Abstract
Neurogranin (Nrgn) is a highly expressed brain-specific protein, which sequesters calmodulin at low Ca2+-levels. We report here on retroviral activation of the Nrgn gene in tumors induced by the T-cell lymphomagenic SL3-3 murine leukemia virus. We have performed a systematic expression analysis of Nrgn in various mouse tissues and SL3-3 induced T-cell tumors. This demonstrated that insertional activation of Nrgn increased RNA and protein expression levels to that observed in brain. Furthermore, elevated Nrgn expression was also observed in some T-cell tumors with no detected provirus integrations into this genomic region. The presented data demonstrate that Nrgn can be produced at high levels outside the brain, and suggest a novel oncogenic role in T-cell lymphomas in mice.
Keywords: Neurogranin, brain, murine leukemia virus, insertional mutagenesis, T-cell lymphoma
1. Introduction
Neurogranin (Nrgn, also denoted RC3) belongs to the neuron-specific calpacitin family of proteins, and functions as a calmodulin (CaM) storage protein at low Ca2+ levels (Baudier et al., 1991; Gerendasy, 1999; Prichard et al., 1999; van Dalen et al., 2003). The focus on Nrgn's function has been primarily in brain tissues, and several studies have demonstrated that the protein plays a role in learning and memory (Pak et al., 2000; Huang et al., 2004; Huang et al., 2006). The expression level of Nrgn is highest in cortex, striatum and hippocampus, while lower levels are detected in the olfactory bulb and midbrain as analyzed in rat and human tissue samples (Represa et al., 1990; Watson et al., 1990; Martinez de Arrieta et al., 1997). Additionally, low levels of expression have been reported in rat thymus and spleen (Watson et al., 1990).
Murine Nrgn consists of four exons of which the first two encompass the coding sequence for a 78-amino acid protein. It is located in a gene-dense region on chromosome 9 surrounded by Esam1 (endothelial cell-selective adhesion molecule), Vsig2 (V-set and immunoglobulin domain containing 2 (also known as CTM)), Ysg2 (yolk sac gene 2/ Sialic acid-specific 9-O-acetylesterase (Siae)) and Spa17 (sperm autoantigenic protein 17). While Esam1 and Spa17 have been reported to play a role in cancer development, no carcinogenic role has been correlated with Vsig2 or Ysg2. Esam1 belongs to the immunoglobulin receptor family and may play an important role in pathological angiogenic processes such as tumor growth (Hirata et al., 2001; Ishida et al., 2003). Spa17 is a member of the cancer/testis antigen family and is expressed in various human cancers including multiple myeloma, ovarian cancer and nervous system tumors (Lim et al., 2001a; Lim et al., 2001b; Chiriva-Internati et al., 2002; Grizzi et al., 2006).
Murine leukemia viruses (MLVs) induce hematopoietic tumors when injected into newborn susceptible mice (for recent reviews see (Mikkers and Berns, 2003; Uren et al., 2005)). Tumor induction by non-acutely transforming MLVs is a complex process containing multiple steps of which the activation of cooperating genes by retroviral insertional mutagenesis is believed to play an important role in the clonal expansion of target cells into full-blown tumors (Mikkers and Berns, 2003; Uren et al., 2005). Large-scale screenings in various virus/host systems have identified thousands of insertion sites of which several hundred represent genes or loci with putative oncogenic potential (recently reviewed in (Uren et al., 2005; Touw and Erkeland, 2007)). Many of these sites are accessible online in the Retroviral Tagged Cancer Gene Database RTCGD (http://rtcgd.abcc.ncifcrf.gov/) (Akagi et al., 2004). The murine leukemia virus SL3-3 is a potent inducer of T-cell lymphomas in susceptible mice with a mean latency between two and four months (Sørensen et al., 2004; Ejegod et al., 2009). Previously, we reported on the enhancer mutant SL3-3(turbo) (Ethelberg et al., 1997b; Nielsen et al., 2005). SL3-3(turbo) has an extra LTR repeat in combination with deletion of two binding site sites for nuclear factor 1, which significantly shortened the mean latency time of T-cell lymphoma induction in mice (Ethelberg et al., 1997b).
In the present work, we report on insertional activation of Nrgn as a result of SL3-3(turbo) and SL3-3 wt integration into a novel retroviral insertion site, the gene-dense Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus, and give a comprehensive expression analysis of Nrgn in mouse T-cell tumors as well as in normal tissue.
2. Materials and methods
2.1 Tumor and control material
Tumor material from the SL3-3(turbo)/inbred NMRI model originates from a previous study (Ethelberg et al., 1997b). Tumor material from the SL3-3 wt/BALB/c model originates from a study described in (Glud et al., 2005; Wang et al., 2006). Control tissues were isolated from mock-injected and non-treated NMRI and BALB/c mice, respectively. Mice were kept according to approved regulations and monitored on a daily basis. Upon signs of illness or development of tumors of defined sizes mice were terminated and relevant organs removed and stored at −80° C.
2.2 Extraction of total RNA and genomic DNA
Total RNA and genomic DNA were extracted from frozen control or tumor tissues using the TRIzol® Reagent (InvitrogenTM) or DNeasy® Blood & Tissue Kit (Qiagen), respectively, following the manufacturer's protocol.
2.3 PCR detection of proviral integrations
Identification of SL3-3(turbo) integration sites in NMRI mice was done using the two-step PCR approach described in Sørensen et al. (Sørensen et al., 1993). Screening and validation of Nrgn promoter insertion sites and orientations were done by PCR using the provirus-specific primers 2620 (5′-GATCCCCGGTCATCTGGG-3′; specific for the minus strand of the U3 region of the long terminal repeat) or 6197 (5′-CCCAGATGACCGGGGATC-5′; specific for the plus strand of the U3 region) in combination with either of the Nrgn promoter-specific primers 5′-CTCATAAGCCCCTCCTCTTTCCAT-3′ (plus strand) and 5′-CCCACTCATTCTCCCTTTAACA-3′ (minus strand). PCR amplification products were sequenced (ABI™ BigDye Terminators, Applied Biosystems) with retrovirus primers 793 (5′-CTCTGGTATTTTTCCCATG −3′) or 540 (5′-TCCGAATCGTGGTCTCGCTGATCCTTGG-3′) (Nielsen et al., 2005). Primers were purchased from DNA Technology A/S. PCR was performed with Taq DNA polymerase (Invitrogen™) using reaction conditions as described previously (Nielsen et al., 2005). Provirus 1423 and 3427 were detected by a splinkerette-based PCR method (Mikkers et al., 2002), and is described in (Wang et al., 2006).
2.4 Southern and Northern blot analysis
Southern blot (Ethelberg et al., 1997a) and Northern blot (Rasmussen et al., 2005) analysis were done with random labeled DNA probes as described in (Sørensen et al., 2000). Hybridization conditions were either ULTRAhyb® Ultrasensitive Hybridization Buffer (Ambion) or Na2HPO4/NaH2PO4 buffer (Ethelberg et al., 1997a). Probe A was a PCR product from mouse DNA using primers 5′- CCCACTCATTCTCCCTTTAACA-3′ and 5′- CTCATAAGCCCCTCCTCTTTCCAT-3′. Probe B-D were PCR amplification products from brain total cDNA using the following primers: 5′-GAAAGTGTCTTCTGATTGGCTTCGAG-3′ and 5′-CACAGTAGGGAAGTCTTGTCACTGCG-3′ (Probe B), 5′-CTCTTCAGTCTAACGTGGTCTCCT-3′ and 5′- CGCAGAGATTAAAAACCTTCCAGCCA-3′ (Probe C), 5′ CAACCACCAAGTCCTTTCGT-3′ and 5′-GGTAACATGCACACGCAGAG-3′ (Probe D). Northern-blot hybridization of the multiple-tissue Northern filter containing poly(A)+ RNA (Clontech Laboratories, Inc.) was performed according to the manufacturer's protocol.
2.5 Reverse transcriptase PCR
For each reverse transcriptase PCR (RT-PCR) reaction, cDNA (First-Strand CDNA Synthesis Kit (GE Healthcare)) originating from 1.5 ng of total RNA was used as template. The different RT-PCR products (Fig. 1A) were amplified with Taq DNA polymerase (Invitrogen™) using the following primers: 5′-GCTCAAAGTGCTGGTTCCTC-3′ and 5′-GAGACACTGGGTGTGGGAGT-3′ (Esam1), 5′-ACTGGGACCTACCTCTGCAA-3′ and 5′-CATCCTCCCGAAGGTCACTA-3′ (Vsig2), 5′-CCCTGAGCTGCCACCCAGCAT-3′ and 5′-ATCTTCTTCCTCGCCATGTG-3′ (Nrgn), 5′-CAACCACCAAGTCCTTTCGT-3′ and 5′-GGTAACATGCACACGCAGAG -3′ (Nrgn, acc. no. NM_022029), 5′-ATGTCGATTCCTTTCTCCAACAC-3′ and 5′-GGGGGTAAAACCTGTGGTCT-3′ (Spa17), 5′-GGCCTGTGTTTGGGATAGTG-3′ and 5′-AAAGGACATGAGGACTCCTCAC-3′ (Ysg2), 5′-GAAACCTCTCTTCTGGACAAG-3′ and 5′-AAAGGACATGAGGACTCCTCAC-3′ (AF156856), and 5′-TCAACACCCCAGCCATGTACGTAGCCATCC-3′ and 5′-ACATCTGCTGGAAGGTGGACA-3′ (β-actin, Bact). The integrity and size of the amplification products were validated by agarose gel electrophoreses and sequencing. Prior to sequencing using the employed PCR primers and ABI™ BigDye Terminators (Applied Biosystems), amplicons were excised from agarose gels and purified using GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare).
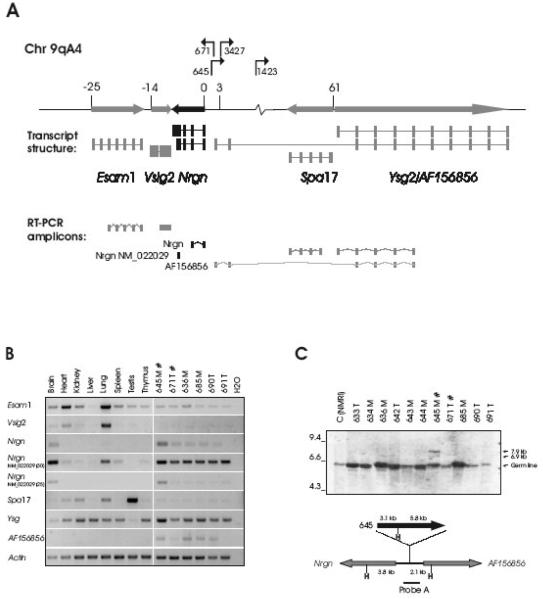
Fig. 1. Proviral integrations in the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus activate Nrgn expression in T-cell tumors.
A. The proviral positions and transcriptional orientations in the locus are shown with arrows. The relative position of the transcription start sites of the genes are given in kb and a schematic mRNA structure is depicted with exons as bars. Both a long (NM_022029) and a short (BC061102) transcript form of Nrgn is shown in black. The RT-PCR amplicons are drawn below. B. RT-PCR on RNA from a panel of tissues from non-infected mice (lanes 1-8) as well as six independent mesenteric (‘M’) and thymic (‘T’) tumors induced by SL3-3(turbo) (lanes 9-14). Tumors with integration into the Nrgn locus are indicated with ‘#’. For the ‘NM_022029’ amplicon PCR amplification products with 25 and 30 cycles are shown. C. Southern blotting on HindIII-digested tumor DNA using Probe A (top panel). Positions of the germ line band as well as the sizes of the expected rearranged bands (6.9 kb and 7.9 kb) are indicated with arrows. The position of Probe A across the integration sites between Nrgn and AF156856 is depicted schematically together with HindIII sites (‘H’), and the distances in kb between HindIII positions and the integration site (bottom panel). For clarity, only the clonal provirus in tumor 645 is depicted. ‘T’ and ‘M’ designates a thymic or mesenteric lymph node tumor, respectively.
2.6 Quantitative real-time PCR
For each quantitative real-time PCR (qRT-PCR) reaction, cDNA (First-Strand cDNA Synthesis Kit (GE Healthcare)) originating from 1.5 ng of total RNA was used as template. qRT-PCR was performed on a Stratagene MX3005 apparatus (AH Diagnostics), using TaqMan probes, assays-on-DemandTM (Applied Biosystems) (Nrgn: Mm00480741_m1, exon 1-2 and UBC: Mm01201237_m1, exon 1-2), and run in 20 μl using TaqMan Universal PCR Master Mix as specified by the manufacturer. Relative quantification was performed using a standard curve method on cDNA isolated from wild-type mouse brain (Nrgn amplifications) and thymus (UBC amplifications), and presented as normalized to Ubiquitin C (UBC) signal. All samples were performed in duplicate. The amplification PCR program was: 95°C for 10 min (1 cycle), 95°C for 15 sec and 60°C for 1 min (40 cycles). Data were analyzed by using Mx3005 software.
2.7 Purification of proteins and Western blot analysis
Whole-cell extracts were isolated from frozen tissue samples by lysis in 360 μl lysis buffer (0.1 M NaCl, 0.01 M Tris-HCl (pH 8.0), 0.5 mM EDTA (pH 8.0) and 0.5 mM PMSF) followed by 30 minutes of incubation on ice and 10 minutes of centrifugation at 20,000 × g. Samples equivalent to 10 μg of total proteins (BCATM Protein Assay Kit, Pierce Biotechnology) were resolved on a 12.5% polyacrylamide gel and transferred to an ImmobilonTM-P transfer membrane (Millipore A/S). The Western blot was probed with primary antibodies Anti-neurogranin (Upstate, catalogue number 07-425) and Anti-actin (I-19) (Santa Cruz Biotechnology, catalogue number sc-1616), both in 1:5000 dilution (3% BSA, 1× TBS-T). Subsequently, the membrane was probed with the HRP-conjugated secondary antibodies goat anti-rabbit IgG (Upstate, catalogue number 12-348) and rabbit anti-goat IgG (DAKO, catalogue number 0449), respectively, both in 1:6666 dilution (3% BSA, 1× TBS-T) and developed using ECL Plus Western Blotting Detection Reagents (GE Healthcare).
3. Results
3.1 Proviral insertions into the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus
T-cell lymphomas were induced in twelve inbred NMRI mice with a mean latency period of 51 days upon inoculation with SL3-3(turbo) MLV (Ethelberg et al., 1997b). All tumors were oligoclonal T-cell lymphomas as revealed by Southern blot analysis of T-cell receptor and immunoglobulin κ-chain rearrangements (data not shown). Tumor DNA was extracted and a total of 45 retrovirus integration sites (RIS) were isolated using the 2-step PCR method described in (Sørensen et al., 1993). PCR products were sequenced and the position and orientation of the integrations were plotted onto the mouse genome (February 2006 UCSC assembly (http://genome.ucsc.edu/)). Forty-four of the sequences could be unambiguously mapped to a RefSeq gene (Table 1). Many integrations map to well known integration sites, such as Myc, Ccnd3, Ccnd1, Rras2, Evi5, Runx1 and Rasgrp1, whereas other RISs have not previously been reported. Upon screening of the tumors by PCR using virus and gene-specific primers, we confirmed the integration in the promoter of Nrgn in tumor 645, and detected a novel integration site at the locus in tumor 671. The two integrations are situated 662 bp apart (Fig. 1A).
Table 1.
RefSeq genes associated with SL3-3(turbo) RISs.
| Mouse ID |
Latency period (days) |
RIS-associated RefSeq a |
|---|---|---|
| 633 | 49 | Rrs1 b |
| 634 | 47 | Myc |
| 635 | 47 | Ccnd3 |
| 636 | 47 | Myc, Wfikkn2 |
| 642 | 49 | Myc (3) c, Pbrm1, Fam169b, Abcb9, Kpn1 |
| 643 | 49 | Myc, Evi5, Sh3pxd2a |
| 644 | 51 | Ganab, Rras2, Set, Plac |
| 645 | 51 | Myc, Zfp507, Nrgn, Evi5, Gmn, Vdac1, Mcl1, Runx1 |
| 671 | 54 | OTTMUSG00000009322, Rassf2, Csrp2bp, Rasgrp1, Naif1, Runx1 |
| 685 | 56 | Myc (2), Rras2 |
| 690 | 57 | Myc (2), Sept7, Ppcdc, Ahi1, Srp68, Pik3r1 |
| 691 | 57 | Ccnd1 |
February 2006 UCSC assembly (http://genome.ucsc.edu/).
Gene names in bold are novel RISs according to the RTCG database (http://rtcgd.abcc.ncifcrf.gov/).
Several independent Myc integrations within one tumor sample.
3.2 The Nrgn gene is the main target of proviral deregulation
Proviruses can disturb the regulation of cellular genes over hundreds of kilobases (Lazo et al., 1990). Thus, in order to point out specific host genes of the locus that may be affected by provirus integration into the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus, semiquantitative reverse transcriptase PCRs (RT-PCR) were performed (Fig. 1B). For comparison, we included tumors from the same panel in which no integration in the locus had been identified in addition to total RNA from various tissues from non-injected animals. All amplicons except amplicon ‘Nrgn (NM_022029)’ spanned at least one intron to rule out amplification of genomic DNA. The identity of the amplification products was determined by sequencing of the PCR fragments.
As it appears in Fig. 1B, the gene of the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus that most clearly seemed to be affected by an integrated provirus was Nrgn. When using an exon 1 to exon 2 amplicon (amplicon ‘Nrgn’) we observed, as expected, expression of Nrgn in brain tissue. Remarkably, however, in most MLV-induced tumor tissues expression of Nrgn was detected. Moreover, in tumor 645, but not tumor 671, both of which harbor provirus integration at the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus, the Nrgn expression level was comparable to that found in brain tissue. We employed an amplicon specific for a longer Nrgn mRNA species (amplicon ‘Nrgn(NM_022029)’), which presumably arises as a result of alternative (downstream) polyadenylation site usage. With this amplicon we observed a pattern similar to that found with the exon 1 to exon 2 amplicon regarding expression in the MLV induced tumors. When we decreased the number of amplification cycles a clear signal in brain and tumor 645 was evident while faint or no bands were observed in the remaining tissues. This made us confident that the signal primarily derived from amplification of cDNA and not from possible carrier-over DNA. Additionally, low levels of expression were seen in lung and spleen.
For the remaining genes at the locus, the expression in the different tissues from non-treated animals in essence correlated with previously published observations (Esam1 (Hirata et al., 2001), Vsig2 (Chretien et al., 1998), Spa17 (Kong et al., 1995; Frayne and Hall, 2002), Ysg2 (Takematsu et al., 1999). Expression of Esam1 and the lysosomal isoform of Ysg2 (Ysg) was observed in the different tumor tissues, but at levels almost similar to those seen in the comparable normal tissues (spleen and thymus). Also, there seemed to be no clear deregulation of these two genes due to proviral integrations in the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus. Notably, expression of the cytosolic Ysg2 variant AF156856, which initiates from an alternative promoter region close to Nrgn (data from UCSC genome browser, see Fig. 1B) is absent in all control tissue while present in four out of six tumor sample. In summary, Nrgn seems to be the main proviral target gene at this locus, but effects of the integrated proviruses on expression of the other genes cannot be ruled out.
The specific increase in Nrgn mRNA expression observed in tumor 645 but not in tumor 671 could suggest differences in clonality status of the tumors with respect to Esam1/Vsig2/Nrgn/Ysg2/Spa17 insertion. In order to address this, Southern blot analysis was performed on genomic DNA from the SL3-3(turbo) induced tumors in NMRI mice. The blot was hybridized with Probe A, which spans the integration-site positions of the inserted proviruses in tumor 645 and 671 (Fig. 1C). In tumor 645 rearranged bands were detected that corresponded to the expected 5′LTR (6.9 kb) and 3′LTR (7.9 kb)-containing fragments (Fig. 1C), supporting a clonal tumor. In contrast, no rearranged fragments were observed in tumor 671 (expected band sizes of 4.7 kb and 10 kb for 5′- and 3′LTR-containing fragments, respectively), indicating a low fraction of cells in the tumor sample containing this particular integration.
3.3 Nrgn is highly expressed in mouse brain
The tissue distribution of Nrgn transcripts has predominantly been examined in human and rat samples (Represa et al., 1990; Watson et al., 1990; Martinez de Arrieta et al., 1997; Pak et al., 2000). In order to address this thoroughly in mouse tissues, we carried out Northern blot, quantitative real-time PCR (qRT-PCR), and Western blot analyses, the results of which are summarized in Fig. 2.
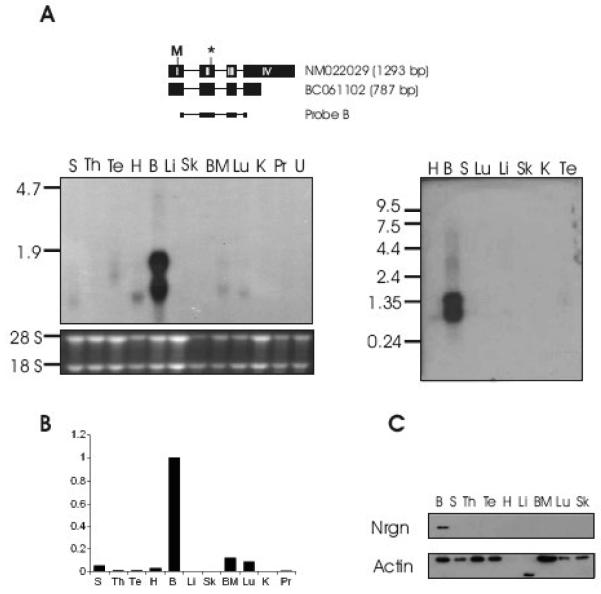
Fig. 2. Expression of Nrgn is highest in brain.
A. Northern blotting using Probe B positioned on the long and short Nrgn mRNA as shown schematically (top panel). The extent of CDS is indicated by start (‘M’) and termination (‘*’) codon. Northern blotting was performed on a mRNA Multiple Northern Blot (Clontech) (right panel) as well as on a membrane containing total RNA from various organs from BALB/c mice (left panel). H, heart; B, brain; S, spleen; Lu, lung; Li, liver; Sk, skeletal muscle; K, kidney; Te, testis; Th, thymus; BM, bone marrow; Pr, prostate; U, uterus. B. Relative Nrgn levels as measured by quantitative real-time PCR. C. Nrgn Western blotting on protein isolated from a panel of mouse organs.
Northern blot analysis was performed on total RNA extracted from a panel of BALB/c mouse tissues (Fig. 2A, left panel). In an effort to detect all transcript forms that may be present in the various tissues, the membrane was probed with a cDNA probe covering exon 1 to exon 4 of Nrgn (Probe B, Fig. 2). This probe detects two transcripts (Fig 2A, left panel), which most likely correspond to the published mRNAs NM_022029 (1293 bp) and BC061102 (815 bp). As expected, very high expression of Nrgn was seen in brain. Additionally, weak expression of a 0.8 kb transcript was observed in spleen, heart, bone marrow and lung, while an intermediate form was detected in testis. Upon hybridization with the same probe to a Northern blot membrane containing mouse poly(A)+ RNA isolated from different tissues (Fig 2A, right panel), we again find high expression of the 1.3 kb and 0.8 kb transcripts in brain.
To further elucidate on the RNA expression pattern, qRT-PCR was performed on tissue RNA from BALB/c mice, employing an Nrgn TaqMan amplicon spanning exon 1 to 2 (Fig. 2B). The results from this assay were in accordance with those obtained from the Northern blot analysis as well as with the RT-PCR data (Fig. 1B).
Finally, we wanted to determine if the high expression of Nrgn RNA in brain is paralleled by an elevated amount of protein and hence, Western blot analysis was performed on whole-cell extracts from different tissues employing antibodies recognizing the C-terminus of Nrgn (Fig. 2C). Nrgn was clearly detected in brain tissues, thus reflecting the RNA expression pattern. We did not, however, detect Nrgn in the low expressing tissues spleen, heart, bone marrow and lung.
3.4 Provirus insertions at the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus upregulate Nrgn RNA and protein
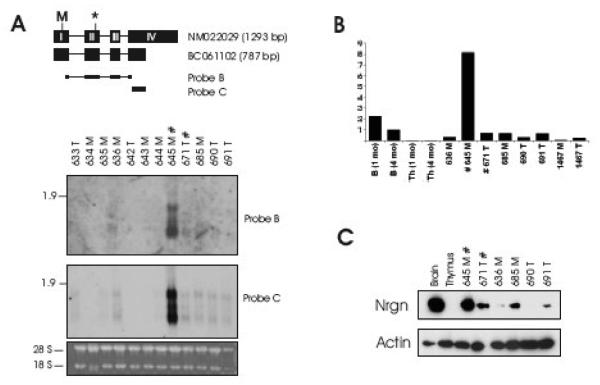
Having established the expression pattern of Nrgn in mouse tissues we next wanted to examine Nrgn mRNA and protein levels in the MLV-induced tumors. Northern blot, qRT-PCR, and Western blot analyses were performed on T-cell lymphomas from thymic and mesenteric lymph nodes of the SL3-3(turbo) infected NMRI mice (Fig. 3). In accordance with the RT-PCR results (Fig. 1B), Northern blot analysis using Probe B detected the 1.3 kb and 0.8 kb transcripts in tumor 645 and, at lower levels, in about half of the other tumor samples. Additionally, an intermediately sized transcript was observed in tumor 645. Upon longer exposure time this mRNA species also appeared in tumors without proviral integration in the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus (data not shown). Hence, the transcript is not directly related to the presence of a nearby provirus insertion, but may represent a novel Nrgn mRNA isoform. We note that an intermediatesize transcript also was observed in testis (Fig. 2A). When we employed a probe situated outside the coding region of Nrgn (Probe C) a similar band pattern appeared.
Fig. 3. Neurogranin expression in SL3-3(turbo) MLV-induced T-cell lymphomas equals that of brain tissue.
Northern blot analysis (A), Quantitative real-time PCR (B) and Western blotting (C) on tumors from SL3-3(turbo)-infected mice. Northern blotting was done using the indicated probes. In (B) brain and thymus from 1 month and 4 month old non-infected mice were included for comparison. In (C) brain and thymus from 1 month old non-infected mice were included. Legend is as in Fig. 1.
Subsequently, Nrgn expression levels were investigated by qRT-PCR using the same TaqMan amplicon as in Fig. 2B, and this analysis confirmed a high Nrgn expression level in tumor 645 exceeding that found in brain (Fig. 3B). Furthermore, in the subclonal tumor 671 as well as in tumors without integration in the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus, Nrgn levels were several fold higher than that found in normal thymus tissue from 1-month and 4-month old mice. Western blot analysis (Fig. 3C) confirmed that high Nrgn RNA levels result in high Nrgn protein levels in MLV-induced tumor tissue harboring the clonal provirus insertion near Nrgn, although seemingly lower than those of brain tissue. We believe that a likely explanation for the discrepancies between protein and RNA levels between brain and tumor 645 is differences in post-transcriptional processes in the two tissues. Nrgn was also evident in the subclonal tumor 671 as well as in half of tumors without integration in the locus. The relative protein level between tumors paralleled the mRNA levels.
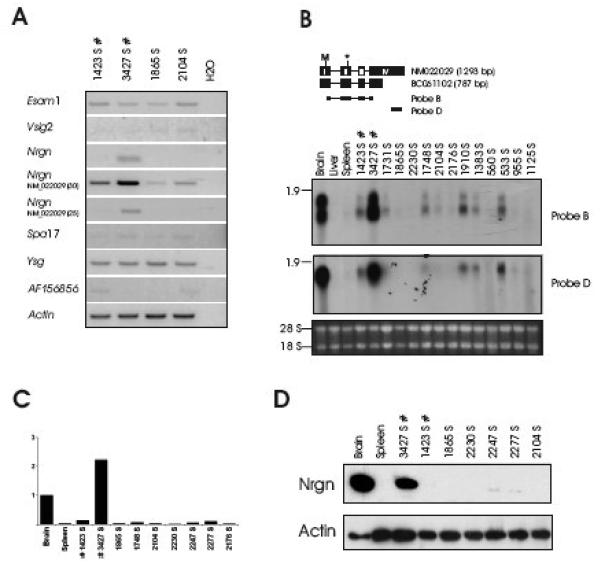
The finding that 2 out of 12 tumors harboring integration in the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus but not in any of the many models listed in the RTCG database (http://rtcgd.abcc.ncifcrf.gov/) (Akagi et al., 2004) suggests a highly model-specific role of Nrgn in T-cell lymphomagenesis, possibly specific to either SL3-3 MLV or the NMRI background, or a combination of both. Interestingly, we subsequently identified two integrations (in tumor 1423 and 3427) in the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus (Fig. 1A) from an independent retroviral tagging screen utilizing wild type SL3-3 in 1767 BALB/c mice (Glud et al., 2005; Wang et al., 2006). The proviruses were inserted 3.1 kb (tumor 3427) and 10.1 kb (tumor 1423) from Nrgn and in opposite transcriptional orientation as Nrgn. RT-PCR, Northern blotting, qRT-PCR and western blot analysis on tumor samples from this tumor panel suggested increased Nrgn expression in tumor 3427, whereas in approximately half of the other tumors - including tumor 1423 - moderate Nrgn mRNA levels were detected (Fig. 4). Upon long exposure time faint levels of Nrgn protein was detected in tumors 1423, 2247 and 2277 (data not shown). In contrast to the SL3-3(turbo)/NMRI model we observed the 0.8 and 1.3 kb but not the intermediate mRNA band. A probe specific to the longer transcript (NM022029) (Probe D) detected only the 1.3 kb transcript supporting the notion that the 0.8 kb Nrgn transcript is generated from alternative polyadenylation within exon 4 (Fig. 4A).
Fig. 4. Nrgn is expressed in SL3-3 wt MLV-induced T-cell lymphomas.
(A) RT-PCR analysis using gene-specific primers as in Fig. 1 is shown. ‘S’ designates splenic tumor. Northern blotting with the indicated probes (B), Quantitative real-time PCR (C) and Western blotting (D) was performed as described for Fig. 3.
4. Discussion
In this work, we have for the first time systematically examined the expression of Nrgn in various mouse tissues as well as in T-cell lymphomas induced by SL3-3 MLV. In accordance with previous observations in rat and human (Represa et al., 1990; Watson et al., 1990; Martinez de Arrieta et al., 1997), our analysis of mouse tissues showed Nrgn to be expressed predominantly in brain tissues. However, we also found low mRNA expression in spleen, heart, bone marrow, lung and testis although we were unable to detect Nrgn protein in these tissues by Western blotting.
The Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus is a novel retroviral target region. Initially, we found integration into the locus in tumors from two out of twelve NMRI mice injected with SL3-3(turbo) MLV (tumors 645 and 671). This virus is highly potent and induces T-cell lymphomas in inbred NMRI mice significantly faster than does wild-type SL3-3 (Ethelberg et al., 1997b). Subsequently, two additional insertions were isolated by retrovirus tagging in a separate study originated from a larger cohort involving 1767 mice injected with wild type SL3-3 (Glud et al., 2005; Wang et al., 2006) (tumors 1423 and 3427). While the SL3-3(turbo) proviruses were inserted within a 662-bp narrow region of the Nrgn promoter region, proviruses in tumor 1423 and 3427 were found in intron sequences of the upstream-located Ysg2 gene. In targeting Nrgn the orientation of the provirus in tumor 671 predicts a promoter activation mechanism and the remaining three proviruses enhancer activation. It is notable that in these studies the utilized viruses were SL3-3 wild-type and a derivative hereof.
All four tumors with proviral integrations in the Esam1/Vsig2/Nrgn/Ysg2/Spa17-locus showed elevated expression levels compared to normal thymus and spleen tissue. In two of these, Nrgn RNA and protein expression levels were as high as in brain. The variable expression levels may reflect the fraction of cells in tumor tissues with provirus insertion at this locus, however we only addressed this specifically in the SL3-3(turbo)-model by Southern blot analysis. Elevated expression of Nrgn was detected in some control tumors in which we have not identified proviral integration. This may result from indirect effects of cellular signaling cascades activated by proviruses inserted at other loci. Alternatively, it might be a direct effect of proviruses positioned in other parts at the locus in question not revealed by our analysis. Finally, it is formally possible that a specific subset of T cells targeted by SL3-3 naturally express Nrgn, although in both 1-month and 4-month old mice thymic expression of Nrgn was barely detectable.
By means of Northern blot analysis employing a full-length probe three different transcripts were detected in T-cell tumors isolated from the NMRI mouse-strain background in contrast to two messengers only in the BALB/c background. While the overall levels of Nrgn RNA varied among tumors, the relative abundance of the individual RNA forms appeared similar within tumors of each of the two models. The BALB/c mRNA species as well as the small and large mRNAs in the NMRI correlate with the two major bands observed in brain. The larger of these corresponds to the 1.35 kb Refseq Nrgn mRNA (acc. no. NM_022029). Using a probe situated in the 3′ terminal end of exon 4 we found only the longer transcript, thus indicating the shorter 0.8 kb to result from premature alternative polyadenylation within exon 4 (e.g. transcript BC061102). Although differential expression of the long and short Nrgn mRNA remains purely speculative, the extended 3′UTR of Nrgn is indicative of miRNA regulation. A search on miRDB (http://mirdb.org/) revealed two miRNAs, miR-423-5p and miR-705, which potentially target exon 4 of Nrgn. Interestingly, while the shorter Nrgn mRNA species is putatively targeted by both miRs, only miR-423-5p targets the longer mRNA. The presented data support a role for Nrgn outside the nervous system. Indeed, in contrast to most neuronal-restricted genes like synapsin and type II Na+ channel genes (Kraner et al., 1992; Li et al., 1993), the promoter of Nrgn does not appear to harbor a brain-specific silencer element (Sato et al., 1995). Previous observations suggest Nrgn to display pro-apoptotic capacity upon interleukin-2 deprivation in T-cells, and cell death induced by the NO-donor sodium nitroprusside in a stable Nrgn-expressing neuroblastoma cell line (Chakravarthy et al., 1999; Devireddy and Green, 2003; Gui et al., 2007). In that respect, it is interesting that the expression of NRGN has been observed to be downregulated in human malignant glial neoplasms as compared to normal brain samples (Yokota et al., 2006). In contrast, we observed relative elevated expression of Nrgn in the T-cell tumors, which might imply that Nrgn displays a dual function in proliferation and apoptosis with the route of action dependent on other signals activated in the cell as reported for other cancer-related genes such as Myc and TGFβRII (Dang et al., 1999; Nasi et al., 2001; Dennler et al., 2002; Roberts and Wakefield, 2003). Nrgn is a redox-sensitive phosphoprotein that does not possess any known enzymatic activity (Sheu et al., 1995; Mahoney et al., 1996; Sheu et al., 1996; Li et al., 1999; Miao et al., 2000), and has only been demonstrated to interact with CaM and phosphatidic acid (PA) (Dominguez-Gonzalez et al., 2007). Based on the CaM-sequestering function in neurons, elevated levels of Nrgn in T-cells may perturb delicate Ca2+ and Ca2+-CaM-dependent pathways (Pak et al., 2000). Regulation of the transcription factor families NFAT, NFkB and AP-1, which are central for transcriptional activity and proliferation of T-cells, is highly dependent on the rise in intracellular [Ca2+] (for review see (Quintana et al., 2005)). Hence, aberrant amplitudes and kinetics of Ca2+-signals caused by high Nrgn expression in lymphoid tissue as reported here may participate in tumorigenesis by disrupting normal cell homeostasis.
Acknowledgment
Technical assistance from Astrid van der Aa Kühle and help from Randi Jessen are gratefully acknowledged.
This work was supported by the Danish Agency for Science, Technology and Innovation, the Danish Cancer Society, the Novo Nordic Foundation, the Danish Cancer Foundation, the Karen Elise Jensen Foundation, and NIH grant R01AI41570.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucl. Acids. Res. 2004;32:D523–527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudier J, Deloulme JC, Van Dorsselaer A, Black D, Matthes HW. Purification and characterization of a brain-specific protein kinase C substrate, neurogranin (p17). Identification of a consensus amino acid sequence between neurogranin and neuromodulin (GAP43) that corresponds to the protein kinase C phosphorylation site and the calmodulin-binding domain. J Biol Chem. 1991;266:229–37. [PubMed] [Google Scholar]
- Chakravarthy B, Morley P, Whitfield J. Ca2+-calmodulin and protein kinase Cs: a hypothetical synthesis of their conflicting convergences on shared substrate domains. Trends Neurosci. 1999;22:12–6. doi: 10.1016/s0166-2236(98)01288-0. [DOI] [PubMed] [Google Scholar]
- Chiriva-Internati M, Wang Z, Salati E, Timmins P, Lim SH. Tumor vaccine for ovarian carcinoma targeting sperm protein 17. Cancer. 2002;94:2447–53. doi: 10.1002/cncr.10506. [DOI] [PubMed] [Google Scholar]
- Chretien I, Marcuz A, Courtet M, Katevuo K, Vainio O, Heath JK, White SJ, Du Pasquier L. CTX, a Xenopus thymocyte receptor, defines a molecular family conserved throughout vertebrates. Eur J Immunol. 1998;28:4094–104. doi: 10.1002/(SICI)1521-4141(199812)28:12<4094::AID-IMMU4094>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- Dennler S, Goumans MJ, Ten Dijke P. Transforming growth factor beta signal transduction. J Leukoc Biol. 2002;71:731–40. [PubMed] [Google Scholar]
- Devireddy LR, Green MR. Transcriptional program of apoptosis induction following interleukin 2 deprivation: identification of RC3, a calcium/calmodulin binding protein, as a novel proapoptotic factor. Mol Cell Biol. 2003;23:4532–41. doi: 10.1128/MCB.23.13.4532-4541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Gonzalez I, Vazquez-Cuesta SN, Algaba A, Diez-Guerra FJ. Neurogranin binds to phosphatidic acid and associates to cellular membranes. Biochem J. 2007;404:31–43. doi: 10.1042/BJ20061483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejegod D, Sorensen KD, Mossbrugger I, Quintanilla-Martinez L, Schmidt J, Pedersen FS. Control of pathogenicity and disease specificity of a Tlymphomagenic gammaretrovirus by E-box motifs but not by an overlapping glucocorticoid response element. J Virol. 2009;83:336–46. doi: 10.1128/JVI.01368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethelberg S, Hallberg B, Lovmand J, Schmidt J, Luz A, Grundstrom T, Pedersen FS. Second-site proviral enhancer alterations in lymphomas induced by enhancer mutants of SL3-3 murine leukemia virus: negative effect of nuclear factor 1 binding site. J Virol. 1997a;71:1196–206. doi: 10.1128/jvi.71.2.1196-1206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethelberg S, Sørensen AB, Schmidt J, Luz A, Pedersen FS. An SL3-3 murine leukemia virus enhancer variant more pathogenic than the wild type obtained by assisted molecular evolution in vivo. J Virol. 1997b;71:9796–9. doi: 10.1128/jvi.71.12.9796-9799.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frayne J, Hall L. A re-evaluation of sperm protein 17 (Sp17) indicates a regulatory role in an A-kinase anchoring protein complex, rather than a unique role in sperm-zona pellucida binding. Reproduction. 2002;124:767–74. doi: 10.1530/rep.0.1240767. [DOI] [PubMed] [Google Scholar]
- Gerendasy D. Homeostatic tuning of Ca2+ signal transduction by members of the calpacitin protein family. J Neurosci Res. 1999;58:107–19. [PubMed] [Google Scholar]
- Glud SZ, Sørensen AB, Andrulis M, Wang B, Kondo E, Jessen R, Krenacs L, Stelkovics E, Wabl M, Serfling E, Palmetshofer A, Pedersen FS. A tumor-suppressor function for NFATc3 in T-cell lymphomagenesis by murine leukemia virus. Blood. 2005;106:3546–52. doi: 10.1182/blood-2005-02-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grizzi F, Gaetani P, Franceschini B, Di Ieva A, Colombo P, Ceva-Grimaldi G, Bollati A, Frezza EE, Cobos E, Rodriguez y Baena R, Dioguardi N, Chiriva-Internati M. Sperm protein 17 is expressed in human nervous system tumours. BMC Cancer. 2006;6:23. doi: 10.1186/1471-2407-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J, Song Y, Han NL, Sheu FS. Characterization of transcriptional regulation of neurogranin by nitric oxide and the role of neurogranin in SNP-induced cell death: implication of neurogranin in an increased neuronal susceptibility to oxidative stress. Int J Biol Sci. 2007;3:212–24. doi: 10.7150/ijbs.3.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata K, Ishida T, Penta K, Rezaee M, Yang E, Wohlgemuth J, Quertermous T. Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells. J Biol Chem. 2001;276:16223–31. doi: 10.1074/jbc.M100630200. [DOI] [PubMed] [Google Scholar]
- Huang FL, Huang KP, Wu J, Boucheron C. Environmental enrichment enhances neurogranin expression and hippocampal learning and memory but fails to rescue the impairments of neurogranin null mutant mice. J Neurosci. 2006;26:6230–7. doi: 10.1523/JNEUROSCI.1182-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KP, Huang FL, Jager T, Li J, Reymann KG, Balschun D. Neurogranin/RC3 enhances long-term potentiation and learning by promoting calcium-mediated signaling. J Neurosci. 2004;24:10660–9. doi: 10.1523/JNEUROSCI.2213-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kundu RK, Yang E, Hirata K, Ho YD, Quertermous T. Targeted disruption of endothelial cell-selective adhesion molecule inhibits angiogenic processes in vitro and in vivo. J Biol Chem. 2003;278:34598–604. doi: 10.1074/jbc.M304890200. [DOI] [PubMed] [Google Scholar]
- Kong M, Richardson RT, Widgren EE, O'Rand MG. Sequence and localization of the mouse sperm autoantigenic protein, Sp17. Biol Reprod. 1995;53:579–90. doi: 10.1095/biolreprod53.3.579. [DOI] [PubMed] [Google Scholar]
- Kraner SD, Chong JA, Tsay HJ, Mandel G. Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- Lazo P, Lee J, Tsichlis P. Long-Distance Activation of the Myc Protooncogene by Provirus Insertion in Mlvi-1 or Mlvi-4 in Rat T-Cell Lymphomas. PNAS. 1990;87:170–173. doi: 10.1073/pnas.87.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Pak JH, Huang FL, Huang KP. N-methyl-D-aspartate induces neurogranin/RC3 oxidation in rat brain slices. J Biol Chem. 1999;274:1294–300. doi: 10.1074/jbc.274.3.1294. [DOI] [PubMed] [Google Scholar]
- Li L, Suzuki T, Mori N, Greengard P. Identification of a functional silencer element involved in neuron-specific expression of the synapsin I gene. Proc Natl Acad Sci U S A. 1993;90:1460–4. doi: 10.1073/pnas.90.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SH, Bumm K, Chiriva-Internati M, Xue Y, Wang Z. MAGE-C1 (CT7) gene expression in multiple myeloma: relationship to sperm protein 17. Eur J Haematol. 2001a;67:332–4. doi: 10.1034/j.1600-0609.2001.00552.x. [DOI] [PubMed] [Google Scholar]
- Lim SH, Wang Z, Chiriva-Internati M, Xue Y. Sperm protein 17 is a novel cancer-testis antigen in multiple myeloma. Blood. 2001b;97:1508–10. doi: 10.1182/blood.v97.5.1508. [DOI] [PubMed] [Google Scholar]
- Mahoney CW, Pak JH, Huang KP. Nitric oxide modification of rat brain neurogranin. Identification of the cysteine residues involved in intramolecular disulfide bridge formation using site-directed mutagenesis. J Biol Chem. 1996;271:28798–804. doi: 10.1074/jbc.271.46.28798. [DOI] [PubMed] [Google Scholar]
- Martinez de Arrieta C, Perez Jurado L, Bernal J, Coloma A. Structure, organization, and chromosomal mapping of the human neurogranin gene (NRGN) Genomics. 1997;41:243–9. doi: 10.1006/geno.1997.4622. [DOI] [PubMed] [Google Scholar]
- Miao HH, Ye JS, Wong SL, Wang BX, Li XY, Sheu FS. Oxidative modification of neurogranin by nitric oxide: an amperometric study. Bioelectrochemistry. 2000;51:163–73. doi: 10.1016/s0302-4598(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Mikkers H, Allen J, Berns A. Proviral activation of the tumor suppressor E2a contributes to T cell lymphomagenesis in EmuMyc transgenic mice. Oncogene. 2002;21:6559–66. doi: 10.1038/sj.onc.1205930. [DOI] [PubMed] [Google Scholar]
- Mikkers H, Berns A. Retroviral insertional mutagenesis: tagging cancer pathways. Adv Cancer Res. 2003;88:53–99. doi: 10.1016/s0065-230x(03)88304-5. [DOI] [PubMed] [Google Scholar]
- Nasi S, Ciarapica R, Jucker R, Rosati J, Soucek L. Making decisions through Myc. FEBS Lett. 2001;490:153–62. doi: 10.1016/s0014-5793(01)02118-4. [DOI] [PubMed] [Google Scholar]
- Nielsen AA, Sørensen AB, Schmidt J, Pedersen FS. Analysis of wild-type and mutant SL3-3 murine leukemia virus insertions in the c-myc promoter during lymphomagenesis reveals target site hot spots, virus-dependent patterns, and frequent error-prone gap repair. J Virol. 2005;79:67–78. doi: 10.1128/JVI.79.1.67-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak JH, Huang FL, Li J, Balschun D, Reymann KG, Chiang C, Westphal H, Huang KP. Involvement of neurogranin in the modulation of calcium/calmodulin-dependent protein kinase II, synaptic plasticity, and spatial learning: a study with knockout mice. Proc Natl Acad Sci U S A. 2000;97:11232–7. doi: 10.1073/pnas.210184697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard L, Deloulme JC, Storm DR. Interactions between neurogranin and calmodulin in vivo. J Biol Chem. 1999;274:7689–94. doi: 10.1074/jbc.274.12.7689. [DOI] [PubMed] [Google Scholar]
- Quintana A, Griesemer D, Schwarz EC, Hoth M. Calcium-dependent activation of T-lymphocytes. Pflugers Arch. 2005;450:1–12. doi: 10.1007/s00424-004-1364-4. [DOI] [PubMed] [Google Scholar]
- Rasmussen MH, Sørensen AB, Morris DW, Dutra JC, Engelhard EK, Wang CL, Schmidt J, Pedersen FS. Tumor model-specific proviral insertional mutagenesis of the Fos/Jdp2/Batf locus. Virology. 2005;337:353–64. doi: 10.1016/j.virol.2005.04.027. [DOI] [PubMed] [Google Scholar]
- Represa A, Deloulme JC, Sensenbrenner M, Ben-Ari Y, Baudier J. Neurogranin: immunocytochemical localization of a brain-specific protein kinase C substrate. J Neurosci. 1990;10:3782–92. doi: 10.1523/JNEUROSCI.10-12-03782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci U S A. 2003;100:8621–3. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Xiao DM, Li H, Huang FL, Huang KP. Structure and regulation of the gene encoding the neuron-specific protein kinase C substrate neurogranin (RC3 protein) J Biol Chem. 1995;270:10314–22. doi: 10.1074/jbc.270.17.10314. [DOI] [PubMed] [Google Scholar]
- Sheu FS, Huang FL, Huang KP. Differential responses of protein kinase C substrates (MARCKS, neuromodulin, and neurogranin) phosphorylation to calmodulin and S100. Arch Biochem Biophys. 1995;316:335–42. doi: 10.1006/abbi.1995.1045. [DOI] [PubMed] [Google Scholar]
- Sheu FS, Mahoney CW, Seki K, Huang KP. Nitric oxide modification of rat brain neurogranin affects its phosphorylation by protein kinase C and affinity for calmodulin. J Biol Chem. 1996;271:22407–13. doi: 10.1074/jbc.271.37.22407. [DOI] [PubMed] [Google Scholar]
- Sørensen AB, Duch M, Jørgensen P, Pedersen FS. Amplification and sequence analysis of DNA flanking integrated proviruses by a simple two-step polymerase chain reaction method. J Virol. 1993;67:7118–24. doi: 10.1128/jvi.67.12.7118-7124.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen AB, Lund AH, Ethelberg S, Copeland NG, Jenkins NA, Pedersen FS. Sint1, a common integration site in SL3-3-induced T-cell lymphomas, harbors a putative proto-oncogene with homology to the septin gene family [In Process Citation] J Virol. 2000;74:2161–8. doi: 10.1128/jvi.74.5.2161-2168.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen KD, Quintanilla-Martinez L, Kunder S, Schmidt J, Pedersen FS. Mutation of all Runx (AML1/core) sites in the enhancer of T-lymphomagenic SL3-3 murine leukemia virus unmasks a significant potential for myeloid leukemia induction and favors enhancer evolution toward induction of other disease patterns. J Virol. 2004;78:13216–31. doi: 10.1128/JVI.78.23.13216-13231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takematsu H, Diaz S, Stoddart A, Zhang Y, Varki A. Lysosomal and cytosolic sialic acid 9-O-acetylesterase activities can Be encoded by one gene via differential usage of a signal peptide-encoding exon at the N terminus. J Biol Chem. 1999;274:25623–31. doi: 10.1074/jbc.274.36.25623. [DOI] [PubMed] [Google Scholar]
- Touw IP, Erkeland SJ. Retroviral insertion mutagenesis in mice as a comparative oncogenomics tool to identify disease genes in human leukemia. Mol Ther. 2007;15:13–9. doi: 10.1038/sj.mt.6300040. [DOI] [PubMed] [Google Scholar]
- Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–72. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- van Dalen JJ, Gerendasy DD, de Graan PN, Schrama LH, Gruol DL. Calcium dynamics are altered in cortical neurons lacking the calmodulin-binding protein RC3. Eur J Neurosci. 2003;18:13–22. doi: 10.1046/j.1460-9568.2003.02720.x. [DOI] [PubMed] [Google Scholar]
- Wang CL, Wang BB, Bartha G, Li L, Channa N, Klinger M, Killeen N, Wabl M. Activation of an oncogenic microRNA cistron by provirus integration. Proc Natl Acad Sci U S A. 2006;103:18680–4. doi: 10.1073/pnas.0609030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JB, Battenberg EF, Wong KK, Bloom FE, Sutcliffe JG. Subtractive cDNA cloning of RC3, a rodent cortex-enriched mRNA encoding a novel 78 residue protein. J Neurosci Res. 1990;26:397–408. doi: 10.1002/jnr.490260402. [DOI] [PubMed] [Google Scholar]
- Yokota T, Kouno J, Adachi K, Takahashi H, Teramoto A, Matsumoto K, Sugisaki Y, Onda M, Tsunoda T. Identification of histological markers for malignant glioma by genome-wide expression analysis: dynein, alpha-PIX and sorcin. Acta Neuropathol. 2006;111:29–38. doi: 10.1007/s00401-005-1085-6. [DOI] [PubMed] [Google Scholar]