Abstract
Tryptophan hydroxylase-2 (TPH2) is a recently identified TPH isoform responsible for neuronal serotonin (5-HT) synthesis, and TPH2 polymorphisms are associated with a range of behavioral traits and psychiatric disorders. This study characterized cis-acting elements and three common polymorphisms (−703G/T, −473T/A, and 90A/G) in the 5′ regulatory region of human TPH2 by using luciferase reporter assay, quantitative real-time PCR, and electrophoretic mobility shift assay (EMSA). The core promoter of human TPH2 was localized to the region between −107 and +7, and the segment of +8 to +53 within the 5′-UTR was found to exert a potent inhibitory effect on gene expression at both transcriptional and post-transcriptional levels. In both RN46A and HEK-293 cell lines, the TTA (−703T/−473T/90A) haplotype of the three polymorphisms showed the lowest gene expression compared with other haplotypes, and the −703G/T and −473T/A polymorphisms tended to exert a synergic effect on gene expression dependent upon the sequence of the 5′-UTR. In RN46A, the 90A/G polymorphism significantly increased luciferase activity and mRNA level irrespective of the other two polymorphisms, while in HEK-293 cells the effect of 90A/G was dependent on the alleles at loci −703 and −473. EMSA showed that all the three polymorphisms potentially alter DNA–protein interactions, while the 90A/G polymorphism predictably alters the 5′-UTR secondary structure of mRNA and influences RNA–protein interactions. In conclusion, our present study demonstrates that both the 5′-UTR and common polymorphisms (especially the 90A/G) in the 5′ regulatory region of human TPH2 have a significant impact on gene expression.
Introduction
Serotonin (5-HT) is a major central neurotransmitter involved in many brain functions and neuropsychiatric disorders, and pharmaceuticals that target the 5-HT system are widely used for the treatment of various psychiatric diseases. Tryptophan hydroxylase (TPH) is the first and rate-limiting enzyme for 5-HT synthesis. It was thought that TPH was derived from a single gene (now referred to as TPH1) until a second TPH isoform (TPH2) was recently described (Walther et al. 2003a, 2003b; Côté et al. 2003). While the classical TPH1 is primarily expressed in the periphery, TPH2 is predominantly expressed in the brain by a distinct TPH2 gene and is responsible for neuronal 5-HT synthesis (Walther et al. 2003b; Côté et al. 2003; Zhang et al. 2004; Zill et al. 2004b). Hence, TPH2 has received much attention for its role in the regulation of 5-HT neurotransmission, as well as in the pathophysiology of various neuropsychiatric disorders related to 5-HT dysfunction.
The association of TPH2 genetic variance with neuropsychiatric disorder incidence has been widely investigated. Specific polymorphisms in human TPH2 have reported association with major depression (Van Den Bogaert et al. 2006; Zhang et al. 2005; Zhou et al. 2005a; Zill 2004a), affective disorders (Harvey et al. 2004; Lopez et al. 2007), suicidality (Van Den Bogaert et al. 2006; Lopez et al. 2007; Lopez de Lara et al. 2007; Jollant et al. 2007; Ke et al. 2006; Zill et al. 2004c), autism (Coon et al. 2005), early onset obsessive-compulsive disorder (Mossner et al. 2006), attention deficit hyperactivity disorder (ADHD; Sheehan et al. 2005; Walitza et al. 2005), panic disorder (Maron et al. 2007), chronic fatigue syndrome (Goertzel et al. 2006; Smith et al. 2006), and Tourette syndrome (Mossner et al. 2007). Of the previously described polymorphisms, a nonsynonymous single nucleotide polymorphism (SNP) named 1463G/A (441R/H) was a focus of investigation because it causes ∼80% loss of 5-HT production in vitro and correlates to unipolar major depression (Zhang et al. 2005); However, this SNP is extremely rare and has not yet been replicated by subsequent studies in large populations (Bicalho et al. 2006; Blakely et al. 2005; Delcorme et al. 2006; Glatt et al. 2005; Van Den Bogaert et al. 2005; Zhou et al. 2005b). Soon after, a common promoter polymorphism −703G/T (rs4570625) was reported to be associated with ADHD (Walitza et al. 2005), amygdala responsiveness (Brown et al. 2005; Canli et al. 2005), emotional processing (Herrmann et al. 2007), personality traits and disorders related to emotional dysregulation (Gutknecht et al. 2007; Reuter et al. 2007), while another common polymorphism −473T/A (rs11178997) in the 5′-regulatory region was associated with ADHD (Walitza et al. 2005), and unipolar and bipolar disorder (Van Den Bogaert et al. 2006). Accordingly, genetic variance in the 5′-regulatory region may play an important contributory role across a wide spectrum of neuropsychiatric disorders.
The expression of TPH2 in brain exhibits a circadian rhythm and is influenced by specific stressors and hormones (Brown et al. 2006; Chamas et al. 2004; Clark et al. 2005; Hiroi et al. 2006; Liang et al. 2004; Malek et al. 2005, 2007; Sanchez et al. 2005), suggesting that TPH2 gene expression may be frequently and subtly regulated under specific physiological or stress conditions. This characteristic feature of TPH2 supports the notion that 5-HT is involved in the function of the hypothalamic–pituitary–adrenal (HPA) axis (Dinan et al. 1996; Leonard et al. 2005; Lowry et al. 2002), a critical neuroendocrine system that responds to stress and also shows a circadian rhythm. The dysfunction of this system is implicated in numerous psychiatric diseases. Thus, genetic polymorphisms or other cis-acting factors affecting TPH2 gene expression might result in the alteration of HPA axis function or other physiological processes related to 5-HT. Indeed, we have previously reported that functional polymorphisms in rhesus monkey TPH2 3′-UTR are strikingly associated with HPA axis function, whereas a common polymorphism 1952G/A (rs17110747) in the human TPH2 3′-UTR does not alter in vitro gene expression (Chen et al. 2006). Recently, two papers regarding TPH2 gene expression regulation were published, of which one reported a functional significance of the −473T/A polymorphism (Scheuch et al. 2007), while the other characterized a negative motif for REST/NRSF in the upstream of the 5′-UTR (Patel et al. 2007). In the present study, we characterized cis-acting elements in the human TPH2 5′ regulatory region, as well as the functional significance of three previously described common SNPs in this region, including the two above-mentioned promoter SNPs (−703G/T and −473T/A) and another previously reported 90A/G SNP (rs11178998) in the 5′-UTR.
Materials and methods
Construction of reporter plasmids for assessing the TPH2 5′ regulatory region
Primers for cloning and mutagenesis (Table 1) were designed based on the genomic and mRNA sequences of human TPH2 (GenBank accession no. AC090109 and NM_173353, respectively). A 1619-bp fragment upstream from the ATG initiation codon was first amplified from human genomic DNA (Promega, Madison, WI, USA) using KpnI-hTPH2(−1478)F and XhoI-hTPH2(+141)R primers. Following sequential digestion by KpnI and XhoI, the PCR product was cloned into the pGL4.14 reporter vector (Promega) and the resultant construct was then used as the template to generate other constructs containing serial 5′ and 3′ deletion or specific mutations. Primers with various 5′ start and 3′ end sites were used for the cloning of serial deletion fragments. The −703G/T, −473T/A and 90A/G variants were generated by using a QuickChange® Site-Directed Mutagenesis Kit (Stratagene, LA Jolla, CA, USA). For the three polymorphisms, all eight theoretic haplotypes (TTA, GAA, GTA, TAA, TTG, GAG, GTG and TAG, named H1 to H8 respectively for abbreviation, shown in detail in Fig. 2a) with different alleles at the polymorphic sites were constructed by using the insert spanning −1478 to +141. All constructs were sequence-verified and correct orientation was confirmed.
Table 1.
Primers used for cloning and mutagenesis
| Name | Sequence |
|---|---|
| Cloninga | |
| KpnI-hTPH2(−1478)F | 5′-gcctggtacctgattagagctgacccttacaga-3′ |
| KpnI-hTPH2(−792)F | 5′-cagaggtacctgcatagaggcatcacagga-3′ |
| KpnI-hTPH2(−615)F | 5′-gataggtaccagtgttcgggagcacaata-3′ |
| KpnI-hTPH2(−219)F | 5′-agatggtaccttgtacaaataacaccccct-3′ |
| KpnI-hTPH2(−135)F | 5′-atcaggtaccgagcacagataaccccaggc-3′ |
| KpnI-hTPH2(−107)F | 5′-gcttggtacctgtaatctgactgtggccatc-3′ |
| KpnI-hTPH2(+1)F | 5′-ggagggtacccattgctcttcagcaccag-3′ |
| XhoI-hTPH2(−67)R | 5′-gagtctcgagaaactcatttctggttgccg-3′ |
| XhoI-hTPH2(+7)R | 5′-tgtgctcgagagcaatgcgaatcctcccat-3′ |
| XhoI-hTPH2(+53)R | 5′-agagctcgaggatcagctgcctgcttgg-3′ |
| XhoI-hTPH2(+141)R | 5′-ctggctcgagggatcccggtgtaatattctttc-3′ |
| hTPH2(+1)F | 5′-cattgctcttcagcaccagg-3′ |
| hTPH2(+61)F | 5′-ccttcctctcaatctccg-3′ |
| hTPH2(+141)R | 5′-ggatcccggtgtaatattctttc-3′ |
| Mutagenesis | |
| hTPH2(−703T)-F | 5′-cacacatttgcatgcacaaaattaTaatatgtcaagtcag-3′ |
| hTPH2(−703T)-R | 5′-ctgacttgacatattAtaattttgtgcatgcaaatgtgtg-3′ |
| hTPH2(−473A)-F | 5′-ccttatttgatcattacacaAtgtacgcttgtgtc-3′ |
| hTPH2(−473A)-R | 5′-gacacaagcgtacaTtgtgtaatgatcaaataagg-3′ |
| hTPH2(+90G)-F | 5′-aatctccgccagcgctgctGctgcccctctagtacc-3′ |
| hTPH2(+90G)-R | 5′-ggtactagaggggcagCagcagcgctggcggagatt-3′ |
Underlined nucleotides were designed to introduce a restriction site of KpnI or XhoI; upper case denotes variant nucleotide
F forward, R reverse
Numbers in parentheses represent the start sites of cloning primers or polymorphic sites for mutagenesis oligos
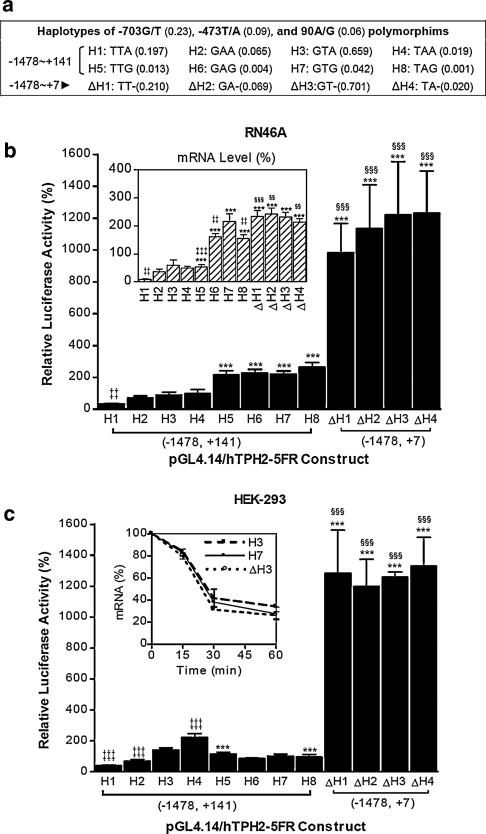
Fig. 2.
Haplotypes of the common polymorphisms in human TPH2 5′ regulatory region (a) and their effect on reporter gene expression in RN46A (b) and HEK−293 (c). Numbers in parentheses following the polymorphisms and haplotypes represent the documented variant allele frequency (Zhou et al. 2005) and expected incidence in the population, respectively. Expected incidences of the haplotypes were calculated regardless of linkage disequilibrium. Relative luciferase activities driven by the haplotypes are shown as means ± SE. The relative luciferase mRNA levels in RN46A are shown as means ± SE (inset in b). ***P < 0.001 compared with the 90A haplotype with identical allele at −703 and −473 loci (KWG vs. KWA, K-G/T, W–T/A); ‡‡P < 0.01, ‡‡‡P < 0.001 compared with the −703G/−473T haplo-type with the identical allele at locus 90 (KWA vs. GTA or KWG vs. GTG); §§§P < 0.001 compared with the 90G haplotype with identical allele at −703 and −473 loci (KW- vs. KWG). Repeated measures ANOVA was used for the comparisons. For the analysis of mRNA stability, firefly luciferase mRNA levels of three representative haplotype constructs (H3, H7 and ΔH3) in HEK-293 were measured at 0, 15, 30 and 60 min after treatment with 100 μM of DRB, and are expressed relative to the value at 0 min (inset in c)
Cell Culture
RN46A and HEK-293 cells were employed in the present study. RN46A is a serotonergic cell line derived from embryonic day 13 rat medullary raphe cells (White et al. 1994). The HEK-293 cell line, which is derived from primary cultures of human embryonic kidney cells, encodes many proteins normally expressed in neuronal lineage cells and reportedly displays 5-HT immunoreactivity (Shaw et al. 2002). By using real-time PCR, we found that TPH2 mRNA is expressed in RN46A, but not in HEK-293 (data not shown). HEK-293 cells were cultured in Dulbecco's Modified Eagle's Medium containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg streptomycin and 0.1 mM non-essential amino acids in an atmosphere of 5% CO2 at 37°C, while RN46A cells were maintained in Neurobasal™ Medium (Invitrogen, Carlsbad, CA, USA) complemented with 8% heat-inactivated FBS and 0.05% l-glutamine at 33°C in a 5% CO2 incubator.
Transient transfection and luciferase assays
The day before transfection, the cells were seeded in 24-well plates at approximately 2 × 105 cells/well. Transfections for RN46A and HEK-293 cells were performed by using Lipofectamine™ 2000 (Invitrogen) and ProFection® Mammalian Transfection System (Promega) respectively, according to the manufacturer's protocol. For each well, 0.8 μg of the construct was transfected along with 0.2 μg of pRL-TK renilla luciferase reporter vector as the control to determine transfection efficiency. The transfected cells were maintained in culture and the growth medium was changed 4−6 h after transfection. The cells were harvested 24 h after transfection, and the activity of firefly and renilla luciferase was measured using Dual Luciferase Reporter Assay System (Promega) on a Victor3™V Multilabel Counter (Wallac-PerkinElmer, Turku, Finland). Transfections and assays were performed in triplicate at least three times, with different DNA preparations of the same clone to ensure validity. The normalized luciferase data (firefly/renilla) were used to perform the statistics and are expressed relative to pGL4.14.
Quantative real-time PCR (qRT-PCR) for mRNA analysis
Cells were seeded in 6-well plates at approximately 5 × 105 cells/well and transfections were performed the next day using 2.4 μg of construct plus 0.6 μg of pRL-TK per well. For the comparisons of mRNA level between haplotypes, the cells were lysed at 24 h after transfection by the addition of 800 μl of TRIzol Reagent (Invitrogen) per well and total RNA was isolated. For the analysis of mRNA stability, 5,6-dichlororibofuranosyl benzimidazole (DRB, 100 μM; Sigma-Aldrich, St Louis, MO, USA) was added at 24 h after transfection and cells were lysed at 0, 15, 30 and 60 min after DRB treatment, followed by isolation of total RNA. Total RNA was then reverse transcribed into cDNA using Superscript™ III reverse transcriptase and oligo-dTs (Invitrogen). To avoid DNA contamination, RNA samples were treated with RQ1 RNase-free DNase I (Promega) for 1 h at 37°C. The synthesized cDNA was diluted to 50 ng/μl for further use. Real-time PCR was performed on a Roche LightCycler 2.0 system (Roche Diagnostics, Indianapolis, IN, USA) using 50 ng of cDNA in each reaction. A Taqman® Master kit in combination with the Universal Probe Library (Human) was used to assess firefly gene expression (generously supplied by Roche Diagnostics, Indianapolis, IN, USA and Penzberg, Germany). Primer set Luc2-#20F/R (Table 2) for the qRT-PCR was designed using the Probe Library Assay Design Center (http://www.roche-applied-science.com/sis/rtpcr/upl/adc.jsp). Quantification of the firefly luciferase mRNA was based on an external standard curve using serial 10-fold dilutions (107−100 copies) of pGL4.14. The co-transfected renilla luciferase mRNA levels were determined by SYBR Green RT-PCR using primer set Rluc-F1/R1 (Table 2) with serial dilutions of pRL-TK as the external standard. The PCR reactions were run in triplicate and the entire experiment was replicated. For the comparisons of gene expression between haplotypes, normalized mRNA levels (firefly/renilla) were used to perform the statistics and then expressed relative to pGL4.14, while for the mRNA stability analysis, the firefly mRNA levels were expressed relative to the values without DRB treatment.
Table 2.
Oligos used for EMSA and qRT-PCR
| Name | Position | Sequence |
|---|---|---|
| aEMSA (probes and competitors) | ||
| hTPH2(+90)A/G-F | +72 to +106 | 5′-aatctccgccagcgctgctA/Gctgcccctctagtacc-3′ |
| hTPH2(+90)A/G-R | +106 to +72 | 5′-ggtactagaggggcagT/Cagcagcgctggcggagatt-3′ |
| hTPH2(−473)T/A-F | −494 to −451 | 5′-accttatttgatcattacacaT/Atgtacgcttgtgtcaaaatatc-3′ |
| hTPH2(−473)T/A-R | −451 to −494 | 5′-gatattttgacacaagcgtacaA/Ttgtgtaatgatcaaataaggt-3′ |
| hTPH2(−703)G/T-F | −725 to −681 | 5′-cacatttgcatgcacaaaattaG/Aaatatgtcaagtcagaaaaagc-3′ |
| hTPH2(−703)G/T-R | −681 to −725 | 5′-gctttttctgacttgacatattC/Ttaattttgtgcatgcaaatgtg-3′ |
| qRT-PCR (primers) | ||
| bLuc2-#20F | 1575−1594 | 5′-catgaccgagaaggagatcg-3′ |
| bLuc2-#20R | 1635−1617 | 5′-cagcttcttggcggttgta-3′ |
| Rluc-F1 | 1075−1094 | 5′-gataactggtccgcagtggt-3′ |
| Rluc-R1 | 1297−1278 | 5′-accagatttgcctgatttgc-3′ |
The complementary single-strand oligos (end with F and R, respectively) were annealed to generate double-strand probes or competitors. Upper case nucleotides following the polymorphic sites represent either allele. Positions of the qRT-PCR primers for firefly (luc2) and renilla (Rluc) luciferase mRNA quantifications are based on the sequences of pGL4.14 (AY864928) and pRL-TK (AF025846), respectively
Each probe and its corresponding competitor for the same EMSA reaction share the same sequence, but differ in whether or not the 3′-end is biotinylated
Number following the “#” represents the probe number according to the Universal ProbeLibrary for Human (Roche)
In vitro transcription
The full-length (+1 to+141) and partial (+61 to +141) TPH2 5′-UTR constructs containing 90A/G alleles were amplified by using primer sets hTPH2(+1)F/hTPH2 (+141)R and hTPH2(+61)F/hTPH2(+141)R (Table 1), respectively, and the purified PCR products were then cloned into pGEM-T (Promega). Following the verification of the correct direction by sequencing, the constructs were linearized by NotI. In vitro transcription reactions with the purified linearized templates were carried out with a MAXIscript® T7 Kit (Ambion, Austin, TX, USA) in the presence or absence of biotinylated UTP (Ambion) according to the manufacturer's instructions. The mRNA products were purified with Ambion's MEGAclear™ kit and an aliquot of each was run on 5% native or denaturing (8 M urea) acrylamide gels in 0.5 × TBE, followed by detection with ethidium bromide (unlabeled) or chemiluminescence (biotinylated).
Electrophoretic mobility shift assay
Nuclear and cytoplasmic extracts from RN46A and HEK-293 cells were prepared by using NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Pirece, Rockford, IL, USA). For DNA-EMSA (DNA-electrophoretic mobility shift assay), both 3′-biotinylated probes and unlabeled competitors (Table 2) were synthesized by Alpha DNA (Montreal, Canada). To make double-stranded probes and competitors, complementary oligos of equal amount were heated at 95°C for 5 min and then annealed for 1 h at room temperature (RT). Binding reactions were carried out for 20 min at RT in the presence of 50 ng/μl poly(dI-dC), 0.05% NP-40, 5 mM MgCl2, 5 mM EDTA, and 2.5% glycerol in 1 × binding buffer (LightShift™ chemiluminescent EMSA kit, Pierce) using 0.05−0.20 pmol of probes and ∼20 μg of nuclear or cytoplasmic extract. For the competition experiments, unlabeled 100× competitor was incubated with the nuclear extract for 10 min before the addition of the labeled probe. Assays were loaded onto 6% native acrylamide gels in 0.5× TBE buffer and electrophoresed at 80 V before being transferred onto a ZetaProbe® membrane (Bio-Rad Laboratories, Hercules, CA, USA). Transferred DNAs were cross-linked to the membrane and detected using horseradish peroxidase-conjugated streptavidin and an LAS-3000 imaging system (Fujifilm Life Science, New Haven, CT, USA).
RNA-EMSA was performed as previously described with minor modification (Li et al. 2004). Reaction mixtures (20 μl) contained 50 mM Tris–HCl of pH 7.0, 150 mM NaCl, 0.25 mg/ml tRNA, 0.25 mg/ml bovine serum albumin, 100 fmol of labeled mRNA probe, 4 U of SuperRase-In (Ambion), and 10 μg of cytoplasmic extracts, in the absence or presence of 20× unlabeled mRNA competitor. The mixtures were incubated at 37°C for 10 min. Following the incubation, 4 μl of a dye mixture (Gel Loading buffer II, Ambion) was added and the samples were immediately loaded on a 5% native acrylamide gel in 0.5× TBE buffer and electrophoresed at 120 V at RT. The RNAs were then transferred, cross-linked and detected as described for DNA-EMSA.
Bioinformatics and data analysis
Transcription factor (TF) binding sites were predicted by the MatInspector (http://www.genomatix.de/products/MatInspector/index.html) and Match™ programs (http://www.gene-regulation.com/pub/programs.html). TF binding-sites altered by the polymorphisms were described by the ElDorado database (http://www.genomatix.de/products/ElDorado/). Prediction of the mRNA secondary structure was performed by Mfold program (http://www.bioinfo.rpi.edu/applications/mfold; Zuker et al. 2003). Statistics were carried out using StatView 5.0 software (SAS Institute Inc., USA). Comparisons of the luciferase activities and mRNA levels between the constructs were performed by an analysis of variance (ANOVA) or t test as indicated.
Results
Identification of the TPH2 core promoter region and inhibitory effect of the TPH2 5′-UTR on gene expression
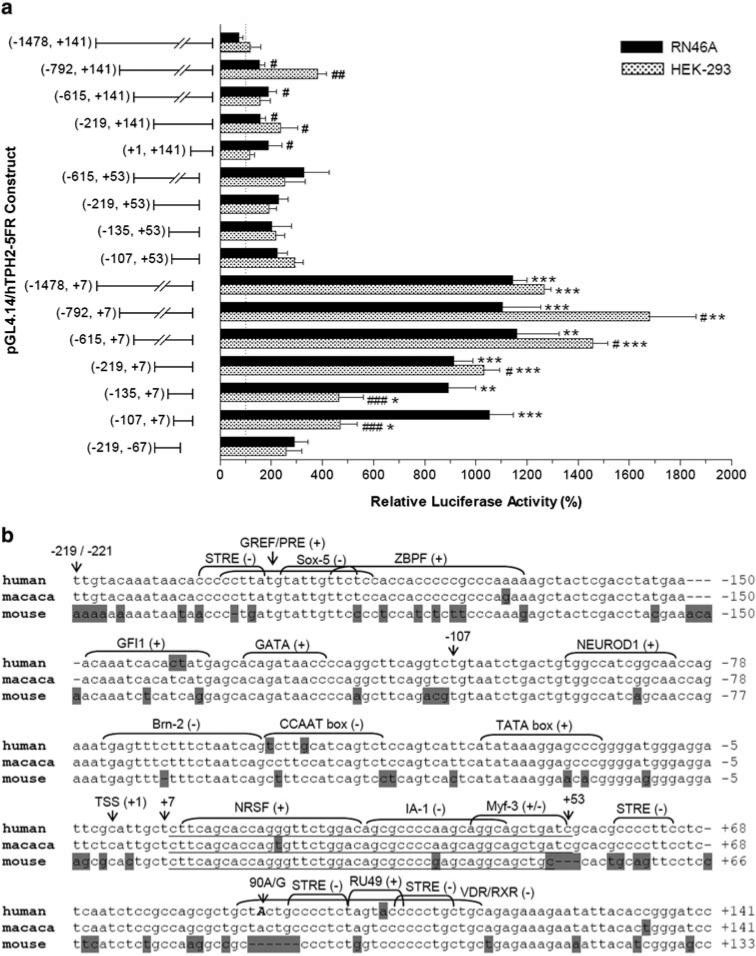
A series of 5′ and 3′ truncated fragments of the TPH2 5′ regulatory region between +141 (position immediately upstream the start codon) and −1478 were cloned into pGL4.14, and the expressed luciferase activities driven by these fragments were measured following transient transfection in RN46A and HEK-293 cells. As shown in Fig. 1a, the 114-bp fragment between −107 and +7 can efficiently drive luciferase expression about 5 and 10-fold over the negative pGL4.14 control in HEK-293 and RN46A cells, respectively, suggesting that the core promoter is located in this region. Interestingly, constructs containing the whole 5′-UTR (+1 to +141) displayed very low luciferase expression independent of the cell line (averaging 1.3 and 2.0-fold of pGL4.14 in RN46A and HEK-293 cells, respectively), and deletion of the region +8 to +141 (inserts end with +7) but not +54 to+141 (inserts end with +53) strikingly increased the luciferase expression by an average of ∼10-fold over pGL4.14. Accordingly, the segment +8 ∼ +53 in TPH2 5′-UTR has a potent inhibitory effect on gene expression, regardless of the upstream sequence or cell line.
Fig. 1.
Effect of serial 5′ and 3′ deletion of the human TPH2 5′ regulatory region on reporter gene expression (a) and analysis of the proximal 5′ region of human, macaca and mouse TPH2 (b). Relative luciferase activities (%) driven by the constructs are shown as Mean ± SE. *P < 0.05, **P < 0.01, ***P < 0.001 compared with constructs with identical 5′ start but different 3′ end; #P < 0.05, ##P < 0.01, ###P < 0.001 compared with the longest construct with the identical 3′ end. Data were analyzed by t test or ANOVA followed by Fisher's protected least-squares difference test. TPH2 sequences for human (AC090109) and mouse (AC153497) were obtained from GenBank, while the macaca TPH2 sequence was identified by our previous study (Chen et al. 2006), which shows 100% identity with 10637260−10637619 of chromosome 11 genomic contig (Mmu11_WGA17192_1). Nucleotides are numbered relative to the transcription start site (TSS, +1), with the position immediately upstream of the TSS as −1. Non-identical nucleotides between the species are shadowed and putative TF binding sites are indicated, with the symbols “+” and “−” representing the sense and anti-sense strand, respectively. The 90A/G polymorphic site is shown in upper case and indicated by an arrow. Borders of the core promoter (−107, +7) are also indicated, and the negative segment (+8, +53) is underlined
As shown in Fig. 1b, the proximal 5′ regulatory region (especially the inhibitory segment +8 to +53) shares a high homology between human, macaca and mouse TPH2. Putative TF binding sites for NEUROD1 (Beta-2/E47 dimer), Brn-2, and CEBP (CCAAT enhancer binding protein) were predicted in the core promoter, while the inhibitory region (+8 ∼ +53) contains putative sites for three TFs (NRSF, IA-1 and Myf-3), among which NRSF has been recently identified to confer transcriptional repression of TPH2 gene expression (Patel et al. 2007), while IA-1 can also function as a transcriptional repressor (Breslin et al. 2002). There are also several putative TF binding sites spanning or neighboring the 90A/G SNP, including two STRE (stress-response element) sites.
The effect of common polymorphisms and haplotypes of TPH2 5′ regulatory region on gene expression
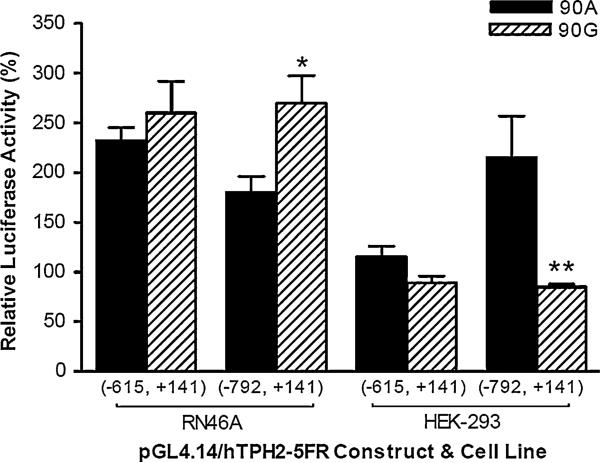
To investigate the effect of common polymorphisms and haplotypes of the human TPH2 5′ regulatory region on gene expression, we constructed all the eight theoretic haplotypes (H1–H8, shown in Fig. 2a) of the −703G/T, −473T/A, and 90A/G polymorphisms. Considering the potent suppression of gene expression by the 5′-UTR, we also made another four 5′-UTR-absent haplotypes (ΔH1–ΔH4, insert spanning −1478 to +7) of −703G/T and −473T/A. The expressed luciferase activities of these constructs are shown in Fig. 2b and c. No significant difference was observed between the four haplotypes with 5′-UTR absent; however, of the haplotypes containing the whole 5′-UTR, the TTA (H1) haplotype showed the lowest luciferase activity in both RN46A and HEK-293 cells (P < 0.01 compared with H2–H4). In RN46A cells, all four 90G-containing haplotypes (H5–H8) showed significantly higher luciferase activity than the four 90A-containing haplotypes (H1–H4; all P < 0.001). In HEK-293 cells, the 90G-containing constructs expressed significantly higher luciferase activity than 90A when the haplotype contained −703T/−473T alleles (H5 vs. H1; TTG > TTA, P < 0.001), but expressed lower luciferase activity when the construct contained −703T/−473A alleles (H8 vs. H4; TAG < TAA, P < 0.001). We also examined the effect of 90A/G on gene expression in another two 5′ deletion constructs (spanning −615 to +141 and that in the −792 to +141, respectively), and found −792 to +141 construct, luciferase expression was also up-regulated by 90A/G transition in RN46A, but down-regulated in HEK-293 (Fig. 3).
Fig. 3.
The effect of the 90A/G polymorphism on reporter gene expression in another two 5′ truncated constructs (−615 to +141 and −792 to +141, respectively), which contain wild-type alleles at −703 and −473T and −473 loci (−703/−473T) if applicable. Relative luciferase activities are shown as means ± SE. *P < 0.05, **P < 0.01 compared with the construct containing the 90A-allele. Repeated measures ANOVA was used for the comparisons
To clarify whether transcriptional or post-transcriptional mechanisms are involved in the regulation of luciferase expression, we assessed luciferase mRNA levels by using quantative RT-PCR. As shown in Fig. 2b inset, constructs containing the wild-type (90A) 5′-UTR expressed very low luciferase mRNA levels, among which the TTA (H1) haplotype showed the lowest mRNA level (P < 0.01 compared with H2–H4). Just as in the case for luciferase activity, the 90A-containing constructs showed significantly lower luciferase mRNA expression compared with the corresponding 90G-containing and 5′-UTR-free constructs that have identical alleles at loci −703 and −473. We also examined the luciferase mRNA stability for three representative constructs (H3, H7 and ΔH3) in HEK-293 cells, but found no difference in mRNA degradation rate between these constructs (Fig. 2c inset). Despite the general consistency with the data of luciferase activity that reflects the protein yields, it is noteworthy that for some haplotypes the mRNA level did not parallel the luciferase activity reflecting the final protein production. Particularly, mRNA level of the haplotypes lacking 5′-UTR (ΔH1–ΔH4) showed only an average of ∼2.5-fold over pGL4.14, while the luciferase activity of these haplotypes displayed ∼12-fold over pGL4.14. This implies that the TPH2 5′-UTR might exert a striking inhibitory effect on the post-transcriptional process of mRNA. In summary, these observations indicate that transcriptional mechanisms contribute primarily to the effect of polymorphisms on gene expression, while the regulation of gene expression by the 5′-UTR involves both transcriptional and post-transcriptional mechanisms, with the latter likely being more important.
The effect of common polymorphisms in human TPH2 5′ regulatory region on potential DNA–protein interaction
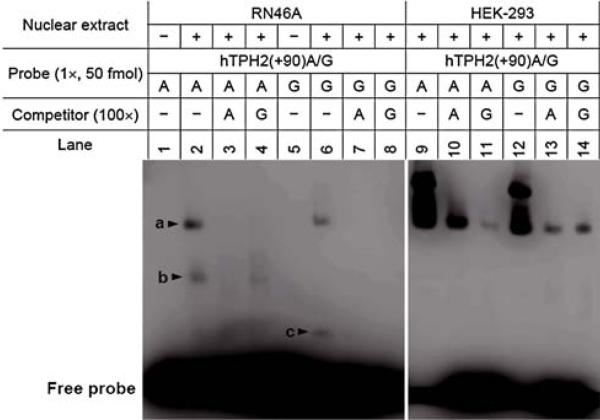
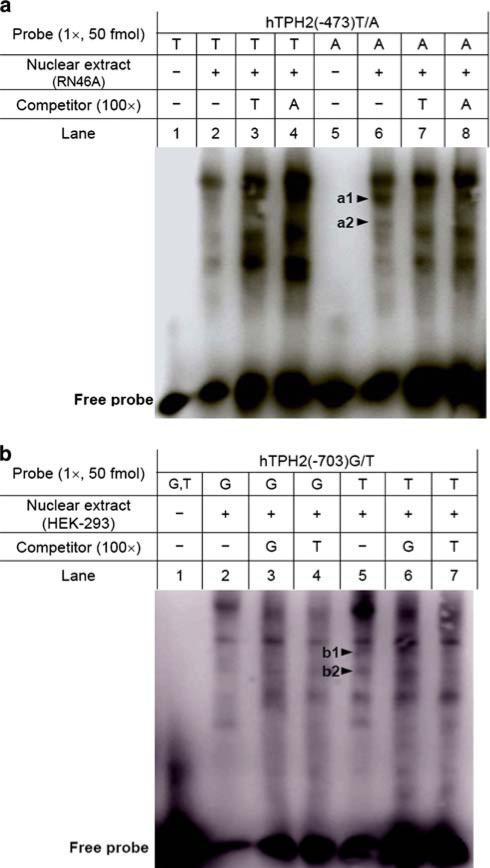
As −703G/T, −473T/A and 90A/G are all predictive of alteration in TF-binding sites (Table 3), we performed DNA-EMSA with allele-specific probes to detect whether these SNPs could affect any DNA–protein interaction. As shown in Fig. 4, EMSA with RN46A nuclear extract revealed that a specific complex (b) was formed by the 90A, but not by the 90G allele, and this complex could be completely abolished by 100× 90A competitor but not by 90G competitor, indicating that this DNA–protein binding is likely specific for the 90A allele and that 90A/G transition might eliminate this binding site. Another complex c is obvious for 90G probe, but it is hardly observed for 90A probe. No allele-specific complex for 90A/G was found by EMSA with HEK-293 extract, but the possibility of a difference in binding affinity for one complex (a in lane 9−14) between the two alleles cannot be excluded.
Table 3.
Prediction of transcription factor binding-site alteration caused by common SNPs in human TPH2 5′ regulatory region (predicted by the ElDorado database)
| SNP | Type | Matrix name | Stranda | Core match | Matrix match |
|---|---|---|---|---|---|
| −703G/T | New | V$FKHD/FHXB.01 | + | 1.000 | 0.850 |
| Lost | V$HOMF/MSX.01 | − | 1.000 | 0.989 | |
| New | V$SATB/SATB1.01 | + | 1.000 | 0.953 | |
| −473T/A | New | V$CEBP/CEBPB.01 | − | 0.940 | 0.968 |
| New | V$CREB/E4BP4.01 | − | 1.000 | 0.852 | |
| New | V$PARF/TEF_HLF.01 | + | 1.000 | 0.834 | |
| New | V$PARF/HLF.01 | − | 1.000 | 0.939 | |
| 90A/G | New | V$MYOD/MYF5.01 | − | 1.000 | 0.917 |
New binding site generated due to SNP, lost binding site lost due to SNP
The symbols “+” and “−” represent the sense and antisense strands, respectively
Fig. 4.
DNA-EMSA for the 90A/G polymorphism using both RN46A and HEK-293 nuclear extracts. The allele-specific probes hTPH2(+90)A and hTPH2(+90)G spanning +72 to +106 were used to perform the EMSA. All samples were run on the same gel, but were separated for RN46A and HEK-293, respectively, so that the bands for RN46A could be made more distinct by adjusting the brightness and contrast. Three specific DNA–protein complexes (a, b and c) were observed with the RN46A nuclear extract, among which complex b was formed by the 90A probe, but not by the 90G probe, and this complex was completely abolished by the 100× 90A competitor but not by the 90G competitor
DNA-EMSA for the −473T/A and −703/T are shown in Fig. 5a and b, respectively. With RN46A nuclear extract, two complexes (a1 and a2 in Fig. 5a) were formed by −473A, but were invisible for −473T, of which the a2 complex was abolished by both allele competitors, suggesting that −473T/A might change DNA–protein binding affinity. EMSA for −703G/T using HEK-293 nuclear extract showed that a complex (b1 in Fig. 5b) was formed by −703T, but not for −703G, and this complex was abolished by the −703T competitor, but not by the −703G competitor, implying that −703G/T might introduce a new DNA–protein interaction. In addition, another complex (b2 in Fig. 5b) formed by −703T was largely reduced by −703T competitor, but not altered by the −703G competitor, suggesting that −703G/T might also change the binding affinity for a specific DNA–protein interaction.
Fig. 5.
DNA-EMSA for −473T/A (a) and −703G/T (b) using nuclear extracts and allele-specific probes. The allele-specific probes hTPH2(−473)T and hTPH2 (−473)A spanning −494 to −451, and hTPH2(−703)G and hTPH2(−703)T spanning −725 to −681 were used to perform the EMSA. For the −473T/A, two complexes (a1 and a2) were formed by −473A but not by −473T, of which the complex a2 was abolished by competitors of both alleles. For the −703G/T, one complex (b1) was formed by the −703T, but not the −703G probe, and it was abolished by the −703T but not the −703G competitor. Another complex (b2) formed by −703T was largely reduced by the −703T competitor but not altered by the −703G competitor
Potential effect of 90A/G polymorphisms on the 5′-UTR secondary structure of TPH2 mRNA and RNA–protein interaction
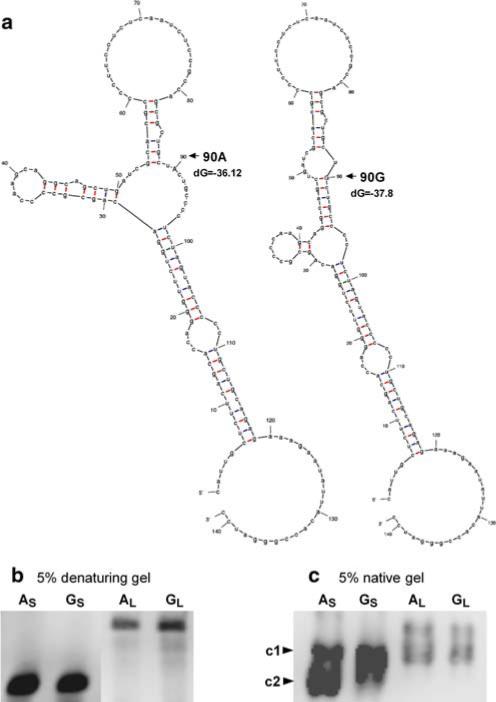
Because the 90A/G polymorphism in 5′-UTR appeared to be functionally significant, independent of the two other SNPs, we further characterized this SNP for its effect on RNA secondary structure and potential RNA–protein interaction. Prediction of the 5′-UTR secondary structure of human TPH2 mRNA by Mfold program showed that the 90A/G may alter the mRNA structure (Fig. 6a). We generated full-length (+1 to +141) and partial (+61 to +141) 5′-UTR RNA containing 90A and 90G alleles, and an aliquot of each product was subjected to electrophoresis on both native and denaturing acrylamide gels. The typical patterns of electrophoresis on denaturing and native gels are shown in Fig. 6b and c, respectively. Both the partial (S) and full-length (L) 5′-UTR transcripts showed no shift difference between 90A and 90G on a denaturing gel; however, when run on a native acrylamide gel, the partial 5′-UTR RNA showed shift differences between 90A and 90G, with two major bands (c1 and c2) being observed for 90A, while only one major band (c1) was observed for 90G.
Fig. 6.
The potential effect of the 90A/G polymorphism on 5′-UTR secondary structure of mRNA (a) and the electrophoretic patterns of in vitro transcribed 5′-UTR RNA probes on denaturing (b) and native (c) gels. Mfold program (Zuker et al. 2003) was employed to predict the mRNA secondary structure for 90A and 90G, respectively, and the free energy (dG, kcal/mol) is also shown. In vitro transcribed RNA probes containing 90A and 90G alleles were run on 5% native and denaturing (8 M urea) acrylamide gels, respectively, and were detected by chemiluminescence. AS and AL represent the partial (+61 to +141) and full-length (+1 to +141) of 5′-UTR RNA probes containing 90A, respectively, while GS and GL represent the corresponding RNA probes containing 90G. No shift difference was observed on the denaturing gel between the 90A and 90G probes. When running on the native gel, AS showed two major bands (c1 and c2, with c2 as the predominant), while GS showed only one major band (corresponding to c1)
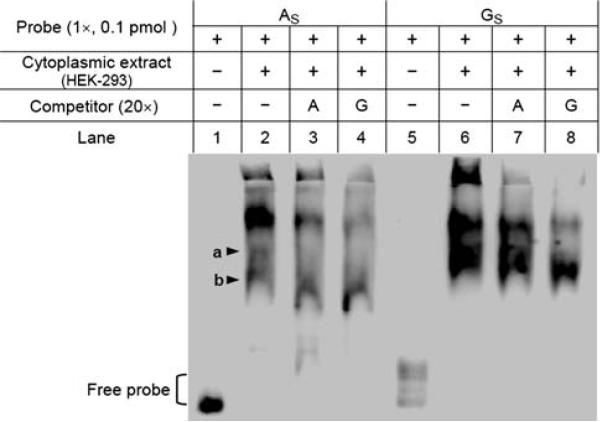
We also performed RNA-EMSA using the partial 5′-UTR mRNAs and cytoplasmic extracts to check whether this polymorphism would alter any RNA–protein interaction. With HEK-293 cytoplasmic extract (Fig. 7), we found that one complex (b1) was formed by both 90A and 90G probe, but the 90A-complex was abolished by 20× competitors while the 90G-complex was not, implying that 90G probe might bind more potently to the specific protein. In addition, a complex (b2) was observed only for 90A, but not for 90G. Although the possible structure difference between the 90A and 90G probes might confound the RNA-EMSA, the data do suggest the possibility that 90A/G polymorphism might have a profound effect on the mRNA structure and/or RNA–protein interaction and thereby affect TPH2 gene expression at the post-transcriptional level.
Fig. 7.
The potential effect of the 90A/G polymorphism on RNA–protein interaction. RNA-EMSA was performed using the partial 5′-UTR RNA probes (AS and GS) and HEK-293 cytoplasmic extracts. One complex (a) was observed for both AS and GS probes, but the AS-complex (a) was abolished by both competitors (lanes 3 and 4), while the GS-complex (a) was not. Another complex (b) was formed by the AS but not by the GS probe
Discussion
The recent identification of brain-specific TPH2 has opened up a new area of investigation on the regulation of the central 5-HT system. As 5-HT has been well documented to modulate HPA axis function, it is noteworthy that TPH2 expression exhibits a circadian rhythm and is influenced by specific stressors and hormones. Our previous study in rhesus macaques revealed a striking association between functional TPH2 polymorphisms and HPA axis activity (Chen et al. 2006). It can thus be inferred that alteration in TPH2 gene expression might result in HPA axis dysfunction, which may in turn lead to abnormality of behavior and increased susceptibility to psychiatric disorders. Hence, knowledge of TPH2 gene expression regulation can advance our understanding of the mechanisms by which the HPA axis is precisely modulated, as well as the pathogenesis of various psychiatric disorders.
It has been well documented that 5′-UTR plays an important role in the regulation of gene expression, primarily due to its effect on the post-transcriptional process, including the initiation and efficiency of translation, as well as the stability of mRNA (Van der Velden and Thomas 1999; Pickering and Willis 2005; Derrigo et al. 2000; Wilkie et al. 2003); however, since the basal transcription apparatus also contains partial sequence downstream of the transcription start site (TSS), it can be inferred that the 5′-UTR might also affect gene expression via a transcriptional mechanism, as has already been reported for some genes (Berardi et al. 2003; Coppotelli et al. 2006; Minet et al. 1999; Singh et al. 2002). In the present study, the segment +8 to +53 of the TPH2 5′-UTR decreased luciferase activity and mRNA level strikingly, yet had a negligible effect on mRNA stability, suggesting that transcriptional mechanisms may be involved in the repression of gene expression by this region and that negative cis-element(s) should exist in this region. In accordance with these findings, a bipartite REST/NRSF binding motif that mediates transcriptional repression was recently identified in TPH2 5′-UTR between +9 to +35 (Patel et al. 2007), just within the suppressive region (+8 to +53) revealed by our present study. In addition, our current preliminary data indicates that additional TF binding motifs in this region might also contribute to the transcriptional suppression of gene expression (data not shown).
However, it deserves our attention that for the 5′-UTR-absent constructs (ΔH1–ΔH4) there is a remarkable discrepancy between the luciferase activity and mRNA level, which are about 12 and 2.5-fold over pGL4.14, respectively. This indicates that the segment +8 to +141 of TPH2 5′-UTR may also have a strong inhibitory effect on translation or other post-transcriptional processes of mRNA, and that deletion of this region could greatly abolish post-transcriptional repression. Interestingly, the human TPH2 5′-UTR has a GC content of ∼60% (even as high as 68% between +11 and +50) and predicted free energy of −36.12 kcal/mol, making the mRNA transcript likely to be poorly translated (Davuluri et al. 2000). Nevertheless, further studies are required to elucidate the mechanisms underlying the regulation of gene expression by TPH2 5′-UTR.
Based on our experimental data on luciferase activity, mRNA level and EMSA, as well as the prediction of mRNA secondary structure, it is likely that the 90A/G polymorphism alone can affect (in most cases up-regulate) gene expression at both transcriptional and post-transcriptional levels. The regulation of gene expression by this SNP is apparently cell-specific. Since there are putative TF binding sites spanning or neighboring this locus and also because the EMSA for this SNP showed a cell-dependent profile of potential DNA–protein interaction, it is possible that 90A/G affects gene transcription in a cell-specific manner via trans-acting factors. Moreover, 90A/G might also change mRNA secondary structure and RNA–protein interaction, making it reasonable that it may also influence gene expression via post-transcriptional mechanism(s). It is interesting that the effect of 90A/G is significant in constructs starting at −1478 and −792, but not significant in construct at −615, implying an interaction between the promoter and 5′-UTR. As for the −703G/T and −473T/A polymorphisms, comparisons of the luciferase expression between TTA, GTA and TAA haplotypes suggest that in vitro gene expression is down-regulated by −703G/T, but up-regulated by 473T/A, albeit being inconsistent with the comparisons between GAA, GTA and TAA haplotypes. The difference between the haplotypes disappears when the G allele is present at locus 90, or the 5′-UTR is truncated. Accordingly, these data suggest that −703G/T and −473T/ A might exert a synergic effect on gene expression, and the synergic effect interacts with other elements in the 5′-UTR. Considering the natural existence of 5′-UTR in human TPH2 and the common frequency (expected to be ∼20% regardless of linkage disequilibrium) of the TTA haplotype (Zhou et al. 2005), it is possible that human TPH2 normally shows a relatively low constitutive expression due to both transcriptional and post-transcriptional repression by the 5′-UTR, and that haplotypes of the TPH2 5′ flanking region may underlie differential regulation of TPH2 gene expression, thereby conferring individual variation in behavioral traits and/or disease susceptibility. In this regard, our findings are well supported by the association of −703G/T and −473T/A with accumulating behavioral traits and psychiatric disorders (Walitza et al. 2005; Brown et al. 2005; Canli et al. 2005; Herrmann et al. 2007; Gutknecht et al. 2007; Reuter et al. 2007).
Our findings are somewhat in conflict with the recent study of Scheuch et al., which identified −473T/A, but not −703G/T and 90A/G, as exerting an effect on gene expression (Scheuch et al. 2007). It is noteworthy that the methodology used in the study of Scheuch et al. differs from ours in several aspects: (1) primary serotonergic neurons and SHP-77 cell lines, but not RN46A and HEK-293 cell lines, were used for transfection; (2) the pGL3-basic and pRL-null (promoterless) vectors, which provide different background expression compared with pGL4.14 and pRLTK (reference: Promega technical manuals), were used for the reporter and internal control; (3) the 5′ flanking region spanning −995 to +162, which includes partial sequence (21-bp length) of exon 1, was cloned for reporter gene assays; (4) the −473T/A polymorphism was focused and intensively investigated, while the functional significance of the other two SNPs was only judged by luciferase assay. In accordance with our data, a recent study confers that both −703G/T and −473T/A affect gene expression, yet the significance of 90A/G was not assessed (Lin et al. 2007). Despite the discrepancies between studies, however, all these studies suggest that the 5′ regulatory region plays an important role in the regulation of TPH2 gene expression, and our study clearly suggests that 90A/G should be further examined and investigated in association studies. Nevertheless, as in vitro findings may not always and exactly reflect what occurs in vivo, the functional significance of these SNPs need to be further verified by in vivo evidence, notably the alteration in the levels of TPH2 mRNA and protein, as well as serotonin turnover in the brain. Interestingly, our current preliminary data indicates that polymorphisms in the 5′ regulatory region of rhesus monkey TPH2 are also functionally significant (unpublished), supporting the feasibility of using the naturally occurring rhesus monkey polymorphisms as models to further understand the mechanisms by which comparable functional SNPs can affect behavioral traits and disease susceptibility.
In summary, our present study reveals an inhibitory effect of the human TPH2 5′-UTR on gene expression and provides evidence for the functional significance of common polymorphisms and haplotypes of the human TPH2 5′ regulatory regions. Our findings promote a better understanding of the regulation of TPH2 gene expression and further advance our knowledge of the brain functions involved in psychiatric disorders and behavioral traits, related to the 5-HT system.
Acknowledgments
This study was supported by AA016194 (GMM), DA016606 (GMM), DA06303 (GMM), DA021180 (GMM), MH082507 (EJV) and RR00168.
References
- Berardi P, Meyyappan M, Riabowol KT. A novel transcriptional inhibitory element differentially regulates the cyclin D1 gene in senescent cells. J Biol Chem. 2003;278:7510–7519. doi: 10.1074/jbc.M210864200. [DOI] [PubMed] [Google Scholar]
- Bicalho MA, Pimenta GJ, Neves FS, Correa H, de Moraes EN, De Marco L, Romano-Silva MA. Genotyping of the G1463A (Arg441His) TPH2 polymorphism in a geriatric population of patients with major depression. Mol Psychiatry. 2006;11:799–800. doi: 10.1038/sj.mp.4001861. [DOI] [PubMed] [Google Scholar]
- Blakely RD. Overview: a rare opportunity or just one less reason to be depressed. Neuron. 2005;48:701–702. doi: 10.1016/j.neuron.2005.11.029. [DOI] [PubMed] [Google Scholar]
- Breslin MB, Zhu M, Notkins AL, Lan MS. Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence. Nucleic Acids Res. 2002;30:1038–1045. doi: 10.1093/nar/30.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown HJ, Henderson LA, Keay KA. Hypotensive but not normotensive haemorrhage increases tryptophan hydroxylase-2 mRNA in caudal midline medulla. Neurosci Lett. 2006;398:314–318. doi: 10.1016/j.neulet.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Brown SM, Peet E, Manuck SB, Williamson DE, Dahl RE, Ferrell RE, Hariri AR. A regulatory variant of the human tryptophan hydroxylase-2 gene biases amygdala reactivity. Mol Psychiatry. 2005;10:884–888. doi: 10.1038/sj.mp.4001716. [DOI] [PubMed] [Google Scholar]
- Canli T, Congdon E, Gutknecht L, Constable RT, Lesch KP. Amygdala responsiveness is modulated by tryptophan hydroxylase-2 gene variation. J Neural Transm. 2005;112:1479–1485. doi: 10.1007/s00702-005-0391-4. [DOI] [PubMed] [Google Scholar]
- Chamas FM, Underwood MD, Arango V, Serova L, Kassir SA, Mann JJ, Sabban EL. Immobilization stress elevates tryptophan hydroxylase mRNA and protein in the rat raphe nuclei. Biol Psychiatry. 2004;55:278–283. doi: 10.1016/s0006-3223(03)00788-1. [DOI] [PubMed] [Google Scholar]
- Chen GL, Novak MA, Hakim S, Xie Z, Miller GM. Tryptophan hydroxylase-2 gene polymorphisms in rhesus monkeys: association with hypothalamic-pituitary-adrenal axis function and in vitro gene expression. Mol Psychiatry. 2006;11:914–928. doi: 10.1038/sj.mp.4001870. [DOI] [PubMed] [Google Scholar]
- Clark JA, Pai LY, Flick RB, Rohrer SP. Differential hormonal regulation of tryptophan hydroxylase-2 mRNA in the murine dorsal raphe nucleus. Biol Psychiatry. 2005;57:943–946. doi: 10.1016/j.biopsych.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Coon H, Dunn D, Lainhart J, Miller J, Hamil C, Battaglia A, Tancredi R, Leppert MF, Weiss R, McMahon W. Possible association between autism and variants in the brain-expressed tryptophan hydroxylase gene (TPH2). Am J Med Genet B Neuropsychiatr Genet. 2005;135:42–46. doi: 10.1002/ajmg.b.30168. [DOI] [PubMed] [Google Scholar]
- Coppotelli G, Summers A, Chidakel A, Ross JM, Celi FS. Functional characterization of the 258 A/G (D2-ORFa-Gly3Asp) human type-2 deiodinase polymorphism: a naturally occurring variant increases the enzymatic activity by removing a putative repressor site in the 5′ UTR of the gene. Thyroid. 2006;16:625–632. doi: 10.1089/thy.2006.16.625. [DOI] [PubMed] [Google Scholar]
- Côté F, Thévenot E, Fligny C, Fromes Y, Darmon M, Ripoche MA, Bayard E, Hanoun N, Saurini F, Lechat P, Dandolo L, Hamon M, Mallet J, Vodjdani G. Disruption of the nonneuronal tph1 gene demonstrates the importance of peripheral serotonin in cardiac function. Proc Natl Acad Sci USA. 2003b;100:13525–13530. doi: 10.1073/pnas.2233056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Zhang MQ. CART classification of human 5′ UTR sequences. Genome Res. 2000;10:1807–1816. doi: 10.1101/gr.gr-1460r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme R, Durand CM, Betancur C, Wagner M, Ruhrmann S, Grabe HJ, Nygren G, Gillberg C, Leboyer M, Bourgeron T, Courtet P, Jollant F, Buresi C, Aubry JM, Baud P, Bondolfi G, Bertschy G, Perroud N, Malafosse A. No human tryptophan hydroxylase-2 gene R441H mutation in a large cohort of psychiatric patients and control subjects. Biol Psychiatry. 2006;60:202–203. doi: 10.1016/j.biopsych.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Derrigo M, Cestelli A, Savettieri G, Di Liegro I. RNA–protein interactions in the control of stability and localization of messenger RNA (review). Int J Mol Med. 2000;5:111–123. [PubMed] [Google Scholar]
- Dinan TG. Serotonin and the regulation of hypothalamic–pituitary–adrenal axis function. Life Sci. 1996;58:1683–1694. doi: 10.1016/0024-3205(96)00066-5. [DOI] [PubMed] [Google Scholar]
- Glatt CE, Carlson E, Taylor TR, Risch N, Reus VI, Schaefer CA. Response to Zhang et al. (2005): loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron. 2005;48:704–705. doi: 10.1016/j.neuron.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Goertzel BN, Pennachin C, de Souza Coelho L, Gurbaxani B, Maloney EM, Jones JF. Combinations of single nucleotide polymorphisms in neuroendocrine effector and receptor genes predict chronic fatigue syndrome. Pharmacogenomics. 2006;7:475–483. doi: 10.2217/14622416.7.3.475. [DOI] [PubMed] [Google Scholar]
- Gutknecht L, Jacob C, Strobel A, Kriegebaum C, Müller J, Zeng Y, Markert C, Escher A, Wendland J, Reif A, Mössner R, Gross C, Brocke B, Lesch KP. Tryptophan hydroxylase-2 gene variation influences personality traits and disorders related to emotional dysregulation. Int J Neuropsychopharmacol. 2007;10:309–320. doi: 10.1017/S1461145706007437. [DOI] [PubMed] [Google Scholar]
- Harvey M, Shink E, Tremblay M, Gagne B, Raymond C, Labbe M, Walther DJ, Bader M, Barden N. Support for the involvement of TPH2 gene in affective disorders. Mol Psychiatry. 2004;9:980–981. doi: 10.1038/sj.mp.4001557. [DOI] [PubMed] [Google Scholar]
- Herrmann MJ, Huter T, Müller F, Mühlberger A, Pauli P, Reif A, Renner T, Canli T, Fallgatter AJ, Lesch KP. Additive effects of serotonin transporter and tryptophan hydroxylase-2 gene variation on emotional processing. Cereb Cortex. 2007;17:1160–1163. doi: 10.1093/cercor/bhl026. [DOI] [PubMed] [Google Scholar]
- Hiroi R, McDevitt RA, Neumaier JF. Estrogen selectively increases tryptophan hydroxylase-2 mRNA expression in distinct subregions of rat midbrain raphe nucleus: association between gene expression and anxiety behavior in the open Weld. Biol Psychiatry. 2006;60:288–295. doi: 10.1016/j.biopsych.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Jollant F, Buresi C, Guillaume S, Jaussent I, Bellivier F, Leboyer M, Castelnau D, Malafosse A, Courtet P. The influence of four serotonin-related genes on decision-making in suicide attempters. Am J Med Genet B Neuropsychiatr Genet. 2007;144:615–624. doi: 10.1002/ajmg.b.30467. [DOI] [PubMed] [Google Scholar]
- Ke L, Qi ZY, Ping Y, Ren CY. Effect of SNP at position 40237 in exon 7 of the TPH2 gene on susceptibility to suicide. Brain Res. 2006;1122:24–26. doi: 10.1016/j.brainres.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Leonard BE. The HPA and immune axes in stress: the involvement of the serotonergic system. Eur Psychiatry. 2005;20(Suppl 3):S302–S306. doi: 10.1016/s0924-9338(05)80180-4. [DOI] [PubMed] [Google Scholar]
- Li Y, Jiang Z, Chen H, Ma WJ. A modified quantitative EMSA and its application in the study of RNA–protein interactions. J Biochem Biophys Methods. 2004;60:85–96. doi: 10.1016/j.jbbm.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Liang J, Wessel JH, 3rd, Iuvone PM, Tosini G, Fukuhara C. Diurnal rhythms of tryptophan hydroxylase 1 and 2 mRNA expression in the rat retina. Neuroreport. 2004;15:1497–1500. doi: 10.1097/01.wnr.0000131007.59315.66. [DOI] [PubMed] [Google Scholar]
- Lin YM, Chao SC, Chen TM, Lai TJ, Chen JS, Sun HS. Association of functional polymorphisms of the human tryptophan hydroxylase 2 gene with risk for bipolar disorder in Han Chinese. Arch Gen Psychiatry. 2007;64:1015–1024. doi: 10.1001/archpsyc.64.9.1015. [DOI] [PubMed] [Google Scholar]
- Lopez de Lara C, Brezo J, Rouleau G, Lesage A, Dumont M, Alda M, Benkelfat C, Turecki G. Effect of tryptophan hydroxylase-2 gene variants on suicide risk in major depression. Biol Psychiatry. 2007;62:72–80. doi: 10.1016/j.biopsych.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Lopez VA, Detera-Wadleigh S, Cardona I, Kassem L, The National Institute of Mental Health Genetics Initiative Bipolar Disorder consortium. McMahon FJ. Nested association between genetic variation in tryptophan hydroxylase II, bipolar affective disorder, and suicide attempts. Biol Psychiatry. 2007;61:181–186. doi: 10.1016/j.biopsych.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Lowry CA. Functional subsets of serotonergic neurones: implications for control of the hypothalamic–pituitary–adrenal axis. J Neuroendocrinol. 2002;14:911–923. doi: 10.1046/j.1365-2826.2002.00861.x. [DOI] [PubMed] [Google Scholar]
- Malek ZS, Dardente H, Pevet P, Raison S. Tissue-specific expression of tryptophan hydroxylase mRNAs in the rat mid-brain: anatomical evidence and daily profiles. Eur J Neurosci. 2005;22:895–901. doi: 10.1111/j.1460-9568.2005.04264.x. [DOI] [PubMed] [Google Scholar]
- Malek ZS, Sage D, Pevet P, Raison S. Daily rhythm of tryptophan hydroxylase-2 mRNA within Raphe neurons is induced by corticoid daily surge and modulated by enhanced locomotor activity. Endocrinology. 2007;148(11):5165–5172. doi: 10.1210/en.2007-0526. [DOI] [PubMed] [Google Scholar]
- Maron E, Tõru I, Must A, Tasa G, Toover E, Vasar V, Lang A, Shlik J. Association study of tryptophan hydroxylase 2 gene polymorphisms in panic disorder. Neurosci Lett. 2007;411:180–184. doi: 10.1016/j.neulet.2006.09.060. [DOI] [PubMed] [Google Scholar]
- Minet E, Ernest I, Michel G, Roland I, Remacle J, Raes M, Michiels C. HIF1A gene transcription is dependent on a core promoter sequence encompassing activating and inhibiting sequences located upstream from the transcription initiation site and cis elements located within the 5′UTR. Biochem Biophys Res Commun. 1999;261:534–540. doi: 10.1006/bbrc.1999.0995. [DOI] [PubMed] [Google Scholar]
- Mossner R, Muller-Vahl KR, Doring N, Stuhrmann M. Role of the novel tryptophan hydroxylase-2 gene in Tourette syndrome. Mol Psychiatry. 2007;12:617–619. doi: 10.1038/sj.mp.4002004. [DOI] [PubMed] [Google Scholar]
- Mössner R, Walitza S, Geller F, Scherag A, Gutknecht L, Jacob C, Bogusch L, Remschmidt H, Simons M, Herpertz-Dahlmann B, Fleischhaker C, Schulz E, Warnke A, Hinney A, Wewetzer C, Lesch KP. Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in children and adolescents with obsessive-compulsive disorder. Int J Neuropsychopharmacol. 2006;9:437–442. doi: 10.1017/S1461145705005997. [DOI] [PubMed] [Google Scholar]
- Patel PD, Bochar DA, Turner DL, Meng F, Mueller HM, Pontrello CG. Regulation of tryptophan hydroxylase-2 gene expression by a bipartite REST/NRSF binding motif. J Biol Chem. 2007;282(37):26717–26724. doi: 10.1074/jbc.M705120200. [DOI] [PubMed] [Google Scholar]
- Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Reuter M, Kuepper Y, Hennig J. Association between a polymorphism in the promoter region of the TPH2 gene and the personality trait of harm avoidance. Int J Neuropsychopharmacol. 2007;10:401–404. doi: 10.1017/S1461145706007073. [DOI] [PubMed] [Google Scholar]
- Sanchez RL, Reddy AP, Centeno ML, Henderson JA, Bethea CL. A second tryptophan hydroxylase isoform, TPH-2 mRNA, is increased by ovarian steroids in the raphe region of macaques. Brain Res Mol Brain Res. 2005;135:194–203. doi: 10.1016/j.molbrainres.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Scheuch K, Lautenschlager M, Grohmann M, Stahlberg S, Kirchheiner J, Zill P, Heinz A, Walther DJ, Priller J. Characterization of a functional promoter polymorphism of the human tryptophan hydroxylase 2 gene in serotonergic raphe neurons. Biol Psychiatry. 2007 doi: 10.1016/j.biopsych.2007.01.015. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002;16:869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- Sheehan K, Lowe N, Kirley A, Mullins C, Fitzgerald M, Gill M, Hawi Z. Tryptophan hydroxylase 2 (TPH2) gene variants associated with ADHD. Mol Psychiatry. 2005;10:944–949. doi: 10.1038/sj.mp.4001698. [DOI] [PubMed] [Google Scholar]
- Singh IS, He JR, Calderwood S, Hasday JD. A high affnity HSF-1 binding site in the 5′-untranslated region of the murine tumor necrosis factor-alpha gene is a transcriptional repressor. J Biol Chem. 2002;277:4981–4988. doi: 10.1074/jbc.M108154200. [DOI] [PubMed] [Google Scholar]
- Smith AK, White PD, Aslakson E, Vollmer-Conna U, Rajeevan MS. Polymorphisms in genes regulating the HPA axis associated with empirically delineated classes of unexplained chronic fatigue. Pharmacogenomics. 2006;7:387–394. doi: 10.2217/14622416.7.3.387. [DOI] [PubMed] [Google Scholar]
- Van Den Bogaert A, De Zutter S, Heyrman L, Mendlewicz J, Adolfsson R, Van Broeckhoven C, Del-Favero J. Response to Zhang et al. (2005): loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron. 2005;48:704. doi: 10.1016/j.neuron.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Van Den Bogaert A, Sleegers K, De Zutter S, Heyrman L, Norrback KF, Adolfsson R, Van Broeckhoven C, Del-Favero J. Association of brain-specific tryptophan hydroxylase, TPH2, with unipolar and bipolar disorder in a Northern Swedish, isolated population. Arch Gen Psychiatry. 2006;63:1103–1110. doi: 10.1001/archpsyc.63.10.1103. [DOI] [PubMed] [Google Scholar]
- van der Velden AW, Thomas AA. The role of the 5′ untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- Walitza S, Renner TJ, Dempfle A, Konrad K, Wewetzer Ch, Halbach A, Herpertz-Dahlmann B, Remschmidt H, Smidt J, Linder M, Flierl L, Knölker U, Friedel S, Schäfer H, Gross C, Hebebrand J, Warnke A, Lesch KP. Transmission disequilibrium of polymorphic variants in the tryptophan hydroxylase-2 gene in attention-deficit/hyperactivity disorder. Mol Psychiatry. 2005;10:1126–1132. doi: 10.1038/sj.mp.4001734. [DOI] [PubMed] [Google Scholar]
- Walther DJ, Bader M. A unique central tryptophan hydroxylase isoform. Biochem Pharmacol. 2003a;66:1673–1680. doi: 10.1016/s0006-2952(03)00556-2. [DOI] [PubMed] [Google Scholar]
- Walther DJ, Peter JU, Bashammakh S, Hortnagl H, Voits M, Fink H, Bader M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science. 2003b;299:76. doi: 10.1126/science.1078197. [DOI] [PubMed] [Google Scholar]
- White LA, Eaton MJ, Castro MC, Klose KJ, Globus MY, Shaw G, Whittemore SR. Distinct regulatory pathways control neurofilament expression and neurotransmitter synthesis in immortalized serotonergic neurons. J Neurosci. 1994;14:6744–6753. doi: 10.1523/JNEUROSCI.14-11-06744.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Zhang X, Beaulieu JM, Sotnikova TD, Gainetdinov RR, Caron MG. Tryptophan hydroxylase-2 controls brain serotonin synthesis. Science. 2004;305:217. doi: 10.1126/science.1097540. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gainetdinov RR, Beaulieu JM, Sotnikova TD, Burch LH, Williams RB, Schwartz DA, Krishnan KR, Caron MG. Loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron. 2005;45:11–16. doi: 10.1016/j.neuron.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Peters EJ, Hamilton SP, McMahon F, Thomas C, McGrath PJ, Rush J, Trivedi MH, Charney DS, Roy A, Wisniewski S, Lipsky R, Goldman D. Response to Zhang et al. (2005): loss-of-function mutation in tryptophan hydroxylase-2 identified in unipolar major depression. Neuron. 2005a;48:702–703. doi: 10.1016/j.neuron.2005.11.018. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Roy A, Lipsky R, Kuchipudi K, Zhu G, Taubman J, Enoch MA, Virkkunen M, Goldman D. Haplotype-based linkage of tryptophan hydroxylase 2 to suicide attempt, major depression, and cerebrospinal fluid 5-hydroxyindoleacetic acid in 4 populations. Arch Gen Psychiatry. 2005b;62:1109–1118. doi: 10.1001/archpsyc.62.10.1109. [DOI] [PubMed] [Google Scholar]
- Zill P, Baghai TC, Zwanzger P, Schule C, Eser D, Rupprecht R, Moller HJ, Bondy B, Ackenheil M. SNP and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene provide evidence for association with major depression. Mol Psychiatry. 2004a;9:1030–1036. doi: 10.1038/sj.mp.4001525. [DOI] [PubMed] [Google Scholar]
- Zill P, Buttner A, Eisenmenger W, Bondy B, Ackenheil M. Regional mRNA expression of a second tryptophan hydroxylase isoform in postmortem tissue samples of two human brains. Eur Neuropsychopharmacol. 2004b;14:282–284. doi: 10.1016/j.euroneuro.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Zill P, Buttner A, Eisenmenger W, Moller HJ, Bondy B, Ackenheil M. Single nucleotide polymorphism and haplotype analysis of a novel tryptophan hydroxylase isoform (TPH2) gene in suicide victims. Biol Psychiatry. 2004c;56:581–586. doi: 10.1016/j.biopsych.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]