Abstract
Understanding the energetics of molecular interactions is fundamental to all of the central quests of structural biology including structure prediction and design, mapping evolutionary pathways, learning how mutations cause disease, drug design, and relating structure to function. Hydrogen-bonding is widely regarded as an important force in a membrane environment because of the low dielectric constant of membranes and a lack of competition from water1–6. Indeed, polar residue substitutions are the most common disease-causing mutations in membrane proteins6, 7. Because of limited structural information and technical challenges, however, there have been few quantitative tests of hydrogen-bond strength in the context of large membrane proteins. Here we show, by using a double-mutant cycle analysis, that the average contribution of eight interhelical side-chain hydrogen-bonding interactions throughout bacteriorhodopsin is only 0.6 kcalmol−1. In agreement with these experiments, we find that 4% of polar atoms in the non-polar core regions of membrane proteins have no hydrogen-bond partner and the lengths of buried hydrogen bonds in soluble proteins and membrane protein transmembrane regions are statistically identical. Our results indicate that most hydrogen-bond interactions in membrane proteins are only modestly stabilizing. Weak hydrogen-bonding should be reflected in considerations of membrane protein folding, dynamics, design, evolution and function.
The few evaluations of hydrogen-bond contributions in membrane proteins have tested the effect of single point mutants on either the free energy of unfolding or the free energy of dissociation4, 8, 9. However, these measurements combine hydrogen-bond contributions with desolvation and many other factors10, so the hydrogen-bond contribution cannot necessarily be extracted without the incorporation of correction factors11 that are particularly uncertain for membrane proteins.
The energetic complexities of single side-chain alterations can be illustrated by mutations in bacteriorhodopsin residues T90 and D115 that make two hydrogen bonds near the centre of the membrane (Fig. 1). We eliminated the hydrogen bonds by making T90A and D115A mutations and measured the change in the free energy of unfolding with an SDS unfolding assay9. The T90A mutation decreases stability by 1.3 ± 0.1 kcal mol−1, whereas the D115A mutant increases stability by 0.5 ± 0.1 kcal mol−1. The large variation suggests that hydrogen-bonding alone does not dominate the stability effects, and other energetic contributions must be accounted for. Below we present evidence that a principal factor is changes in solvation free energy in the unfolded protein.
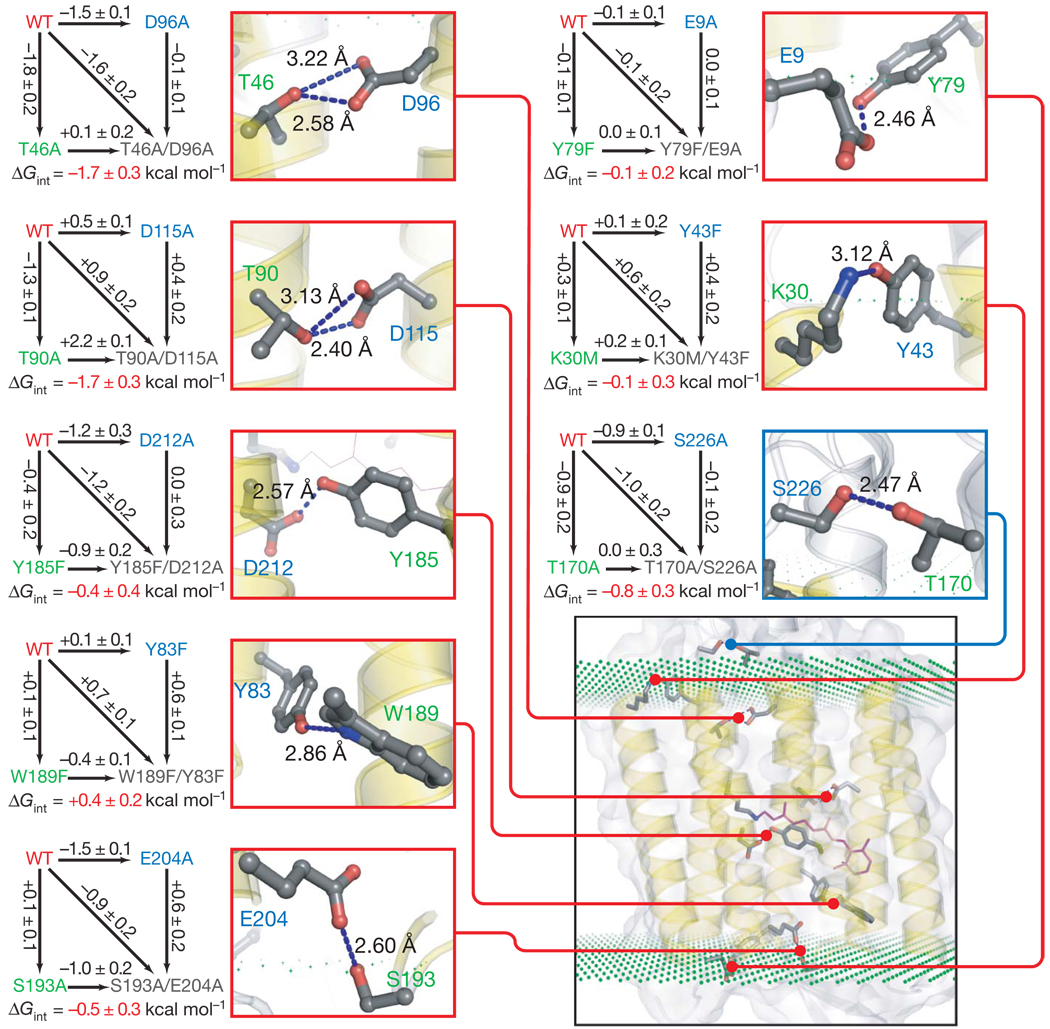
Figure 1. Double-mutant cycles for hydrogen-bonding interactions in bacteriorhodopsin.
For each cycle shown, the difference in free energies of unfolding (black number by the arrow) was measured for the pair of proteins connected by the arrow. Free energies of unfolding are compared at an SDS concentration at which the wild-type protein (WT) is 50% unfolded to minimize extrapolations needed. Errors are s.d. for three separate measurements. Next to each double-mutant cycle is a close-up view of the relevant hydrogen bond shown as blue dotted line between the altered side chains along with the heavy atom donor–acceptor distance. Donor and acceptor residues are labelled in green and blue, respectively.
Donor–acceptor distinction in the two strongest interactions was arbitrary. On the basis of hydrogen-bonding patterns and nearest neighbours, it seems that all the potentially charged residues are the neutral species. The inset (bottom right) shows the location of each interaction in the context of the protein (PDB ID 1C3W). The planes of green dots indicate the estimated position of the edge of the hydrocarbon region of the bilayer as defined previously28. Any interaction mediated by the residues that contain at least one atom in the hydrocarbon region is mapped with the red line, and the interaction in the lipid/water interface region is mapped with a blue line.
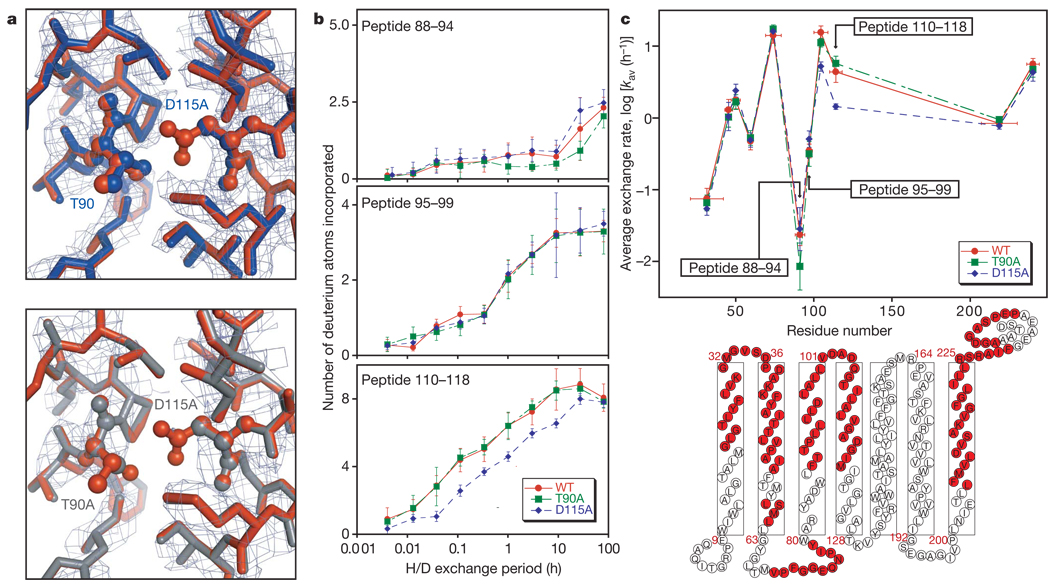
To examine the effects of the T90A and D115A mutations on the folded state of bacteriorhodopsin, we solved the structures of the D115A mutant and a T90A/D115A double mutant (T90A proved too unstable to crystallize). We were unable to detect any structural changes in the mutant proteins that would obviously explain the contrasting energetic consequences, beyond the loss of density around the deleted side chains (see Fig. 2a).
Figure 2. Characterization of the T90A, D115A and T90A/D115Amutants.
a, Omit electron density maps and overlay of refined mutant and wild-type structures for D115A (top) and T90A/D115A (bottom) mutants. The wild-type structure (PDB ID 1PY6) is shown in rust, D115A in blue and T90A/D115A in grey. The mutated side chains are shown in ball-and-stick representation and labelled. The side chains of all residues within 4Å of T90 and D115 of the wild-type (WT) structure were eliminated during refinement for the omit map and are shown here with the exception of W182, which was left out for clarity. The electron density map is contoured at 1.0σ and 1.5σ for D115A and T90A/D115A, respectively. b, Plot of the number of hydrogens exchanged in the denatured state against time for peptides overlapping the T90A mutation (top), a region between T90A and D115A mutation (middle) and the D115A mutation (bottom). In brief, wild-type and mutant proteins were unfolded in SDS and incubated in D2O; the exchange reaction was quenched by rapidly lowering the temperature and pH. The proteins were then digested with pepsin, distinct peptides were separated chromatographically and the change in the mass envelope was measured by electrospray ionization-mass spectroscopy. The maximum scale on the y axis is the maximum number of exchangeable backbone amide hydrogens. Error bars are s.d. estimated with results from triplicate experiments. c, A plot of average exchange rates for peptides throughout the protein (top) and a schematic illustration of the bacteriorhodopsin structure (bottom) showing the sequences covered by the deuterium exchange experiment in light red. Error bars on the x axis reflect the range of the peptic peptides, and those on the y axis are s.d. for ten simulated data sets incorporating the experimental errors observed in the exchange time courses (see Methods).
To probe the consequences of the mutations on the unfolded state, we developed a hydrogen-exchange assay. Unfolded-state backbone hydrogens that are shielded from solvent by burial in the detergent micelle will exchange at a slower rate than backbone hydrogens exposed to the aqueous phase12, 13. Figure 2b shows the detailed time course of exchange for the unfolded state of the wild-type and mutant proteins at three regions, one resolved by the peptide overlapping the site of the T90A mutation, the second overlapping a region in between the sites of the T90A and D115A mutations, and the third overlapping the site of the D115A mutation. Figure 2c summarizes the average exchange rates of peptides throughout the unfolded states.
The T90A mutation modestly slows the exchange in the vicinity of position 90, whereas D115A markedly slows exchange in the vicinity of position 115. Although the sequence effects on intrinsic exchange rates14 are uncertain in an SDS environment15, the results suggest that the polar to non-polar substitutions alter the unfolded state by increasing burial in the detergent micelle at the sites of mutation. The larger change in polarity in D115A than in T90A is consistent with the larger effect on exchange rate and probably explains the stabilizing effect of the D115A mutation. In particular, the loss of the favourable escape of D115 to solvent could increase the free energy of the unfolded state in the D115A mutant, compensating for the increased free energy of the folded state. Thus, solvation effects in the unfolded state may mask the hydrogen-bond contribution that we wish to measure.
In an effort to obtain side-chain interaction energies within the folded state, we turned to double-mutant cycle analysis. Double-mutant cycle analysis has the potential to measure the free energy of side-chain interaction directly in the context of the folded protein by cancelling out energetic perturbations in both the folded and unfolded states that are not due to the interactions between the side chains16, 17. Thus, desolvation contributions and any other new interactions made in the unfolded state can be eliminated. As a result, double-mutant cycle analysis can be interpreted as reporting the contribution of the hydrogen-bonded interaction to the free energy of the folded state, not the difference in free energy between the folded and unfolded states. The unfolded state becomes simply a common reference state in which the interaction of interest is broken (see Supplementary Methods).
We were able to express and purify complete single-mutant and double-mutant sets for eight interhelical hydrogen-bonding interactions as shown in Fig. 1. Four of the hydrogen bonds are in the middle of the hydrocarbon core region of the bilayer, three are on the edge of the hydrocarbon core and one is in the interfacial region. The strongest interactions were T46–D96 and T90–D115, each contributing −1.7 ± 0.3 kcal mol−1, and T170–S226, contributing −0.8 ± 0.3 kcal mol−1. The strongest interactions, between T46 and D96 and between T90 and D115, both involve two hydrogen bonds, corresponding to about −0.9 kcal mol−1 per hydrogen bond. Y185–D212 and S193–E204 make weaker, but favourable, interactions contributing −0.4 ± 0.4 and −0.5 ± 0.3 kcal mol−1, respectively. The K30–Y43 and E9–Y79 interactions were found to make no measurable contribution to stability, and W189–Y83 was found to be slightly destabilizing, contributing +0.4 ± 0.2 kcal mol−1.
The results of the double-mutant cycles suggest three main conclusions. First, hydrogen-bonded side-chain contributions are quite variable and depend on the characteristics and local environment of each hydrogen bond. Second, the strength of a hydrogen-bonded interaction is not strongly correlated with the location in the protein. For example, the T170–S226 interaction in the interfacial region contributes −0.8 ± 0.3 kcal mol−1, whereas the T185–D212 interaction in the centre of the hydrocarbon core contributes only −0.4 ± 0.4 kcal mol−1. Third, the eight hydrogen-bonding interactions studied here make a remarkably modest average contribution of only about 0.6 kcal mol−1, which corresponds to a roughly threefold effect on an equilibrium constant at room temperature.
Protein folding experiments are complex and not all variables can be eliminated, so we sought an additional, independent evaluation of the hydrogen-bond contribution in membrane proteins. We reasoned that if hydrogen-bond strengths were low, we would see a large number of unsatisfied hydrogen-bonding groups in membrane protein structures18. To test this idea, we examined six membrane protein structures solved at 1.7 Å resolution or better. HBPLUS was used to identify the hydrogen-bonding of all polar atoms within the hydrocarbon core region of the bilayer. Any polar atoms that made no hydrogen bonds were further verified by eye.
The results are summarized in Table 1 and reveal that unsatisfied hydrogen bonds are not rare in the hydrocarbon core region (also see Supplementary Information). Of 2,892 protein donors and acceptors examined in the hydrocarbon core region, 111 have no hydrogen-bonding partner (about 4%). We believe this to be a low estimate of the number of unsatisfied hydrogen bonds because the HBPLUS criteria permit even marginal hydrogen bonds to be counted. Moreover, the crystal structure reports the predominant conformation and does not report the fraction of time for which a hydrogen bond is broken.
Table 1.
Unsatisfied hydrogen-bond donors and acceptors in hydrophobic cores of membrane proteins
| Protein | PDB code | Resolution (Å) | Donor and acceptor population, unsatisfied/total* | Percentage unsatisfied |
|---|---|---|---|---|
| Bacteriorhodopsin | 1C3W | 1.55 | 17/345 | 4.9 |
| Formate dehydrogenase N | 1KQF | 1.60 | 9/288 | 3.1 |
| NH4+transporter Amt B | 1U7G | 1.40 | 17/571 | 3.0 |
| Na+/Cl−-dependent neurotransporter transmitter | 2A65 | 1.65 | 34/699 | 4.9 |
| NH4+ transporter Amt-1 | 2B2H | 1.54 | 24/593 | 4.0 |
| Aquaportin | 2F2B | 1.68 | 10/396 | 2.5 |
| Total | 111/2,892 | 3.8 |
See Supplementary Fig. 2 for further details.
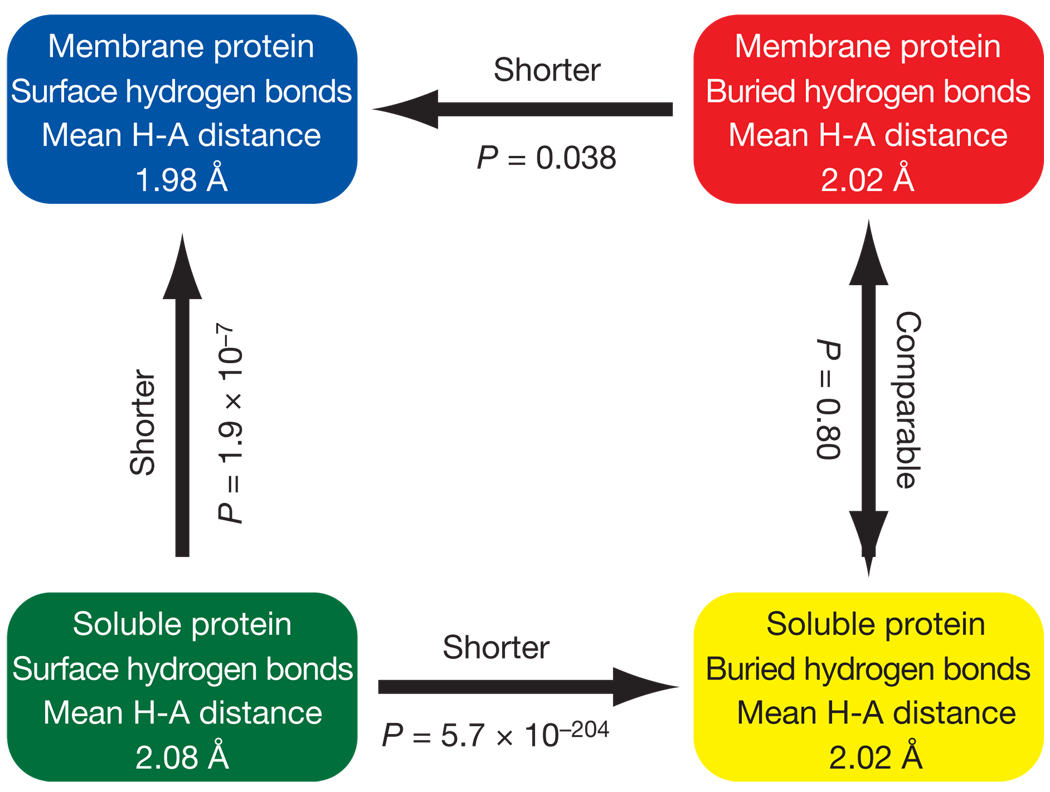
The hydrogen-bonded interaction strengths we measured in a membrane protein are very similar to hydrogen-bond strengths in soluble proteins measured in a variety of double-mutant cycle analyses (see Supplementary Fig. 1). Retrospectively, this finding is not unreasonable because the polarities of the interiors of soluble proteins and membrane proteins are quite similar19, 20. Because folding studies in soluble proteins are well accepted, we decided to validate our findings further by comparing hydrogen-acceptor distance distributions in membrane and soluble proteins. As summarized in Fig. 3, the buried hydrogen-bond distances in the interior of soluble and membrane protein transmembrane regions are statistically indistinguishable (see Supplementary Fig. 3 for full distributions), both averaging 2.02Å. However, the hydrogen-bond distances in surface residues are markedly different. For the transmembrane regions of membrane proteins, the hydrogen-acceptor distances on the surface are slightly shortened to 1.98Å on average, whereas for soluble proteins the average distance lengthens to 2.08Å. These results further validate our results, indicating similar contributions from interior hydrogen bonds in soluble and membrane proteins. It also hints that hydrogen bonds at the surface of membrane proteins may be stronger than what we have measured here for interior ones.
Figure 3. Comparison of average hydrogen-bond distances in different environments.
The arrows point towards the shorter hydrogen bonds. The P value is the probability that the distance distributions are different by random chance based on Student’s t-test. The distributions are shown in Supplementary Information.
Many of the hydrogen-bonded residues we tested are involved in function, so it is possible that they are in a separate class from structural hydrogen-bonded side chains. However, previous work eliminating hydrogen bonds in structural residues is consistent with our findings. For example, a Gln residue that makes two hydrogen bonds across the interface of an OMPLA dimer contributes less than 1 kcal mol−1 to dimerization (less than 0.5 kcal mol−1 per hydrogen bond)21. An Asp to Ala substitution in a designed trans-membrane helix oligomer decreases the free energy of association by 1.8 kcal mol−1 (0.9 kcal mol−1 per potential hydrogen bond)4. A T87A substitution in glycophorin A decreases the free energy of dimerization by 0.9 kcal mol−1 (about 0.5 kcal mol−1 per hydrogen bond in the dimer)22. Mutations in hydrogen-bonding polar residues in the T-cell receptor ζ-subunit dimer affected dimerization by a maximum of 7.7-fold, as indirectly measured by assembly rate, which corresponds to a maximum of about 1.2 kcal mol−1 (0.6 kcal mol−1 per residue in the dimer)8. Analysis of hydrogen-bonding mutations in bacteriorhodopsin (ref. 9) suggest a contribution of about 1 kcal mol−1 (ref. 23). Because double-mutant cycle analysis was not employed in these cases, however, the results combine hydrogen-bond contributions with other effects that could contribute favourably or unfavourably10, 24.
Although our results indicate that most hydrogen-bond interactions observed in membrane proteins make modest energetic contributions, it does not mean that polar interactions cannot be strong. It has been found16 that charge-stabilized salt-bridge interactions can contribute 5.6 kcal mol−1, and mutations in residues that hydrogen-bond to ligands in the β-adrenergic receptor can have marked effects on ligand binding25.
Why, then, are hydrogen-bond interactions not much stronger on average? It is possible that optimal geometries are difficult to achieve, that there are entropic costs to fixing hydrogen-bonded groups, and that polar groups in the protein can increase the local dielectric constant26. In addition to possible physical limitations, there may also be evolutionary pressure favouring weak hydrogen bonds. Evolutionary pressure for weak hydrogen bonds could come in the form of the conformational flexibility needed for protein function6. Moreover, the helical distortions that are common in membrane proteins would be hard to create by random mutation if the breakage of hydrogen bonds presented a large energy barrier. It is also possible that weak hydrogen bonds are more robust evolutionarily. Strong side-chain hydrogen bonds, once established, could no longer be altered by mutation without destroying fitness. Thus, proteins that rely on a strong hydrogen bond for stability would be more likely to be lost from a population than proteins that rely on more broadly distributed stabilizing interactions. Whatever the mechanism, our results indicate that it is not difficult to make and break interactions between polar residues, enabling the structural variation and dynamic flexibility necessary to optimize membrane protein function and folding. The results also suggest that a primary mechanism for the prevalence of disease-causing substitutions of polar residues may not be inappropriate hydrogen-bond formation but, instead, alterations in bilayer partitioning6. However, the loss or gain of even a weak hydrogen bond could tip the balance between biological function and dysfunction.
METHODS SUMMARY
Equilibrium unfolding measurements
Stability measurements were performed essentially as described previously9. In brief, purple membrane was dissolved in a DMPC (1,2-dimyristoyl-sn-glycerol-3-phosphocholine)/CHAPSO (3((3-cholamidopropyl)dimethylammonio)-2-hydroxy-1-propane-sulphonate) mixture and unfolded by adding increasing concentrations of SDS. Unfolding was monitored either by retinal absorbance at 560nm or far-ultraviolet circular dichroism at 228 nm. Unfolding free energies were compared at the SDS concentration at which wild-type bacteriorhodopsin was 50% unfolded, to minimize the extrapolation error due to the varying m values.
X-ray crystallography
Crystals were grown by the bicelle method27 and, diffraction data were phased by molecular replacement.
Deuterium exchange
Unfolded proteins in SDS were deuterium-exchanged for various periods. The exchange reactions were then quenched by rapid cooling and the addition of low-pH buffer containing an acid-labile detergent to maintain solubility during digestion with pepsin. K+ was also included to precipitate SDS. Quenched reactions were flash-frozen and stored at−80 °C. For analysis of deuterium exchange, the quenched reactions were rapidly thawed, treated with pepsin at 0 °C and immediately analysed by liquid chromatography–mass spectrometry (LC–MS).
Structure analysis
To analyse unsatisfied hydrogen-bond donors and acceptors, the transmembrane regions were first identified as described28 and hydrogen bonds were identified with HBPLUS29. To obtain hydrogen-bond distance distributions, hydrogen-acceptor distances of all α-helix backbone–backbone hydrogen bonds in six high-resolution membrane protein structures and 839 unique soluble protein structures were calculated with HBPLUS. The soluble proteins chosen were solved in the same resolution range as the membrane proteins used.
Supplementary Material
Acknowledgments
We thank the staff at the beamlines 8.2.1 and 8.2.2 at the Advanced Light Source; F. Pettit for advice on statistics; M. Philips for assisting with circular dichroism experiments; S. Bassilian for assisting with LC–MS analysis of intact bacteriorhodopsin; Y. Ihm for the identification of transmembrane regions; Z. Zhang for providing deuterium-exchange data reduction software MAGTRAN and LAPLACE; N. L. Kelleher for providing the acid-labile detergent; and M. Chamberlain, H. Cheng, E. Gendel, H. Hong, Y. Ihm, A. D. Meruelo, T. Mitchell and R. Stafford for critically reading the manuscript. This work was supported by National Institutes of Health grant RO1 GM063919 (J.U.B.) and by the National Institutes of Health National Cancer Institute Innovative Molecular Analysis Technologies Program (V.L.W.).
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Coordinates and structure factors for the D115A and T90A/D115A mutant bacteriorhodopsins have been deposited in the Protein Data Bank under accession codes 3COC and 3COD, respectively. Reprints and permissions information is available at www.nature.com/reprints
References
- 1.White SH. How hydrogen bonds shape membrane protein structure. Adv. Protein Chem. 2005;72:157–172. doi: 10.1016/S0065-3233(05)72006-4. [DOI] [PubMed] [Google Scholar]
- 2.Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 3.Zhou FX, Merianos HJ, Brunger AT, Engelman DM. Polar residues drive association of polyleucine transmembrane helices. Proc. Natl Acad. Sci. USA. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc. Natl Acad. Sci. USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adamian L, Liang J. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins. 2002;47:209–218. doi: 10.1002/prot.10071. [DOI] [PubMed] [Google Scholar]
- 6.Partridge AW, Therien AG, Deber CM. Polar mutations in membrane proteins as a biophysical basis for disease. Biopolymers. 2002;66:350–358. doi: 10.1002/bip.10313. [DOI] [PubMed] [Google Scholar]
- 7.Partridge AW, Therien AG, Deber CM. Missense mutations in transmembrane domains of proteins: phenotypic propensity of polar residues for human disease. Proteins. 2004;54:648–656. doi: 10.1002/prot.10611. [DOI] [PubMed] [Google Scholar]
- 8.Call ME, et al. The structure of the ζζtransmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faham S, et al. Side-chain contributions to membrane protein structure and stability. J. Mol. Biol. 2004;335:297–305. doi: 10.1016/j.jmb.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 10.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. Changes in apparent free energy of helix-helix dimerization in a biological membrane due to point mutations. J. Mol. Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers JK, Pace CN. Hydrogen bonding stabilizes globular proteins. Biophys. J. 1996;71:2033–2039. doi: 10.1016/S0006-3495(96)79401-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busenlehner LS, Armstrong RN. Insights into enzyme structure and dynamics elucidated by amide H/D exchange mass spectrometry. Arch. Biochem. Biophys. 2005;433:34–46. doi: 10.1016/j.abb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Busenlehner LS, et al. Stress sensor triggers conformational response of the integral membrane protein microsomal glutathione transferase 1. Biochemistry. 2004;43:11145–11152. doi: 10.1021/bi048716k. [DOI] [PubMed] [Google Scholar]
- 14.Molday RS, Englander SW, Kallen RG. Primary structure effects on peptide group hydrogen exchange. Biochemistry. 1972;11:150–158. doi: 10.1021/bi00752a003. [DOI] [PubMed] [Google Scholar]
- 15.O’Neil JD, Sykes BD. NMR studies of the influence of dodecyl sulfate on the amide hydrogen exchange kinetics of a micelle-solubilized hydrophobic tripeptide. Biochemistry. 1989;28:699–707. doi: 10.1021/bi00428a043. [DOI] [PubMed] [Google Scholar]
- 16.Hong H, Szabo G, Tamm LK. Electrostatic couplings in OmpA ion-channel gating suggest amechanismfor pore opening. Nature Chem. Biol. 2006;2:627–635. doi: 10.1038/nchembio827. [DOI] [PubMed] [Google Scholar]
- 17.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J. Mol. Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 18.Fleming PJ, Rose GD. Do all backbone polar groups in proteins form hydrogen bonds? Protein Sci. 2005;14:1911–1917. doi: 10.1110/ps.051454805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamian L, Nanda V, DeGrado WF, Liang J. Empirical lipid propensities of amino acid residues in multispan alpha helical membrane proteins. Proteins. 2005;59:496–509. doi: 10.1002/prot.20456. [DOI] [PubMed] [Google Scholar]
- 20.Rees D, DeAntonio L, Eisenberg D. Hydrophobic organization of membrane proteins. Science. 1989;245:510–513. doi: 10.1126/science.2667138. [DOI] [PubMed] [Google Scholar]
- 21.Stanley AM, Fleming KG. The role of a hydrogen bonding network in the transmembrane β-barrel OMPLA. J. Mol. Biol. 2007;370:912–924. doi: 10.1016/j.jmb.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Fleming KG, Engelman DM. Specificity in transmembrane helix-helix interactions can define a hierarchy of stability for sequence variants. Proc. Natl Acad. Sci. USA. 2001;98:14340–14344. doi: 10.1073/pnas.251367498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Lear JD, Gratkowski H, Adamian L, Liang J, DeGrado WF. Position-dependence of stabilizing polar interactions of asparagine in transmembrane helical bundles. Biochemistry. 2003;42:6400–6407. doi: 10.1021/bi020573j. [DOI] [PubMed] [Google Scholar]
- 25.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 26.Shan SO, Herschlag D. Hydrogen bonding in enzymatic catalysis: analysis of energetic contributions. Methods Enzymol. 1999;308:246–276. doi: 10.1016/s0076-6879(99)08013-1. [DOI] [PubMed] [Google Scholar]
- 27.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J. Mol. Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 28.Chamberlain AK, Lee Y, Kim S, Bowie JU. Snorkeling preferences foster an amino acid composition bias in transmembrane helices. J. Mol. Biol. 2004;339:471–479. doi: 10.1016/j.jmb.2004.03.072. [DOI] [PubMed] [Google Scholar]
- 29.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.