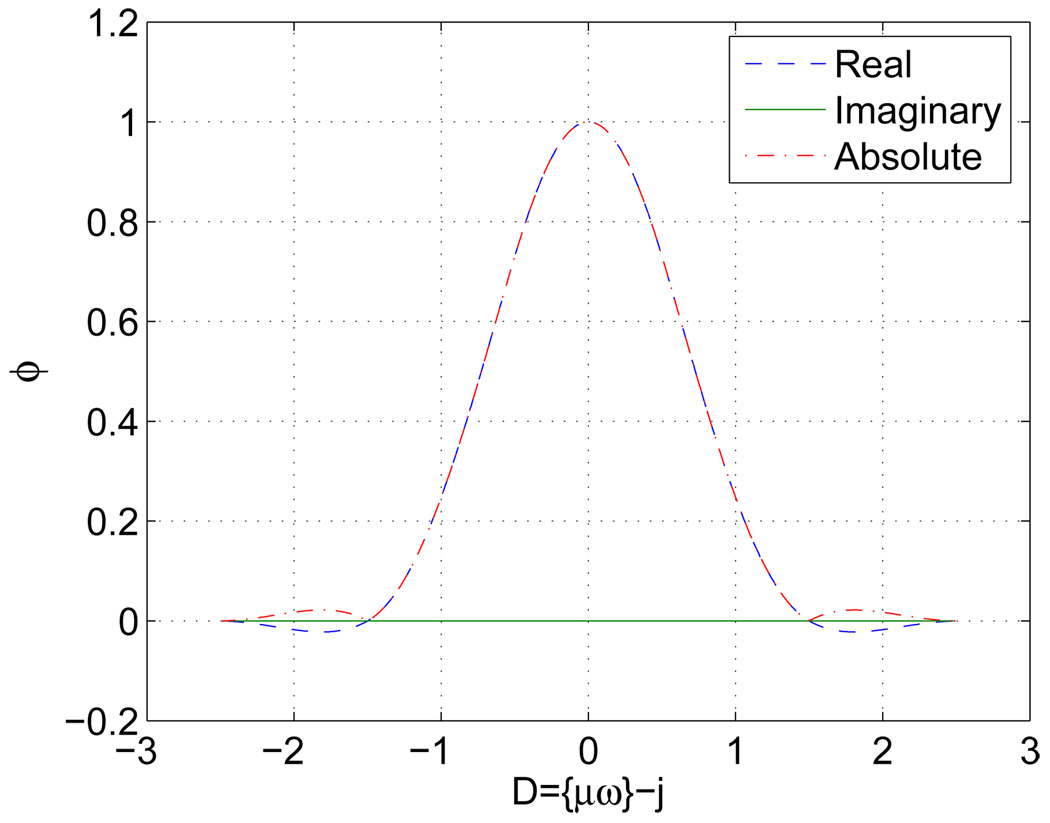
However, it should be point out here that although the LS-NUFFT kernels are “calculated” instead of “defined”, it is not “shift-variant” as claimed in [19]. In fact, α is a function of Dk(υ) = {μNpυ} + q/2 − k with k = 0, 1, 2, …, q. Since F−1 is independent of υ and only manipulates the weights obtained by α, ϕ, it is actually shift-invariant and complex in general. This contradicts with the “shift-variant” conclusion and the associated plots in [19]. In Fig. 1, ϕm of a 1D random sampling trajectory is plotted within D ∈ (−2.5, 2.5) corresponding to a kernel size of 5.
Fig. 1.
1D cosine LS-NUFFT complex kernel of width 5.
B. d-dimensional NUFFT-1 and NUFFT-2
With the calculated Φm, (2) can be computed efficiently with the d-dimensional NUFFT of the 1st-type (NUFFT-1). Here we define and use M consistently as the total number of non-Cartesian samples. The d-dimensional NUFFT-1 procedures are list below:
- Find the new Fourier coefficients on an oversampled uniform grid by kernel convolution
The complexity is O(M(q + 1)d).(10) Use the new Fourier coefficients to calculate the time signal samples hn by regular FFT at the cost of O(μd N log (μd N)).
Scale the resulting signal samples by the accuracy factor h̃n = hn/sn using O(N) operations.
Similarly, with the calculated Φm, the d-dimensional NUFFT of the 2nd-type (NUFFT-2) is evaluated by following steps:
Scale the uniform time signal samples by the accuracy factor g̃n = gn/sn. The complexity is O (N).
Calculate the DFT on an oversampled uniform grid by regular FFT. The complexity associated with this step is O (μd N log (μd N)).
- Find the nonuniform samples using kernel convolution (11) with a complexity of O (M(q + 1)d),
(11)
The conventional gridding method takes similar convolution, FFT and scaling procedures, except that the scaling function is found as the inverse Fourier transform of the defined kernel function. This division is usually called “deapodization” in conventional gridding method.
Since the sampling trajectories are usually repeatedly used in MRI applications, a look up table (LUT) technique is sometimes applied to store and look up the kernel weights for all the non-Cartesian sample points. With the increased interest in high dimension high resolution radial imaging, LUT weights storage implies large memory demands, and as a result, the on-the-fly weights calculation is often preferred. For this reason, the complexity of calculating Φk cannot be excluded. In this case, both NUFFT-1 and NUFFT-2 have a total complexity of O [C1 dM (q + 1) + M (q + 1)d + μd N log (μd N)] if the cosine scaling factor is used, and O [C2 dM N (q + 1) + M (q + 1)d + μd N log (μd N)] otherwise. For Kaiser-Bessel gridding, the complexity is O [C1 dM (q + 1) + M (q + 1)d + μd N log (μd N)]. In the above, C1, C2 and C3 are constants, but C3 involves the computation of a Bessel function so it is considerably larger than C1, C2.
C. Scaling factor and parameter selection
Similar to the conventional gridding method, the LS-NUFFT algorithms proposed also trade off between approximation accuracy and processing speed. In practical applications, this trade-off must be balanced by selecting optimal scaling factor and parameters μ, q.
The scaling factor is analogous to the deapodization function in the conventional gridding method. Therefore, for a particular given kernel function, its inverse Fourier transform could be utilized as the scaling factor. The LS-NUFFT kernel can then be found by least square solution. We compare LS-NUFFT and conventional gridding methods on 1D experiments. Gaussian kernel and Kaiser-Bessel kernel are used as examples. In the Gaussian case, the kernel and its inverse Fourier transform are C(k) = e−bk2 and , respectively. The Kaiser-Bessel kernel and its inverse Fourier transform are given in Eqn.8.
We constructed a 1D random sampling experiment to study the impact of scaling factor as well as the parameters. M = 256 random ωm locations are drawn from a uniform distribution over [−π, π]. The signal intensities are generated complex with the real and imaginary parts drawn from uniform distribution over [0, 1]. Fig. 2 shows the normalized L2 error (defined as where f0 is obtained by DFT) for the conventional gridding methods (shape parameters b = 0.247 and β = 11.441) and LS-NUFFT methods (μ = 2, q = 4). The results show that for a particular kernel function C(k) used in the conventional gridding method, using c(x) as a scaling factor and applying the LS-NUFFT framework will further improve the accuracy. This suggests that LS-NUFFT refines the kernel in an optimal least square sense. It is also noticed that the Kaiser-Bessel kernel with the given optimal β has been excellent in accuracy, so that the LS refinement provides relatively less significant improvement compared to the Gaussian case.
Fig. 2.
Comparison of normalized L2 errors using conventional gridding methods and LS-NUFFT methods.
On the other hand, adjusting parameters μ and q leverages the tradeoff between NUFFT accuracy and processing speed, similar to the conventional gridding. The error decreases exponentially with increasing m and q. However, the complexities of kernel calculation and convolution increases proportionally to (q + 1) and (q + 1)d, and the complexity of FFT increases proportionally to . Therefore, in this work, we use the accuracy-speed efficiency as the criteria to evaluate algorithms.
III. NON-CARTESIAN RECONSTRUCTION AND k-space SIMULATION
It is well known that the spatially encoded MRI signal can be interpreted as a Fourier transform of the spatial distribution of the initial transverse magnetization Mxy in spatial domain x,
| (12) |
Therefore, the image of the object can be reconstructed by taking inverse Fourier transform of the sampled signal in the spatial frequency space k, known as k-space,
| (13) |
Here we consider non-Cartesian acquisition trajectories, so the Fourier transforms above essentially connect the non-Cartesian k-space and Cartesian image space. The procedures of conventional gridding reconstruction methods include density compensation (also known as pre-weighting), convolution with a gridding kernel and resampling in k-space, and deapodization in image space,
| (14) |
where km represents the non-Cartesian samples and kl the regridded Cartesian samples, W(k) is the density compensation function, and C(k), c(x) are convolution kernel functions in k-space and image space, respectively. In this work, we implemented the widely used Kaiser-Bessel gridding method for comparison.
In our approach, we first discretize the integrands (12) and (13) and evaluate their discretized versions numerically using our multi-dimensional NUFFTs. A direct discretization of (12) leads to
| (15) |
where Δxp is the image voxel size in the pth dimension.
Assuming that a variable u exists and satisfies v = f (u), and the corresponding {um} is uniform, a change of variables from k to u is performed to (13) before discretization to minimize the discretization error
| (16) |
| (17) |
where |J(um)| is the determinant of Jacobian matrix and Δup is the sampling interval in the pth dimension.
Equation (15) can be efficiently computed with NUFFT-2 and used for k-space data simulation for computer phantom studies. Due to the negative sign in the exponent, the kernel function Ψm can be found as the conjugate of the one introduced in Section II.A, i.e., . Equation (17) is the non-Cartesian reconstruction formula that can be computed with NUFFT-1. Specifically, in 2D and 3D, the typical radially encoded k-space samples are located uniformly in polar coordinate and spherical coordinates as:
| (18) |
Fig. (3) shows the corresponding radially sampled 2D and 3D k-spaces given by (18). The radial sampling has a degree of freedom in the ray “end point” distribution, which in fact is an important factor for acquisition SNR and sampling efficiency (refer to the discussion section). However, in this work, the patterns described by (18) are chosen to be consistent with the historical PR technique and for the ease of calculating the pre-weighting functions. Therefore, the corresponding pre-weighting functions determined by Jacobian are r and r2 sin θ respectively. These lead (15) and (17) to
| (19) |
for 2D and
| (20) |
for 3D imaging applications.
Fig. 3.
2D and 3D radial sampling k-space samples.
IV. RECONSTRUCTION RESULTS
To test our multi-dimensional NUFFT reconstruction on radially sampled MR images, we first compared the method with the conventional Kaiser-Bessel gridding method on 2D and 3D computer-simulated phantoms and scanned phantoms. Then, we applied the proposed method to 2D and 3D radially encoded MRI scans of small animals.
A. Computer Simulation
For the computer phantom study, we adopted the Shepp-Logan phantom that is widely used by CT reconstruction researchers. The radial k-space data within this acquisition domain is simulated using (15) through NUFFT-2. Although theoretically the k-space is infinite, only a portion of it can be acquired in practice. The size of the acquisition domain (kmax) is decided directly by the desired image resolution as Δl = 1/kmax. To be consistent with Cartesian MRI, the number of samples in each ray is selected so that it satisfies the desired image field of view (FOV) according to Nr = kmax·FOV. The number of rays is typically selected such that Nyquist’s criterion is satisfied at the largest sample distance. This corresponds to Nθ = π Nr in 2D and in 3D. However, undersampled radial MR imaging using a reduced number of rays is of interest due to the emerging application of radial MRI in dynamic imaging, where fast acquisition is required [21][22].
The collected k-samples only occupy a finite region of infinite k-space. Due to the nonuniformity of sampling, a pre-weighting is needed. Truncation and pre-weighting both introduce errors to the reconstructed image. Lauzon et al.[23] described in detail the effects of 2D radial sampling. In this work, however, we only address the computational accuracy of the reconstruction with a given amount of k-space information and after pre-weighting. Therefore, we simulate the k-space data and reconstruct the image using NUDFT by evaluating the summations in (15) and (17) directly to provide the base for comparison. Normalized difference images and errors are defined based on the NUDFT results. However, one should note that the direct summation approach is very expensive at a complexity of O (M N). In the rest of the paper, We use the following abbreviations: the reconstructed image using direct NUDFT (I), simulated k-space data using direct NUDFT (s), NUFFT (·nu), and Kaiser-Bessel (·kb). For example, equations and figures use Inu for the image using the NUFFT method, and Ikb for the image using the Kaiser-Bessel gridding method. Simulated k-space data, s, snu, and skb also use these subscripts. The L2 error and L∞ error, e.g., for NUFFT reconstruction, are defined as
| (21) |
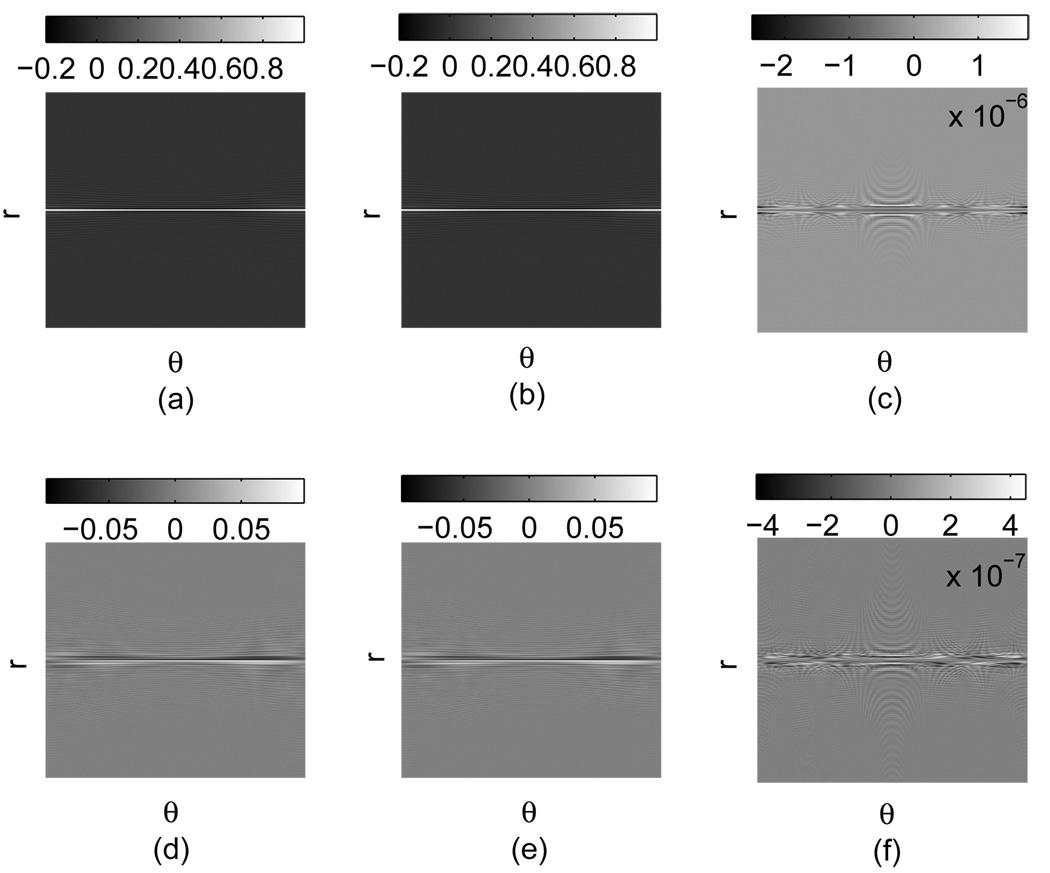
A 2D 128 × 128 Shepp-Logan phantom is first used for simulation. The k-space is sampled by 400 rays with 64 samples in each ray. Fig. 4 shows the comparison of the NUFFT-2 simulated k-space data and the true values. The normalized difference image is defined as the pixel-wise intensity distance between the two images divided by the norm of the comparison base. As a result, the norm of the difference image becomes exactly the L2 error. The simulation is of very high fidelity, for example, the L2 error of this case is 1.62 × 10−6.
Fig. 4.
k-space sample simulation results for a radially encoded Shepp-Logan phantom using NUFFT method with μ = 2 and q = 8. (a) Real part of the true k-space data ℜ(s). (b) Real part of the simulation using NUFFT-2 ℜ(snu). (c) ℜ(snu)−ℜ (s). (d) Imaginary part of the true k-space data 𝔍(s). (e) Imaginary part of the simulation using NUFFT-2 𝔍(snu). (f) 𝔍(snu)−𝔍(s).
With the simulated k-samples, the image is reconstructed using NUDFT, cosine LS-NUFFT, KB LS-NUFFT and conventional Kaiser-Bessel reconstruction methods. The cosine LS-NUFFT is included in the comparison because its speed advantage, although the cosine scaling factor is probably not optimal. According to the 1D study, KB LS-NUFFT has better accuracy but the direct evaluation of α reduces the computational speed.
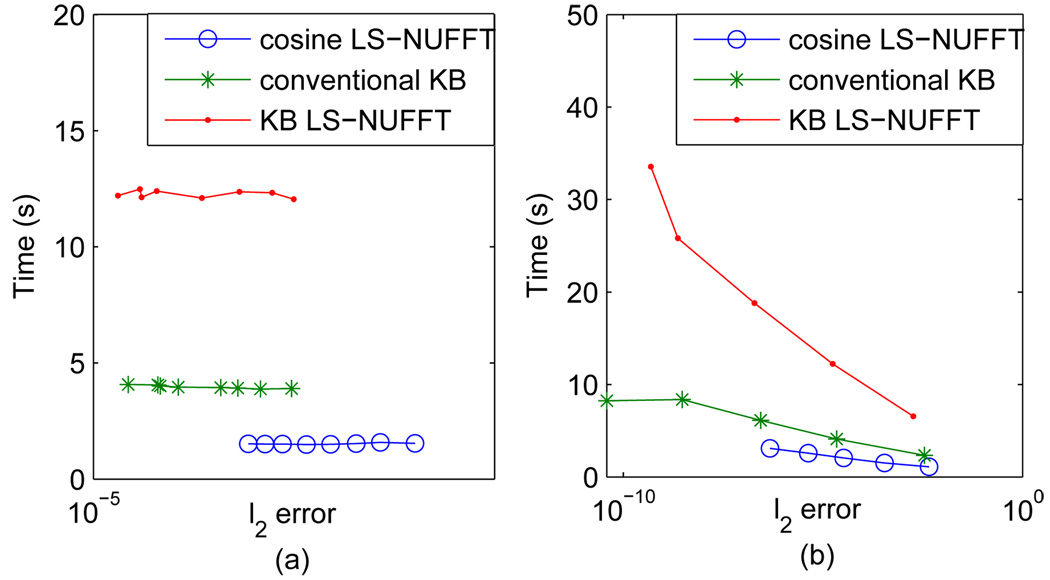
Fig. 5 shows the difference images. Both conventional KB and KB LS-NUFFT methods yield lower L2 error than the cosine LS-NUFFT, while the KB LS-NUFFT is slightly more accurate than the KB method. It should be noted that that the error of image (b) is mainly contributed by a DC offset, while in the other two images, the differences are in the background. On the other hand, a speed measure is also used for a accuracy-speed efficiency study. All of the reconstruction methods are implemented with MATLAB 7.0 on the same computer (CPU: AMD ATHLON 2600+; RAM: 2GB). The study of accuracy-speed efficiency is done by changing the oversampling rate and the kernel size shown by Fig. 6. As window size increases with a fixed oversampling rate of 2, the error in all the three methods decreases. Both charts in Fig. 6 suggest that if a certain error tolerance is set, the cosine LS-NUFFT reconstruction has a relatively higher accuracy-speed efficiency. In other words, it allows a smaller combination of μ and q to reach the criterion than the Kaiser-Bessel method, and consequently requires less processing time and memory.
Fig. 5.
Comparison of reconstruction results for a radially encoded Shepp-Logan phantom using Kaiser-Bessel gridding method and NUFFT method. Both methods use μ = 2 and q = 4 are used. (a) Difference image between DFT reconstruction and conventional KB reconstruction. I−Ikb. (b) Difference image between DFT reconstruction and cosine LS-NUFFT reconstruction. . (c) Difference image between DFT reconstruction and KB LS-NUFFT reconstruction. .
Fig. 6.
Accuracy-error curves of conventional Kaiser-Bessel gridding, cosine LS-NUFFT and KB LS-NUFFT methods. (a) Effect of changing μ and (b) effect of changing q are both provided.
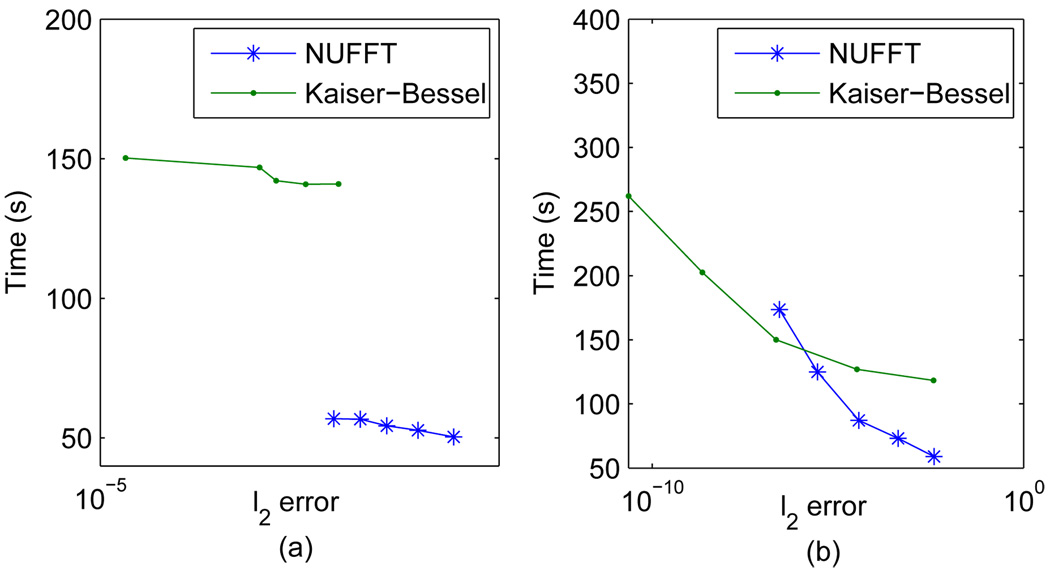
A 3D Shepp-Logan phantom of size 64×64×64 is also simulated. 20000 rays are required by Nyquist’s criterion with 32 samples in each ray. The image size is selected to be relatively small for the test case due to the high computational cost of the direct summation approach that is used for comparison here. However, due to its O (N M) complexity, the computation took a long time even at this small image size. Based on the 2D results, and given that speed will be more concerned in the 3D case, we included only the cosine LS-NUFFT and conventional KB method in the comparison. As in 2D, a few μ and q combinations are tested to study the accuracy-speed efficiency. The results are plotted as accuracy-speed curves in Fig. 7. Effects observed are generally consistent with the 2D study. On the right chart, the two curves cross each other between q = 8 and q = 10. This suggests that the L2 error of the conventional KB decays with q faster than the cosine LS-NUFFT. However, in 3D reconstruction, typically q = 4 is used to avoid high computational cost.
Fig. 7.
Accuracy-error curves of both conventional Kaiser-Bessel gridding and cosine LS-NUFFT methods. (a) Effect of changing μ and (b) effect of changing q.
B. Scanned Phantoms
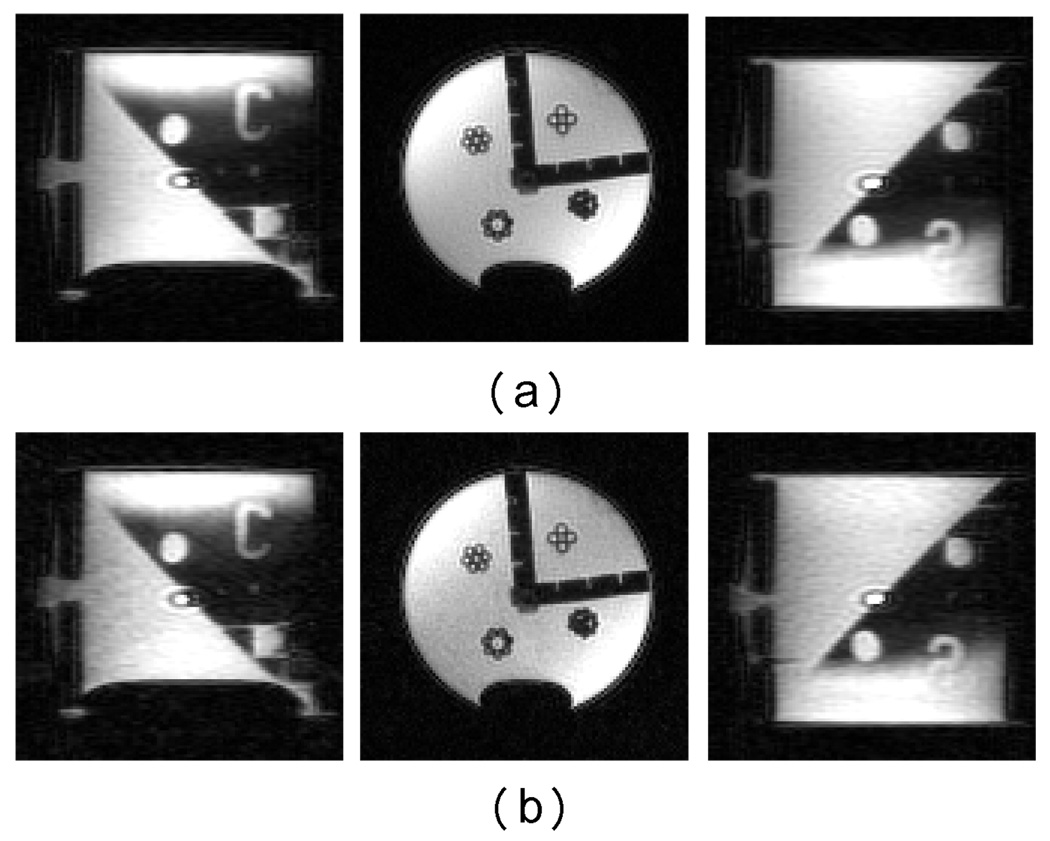
A cylindrical water tube phantom 3.5 cm in diameter and 4 cm in height was scanned on a 2 Tesla Oxford 30 cm horizontal magnet. The custom MRI sequence was driven by a GE EXCITE console. The phantom was filled with 0.05 mmol solution of copper sulfate (CuSO4[H2O]5) for shorter T1 and better proton signal. Both 2D and 3D radially encoded spin echo sequences were run on this phantom. The 2D scan excited a 2 mm slice with a 5 cm field of view. 800 rays with 160 samples in each ray were acquired. A 160×160 image was reconstructed at a spatial resolution of 0.3125 mm using both conventional gridding and cosine LS-NUFFT reconstruction methods. In Fig. 8, the reconstructed images are displayed in a γ-mode, i.e., the image intensity of each pixel is compressed using , as well as the normal mode. Here we use γ = 0.33, so that the noise level is relatively enhanced compared to using a linear colormap, for a better comparison of SNR. Although the 2D computer simulation study concludes that conventional KB yields smaller L2 error in most cases than the cosine LS-NUFFT, the visual qualities do not show much difference. In fact, the SNR is improved by 19.8% in this particular case using the LS-NUFFT reconstruction versus the gridding method. This again suggests that the error of LS-NUFFT may be mostly contributed by the DC offset but not the background. The reconstruction time was 1.61 s without preprocessing for NUFFT method and 3.44 s for gridding method, both of which are significantly shorter than the scan time.
Fig. 8.
2D radial image reconstructions of a water tube phantom scan. Both NUFFT and Kaiser-Bessel reconstruction methods are used with μ = 2 and q = 4. (a) NUFFT reconstruction. (b) γ-mode NUFFT reconstruction (γ = 0.33) (c) Gridding reconstruction. (d) γ-mode Gridding reconstruction (γ = 0.33).
The 3D scan used 40000 rays with 96 samples in each ray. An isotropic field of view (5 cm×5 cm×5 cm) with anisotropic resolution (Rx: Ry: Rz = 2 : 2 : 1) was acquired. To obtain this anisotropic resolution, the z-gradient was adjusted so that the k-space end points of these rays are located on the surface of an ellipsoid instead of a sphere. A 128 × 128 × 64 image was reconstructed using both NUFFT method and gridding method, as displayed in Fig. 9. Again, the SNR is improved by 12.4% using the NUFFT method.
Fig. 9.
3D radial image reconstructions of a water tube phantom scan. Both NUFFT and Kaiser-Bessel reconstruction methods are used with μ = 2 and q = 4. (a) NUFFT reconstruction. (b) Gridding reconstruction.
C. in vivo Radial Imaging
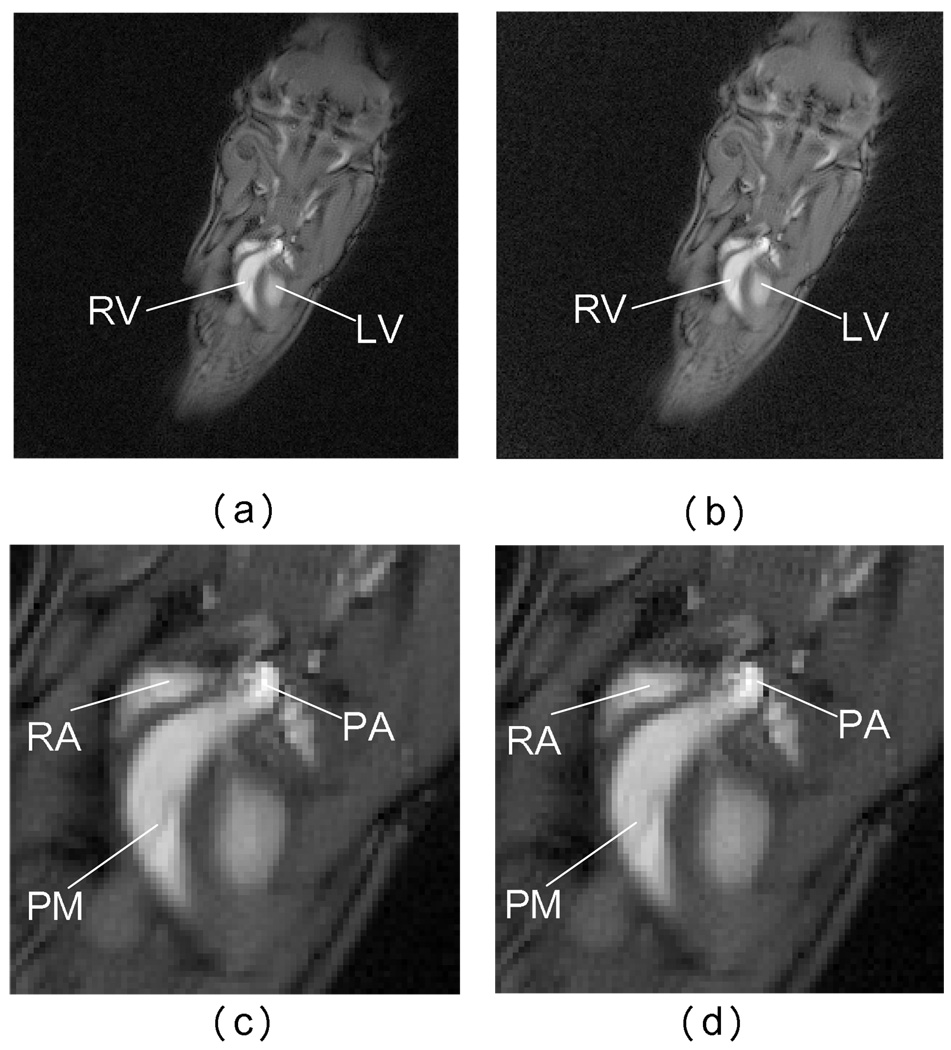
Radial sampling has been popularly used in cardiac imaging because of its robustness against motion and enhanced SNR, due to favorable signal averaging around the center k-space. 2D and 3D radially encoded data sets were acquired of a live mouse using a gated acquisition. Images were acquired on a 7T 21 cm bore Magnex system with a GE EXCIT (11.0) console identical to that on the above 2T system. The 2D radial scan acquired 16000 rays with 256 points on each ray and gave an oblique sagittal view of the whole mouse. The k-space was sampled with a much larger number of rays than required by the Nyquist criterion in order to gain the SNR improvement. The images were reconstructed using both the proposed NUFFT reconstruction and the Kaiser-Bessel gridding method in Fig. 10. In Fig. 10(a) and Fig. 10(b), γ mode (γ = 0.5) is used again to show the SNR benefit of NUFFT reconstruction. Fig. 10(c) and Fig. 10(d) are enlarged images of the heart region. Observe that the NUFFT reconstructed image is less noisy around some regions like the right atrium (RA) and pulmonary artery (PA), but still maintains the detailed structures such as the papillary muscle (PM). In fact, the SNR of Fig. 10(a) is approximately 25% higher than that of Fig. 10(b). Due to the large number of rays used, the pre-processing step that calculates the kernels consumes a relatively significant portion of the total reconstruction time for both methods. Therefore, NUFFT reconstruction is even more speed-advantageous since it uses closed-form solutions. In fact, for this particular example, NUFFT reconstruction method uses roughly 60% less time than the Kaiser-Bessel gridding method.
Fig. 10.
Reconstruction of a 2D radially encoded mouse cardiac image using Kaiser-Bessel gridding method and NUFFT method. In both methods, μ = 2 and q = 4 are used. (a) Image from NUFFT method in γ mode, γ = 0.5. (b) Image from Kaiser-Bessel gridding method in γ mode, γ = 0.5. (c) Zoomed in image of the heart region (using NUFFT reconstruction). (d) Zoomed in image of the heart region (using Kaiser-Bessel gridding method).(RV: right ventricle, LV: left ventricle, RA: right atrium, PA: pulmonary artery, PM: papillary muscle.
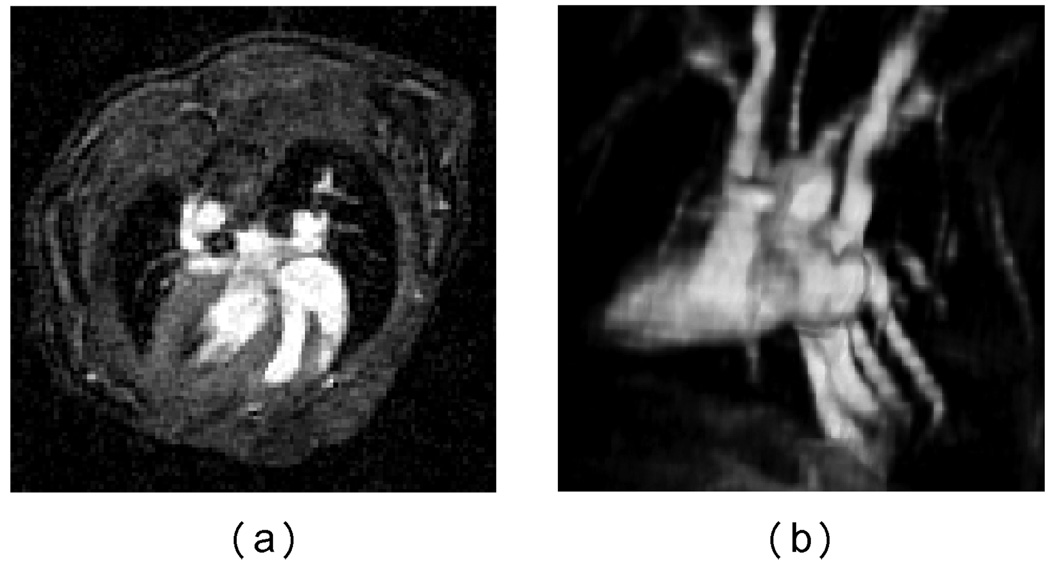
A 3D volume scan of the mouse was also conducted with radial sampling. 46080 rays were acquired and 196 samples were recorded in each ray, of which 64 samples are on the gradient ramp. The image was successfully reconstructed using our NUFFT method and the axial view and 3D volume rendered view highlighting the vasculature are shown in Fig. 11. The reconstructed volume is of size 256 × 256 × 64 with anisotropic resolution of Rx : Ry : Rz = 4 : 4 : 1.
Fig. 11.
NUFFT reconstruction of the 3D cardiac image of a mouse heart. (a) An axial view (b) A volume rendered 3D view of the heart vasculature.
V. DISCUSSION
The present work intends to suggest an alternative MRI reconstruction method for general non-Cartesian problems, and specifically describes its applications to 2D and 3D radially encoded MR imaging. Previously, most non-Cartesian reconstruction has been based on gridding methods using conventional kernels. The proposed LS-NUFFT method is in a framework consistent with the conventional gridding method. The key difference is that the proposed method uses a scaling factor and a least square approach to minimize the gridding error as opposed to the pre-defined kernels in conventional gridding method. It is found that the LS-NUFFT framework can be utilized to further refine those pre-defined kernel functions used in the conventional gridding approach. However, the LS kernel calculation, with many choices of scaling factors, might add on additional complexity. On the other hand, when a cosine scaling factor is used, the proposed method is computationally efficient, because the least square solution of the kernel weights is obtained through closed-form formulas rather than matrix manipulations. The high accuracy-speed efficiency makes the method a potential solution for high-dimensional high resolution non-Cartesian reconstruction problems.
However, we need to point out that both gridding and LS-NUFFT methods only deal with the approximation and fast computation of DFT. Even the DFT reconstructed images significantly suffer from ringing artifacts, known as truncation error or the Gibbs ringing phenomenon. The ringing phenomenon is more severe in low-resolution cases. Although we did not discuss the problem specifically in this paper, the issue has been addressed in a previous work [24] and a ringing reduction technique has been developed in this work based on our NUFFT k-space simulation and reconstruction tools. These ideas are straightforward to extend to 3D radial encoding. In addition, references [25][26] recently proposed a pre-weighting design method in the gridding reconstruction framework to sharpen the point spread function of PR imaging. We feel its integration would potentially improve our method as well.
In addition to reconstruction accuracy, image quality will also be affected by the k-space sampling strategy. The proposed reconstruction method has enabled us to consider more efficient radial sampling strategies beyond the conventional PR sampling distributions [2][3] where samples are constrained on radii as well as on latitudinal rings. Isotropic radial sampling that distributes the ray “end points” uniformly on a sphere has the potential to improve SNR, reduce scan time, and enhance reconstruction performance. Recently, some distributions more isotropic than the conventional PR sampling were explored by adopting geomathematical methods [7][13]. Isotropic sampling pattern that has the potential to reduce the number of rays, which directly corresponds to a reduced scan time, and thus is critical to dynamic imaging. Further investigation is underway.
VI. CONCLUSIONS
Multi-dimensional NUFFT-1 and NUFFT-2 have been developed as fast and accurate evaluations of the NUDFTs using least square kernels. The LS-NUFFT framework is proposed as an alternative approach to conventional gridding method. It calculates kernel functions based on a selected scaling factor, thus has the potential to yield lower L2 error than using pre-defined kernel functions. Choice of scaling factor, oversampling rate and kernel size determines the accuracy-speed efficiency of the algorithm. The cosine LS-NUFFT is efficient to compute and has a favorable accuracy-speed performance under most parameter choices. In this work, MRI k-space data simulation and image reconstruction algorithms are developed. The LS-NUFFT algorithms are implemented in 2D and 3D to facilitate the application of radially encoded MR imaging. The Kaiser-Bessel gridding method is also implemented for comparison. The 2D computer simulation suggests that KB LS-NUFFT yields lowest L2 error among the three methods compared. Both 2D and 3D find improved accuracy-speed performance using the cosine LS-NUFFT reconstruction method. The cosine LS-NUFFT method is successfully applied to 2D and 3D cardiovascular imaging on small animals.
ACKNOWLEDGMENT
The authors would like to thank Dr. Bastiaan Driehuys for providing the 2D in vivo hyperpolarized imaging data and Elizabeth Bucholz for providing the 3D cardiac imaging data. The research was performed at Duke Department of Electrical and Computer Engineering and supported by NIH through grant 5R21CA114680. The MRI scans were performed at the Duke Center for In Vivo Microscopy, an NCRR/NCI National Resource (P41 RR005959/R24 CA092656).
REFERENCES
- 1.Lauterbur PC. Image formations by induced local interactions: examples emplying nuclear magnetic resonance. Nature. 1973;vol. 242:190–191. [PubMed] [Google Scholar]
- 2.Lauterbur PC, Lai CM. Zeugmatography by reconstruction from projections. IEEE Trans. Nuclear Science. 1980;vol. 27:1227. [Google Scholar]
- 3.Lai CM. True three-dimensional nuclear magnetic resonance imaging by fourier reconstruction zeugmatrography. Journal of Applied Physics. 1981;vol. 52:1141–1145. [Google Scholar]
- 4.Golver GH, Pauly JM. Projection reconstruction technique for reduction of motion effects in MRI. Magnetic Resonance in Medicine. 1986;vol. 4:359–376. doi: 10.1002/mrm.1910280209. [DOI] [PubMed] [Google Scholar]
- 5.Gewalt SL, Glover GH, Hedlund LW, Cofer GP, MacFall JR, Johnson GA. MR microscopy of the rat lung using projection reconstruction. Magnetic Resonance in Medicine. 1993;vol. 29:99–106. doi: 10.1002/mrm.1910290117. [DOI] [PubMed] [Google Scholar]
- 6.Cremillieux Y, Briguet A, Deguin A. Projection reconstruction methods: fast imaging sequences and data processing. Magnetic Resonance in Medicine. 1994;vol. 32:23–32. doi: 10.1002/mrm.1910320105. [DOI] [PubMed] [Google Scholar]
- 7.Johnson GA, Cofer GP, Hedlund LW, Maronpot RR, Suddarth SA. Registered 1h and 3he magnetic resonance microscopy of the lung. Magnetic Resonance in Medicine. 2001;vol. 45:365–370. doi: 10.1002/1522-2594(200103)45:3<365::aid-mrm1047>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Schomberg H, Timmer J. The gridding method for image reconstruction by Fourier transformation. IEEE Transactions on Medical Imaging. 1995;vol. 14:596–607. doi: 10.1109/42.414625. [DOI] [PubMed] [Google Scholar]
- 9.Rasche V, Sinkus R, Proska R, Boernert P, Eggers H. Resampling of data between arbitrary grids using convolution interpolations. IEEE Transactions on Medical Imaging. 1999;vol. 18(no 5):385–292. doi: 10.1109/42.774166. [DOI] [PubMed] [Google Scholar]
- 10.Sulllivan JO. A fast sinc function gridding algorithm for Fourier inversion in computed tomography. IEEE Trans. Medical Imaging. 1985;vol. MI-4(no 4):200–207. doi: 10.1109/TMI.1985.4307723. [DOI] [PubMed] [Google Scholar]
- 11.Jackson J, Meyer CH, Nishimura DG, Macovski A. Selection of a convolution function for Fourier inversion using gridding. IEEE Transactions on Medical Imaging. 1991;vol. 10(no 1):473–478. doi: 10.1109/42.97598. [DOI] [PubMed] [Google Scholar]
- 12.Beatty PJ, Nishimura DG, Pauly JM. Rapid gridding reconstruction with a minimal overampling ratio. IEEE Trans. Medical Imaging. 2005;vol. 24(no 6):799–808. doi: 10.1109/TMI.2005.848376. [DOI] [PubMed] [Google Scholar]
- 13.Lethmate R, Wajer F, Cremillieux Y, Ormondt DV, Graveron-Demilly D. Dynamic mr-imaging with radial scanning, a post-acquisition keyhole approach. EURASIP Journal on Applied Signal Processing. 2003;vol. 5:405–412. [Google Scholar]
- 14.Dutt A, Rokhlin V. Fast Fourier transforms for nonequispaced data. SIAM J. Sci. Comp. 1993;vol. 14:1368–1393. [Google Scholar]
- 15.Beylkin G. On the fast Fourier transform of functions with singularities. Appl. Computat. harmonic Anal. 1995;vol. 2:363–382. [Google Scholar]
- 16.Liu QH, Nguyen N. An accurate algorithm for nonuniform fast Fourier transform (NUFFT’s) IEEE Microwave and Guided Wave Letters. 1998;vol. 8(no 1):18–20. [Google Scholar]
- 17.Nguyen N, Liu QH. The regular Fourier matrices and nonuniform fast Fourier transforms. SIAM J. Sci. Compt. 1999;vol. 21(no 1):283–293. [Google Scholar]
- 18.Fessler JA, Sutton BP. Nonuniform fast Fourier transform using min-max interpolation. IEEE Trans. Signal Process. 2003;vol. 51:560–574. [Google Scholar]
- 19.Sha L, Guo H, Song AW. An improved gridding method for spiral MRI using nonuniform fast Fourier transform. J. Mag. Reson. 2003;vol. 162:250–258. doi: 10.1016/s1090-7807(03)00107-1. [DOI] [PubMed] [Google Scholar]
- 20.Sutton BP, Fessler JA, Noll D. A min-max approach to the nonuniform N-D FFT for rapid iterative reconstruction of MR images. Proc. Intl. Soc. Mag. Res. Med. 2001:763. [Google Scholar]
- 21.Vigen KK, Peters DC, Grist TM, Block WF, Mistretta CA. Undersampled projection-reconstruction image for time-resolved contrast-enhanced imaging. Magnetic Resonance in Medicine. 2000;vol. 43:170–196. doi: 10.1002/(sici)1522-2594(200002)43:2<170::aid-mrm2>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 22.Peters DC. Undersampled projection reconstruction applied to MR angiography. Magnetic Resonance in Medicine. 2000;vol. 43:91–101. doi: 10.1002/(sici)1522-2594(200001)43:1<91::aid-mrm11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 23.Lauzon ML, Rutt BK. Effects of polar sampling in k-space. Mag. Res. Med. 1996;vol. 36:940–949. doi: 10.1002/mrm.1910360617. [DOI] [PubMed] [Google Scholar]
- 24.Song J, Liu QH. Improving non-cartesian mri reconstruction through discontinuity subtraction. Intl J. Biomed. Imag. 2006 doi: 10.1155/IJBI/2006/87092. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pipe JG. Sampling density compensation in MRI: rationale and an iterative numberical solution. Magnetic Resonance in Medicine. 1999;vol. 41:179–186. doi: 10.1002/(sici)1522-2594(199901)41:1<179::aid-mrm25>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 26.Pipe JG. Reconstructing MR images from undersampled data: data-weighting considerations. Magnetic Resonance in Medicine. 2000;vol. 43:867–875. doi: 10.1002/1522-2594(200006)43:6<867::aid-mrm13>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]