Abstract
Background
Molecular adaptations are believed to contribute to the mechanism of action of antipsychotic drugs (APDs). Here we try to establish common gene regulation patterns induced by chronic treatment with APDs.
Methods
Gene expression analysis was performed with the Affymetrix U34A array in the frontal cortex (FC) and the striatum of rats chronically treated with two different concentrations of either clozapine or haloperidol. Key data were verified with real-time quantitative PCR.
Results
Many genes in the FC affected by APD-treatment contribute to similar functions. mRNAs coding for synaptic vesicle docking- and microtubule-associated proteins were upregulated; mRNAs for serine-threonine protein phosphatases were downregulated, whereas the serine-threonine kinases protein kinase A, protein kinase C and calcium/calmodulin kinase II alpha and IV, were upregulated, indicating increased potential for protein phosphorylation. In the striatum, altered gene expression was less focused on genes of particular function or location, and the high concentration of haloperidol had a different gene expression profile than any of the other APD treatments.
Conclusion
We found an increase in the transcription of genes coding for proteins involved in synaptic plasticity and synaptic activity in the FC. We furthermore found that the gene expression profile of APDs is different between FC and striatum.
Keywords: antipsychotic drugs, clozapine, haloperidol, frontal cortex, striatum, neuroplasticity
Introduction
Schizophrenia is a chronic and disabling brain disorder that affects approximately 1 percent of the world population (Andreasen 1995; Carpenter and Buchanan 1994). Antipsychotic drugs (APDs) can substantially improve the symptoms but they do not generally cure the disease (Frankenburg 1999; Kane et al 2003; Kane and Marder 1993). APDs take time to exhibit their full therapeutic effect, and it has been suggested that molecular adaptations contribute to their mechanism of action (Hyman and Nestler 1996). Indeed, APDs induce gene expression and activate transcription factors (Dragunow et al 1990; Konradi and Heckers 1995; Konradi and Heckers 2001; Robertson and Fibiger 1992). APDs are classified into conventional and atypical APDs. A commonly used conventional APD is haloperidol; the prototypical atypical APD is clozapine (Frankenburg 1999). Haloperidol and clozapine differ in their pharmacological profile and their side-effect profile. Whereas haloperidol is predominantly an inhibitor of dopamine D2 receptors (Creese et al 1976), clozapine has wide-spread, and not particularly strong, effects at dopamine receptors, serotonin receptors, cholinergic receptors and others (Meltzer 1994; Remington and Chong 1999). Extrapyramidal side effects (tardive dyskinesia) are common during treatment with haloperidol, particularly if haloperidol is administered at higher concentrations and over extended periods of time (Casey 1999). Clozapine does not cause tardive dyskinesia but can cause other side effects including potentially life-threatening agranulocytosis (Casey 1996; Iqbal et al 2003). Animal research has shown that haloperidol affects gene expression predominantly in the striatum, while clozapine affects gene expression in other brains areas such as the frontal cortex (FC) and nucleus accumbens, in addition to the striatum (Nguyen et al 1992; Robertson and Fibiger 1992; Robertson et al 1994).
We have treated rats for 26 days in single, daily injections with APDs, and compared the gene expression profile of APDs in the FC and striatum to vehicle-treated controls. We used Affymetrix U34A gene arrays, which examine the expression of 8,800 genes simultaneously. In four treatment groups, rats received one of two doses of haloperidol or clozapine. Data of these four groups were compared to gene expression levels of a control group. We were interested in significant gene regulations common to all treatment groups, with the assumption that gene expression changes responsible for side effects should be restricted to drug type and/or drug dose and should be diluted out by the other treatments.
Methods and Materials
Animals
Male Sprague-Dawley rats (Taconic Farms, Germantown, NY) weighing 200 g at the beginning of the experiment were housed four to a cage on a 12 hr light/dark cycle. The animals were allowed one week for habituation to the colony prior to the first drug administration. All rats received single, daily injections, administered intraperitoneally. Animal care and experimental procedures conformed to PHS Policy on Humane Care and Use of Laboratory Animals.
Drug treatment
Animals were assigned randomly to the following groups: controls (vehicle-treated; n=5), clozapine-L (8 mg/kg/day; n=6), clozapine-H (20 mg/kg/day; n=5), haloperidol-L (0.2 mg/kg/day; n=6), haloperidol-H (0.5 mg/kg/day; n=6). Haloperidol and Clozapine were purchased from Sigma (St. Louis, MO) and Tocris Cookson, Inc. (Ellisville, MO), respectively, and dissolved in 0.1N acetic acid, pH 5–6. The injections were administered i.p. once per day for twenty-six consecutive days. Vehicle injections consisted of 0.1N acetic acid, pH 5–6. Animals were sacrificed by rapid decapitation twenty-four hours after the final injection. Tissue samples from frontal cortex (FC), and striatum were dissected and frozen immediately. FC was defined as the cortical area rostral to bregma 4.0 mm, which includes orbital- and medial prefrontal cortex as well as a part of the secondary motor cortex (McAlonan and Brown 2003; Paxinos and Watson 1998; Uylings et al 2003). Striatum included caudate-putamen and nucleus accumbens (Paxinos and Watson 1998). All samples were stored at −70°C prior to RNA preparation.
Sample preparation: Sample and array processing
RNA was extracted from approximately 20–30 mg tissue using the RNAgent kit (Promega, Madison WI). RNA quality was assessed in an analytical gel, and 7 µg total RNA was used for cDNA synthesis with the SuperScript double-stranded cDNA synthesis kit (Invitrogen Corp., Carlsbad, CA). In vitro transcription was performed with the Enzo-IVT kit (Enzo Biochem, Farmingdale, NY). Biotinylated RNA was hybridized to the RG-U34A array (Affymetrix, Santa Clara, CA), and washing and staining was carried out according to company protocol (www.Affymetrix.com). Samples from individual rats were hybridized to individual arrays. The Affymetrix RG-U34A array contains 8800 genes; each gene is represented by 16 perfectly matched 25-mer oligonucleotides, and the same number of one-mismatch oligonucleotides to provide values for non-specific binding.
Quality control criteria
Tissue preparation and RNA extractions were performed in a single batch by the same investigators to limit experimental variability. All samples yielded equal amounts of RNA and of biotinylated RNA. An average of 80 ± 13 µg (mean ± STDEV) of biotinylated RNA was obtained from the FC for the in vitro transcription, and 92 ± 15 µg biotinylated RNA from the striatum. All quality control criteria defined by Affymetrix were met by the samples, and no differences between the experimental groups were observed. The average percent ‘present’ call across all FC arrays was 45.7 ± 1.8%, across all striatum arrays was 45.8 ± 2.6%, and the 3’/5’ GAPDH and β-actin ratios were 2.0 ± 1.2, and 1.5 ± 0.3 for FC, and 1.5 ± 0.8, and 1.5 ± 0.5 for the striatum (all average ± STDEV). Note that GAPDH mRNA was upregulated by APD treatment in the FC. Background (44.2 ± 3.5, FC; 50.5 ± 7.5, striatum) and Noise (1.5 ± 0.2, FC; 1.8 ± 0.4, striatum) were comparable between all treatment groups.
Data Analysis
Four different programs were used for data analysis: DNA-Chip Analyzer using the PM/MM difference model (dChip version 1.3, http://www.biostat.harvard.edu/complab/dchip/, see also Li and Hung Wong 2001; Li and Wong 2001), Microarray Suite 5.0 (Affymetrix), RMAExpress (Bolstad et al 2003; Irizarry et al 2003), and Gene Microarray Pathway Profiler (http://www.genmapp.org/, Dahlquist et al 2002; Doniger et al 2003). Hierarchical clustering was performed with the dChip program, which bases hierarchical clustering on previously published algorithms (Eisen et al 1998; Golub et al 1999). Redundant probe sets were excluded from the clustering analyses (figure 1, figure 10, figure 12).
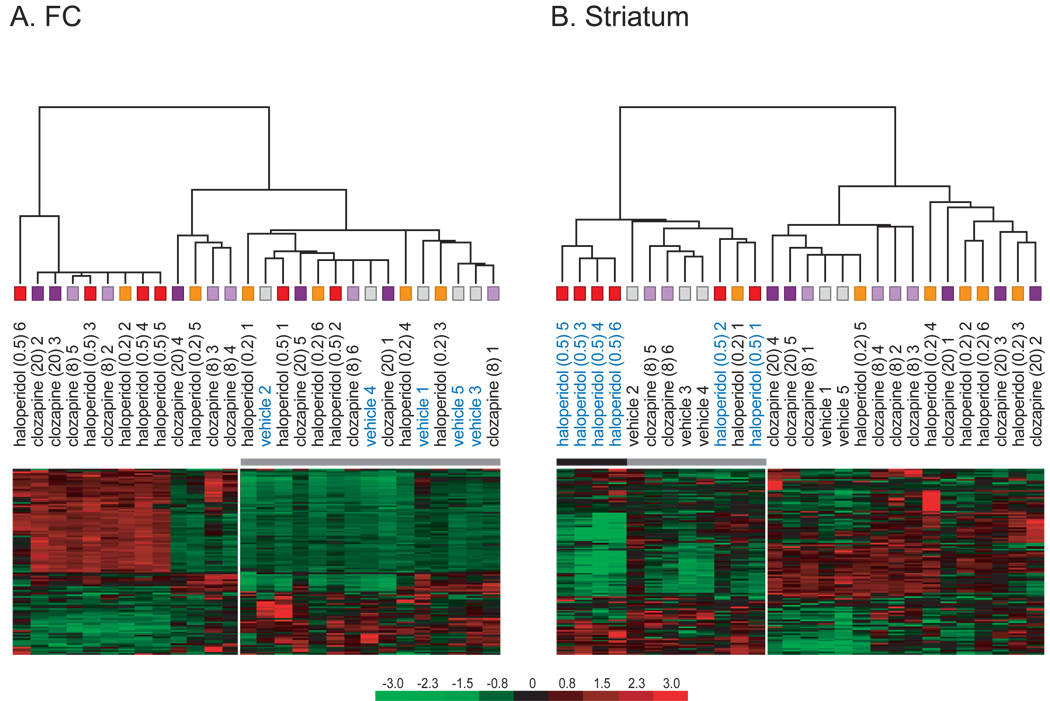
Figure 1. Unsupervised, hierarchical clustering of samples in the FC and the striatum.
Clustering analysis was performed in the FC (A) and the striatum (B), using 120 genes with the highest variation across treatment groups in each brain area, which were expressed in at least 60% of all samples. In the FC, vehicle samples clustered together (p=0.03), whereas haloperidol-H samples clustered in the striatum (p=0.02). No other significant sample clustering was observed.
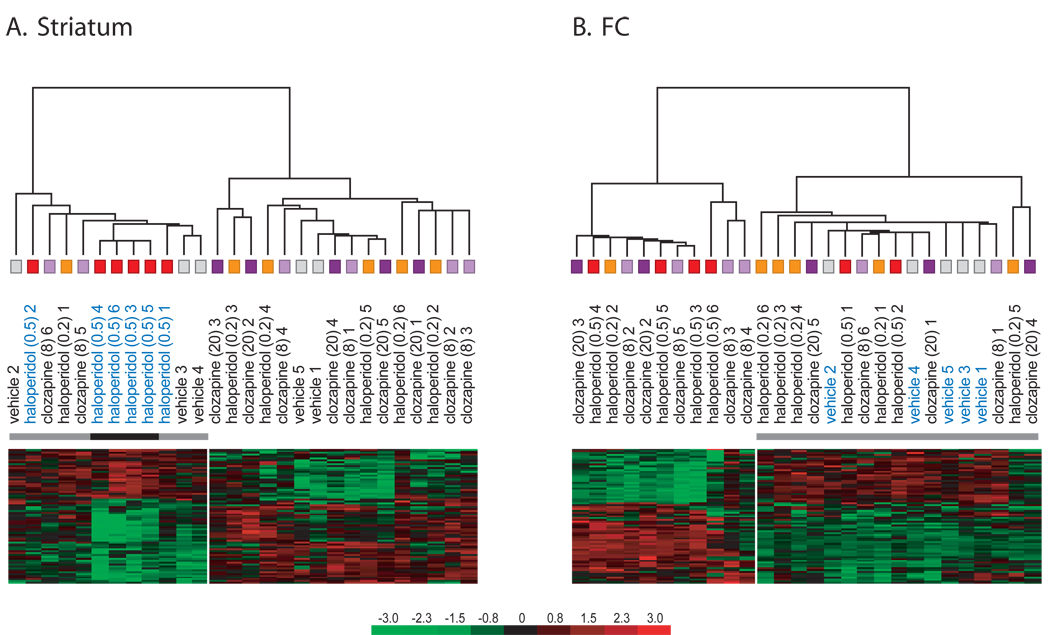
Figure 10. Sample clustering demonstrates a unique molecular profile induced by high doses of haloperidol in the striatum.
The 100 genes with the biggest difference in expression levels between haloperidol-H and vehicle (p-level of <=0.05; ‘present’ in >=60 % of all samples) within each brain area were used for hierarchical clustering in the striatum (A) and the FC (B). In the striatum, these genes clustered haloperidol-H samples (p<0.001; A). In contrast, in the FC, haloperidol-H samples were spread between the other APD samples (B), while vehicle samples were clustered.
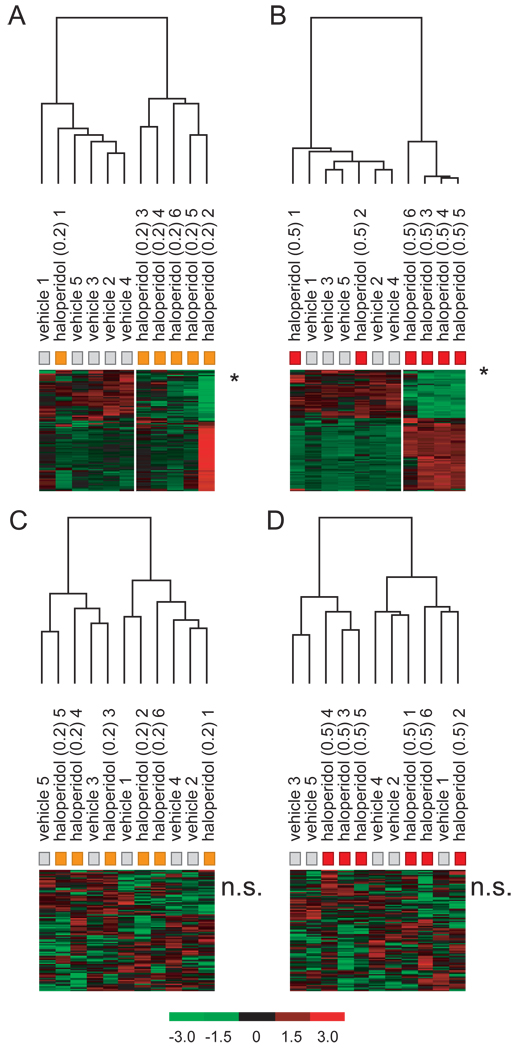
Figure 12. Genes affected by clozapine-treatment in the FC are predictive of chronic haloperidol exposure.
Onehundred genes with the biggest difference in expression between clozapine (H and L) and vehicle samples in the FC were used for hierarchical clustering of haloperidol samples. A. The ‘clozapine’ genes cluster samples of rats treated with 0.2 mg/kg haloperidol, p = 0.01. B. The ‘clozapine’ genes also cluster samples of rats treated with 0.5 mg/kg haloperidol, p = 0.05. Onehundred genes that were not regulated by clozapine did also not cluster haloperidol 0.2 mg/kg (C) or haloperidol 0.5 mg/kg (D) samples.
GenMAPP was used to examine the biological context of the findings. GenMAPP is designed to visualize gene expression data on MAPPs representing biological pathways or any category defined by the investigator. A MAPP is a file prepared with the GenMAPP program that contains a group of genes assembled by the investigator. We have drawn MAPPs of many different groups of genes (>300 MAPPs) that we consider functionally related, such as second messenger pathways, receptors responsive to the same neurotransmitter (i.e. dopamine, glutamate), kinases, phosphatases, enzymes involved in glycolysis, subunits of the proteasome, pre- and postsynaptic proteins, proteins of the mitochondrial respiratory chain, proteins related to specific neurotransmitters, G-protein-coupled receptors, etc. All MAPPs were established prior to data analysis and were not influenced by the data. MAPP-finder calculates the percentage of genes changed in each map and uses this percentage for a z score based on the mean and the standard deviation of the hypergeometric distribution (see http://www.genmapp.org). Positive z scores indicate that more genes than expected are regulated in a particular MAPP, and higher scores denote higher confidence. We list gene categories that obtained a z score above 3 in the FC, and above 2 in the striatum, and that had significant permuted p-values.
Real-time reverse transcriptase-polymerase chain reaction
For the real-time reverse transcriptase-polymerase chain reaction (Q-PCR) experiments, 12 vehicle-treated control samples were compared to six clozapine-L, seven clozapine-H, five haloperidol-L and five haloperidol-H samples. Complementary DNA was synthesized from 2.5 µg and 3.5 µg of total RNA for FC and striatum respectively, with the SuperScript First-Strand Synthesis System for real-time quantitative PCR (Invitrogen Corp), and oligonucleotide deoxythymidine primer. A primer set for each gene was designed with the Primer3 software (www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi.), for amplicons of 100 to 200 base pairs. Melt curve analysis and polyacrylamide gel electrophoresis were used to confirm the specificity of each primer pair. A Q-PCR kit (Platinum SYBR Green Q-PCR SuperMix UDG; Invitrogen Corp) was used for the experiment which was carried out on a DNA engine Opticon I (MJ Research, Waltham Mass) in a volume of 20 µl, with 4 µl 1:10 diluted cDNA samples and 0.3 µM primers. The PCR cycling conditions were initially 50˚C for 2 minutes followed by 95˚C for 7 minutes followed by 39 cycles of 94˚C for 10 seconds, 55˚C for 15 seconds, and 72˚C for 30 seconds. Data were collected between 72˚C and 82˚C depending on amplicon melt temperature. A melt curve analysis was performed at the end of each Q- PCR experiment. Dilution curves were generated for each primer pair in every experiment by diluting complementary DNA from a vehicle sample to a final concentration of 1.00, 0.2, 0.04, 0.008, and 0.0016. The logarithm of the dilution values was plotted against the cycle values for the standard curve. Opticon Monitor Data Analysis Software version 1.4 (MJ Research, Waltham, Mass) was used to analyze the data. Blanks were run with each dilution curve to control for cross contamination. Dilution curves, blanks and samples were run in duplicate. Reported values were normalized to the internal standard β-actin (Genbank accession number NM_031144). β-Actin was not regulated in the gene-array or Q-PCR analysis.
Results
A triple data analysis was performed with dChip, MAS 5.0 and RMAExpress
Most differences in expression levels of abundantly expressed genes treated with APDs and analyzed with dChip 1.3 were below two-fold. Genes with lower expression levels and high signal-to-noise ratios yielded higher fold-differences. For example, syntaxin 12 was expressed 8-fold higher in the FC of APD treated rats than in vehicle-treated controls, but was above detection level in only 9 of 28 samples (32 % ‘present’ call). We set our parameters to exclude genes that were below expression threshold in more than 50% of all samples, which excluded approximately half of all genes on the array, including syntaxin 12.
We first performed a dual analysis using the dChip 1.3 and MAS 5.0 programs, with the emphasis on dChip 1.3. The programs apply different strategies for normalization and calculation of expression levels, and changes that reach significance in both analyses are of higher confidence. Because both programs calculate expression levels differently, we were predominantly interested in levels of significance and direction of regulation (up versus downregulation), with lesser emphasis on fold-different expression level. MAS 5.0 yielded higher fold-difference levels (1.5 fold higher), but the correlation coefficient between dChip 1.3 and MAS 5.0 for all expression levels of significantly regulated genes presented here (p<=0.05 with dChip 1.3), was 0.97 for the FC dataset and 0.98 for the striatum dataset. The dChip 1.3 program had a higher likelihood than MAS 5.0 to yield significant downregulations in the FC dataset, and not all downregulations in the FC, observed with dChip 1.3, were verified with MAS 5.0 (see for instance figure 2). Because the regulation patterns of multiple copies of the same gene were more consistent with dChip 1.3 than MAS 5.0, and because of our past experience with independent confirmation of results (Konradi et al 2004a; Konradi et al 2004b), all data presented here are based on dChip 1.3. Although regulations in MAS 5.0 were similar, in some cases they did not reach significance. These cases are indicated in the figures by a number sign (#). However, the same gene families reached significance in the MAPPfinder analysis irrespective of their expression calculation with MAS 5.0 or dChip 1.3, yielding comparable z scores, due to the fact that additional genes in the same families reached significance with MAS 5.0. After all data were calculated, we performed a third analysis with RMAExpress. Data calculated with RMAExpress matched data obtained with the dChip 1.3 calculation, although this program uses, again, a different strategy for background adjustment, normalization and data summarization (Bolstad et al 2003; Irizarry et al 2003).
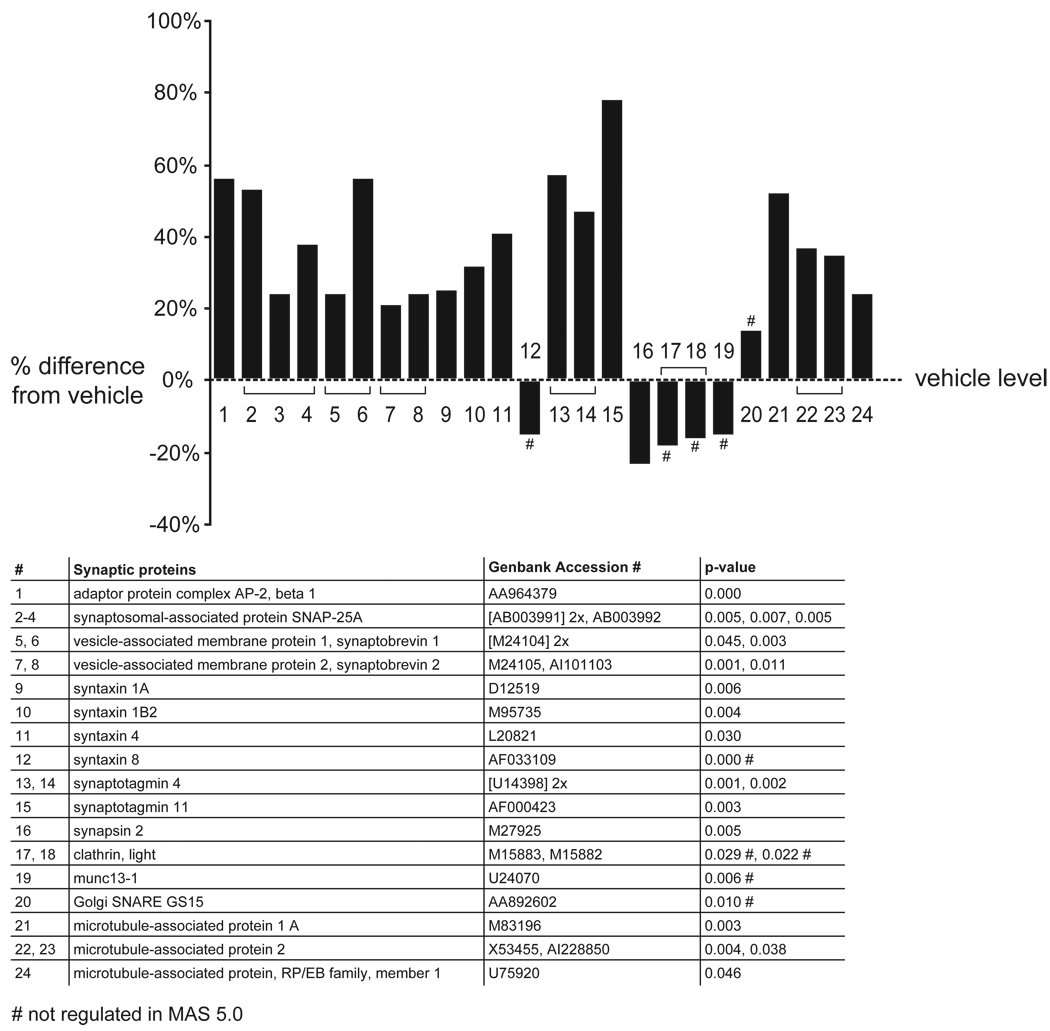
Figure 2. Upregulation of mRNAs for presynaptic proteins in the FC.
The graphs depict the effect of APDs in the FC, comparing vehicle-treated rats to all APD treatments combined. The percentage induction or reduction of mRNAs is shown. Multiple probe sets of the same gene are joined by brackets. Names of regulated genes, Genbank accession numbers and p-values are shown in the tables below the graphs. The number sign (#) denotes genes that were only regulated in dChip 1.3 but not MAS 5.0. The section sign (§) denotes genes that were also significantly regulated in the striatum.
The graph shows the percentage induction or reduction of genes associated with synaptic vesicle function and microtubules. Thirteen genes involved in synaptic vesicle fusion and recycling, or associated with microtubules, were upregulated in the FC. Of four genes that were downregulated with the dChip 1.3 program, three were unchanged in MAS 5.0. Upregulations were concordant between both programs. None of the genes shown were altered in the striatum.
Unsupervised clustering analysis of expression profiles in the FC and striatum
Hierarchical clustering groups samples with similar gene expression levels and presents the results in a dendrogram. Samples within a cluster have more closely related gene expression levels to one another than samples assigned to different clusters. Hierarchical clustering can be used to address the question if variations in gene expression levels can be explained by treatment. For this purpose, genes with high variability, irrespective of their function, are used to cluster samples. Because genes with very small, non-specific signals also have a high variability, these genes have to be first removed from the analysis to reduce noise. We therefore included in the hierarchical clustering analysis only genes that are above background in at least 60% of all samples. Clustering analysis was performed in the FC and the striatum, using 120 genes with the highest variation across all treatment groups, expressed in at least 60% of all samples (only one set of redundant probe sets was included). In this analysis, vehicle samples of the FC clustered together, whereas APD treatment groups were interspersed (figure 1A). In contrast, in the striatum, the haloperidol-H samples clustered together (figure 1B). Thus, in the FC, the vehicle samples were most different from APD samples, while APD, samples were similar to each other. In the striatum, haloperidol-H samples were different from the other APD samples.
Gene expression changes observed in the FC
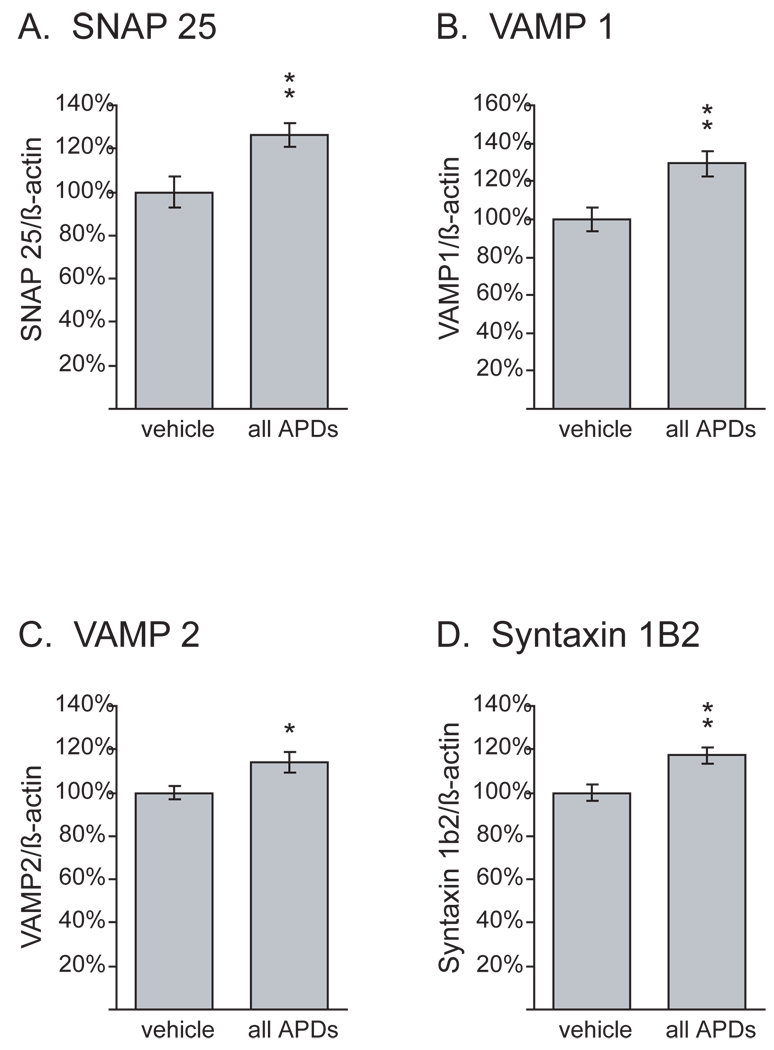
The MAPPfinder analysis of mRNA expression levels in the FC revealed the most significant changes in mRNAs coding for synaptic vesicle docking- and microtubule-associated proteins (figure 2; z score >5; permuted p-value: 0). The regulation of five presynaptic proteins in the FC was further analyzed with Q-PCR. We chose SNAP 25, vesicle associated membrane protein (VAMP) 1 and 2, syntaxin 1B2 and synapsin 2 (not shown) for verification. In the gene array analysis, the former four were upregulated (figure 2), whereas synapsin 2 was downregulated. In the Q-PCR analysis, a significant upregulation was observed with SNAP 25, vesicle associated membrane protein (VAMP) 1 and 2, and syntaxin 1B2 (figure 3), but synapsin 2 was unchanged (not shown). Thus, the upregulations of mRNAs were consistent between dChip 1.3, MAS 5.0 and Q-PCR, but the downregulation of synapsin 2 could not be confirmed.
Figure 3. Verification of upregulation of synaptic proteins in the FC with Q-PCR.
Upregulation of SNAP 25 (A), VAMP1 (B) and 2 (C), and syntaxin 1b2 (D) by APDs in the FC was verified with Q-PCR. Data were normalized to β-actin, which was not regulated in the gene arrays. N=12 vehicle-treated controls, 23 APDs (6 clozapine-L, 7 clozapine-H, 5 haloperidol-L, 5 haloperidol-H). ** p<=0.01, * p<=0.05.
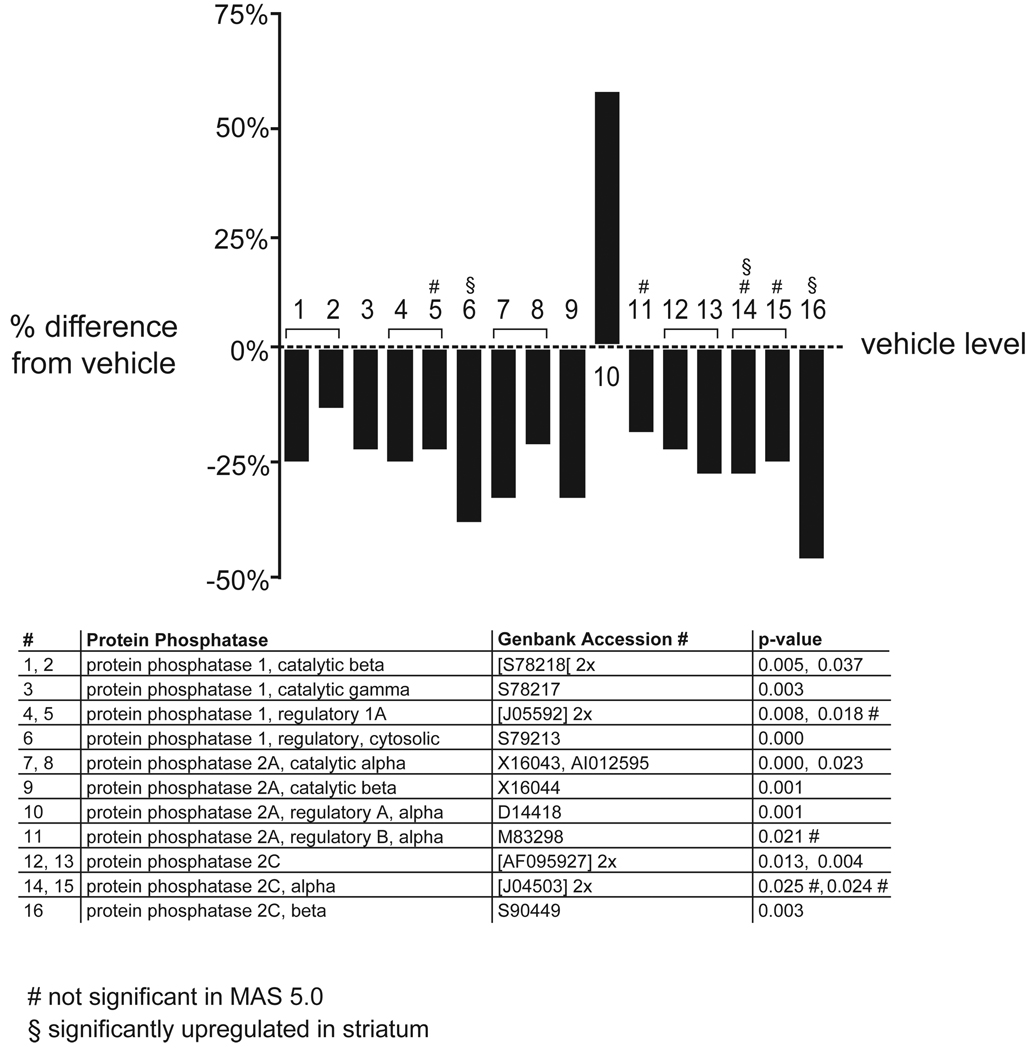
A general downregulation of protein phosphatases was observed in the FC (figure 4; z score >5; permuted p-value: 0). Both, regulatory and catalytic subunits of protein phosphatase (PP) 1 were downregulated, as was PP 2C. Similarly, catalytic subunits for PP 2A and the regulatory B alpha subunit were downregulated, while the regulatory A alpha subunit was upregulated (figure 4). Upregulation of the regulatory subunit should decrease the activity of the catalytic subunit. The downregulation in mRNAs of protein phosphatases was accompanied by an upregulation of various serine-threonine kinases. CaM kinase II alpha, CaM kinase IV, PKC, and the catalytic subunit of PKA were all upregulated, while the regulatory subunit of PKA and the PKA inhibitor, PKI beta (Van Patten et al 1992; Van Patten et al 1991) were downregulated (figure 5; z score >3; permuted p-value: 0.004). CaM kinase II beta and delta were downregulated.
Figure 4. Downregulation of mRNAs for protein phosphatases in the FC.
The graphs depict the effect of APDs in the FC, comparing vehicle-treated rats to all APD treatments combined. The percentage induction or reduction of mRNAs is shown. Multiple probe sets of the same gene are joined by brackets. Names of regulated genes, Genbank accession numbers and p-values are shown in the tables below the graphs. The number sign (#) denotes genes that were only regulated in dChip 1.3 but not MAS 5.0. The section sign (§) denotes genes that were also significantly regulated in the striatum.
PP1 and PP2A exist as multimeric holoenzymes with a limited number of catalytic subunits complexed to a variety of regulatory subunits. PP1 is a heterodimeric complex of catalytic and regulatory subunits, while PP2A forms a heterotrimeric complex between the catalytic subunit and the regulatory A and B subunits (Sim et al 2003). PP2C is a monomeric enzyme (Price and Mumby 1999). A downregulation in mRNA levels for protein phosphatases was observed, except for the regulatory A alpha subunit for PP2A, which was upregulated, in essence causing an inhibition of PP2A. Genes that did not reach significant with MAS 5.0 (denoted with #), still showed a pronounced downregulation in that program. None of the genes presented were similarly regulated in the striatum, although three had a significant opposite regulation (denoted with §).
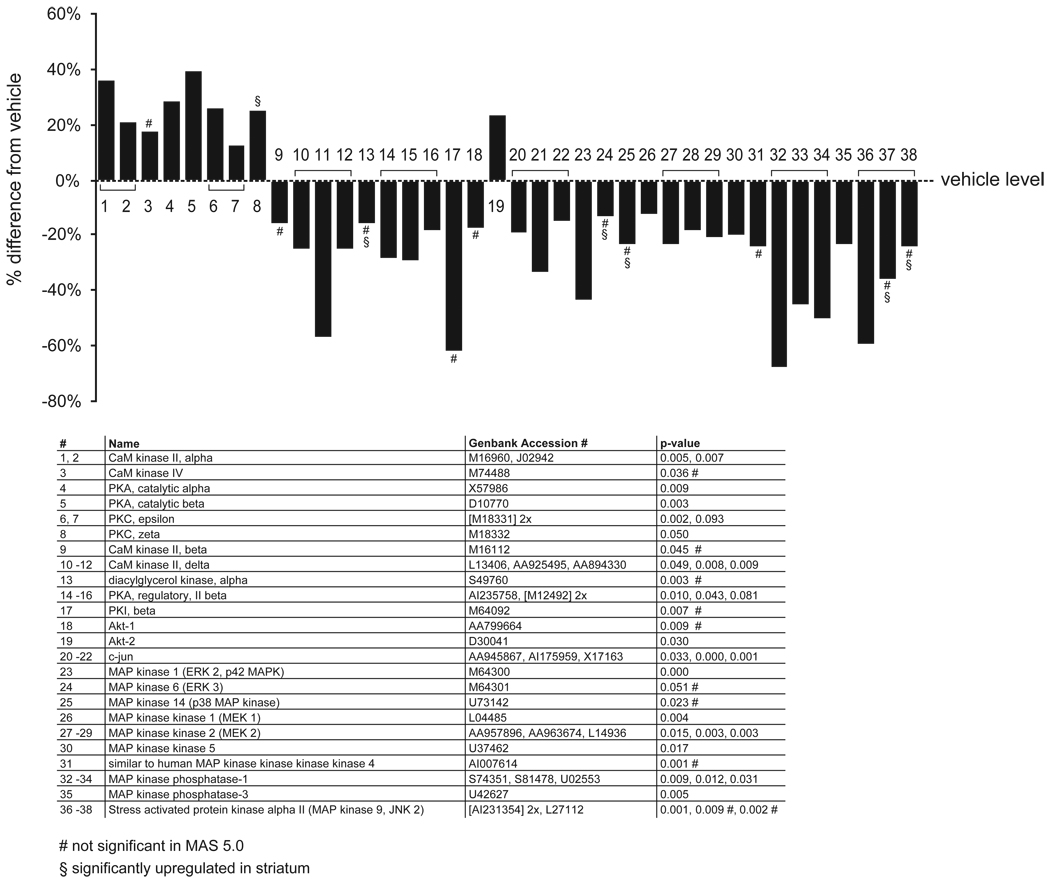
Figure 5. Altered regulation of protein kinase genes.
The graphs depict the effect of APDs in the FC, comparing vehicle-treated rats to all APD treatments combined. The percentage induction or reduction of mRNAs is shown. Multiple probe sets of the same gene are joined by brackets. Names of regulated genes, Genbank accession numbers and p-values are shown in the tables below the graphs. The number sign (#) denotes genes that were only regulated in dChip 1.3 but not MAS 5.0. The section sign (§) denotes genes that were also significantly regulated in the striatum.
Many Ca2+-activated kinases were upregulated in the FC, while members of the MAP kinase pathways were mostly downregulated. Not all regulations were significant in MAS 5.0.
mRNA’s from four separate MAP kinase cascades were downregulated in the FC by APDs (figure 5; z score >3; permuted p-value: 0.004). First, the members of the ERK1/2 cascade, MAP kinase kinase 1 and 2, and MAP kinase 1 (ERK 2), (Grewal et al 1999). The phosphorylation substrates of this cascade, microtubule-associated protein 1A, and microtubule-associated protein 2, were both upregulated (figure 2). The second MAP kinase pathway, the stress-activated c-Jun N-terminal kinase (JNK) pathway (Harper and LoGrasso 2001; Mielke and Herdegen 2000), showed a downregulation of stress-activated protein kinase alpha II (JNK2). The substrate of this pathway, c-jun (Mielke and Herdegen 2000) was also downregulated. The third MAP kinase pathway, the p38 MAP kinase pathway (Harper and LoGrasso 2001; Mielke and Herdegen 2000), was represented with a downregulation of p38 MAP kinase. Finally, a downregulation of MAP kinase kinase 5 was observed. This kinase phosphorylates MAP kinase 5 (Zhou et al 1995).
MRNA levels for the enzymes involved in the dephosphorylation of MAP kinases, MAP kinase phosphatase 1 (Qian et al 1994) and 3 (Mourey et al 1996), were downregulated. MAP kinase phosphatase 2 was below detection level in the samples.
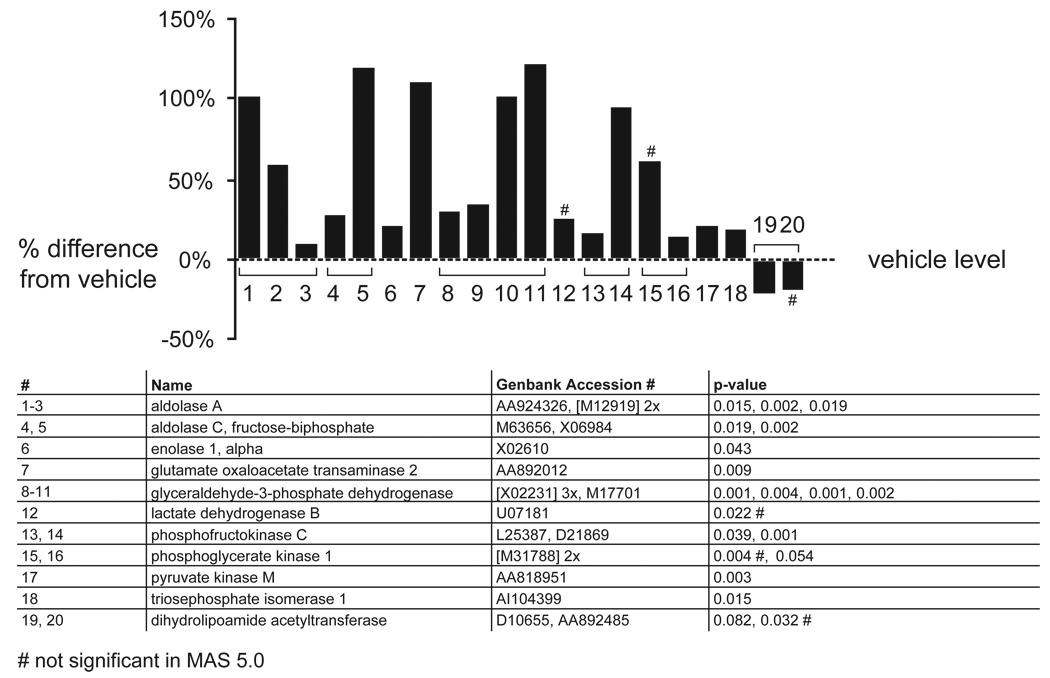
APDs also increased mRNAs of proteins involved in glycolysis and gluconeogenesis (figure 6; z score >4; permuted p-value: 0). The upregulation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was confirmed with Q-PCR (data not shown). Genes involved in energy regulation are also present in the GO database, and received the highest z scores (>6) when the data were analyzed with this database.
Figure 6. Upregulation of genes involved in energy metabolism in the FC.
The graphs depict the effect of APDs in the FC, comparing vehicle-treated rats to all APD treatments combined. The percentage induction or reduction of mRNAs is shown. Multiple probe sets of the same gene are joined by brackets. Names of regulated genes, Genbank accession numbers and p-values are shown in the tables below the graphs. The number sign (#) denotes genes that were only regulated in dChip 1.3 but not MAS 5.0. The section sign (§) denotes genes that were also significantly regulated in the striatum.
A predominant upregulation in mRNA levels for proteins involved in glycolysis was observed with the dChip 1.3 program. None of the genes were changed in the striatum. Note the significant upregulation of GAPDH, a gene frequently used as a normalization control.
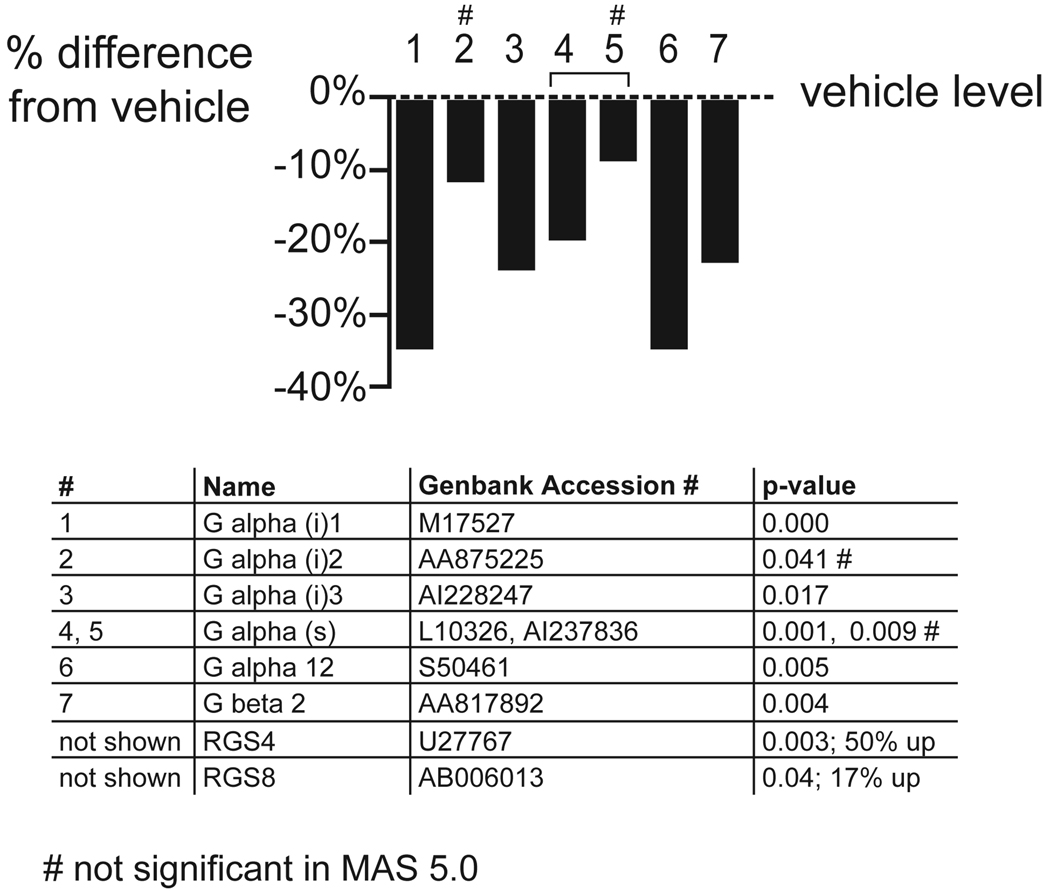
Finally, all three subtypes of the inhibitory G alpha (i) protein and the one stimulatory G alpha (s) protein on the array, were downregulated in the FC after chronic APD treatment (figure 7; z score >3; permuted p-value: 0.008). The regulators of G-protein signaling, RGS 4 and RGS 8, were upregulated after treatment with APDs.
Figure 7. Downregulation of G protein alpha subunits in the FC.
The graphs depict the effect of APDs in the FC, comparing vehicle-treated rats to all APD treatments combined. The percentage induction or reduction of mRNAs is shown. Multiple probe sets of the same gene are joined by brackets. Names of regulated genes, Genbank accession numbers and p-values are shown in the tables below the graphs. The number sign (#) denotes genes that were only regulated in dChip 1.3 but not MAS 5.0. The section sign (§) denotes genes that were also significantly regulated in the striatum.
A downregulation of mRNA levels of G alpha subunits, and of G beta 2, was observed in the FC. RGS4 and RGS8 were upregulated.
Gene expression changes observed in the striatum
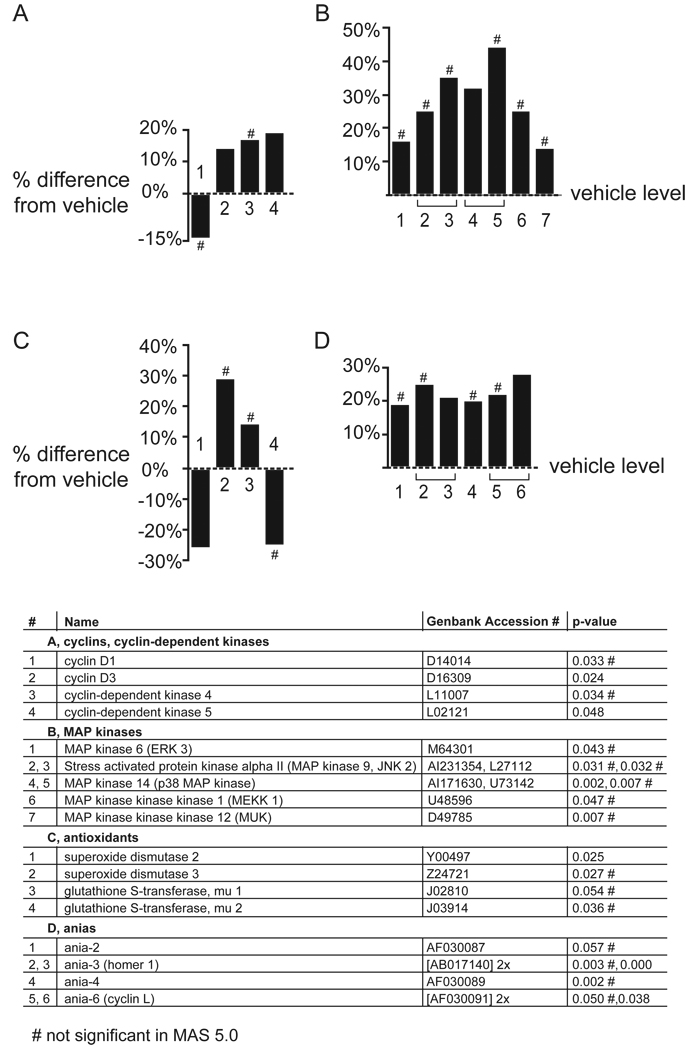
In the striatum, mRNA expression of the gene families affected in the FC was mostly unchanged (figure 2, figure 4–figure 7, section sign). Altered mRNA expression was observed for cyclins and cyclin-dependent kinases (z score >4; permuted p-value: 0.002; figure 8A), the MAP kinase pathway (z score >3; permuted p-value: 0.013; figure 8B), antioxidants (z score >2; permuted p-value: 0.024; figure 8C) and activity and neurotransmitter-induced early genes (ANIAs; z score >3; permuted p-value: 0.013; figure 8D). The gene family representing the Wnt signaling pathway, which included cyclins (figure 8A) and protein kinase C (PKC; figure 9) was upregulated (z score >2; permuted p-value: 0.04). All gene families received lower z scores and p-values than the gene families regulated by APDs in the FC. Although the regulations in MAS 5.0 in the striatum were in the same direction as the regulations observed in dChip 1.3 (correlation coefficient r=0.98), many did not reach significance (p<=0.05) in MAS 5.0.
Figure 8. Gene families regulated by APDs in the striatum.
The four gene families that were most affected by APD treatment in the striatum were (A) cyclins and cyclin-dependent kinases, (B) MAP kinases, (C) antioxidants and (D) ANIAs. Z scores were lower than in the FC, and many of the genes did not reach significance with MAS 5.0 (#).
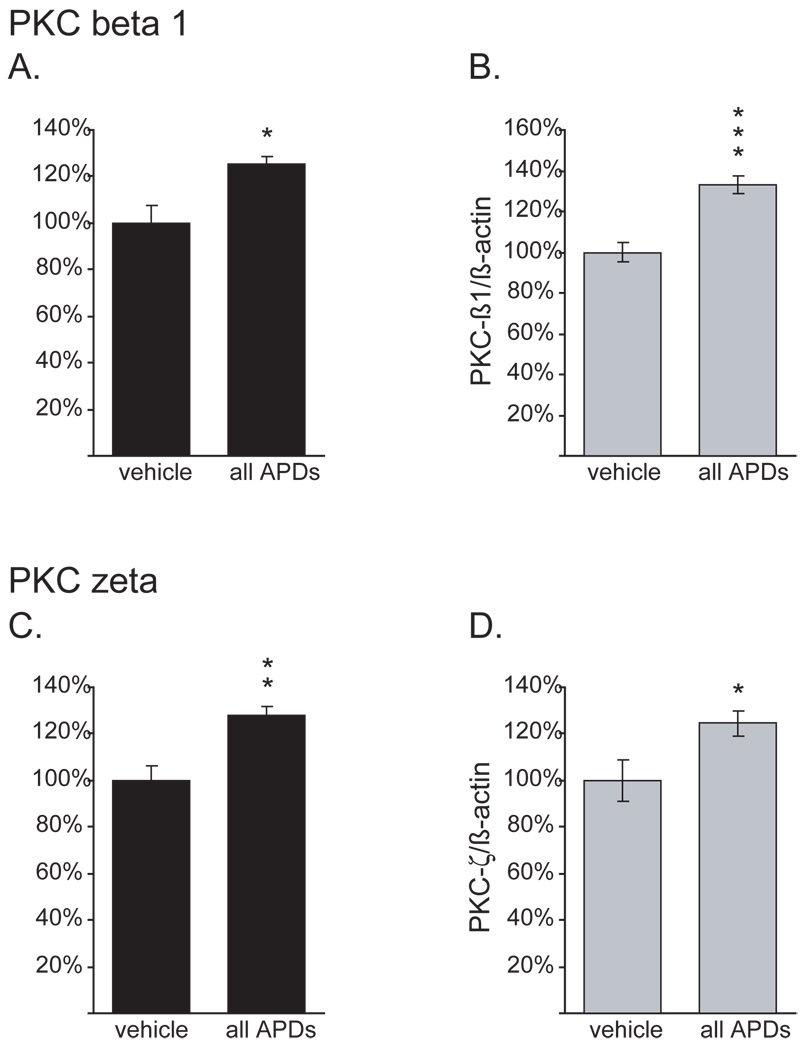
Figure 9. mRNA regulation of PKC beta 1 and PKC zeta by APDs in the striatum.
A, B, PKC beta 1 (K03486), C, D, PKC zeta (M18332). A, C, gene array data, B, D, Q-PCR analysis normalized to β-actin.
Comparison of expression profiles of high dose of haloperidol, lower dose of haloperidol, and clozapine
The unsupervised, hierarchical clustering algorithm demonstrated that treatment with higher doses of haloperidol was associated with a particular molecular profile in the striatum (figure 1B). We used the 100 genes with the biggest difference in expression levels between haloperidol-H and vehicle samples (p-level of <=0.05; ‘present’ in >=60 % of all samples), in the striatum (figure 10A) and the FC (figure 10B), to perform hierarchical clustering. In the striatum, the expression level of these genes was significantly different between haloperidol-H and all the other APDs (p<0.001; figure 10A). In contrast, in the FC, haloperidol-H samples were spread between the other APD samples whereas vehicle samples were clustered (figure 10B), indicating that genes that were differently expressed in haloperidol-H samples were similar across all APD treatments. Therefore, gene expression levels after haloperidol-H were similar to other APDs in the FC, but dissimilar from other APDs in the striatum.
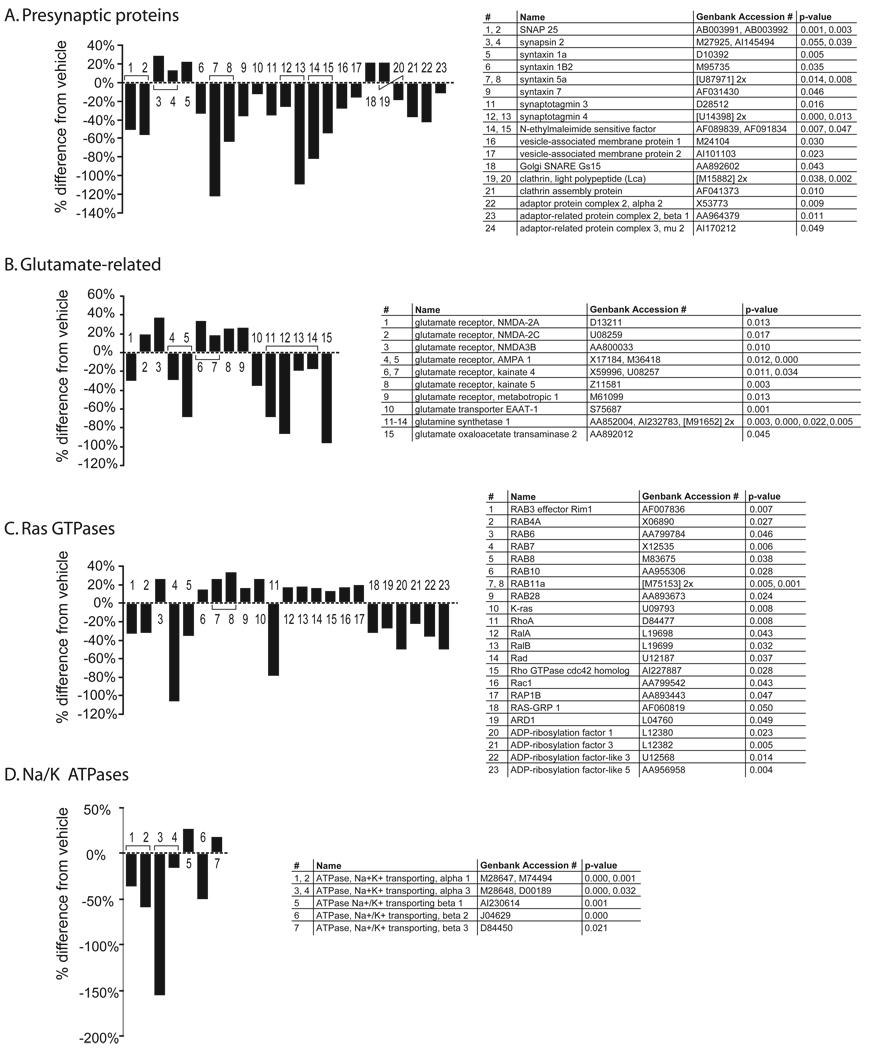
A comparison of haloperidol-H treatment to the combined haloperidol-L and clozapine-H and clozapine-L treatment groups showed a significant difference in the regulation of genes associated with synaptic vesicle proteins (z score >4; figure 11A), glutamate receptors, glutamate synthesis and uptake (z score >3; figure 11B), Ras GTPase superfamily (z score >3; figure 11C) and Na+/K+ ATPases (z score >2; figure 11D). Eight percent of all genes measured in the striatum were regulated differently by haloperidol-H than by all the other APD treatments, a significant difference (at p<0.05 a median false-discovery rate <5%), whereas only 1% of all genes measured in the FC were regulated differently by haloperidol-H than the other APDs, a number that was equal to chance (at p<0.05 a median false discovery rate >100%).
Figure 11. Differential effect of Haloperidol-H on the expression of various gene families in the striatum.
Compared to the other APD treatments, haloperidol-H treatment induced a downregulation of presynaptic proteins (A), a dysregulation of glutamate-related proteins (B), GTPases (C), and a downregulation of Na+/K+ ATPases, in the striatum.
Cluster analysis with the genes of figure 11 showed that they are regulated differently by haloperidol-H in the striatum, but not in the FC (not shown). These data demonstrate that haloperidol-H has a molecular profile in the striatum that is (a) present in the striatum but not the FC, and (b) not associated with APD treatment in general.
Predictive validity of APD treatment in the FC
Because all APDs evoked a similar molecular profile in the FC, we examined if the molecular profile obtained with the two concentrations of clozapine could be used to predict treatment with either concentration of haloperidol. In a comparison of clozapine-treated rats (n=11; 5 treated with 20 mg/kg, 6 treated with 8 mg/kg) to vehicle-treated rats, 100 genes that were present in at least 60% of all samples had a higher than 1.4 fold different expression at a confidence level of p<=0.05 (median false discovery rate < 5%). These genes were used for hierarchical clustering of haloperidol samples, and they clustered both haloperidol-L (figure 12A) and haloperidol-H samples (figure 12B) separate from vehicle samples. In a control experiment, 100 genes that were not affected by clozapine-treatment did also not cluster haloperidol-L (figure 12C) or haloperidol-H (figure 12D) samples.
Discussion
Changes in mRNA levels have been previously correlated with similar changes in protein levels. For example, acute treatment with haloperidol caused upregulations of c-fos, proenkephalin, and synapsin II mRNAs that translated into upregulations of these proteins in the striatum (Chong et al 2002; Dragunow et al 1990; Hong et al 1979; Hong et al 1985; Konradi and Heckers 1995; Konradi et al 1993; Leveque et al 2000). While these findings are encouraging for microarray experiments, it is important to keep in mind that changes in mRNA levels might not always translate into changes in protein levels.
We have previously shown that after 12 days of single, daily i.p. injections of haloperidol, the rat striatum has an altered molecular response to the drug, as demonstrated by the decreased ability to induce c-fos expression, as well as by decreased transcription factor binding to the AP-1 enhancer element (Konradi et al 1993; Leveque et al 2000). Repeated haloperidol-treatment leads furthermore to an upregulation of the proenkephalin gene, which has been shown to be stable during 7 days to 8 months of treatment (Chong et al 2002; Delfs et al 1994; Dragunow et al 1990; Hong et al 1979; Hong et al 1985; Konradi and Heckers 1995; Konradi et al 1993; Leveque et al 2000). We therefore decided that 26 days of APD exposure should be sufficient to obtain the mRNA expression pattern of chronic APD treatment. Although this profile might change further over time, it should provide helpful insights into the molecular consequences of APD exposure. To avoid interference with gene expression due to spikes in APD levels immediately after injection, we sacrificed the rats 24 hours after the final, chronic injection. In past studies, levels of proenkephalin mRNA after 11 days of haloperidol-treatment were similar 30 minutes and 24 hours after the last injection (Konradi et al 1993; Leveque et al 2000). As for drug withdrawal, it has been shown that a single dose of haloperidol has a near-terminal elimination half-live of 6.6 days in rat brain, and is detectable for 21 days after dosing, well beyond the 24 hours we chose (Cohen et al 1992). Interestingly, the elimination half-life in human brain tissue was estimated to be similar, at 6.8 days (Kornhuber et al 1999). Nevertheless, observed fluctuations in the D2 receptor occupancy rate of haloperidol during a 24-hour period (Kapur et al 2000) would suggest that some of the observed effects on mRNA expression are due to falling APD levels.
We chose the conventional APD, haloperidol and the atypical APD, clozapine, because of their broad use in patients and in animal models (Fibiger 1994; Frankenburg 1999; Kane and Marder 1993; Konradi and Heckers 2001). Each APD was used at a concentration that is known to induce gene expression in the rat striatum after acute treatment (haloperidol-H, clozapine-H), (Leveque et al 2000; MacGibbon et al 1994; Robertson and Fibiger 1992), and at a second, lower concentration that is within the range of APD concentrations administered in humans (haloperidol-L and clozapine-L) (Aymard et al 1997; Conley et al 1999; Kane and Marder 1993; Purdon et al 2000). The common denominator of the drugs and drug concentrations used is their antipsychotic action in humans, whereas side effects are dissimilar between both types of drugs and between the high and low drug concentrations (Mortimer 2003; Wirshing et al 2003). By combining the different treatment paradigms and by tracking the common gene regulation pattern across all of them, we hoped to minimize gene expression patterns responsible for side effects. Although the study was carried out in healthy rats, some of the insights gained might be beneficial for our understanding of the molecular effects of APDs in general.
Chronic APD treatment increased mRNA levels for presynaptic proteins in the FC
Many of the upregulated proteins play key roles in neurotransmitter release by mediating the fusion of vesicles with the plasma membrane (Garner et al 2002; Murthy and De Camilli 2003). One of the upregulated genes, VAMP1, is rate-limiting in synaptic vesicle fusion (Hu et al 2002), while three of the upregulated genes, VAMP2, syntaxin 1A and SNAP25 can mediate spontaneous membrane fusion (Hu et al 2003). The upregulation of presynaptic genes, if translated into changes in protein level, could increase the probability of vesicle fusion and neurotransmitter release (Dobrunz and Stevens 1997). Thus, APDs could affect synaptic structure and facilitate synaptic activity in the FC.
Indication of increased protein phosphorylation in the FC, a mechanism that facilitates neuroplasticity
The protein phosphatases PP 1 and PP 2A dephosphorylate most serine and threonine residues in the brain (Price and Mumby 1999). Transcript levels of PPs were downregulated after treatment with APDs.
PPs counteract processes activated by kinases. These include processes of neuronal plasticity, such as synaptic vesicle fusion (Turner et al 1999), gene transcription (Whitmarsh and Davis 2000), and reorganization of the cytoskeleton (Grant and Pant 2000). PPs also counteract long-term potentiation (Winder and Sweatt 2001) and mechanisms of learning and memory. PP 1 facilitates memory decline and promotes forgetting (Genoux et al 2002). Phosphorylation of the transcription factor CREB is a step in memory formation (Frank and Greenberg 1994; Mayr and Montminy 2001), and PP1/2A dephosphorylate CREB (Hagiwara et al 1992; Wadzinski et al 1993). The decrease in PP transcripts was accompanied by an increase in expression of serine-threonine protein kinase transcripts in the FC, suggesting increased activity in cyclic AMP and certain Ca2+ -activated kinases.
The ratio of kinases and phosphatases is important for vesicle fusion, because phosphorylation is crucial for the activity of vesicle proteins (Turner et al 1999). Furthermore, transcription factor activity and histone modification/DNA accessibility depend on phosphorylation mechanisms (Davie and Spencer 1999). Our findings point toward increased levels of phosphorylation of proteins in the FC after treatment with APDs, which could result in higher transcriptional activities and a higher likelihood of vesicle fusion.
APDs decrease transcript expression of members of the MAP kinase family in the FC, but increase them in the striatum
MAP kinase family members regulate diverse biological functions. They mediate stress responses and apoptosis, but also trophic responses (Mielke and Herdegen 2000; Sweatt 2001; Thomas and Huganir 2004). Dependent on mode, strength and duration of activation, MAP kinase pathways can be involved in mechanisms of cell death or mechanisms of neuronal plasticity (Agell et al 2002). MAP kinase cascades are comprised of three serially linked kinases consisting of a MAP kinase kinase kinase which activates a MAP kinase kinase, which activates a MAP kinase. mRNAs of all MAP kinase cascades were downregulated in the FC by APDs, including the mitogenic ERK1/2 pathway (Grewal et al 1999), the stress-activated c-Jun N-terminal kinase (JNK) and p38 MAP kinase pathways (Mielke and Herdegen 2000), and the ERK5 pathway (Cavanaugh et al 2001; Zhou et al 1995). These downregulations were accompanied by a downregulation of the phosphatases that inactivate MAP kinases. Because of the contradictory actions of MAP kinase pathways, it is impossible to deduce the implication of our findings.
Surprisingly, the MAP kinase family was regulated in the opposite direction by APDs in the striatum. The upregulation of MAP kinase family members in the striatum was seen across all treatment groups. Thus, APDs affect transcripts of the MAP kinase pathways strongly and differently in different brain areas.
APDs increase transcript levels of proteins involved in glycolysis in the FC
Transcripts of proteins involved in glycolysis were upregulated by APDs in the FC, but not in the striatum. The increase in synaptic vesicle genes, decrease in protein phosphatases and increase in serine-threonine protein kinases suggest increased synaptic and biochemical activity in the FC, which requires higher expenditure of energy. One of the strongest upregulated genes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), is considered a housekeeping gene and commonly used as normalization control. Like others, we find GAPDH a poor choice for mRNA normalization (Bustin 2002).
In the FC, the different treatment paradigms with APDs yield similar molecular profiles
Hierarchical clustering demonstrated that genes that were affected by clozapine in the FC can be used to reveal exposure to haloperidol. These data demonstrate that APDs leave a characteristic molecular fingerprint in the FC in the rat, which is independent of the pharmacological background of the APD. Moreover, because of the similar molecular pattern by APDs in the FC, but not in the striatum (see below), the results favor the FC as a potential target for therapeutic actions of APDs.
In the striatum, treatment with high concentrations of haloperidol yields a unique molecular profile dissimilar from low concentrations of haloperidol, or from clozapine
Haloperidol-H affected the expression of many transcripts in the striatum different than the other APDs. In rats, prolonged treatment with higher doses of haloperidol can cause striatal cellular stress and extrapyramidal side-effects (EPS) such as vacuous chewing movements (Andreassen and Jorgensen 2000; Roberts et al 1995). The glutamate system has been implicated in EPS (Meshul et al 1996; See and Chapman 1994), and our data show dysregulation of transcripts of the glutamate system. In addition to the changes in the glutamate system our data suggest altered membrane potential (Na+/K+ ATPases), cell stress (Ras/GTPases) and loss of synapses (decrease in synaptic transcripts), which, taken to the extreme, could ultimately lead to EPS. Thus, the molecular profile of haloperidol-H in the striatum might reflect a molecular path toward the side effects that appear after prolonged treatment with high doses of conventional APDs.
Conclusion
The gene array analysis after chronic APD treatment in rats demonstrated a characteristic molecular APD profile in the FC that was exhibited independent of drug or dose. The implication of that profile was of an increase in synaptic strength and activity. In contrast, the APD profile in the striatum was dissimilar between haloperidol-H and the other treatments and indicative of neuronal and synaptic stress inflicted by higher concentrations of conventional APDs.
Acknowledgement
We thank Cheng Li, Ph.D., for his help with the dChip program, and Kristina Hanspers, M.Sc., Bruce Conklin, M.D. and the GenMAPP developing group for help with GenMAPP. We furthermore thank Francine Benes, MD.Ph.D., and Stephan Heckers, M.D., M.Sc, for support. This work was supported by the NIMH Chemical Synthesis and Drug Supply Program, and by MH63266.
References
- Agell N, Bachs O, Rocamora N, Villalonga P. Modulation of the Ras/Raf/MEK/ERK pathway by Ca(2+), and calmodulin. Cell Signal. 2002;14:649–654. doi: 10.1016/s0898-6568(02)00007-4. [DOI] [PubMed] [Google Scholar]
- Andreasen NC. Symptoms, signs, and diagnosis of schizophrenia. Lancet. 1995;346:477–481. doi: 10.1016/s0140-6736(95)91325-4. [DOI] [PubMed] [Google Scholar]
- Andreassen OA, Jorgensen HA. Neurotoxicity associated with neuroleptic-induced oral dyskinesias in rats Implications for tardive dyskinesia? Prog Neurobiol. 2000;61:525–541. doi: 10.1016/s0301-0082(99)00064-7. [DOI] [PubMed] [Google Scholar]
- Aymard N, Baldacci C, Leyris A, Smagghe PO, Tribolet S, Vacheron MN, Viala A, Caroli F. Neuroleptic-resistant schizophrenic patients treated by clozapine: clinical evolution, plasma and red blood cell clozapine and desmethylclozapine levels. Therapie. 1997;52:227–232. [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- Carpenter WT, Jr, Buchanan RW. Schizophrenia. N Engl J Med. 1994;330:681–690. doi: 10.1056/NEJM199403103301006. [DOI] [PubMed] [Google Scholar]
- Casey DE. Side effect profiles of new antipsychotic agents. J Clin Psychiatry. 1996;57:40–45. discussion 46–52. [PubMed] [Google Scholar]
- Casey DE. Tardive dyskinesia and atypical antipsychotic drugs. Schizophr Res. 1999;35:S61–S66. doi: 10.1016/s0920-9964(98)00160-1. [DOI] [PubMed] [Google Scholar]
- Cavanaugh JE, Ham J, Hetman M, Poser S, Yan C, Xia Z. Differential regulation of mitogen-activated protein kinases ERK1/2 and ERK5 by neurotrophins, neuronal activity, and cAMP in neurons. J Neurosci. 2001;21:434–443. doi: 10.1523/JNEUROSCI.21-02-00434.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong VZ, Young LT, Mishra RK. cDNA array reveals differential gene expression following chronic neuroleptic administration: implications of synapsin II in haloperidol treatment. J Neurochem. 2002;82:1533–1539. doi: 10.1046/j.1471-4159.2002.01104.x. [DOI] [PubMed] [Google Scholar]
- Cohen BM, Tsuneizumi T, Baldessarini RJ, Campbell A, Babb SM. Differences between antipsychotic drugs in persistence of brain levels and behavioral effects. Psychopharmacology (Berl) 1992;108:338–344. doi: 10.1007/BF02245121. [DOI] [PubMed] [Google Scholar]
- Conley RR, Tamminga CA, Kelly DL, Richardson CM. Treatment-resistant schizophrenic patients respond to clozapine after olanzapine non-response. Biol Psychiatry. 1999;46:73–77. doi: 10.1016/s0006-3223(99)00029-3. [DOI] [PubMed] [Google Scholar]
- Creese I, Burt DR, Snyder SH. Dopamine receptor binding predicts clinical and pharmacological potencies of antischizophrenic drugs. Science. 1976;192:481–483. doi: 10.1126/science.3854. [DOI] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Davie JR, Spencer VA. Control of histone modifications. J Cell Biochem. 1999;(Suppl):141–148. doi: 10.1002/(sici)1097-4644(1999)75:32+<141::aid-jcb17>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Delfs JM, Yu L, Ellison GD, Reisine T, Chesselet MF. Regulation of mu-opioid receptor mRNA in rat globus pallidus: effects of enkephalin increases induced by short- and long-term haloperidol administration. J Neurochem. 1994;63:777–780. doi: 10.1046/j.1471-4159.1994.63020777.x. [DOI] [PubMed] [Google Scholar]
- Dobrunz LE, Stevens CF. Heterogeneity of release probability, facilitation, and depletion at central synapses. Neuron. 1997;18:995–1008. doi: 10.1016/s0896-6273(00)80338-4. [DOI] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biology. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragunow M, Robertson GS, Faull RL, Robertson HA, Jansen K. D2 dopamine receptor antagonists induce fos and related proteins in rat striatal neurons. Neuroscience. 1990;37:287–294. doi: 10.1016/0306-4522(90)90399-o. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fibiger HC. Neuroanatomical targets of neuroleptic drugs as revealed by Fos immunochemistry. J Clin Psychiatry. 1994;55:33–36. [PubMed] [Google Scholar]
- Frank DA, Greenberg ME. CREB: A mediator of long-term memory from mollusks to mammals. Cell. 1994;79:5–8. doi: 10.1016/0092-8674(94)90394-8. [DOI] [PubMed] [Google Scholar]
- Frankenburg FR. Choices in antipsychotic therapy in schizophrenia. Harv Rev Psychiatry. 1999;6:241–249. doi: 10.3109/10673229909000336. [DOI] [PubMed] [Google Scholar]
- Garner CC, Zhai RG, Gundelfinger ED, Ziv NE. Molecular mechanisms of CNS synaptogenesis. Trends Neurosci. 2002;25:243–251. doi: 10.1016/s0166-2236(02)02152-5. [DOI] [PubMed] [Google Scholar]
- Genoux D, Haditsch U, Knobloch M, Michalon A, Storm D, Mansuy IM. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature. 2002;418:970–975. doi: 10.1038/nature00928. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Grant P, Pant HC. Neurofilament protein synthesis and phosphorylation. J Neurocytol. 2000;29:843–872. doi: 10.1023/a:1010999509251. [DOI] [PubMed] [Google Scholar]
- Grewal SS, York RD, Stork PJ. Extracellular-signal-regulated kinase signalling in neurons. Curr Opin Neurobiol. 1999;9:544–553. doi: 10.1016/S0959-4388(99)00010-0. [DOI] [PubMed] [Google Scholar]
- Hagiwara M, Alberts A, Brindle P, Meinkoth J, Feramisco J, Deng T, Karin M, Shenolikar S, Montminy M. Transcriptional attenuation following cAMP induction requires PP-1 mediated dephosphorylation of CREB. Cell. 1992;70:105–113. doi: 10.1016/0092-8674(92)90537-m. [DOI] [PubMed] [Google Scholar]
- Harper SJ, LoGrasso P. Signalling for survival and death in neurones: the role of stress-activated kinases, JNK and p38. Cell Signal. 2001;13:299–310. doi: 10.1016/s0898-6568(01)00148-6. [DOI] [PubMed] [Google Scholar]
- Hong JS, Yang HY, Gillin JC, Di Giulio AM, Fratta W, Costa E. Chronic treatment with haloperidol accelerates the biosynthesis of enkephalins in rat striatum. Brain Res. 1979;160:192–195. doi: 10.1016/0006-8993(79)90618-8. [DOI] [PubMed] [Google Scholar]
- Hong JS, Yoshikawa K, Kanamatsu T, Sabol SL. Modulation of striatal enkephalinergic neurons by antipsychotic drugs. Fed Proc. 1985;44:2535–2539. [PubMed] [Google Scholar]
- Hu C, Ahmed M, Melia TJ, Sollner TH, Mayer T, Rothman JE. Fusion of cells by flipped SNAREs. Science. 2003;300:1745–1749. doi: 10.1126/science.1084909. [DOI] [PubMed] [Google Scholar]
- Hu K, Carroll J, Fedorovich S, Rickman C, Sukhodub A, Davletov B. Vesicular restriction of synaptobrevin suggests a role for calcium in membrane fusion. Nature. 2002;415:646–650. doi: 10.1038/415646a. [DOI] [PubMed] [Google Scholar]
- Hyman SE, Nestler EJ. Initiation and adaptation: a paradigm for understanding psychotropic drug action. Am J Psychiatry. 1996;153:151–162. doi: 10.1176/ajp.153.2.151. [DOI] [PubMed] [Google Scholar]
- Iqbal MM, Rahman A, Husain Z, Mahmud SZ, Ryan WG, Feldman JM. Clozapine: a clinical review of adverse effects and management. Ann Clin Psychiatry. 2003;15:33–48. doi: 10.1023/a:1023228626309. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kane JM, Leucht S, Carpenter D, Docherty JP. Expert consensus guideline series. Optimizing pharmacologic treatment of psychotic disorders. Introduction: methods, commentary, and summary. J Clin Psychiatry. 2003;64:5–19. [PubMed] [Google Scholar]
- Kane JM, Marder SR. Psychopharmacologic treatment of schizophrenia. Schizophr Bull. 1993;19:287–302. doi: 10.1093/schbul/19.2.287. [DOI] [PubMed] [Google Scholar]
- Kapur S, Wadenberg ML, Remington G. Are animal studies of antipsychotics appropriately dosed? Lessons from the bedside to the bench. Can J Psychiatry. 2000;45:241–246. doi: 10.1177/070674370004500302. [DOI] [PubMed] [Google Scholar]
- Konradi C, Eaton ME, MacDonald ML, Walsh J, Benes FM, Heckers S. Molecular evidence for mitochondrial dysfunction in bipolar disorder. Arch Gen Psychiatry. 2004a;61:300–308. doi: 10.1001/archpsyc.61.3.300. [DOI] [PubMed] [Google Scholar]
- Konradi C, Heckers S. Haloperidol-induced Fos expression in striatum is dependent upon transcription factor cyclic AMP response element binding protein. Neuroscience. 1995;65:1051–1061. doi: 10.1016/0306-4522(94)00546-h. [DOI] [PubMed] [Google Scholar]
- Konradi C, Heckers S. Antipsychotic drugs and neuroplasticity: insights into the treatment and neurobiology of schizophrenia. Biol Psychiatry. 2001;50:729–742. doi: 10.1016/s0006-3223(01)01267-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C, Kobierski LA, Nguyen TV, Heckers S, Hyman SE. The cAMP-response-element-binding protein interacts, but Fos protein does not interact, with the proenkephalin enhancer in rat striatum. Proc Natl Acad Sci U S A. 1993;90:7005–7009. doi: 10.1073/pnas.90.15.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C, Westin J, Carta M, Eaton ME, Kuter K, Dekundy A, Lundblad M, Cenci MA. Transcriptome analysis in a rat model of L-DOPA-induced dyskinesia. Neurobiol Dis. 2004b;17:219–236. doi: 10.1016/j.nbd.2004.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornhuber J, Schultz A, Wiltfang J, Meineke I, Gleiter CH, Zochling R, Boissl KW, Leblhuber F, Riederer P. Persistence of haloperidol in human brain tissue. Am J Psych. 1999;156:885–890. doi: 10.1176/ajp.156.6.885. [DOI] [PubMed] [Google Scholar]
- Leveque JC, Macías W, Rajadhyaksha A, Carlson RR, Barczak A, Kang S, Li XM, Coyle JT, Huganir RL, Heckers S, Konradi C. Intracellular modulation of NMDA receptor function by antipsychotic drugs. J Neurosci. 2000;20:4011–4020. doi: 10.1523/JNEUROSCI.20-11-04011.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Hung Wong W. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biology. 2001;2 doi: 10.1186/gb-2001-2-8-research0032. RESEARCH0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGibbon GA, Lawlor PA, Bravo R, Dragunow M. Clozapine and haloperidol produce a differential pattern of immediate early gene expression in rat caudate-putamen, nucleus accumbens, lateral septum and islands of Calleja. Brain Res Mol Brain Res. 1994;23:21–32. doi: 10.1016/0169-328x(94)90207-0. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- McAlonan K, Brown VJ. Orbital prefrontal cortex mediates reversal learning and not attentional set shifting in the rat. Behav Brain Res. 2003;146:97–103. doi: 10.1016/j.bbr.2003.09.019. [DOI] [PubMed] [Google Scholar]
- Meltzer HY. An overview of the mechanism of action of clozapine. J Clin Psychiatry. 1994;55(Suppl B):47–52. [PubMed] [Google Scholar]
- Meshul CK, Andreassen OA, Allen C, Jorgensen HA. Correlation of vacuous chewing movements with morphological changes in rats following 1-year treatment with haloperidol. Psychopharmacology (Berl) 1996;125:238–247. doi: 10.1007/BF02247334. [DOI] [PubMed] [Google Scholar]
- Mielke K, Herdegen T. JNK and p38 stresskinases--degenerative effectors of signal-transduction-cascades in the nervous system. Prog Neurobiol. 2000;61:45–60. doi: 10.1016/s0301-0082(99)00042-8. [DOI] [PubMed] [Google Scholar]
- Mortimer AM. Antipsychotic treatment in schizophrenia: atypical options and NICE guidance. Eur Psychiatry. 2003;18:209–219. doi: 10.1016/s0924-9338(03)00060-9. [DOI] [PubMed] [Google Scholar]
- Mourey RJ, Vega QC, Campbell JS, Wenderoth MP, Hauschka SD, Krebs EG, Dixon JE. A novel cytoplasmic dual specificity protein tyrosine phosphatase implicated in muscle and neuronal differentiation. J Biol Chem. 1996;271:3795–3802. doi: 10.1074/jbc.271.7.3795. [DOI] [PubMed] [Google Scholar]
- Murthy VN, De Camilli P. Cell biology of the presynaptic terminal. Annu Rev Neurosci. 2003;26:701–728. doi: 10.1146/annurev.neuro.26.041002.131445. [DOI] [PubMed] [Google Scholar]
- Nguyen TV, Kosofsky BE, Birnbaum R, Cohen BM, Hyman SE. Differential expression of c-fos and zif268 in rat striatum after haloperidol, clozapine, and amphetamine. Proc Natl Acad Sci U S A. 1992;89:4270–4274. doi: 10.1073/pnas.89.10.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 4 ed. San Diego: Academic Press; 1998. [Google Scholar]
- Price NE, Mumby MC. Brain protein serine/threonine phosphatases. Curr Opin Neurobiol. 1999;9:336–342. doi: 10.1016/s0959-4388(99)80049-x. [DOI] [PubMed] [Google Scholar]
- Purdon SE, Jones BD, Stip E, Labelle A, Addington D, David SR, Breier A, Tollefson GD. Neuropsychological change in early phase schizophrenia during 12 months of treatment with olanzapine, risperidone, or haloperidol. The Canadian Collaborative Group for research in schizophrenia. Arch Gen Psychiatry. 2000;57:249–258. doi: 10.1001/archpsyc.57.3.249. [DOI] [PubMed] [Google Scholar]
- Qian Z, Gilbert M, Kandel ER. Temporal and spatial regulation of the expression of BAD2, a MAP kinase phosphatase, during seizure, kindling, and long-term potentiation. Learn Mem. 1994;1:180–188. [PubMed] [Google Scholar]
- Remington G, Chong SA. Conventional versus novel antipsychotics: changing concepts and clinical implications. J Psychiatry Neurosci. 1999;24:431–441. [PMC free article] [PubMed] [Google Scholar]
- Roberts RC, Gaither LA, Gao XM, Kashyap SM, Tamminga CA. Ultrastructural correlates of haloperidol-induced oral dyskinesias in rat striatum. Synapse. 1995;20:234–243. doi: 10.1002/syn.890200307. [DOI] [PubMed] [Google Scholar]
- Robertson GS, Fibiger HC. Neuroleptics increase c-fos expression in the forebrain: contrasting effects of haloperidol and clozapine. Neuroscience. 1992;46:315–328. doi: 10.1016/0306-4522(92)90054-6. [DOI] [PubMed] [Google Scholar]
- Robertson GS, Matsumura H, Fibiger HC. Induction patterns of Fos-like immunoreactivity in the forebrain as predictors of atypical antipsychotic activity. J Pharmacol Exp Ther. 1994;271:1058–1066. [PubMed] [Google Scholar]
- See RE, Chapman MA. Chronic haloperidol, but not clozapine, produces altered oral movements and increased extracellular glutamate in rats. Eur J Pharmacol. 1994;263:269–276. doi: 10.1016/0014-2999(94)90722-6. [DOI] [PubMed] [Google Scholar]
- Sim AT, Baldwin ML, Rostas JA, Holst J, Ludowyke RI. The role of serine/threonine protein phosphatases in exocytosis. Biochem J. 2003;373:641–659. doi: 10.1042/BJ20030484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. The neuronal MAP kinase cascade: a biochemical signal integration system subserving synaptic plasticity and memory. J Neurochem. 2001;76:1–10. doi: 10.1046/j.1471-4159.2001.00054.x. [DOI] [PubMed] [Google Scholar]
- Thomas GM, Huganir RL. MAPK cascade signalling and synaptic plasticity. Nature Reviews Neuroscience. 2004;5:173–183. doi: 10.1038/nrn1346. [DOI] [PubMed] [Google Scholar]
- Turner KM, Burgoyne RD, Morgan A. Protein phosphorylation and the regulation of synaptic membrane traffic. Trends Neurosci. 1999;22:459–464. doi: 10.1016/s0166-2236(99)01436-8. [DOI] [PubMed] [Google Scholar]
- Uylings HB, Groenewegen HJ, Kolb B. Do rats have a prefrontal cortex? Behav Brain Res. 2003;146:3–17. doi: 10.1016/j.bbr.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Van Patten SM, Howard P, Walsh DA, Maurer RA. The alpha- and beta-isoforms of the inhibitor protein of the 3',5'-cyclic adenosine monophosphate-dependent protein kinase: characteristics and tissue- and developmental-specific expression. Mol Endocrinol. 1992;6:2114–2122. doi: 10.1210/mend.6.12.1491692. [DOI] [PubMed] [Google Scholar]
- Van Patten SM, Ng DC, Th'ng JP, Angelos KL, Smith AJ, Walsh DA. Molecular cloning of a rat testis form of the inhibitor protein of cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1991;88:5383–5387. doi: 10.1073/pnas.88.12.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadzinski BE, Wheat WH, Jaspers S, Peruski LFJ, Lickteig RL, Johnson GL, Klemm DJ. Nuclear protein phosphatase 2A dephosphorylates protein kinase A-phosphorylated CREB and regulates CREB transcriptional stimulation. Mol Cell Biol. 1993;13:2822–2834. doi: 10.1128/mcb.13.5.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmarsh AJ, Davis RJ. Regulation of transcription factor function by phosphorylation. Cell Mol Life Sci. 2000;57:1172–1183. doi: 10.1007/PL00000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder DG, Sweatt JD. Roles of serine/threonine phosphatases in hippocampal synaptic plasticity. Nat Rev Neurosci. 2001;2:461–474. doi: 10.1038/35081514. [DOI] [PubMed] [Google Scholar]
- Wirshing DA, Pierre JM, Erhart SM, Boyd JA. Understanding the new and evolving profile of adverse drug effects in schizophrenia. Psychiatr Clin North Am. 2003;26:165–190. doi: 10.1016/s0193-953x(02)00035-7. [DOI] [PubMed] [Google Scholar]
- Zhou G, Bao ZQ, Dixon JE. Components of a new human protein kinase signal transduction pathway. J Biol Chem. 1995;270:12665–12669. doi: 10.1074/jbc.270.21.12665. [DOI] [PubMed] [Google Scholar]