Abstract
Protein degradation is increased by both insulin deficiency and insulin resistance in humans and animal models. In skeletal muscle this insulin-dependent increase in protein degradation involves activation of both caspase-3 and the ubiquitin-proteasome system. The influence of abnormal insulin signaling on protein metabolism in cardiac muscle is not well understood; therefore, we measured protein degradation in cardiac muscle of mice with streptozotocin-induced diabetes. Insulin deficiency increased both total muscle proteolysis (measured as tyrosine release in muscle slices or extracts) and the degradation of the myofibrillar protein actin (measured as the appearance of a 14-kDa actin fragment). Expression of ubiquitin mRNA and chymotrypsin-like activity in the proteasome were increased, indicating activation of the ubiquitin-proteasome system in diabetic mouse heart. We also evaluated possible signaling pathways that might regulate cardiac muscle proteolysis. Insulin receptor substrate-1 (IRS-1) tyrosine phosphorylation, and Akt phosphorylation were decreased. Insulin replacement prevented the decrease in IRS-1/Akt phosphorylation, the increase in proteolysis, and attenuated the increase in ubiquitin mRNA. We conclude that insulinopenia accelerates proteolysis in cardiac muscle by reducing IRS-1/Akt signaling, which leads to activation of the ubiquitin-proteasome proteolytic pathway.
DIABETES MELLITUS is associated with a marked increase in the risk of cardiac disease, including a cardiomyopathy that is characterized by impaired ventricular function, even though there is no atherosclerotic coronary heart disease or hypertension (1). The mechanisms underlying this type of heart disease in diabetes are complex. There could be involvement of hyperlipidemia, altered endothelial function, and/or decreased sympathetic neuronal function (2,3). A major component of diabetic cardiomyopathy is cardiac remodeling, which involves increased turnover of cardiac muscle proteins. Destruction of cardiac muscle proteins also could affect contractile function by decreasing the content of myofibrillar protein. It is unclear how diabetes changes the regulation of protein metabolism in the heart.
Insulin is an important regulator of protein turnover in both skeletal and cardiac muscle. It is well established that insulin deficiency or insulin resistance accelerates the degradation of skeletal muscle protein (4,5). The mechanisms underlying this response involve suppressed insulin receptor substrate-1 (IRS-1)/Akt signaling, which causes activation of caspase-3 and stimulation of the ubiquitin-proteasome system (5,6,7). Evidence also suggests that an intact insulin signaling system plays an important role in regulating cardiac size. In mice with a selective knockout of the insulin receptor in cardiomyocytes, the hearts were reduced in size by 20–30% (8). In cardiac muscle, insulin stimulates protein synthesis (9) and exerts an antiapoptotic effect on cardiomyocytes (10). Insulin may also increase cardiac contractility (9).
The importance of the ubiquitin-proteasome system in regulating protein degradation in skeletal muscle in diabetes is well documented. However, little information is known about the regulation of the ubiquitin-proteasome system in the heart (11,12). For example, Kedar et al. (13) reported that the ubiquitin-proteasome system is critical for the degradation of key structural and functional proteins in the heart. This report suggested that the ubiquitin-proteasome system could regulate cardiac function in both normal and stressed situations by changing protein metabolism. In the present study, we evaluated the influence of insulin deficiency on muscle protein metabolism in the heart using a mouse model of acute type 1 diabetes.
In this study, cardiac muscle protein degradation and insulin signaling were examined by several different methods to determine the impact of insulin deficiency on cardiac muscle protein turnover. We investigated four major proteolytic processes: 1) the ATP-dependent, ubiquitin-proteasome system; 2) the lysosomal proteolytic pathway; 3) the calcium-activated proteolytic pathway; and 4) caspase-3-mediated proteolysis. Our results provide evidence that the IRS-1/Akt regulates cardiac muscle mass by modulating the activity of the ubiquitin-proteasome system and caspase-3-mediated proteolysis.
Materials and Methods
Animals
Mice (C57BL/6J) were purchased from The Jackson Laboratory (Bar Harbor, ME). The experiments were approved by the Institutional Animal Care and Use Committee of Emory University. Mice were housed in the animal care facility with a 12-h light, 12-h dark cycle. Control mice were pair fed the same amount of food as the insulin-deficient mice (4). Insulin deficiency was produced by injecting streptozotocin (STZ) (Pfanstiehl Laboratories, Inc., Waukegan, IL), 150 mg/kg body weight [prepared fresh in 0.1 m citrate buffer (pH 4.0)] ip on the first day. On the second day, 75–150 mg STZ/kg was injected depending on the blood glucose level. Control mice were injected with vehicle only.
Some diabetic mice received insulin replacement in the form of a daily sc injection (8 U/100 g body weight) of long-acting bovine Protamine Zine Insulin (BCP Veterinary Pharmacy, Houston, TX), beginning with the initial STZ treatment.
Ten days after STZ injection, diabetic and control mice were anesthetized with 12 mg/kg xylazine and 60 mg/kg ketamine. The heart was removed to measure protein metabolism in cardiac slices. The remaining heart tissue was flash frozen in liquid nitrogen and stored at −80 C for subsequent analyses. In some experiments, plantaris and gastrocnemius muscles were removed to determine protein metabolism in skeletal muscle (14).
Plasma insulin was measured using the 1-2-3 ultrasensitive mouse insulin enzyme immunoassay kit (American Laboratory Products Co., Windham, NH). The blood glucose concentration was measured by the Accu-CHEK advantage blood glucose meter (Roche, Indianapolis, IN).
Cardiac muscle protein degradation
Protein degradation in cardiac muscle was evaluated by three different assays. Total protein degradation was measured in left ventricular slices, whereas breakdown of soluble cardiac muscle protein and myofibrillar proteins were determined in flash-frozen muscles. As described for isolated skeletal muscle (14), we measured the rate of tyrosine release from slices of the left ventricle (∼15 mg/slice). The slices were fixed at resting size (15) on a plastic support and then incubated in Krebs-Ringer bicarbonate buffer (135.5 mm NaCl; 4.7 mm KCl; 24.8 mm NaHCO3; 1 mm MgSO4; 1 mm KH2PO4; 2.5 mm CaCl2; 10 mm glucose) containing 0.5 mm cycloheximide to block tyrosine reutilization. After a 30-min preincubation, the muscle was transferred to a flask containing fresh media and incubated at 37 C for 2 h. All incubation flasks were gassed with 95%/O2/5%/CO2 for 3 min at the beginning of the preincubation and experimental periods. Degradation of proteins was measured by assaying the free tyrosine in the trichloroacetic acid soluble supernatant using a fluorometric technique (16). The influence of diabetes on the different proteolytic pathways was assessed by adding proteolytic inhibitors to the media and measuring tyrosine release (17,18). The ubiquitin-proteasome pathway was specifically inhibited by adding 20 μm epoxomicin (Peptides International Inc., Louisville, KY). Epoxomicin is a cell-permeable, potent, selective, and irreversible inhibitor of proteasome function. It does not inhibit nonproteasomal proteases, including calpains and lysosomal cathepsins, at concentrations less than 50 μm (19). In some experiments, both lysosomal and calcium-activated proteases were blocked. This was accomplished by adding 10 mm methylamine, 200 μm valine, 170 μm leucine, 100 μm isoleucine, and 1 mU/ml insulin to the medium to inhibit lysosomal function, whereas calcium-activated proteases were blocked by deleting calcium from the Krebs-Ringer bicarbonate buffer and adding 50 μm trans-epoxysuccinyl-l-leucylamido-4-guanidino butane (E-64) to the media. E-64 is a potent inhibitor of both the calcium pathway (calpains) and the lysosomal proteases cathepsin B, D, H, and L. At 50 μm, E-64 does not inhibit the proteasome (20).
To assay the degradation of cardiac muscle soluble protein, we used a modification of the proteasomal proteolysis assay reported by Goldberg and colleagues (21). The heart was removed, and, after removal of slices for metabolism tests, two thirds of the remaining heart was freeze clamped in liquid nitrogen. Muscle extracts were prepared by pulverizing cardiac muscle (∼50 mg tissue) in liquid nitrogen and homogenizing the powder in ice-cold harvest buffer [5 mm Tris-HCl (pH 8.8), 1% glycerol, 1 mm EDTA, 1 mm EGTA, freshly constituted 1 mm β-Me, and 50 mm EP-475] (21). The homogenates were centrifuged at 30,000 × g for 30 min, then the soluble fractions were used to measure protein degradation, and the pellets were used to determine actin cleavage (22). For the protein degradation determination, muscle extracts were dialyzed against a basal buffer [20 mm Tris-HCl (pH 7.6) 10% glycerol, 2 mm dithiothreitol (DTT), 10 mm magnesium acetate, and 20 mm potassium chloride] to remove accumulated tyrosine. The protein content was quantified with the DC-AC Protein Assay kit (Bio-Rad Laboratories, Inc., Hercules, CA). Aliquots of extracts were incubated for 2 h at 37 C with or without an ATP-generation system (1 mm ATP, 100 μg/ml creatine kinase, and 10 mm phosphocreatine); ubiquitin (250 μg/ml) was also added. The reaction was stopped with trichloroacetic acid, precipitated proteins were removed by centrifugation, and free tyrosine was measured fluorometrically in a Shimadzu FR-5000 spectrofluorometer (450 nm excitation/550 nm emission; Shimadzu Scientific Instruments, Columbia, MD) to calculate the rate of protein degradation (23).
To assess actin degradation, we measured the level of the characteristic 14-kDa actin fragment that is generated by the cleavage of actomyosin by caspase-3 (22). The pellet fraction of the muscle homogenate was solubilized in 2× Laemmli sample buffer and boiled for 5 min. Proteins were separated by electrophoresis on a 15% sodium dodecyl sulfate polyacrylamide gel. Actin cleavage was detected by Western blot probed with an antibody to the C-terminal 11 amino acids of actin (Sigma-Aldrich Corp., St. Louis, MO).
Antibodies
The levels of signaling proteins were detected in the soluble fraction of the cardiac tissue homogenates by standard Western blotting (24). The primary antibodies used included: anti-IRS-1 (1:1000 dilution; Upstate Biotechnology Inc., Lake Placid, NY); anti-pFOXO-Thr32 (1:500 dilution; Upstate Biotechnology); anti-Akt (1:1000 dilution; Cell Signaling Technologies, Beverly, MA); anti-phospho-Akt (ser473) (1:1000 dilution; Cell Signaling Technologies); and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:1000 dilution; Chemicon International, Inc., Temecula, CA). The IRS-1 tyrosine phosphorylation was measured in muscle extracts by immunoprecipitation with the IRS-1 antibody, followed by Western blot analysis using the PY-20 (antiphosphotyrosine) antibody (1: 2000 dilution, BD Biosciences, San Jose, CA).
Northern blots
Hearts were removed and immediately freeze clamped in liquid nitrogen. Total RNA from the heart was isolated using TriReagent (Molecular Research Center, Inc., Cincinnati, OH), and used in Northern blots for ubiquitin and GAPDH as described (24). Autoradiographic band intensities were quantified by densitometry (Bio-Rad Gel Doc system) using the corresponding GAPDH to correct for variations in RNA loading and transfer.
Proteasome activity
To measure the chymotrypsin-like peptidase activity of the proteasome, frozen pulverized cardiac muscle was homogenized in a proteasome isolation buffer [50 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 250 mm sucrose, 2 mm ATP, 1 mm DTT, 0.5 mm EDTA, and 0.025% digitonin]. After centrifugation at 20,000 × g for 15 min at 4 C, the supernatant was transferred and subjected to centrifugation for 2 h at 300,000 × g. Pellets containing the proteasome were resuspended in assay buffer [50 mm Tris-HCl (pH 7.5); 5 mm MgCl2, 40 mm KCl, 2 mm ATP, 1 mm DTT, and 0.5 mg/ml BSA]. Proteasome chymotrypsin-like activity was measured as the release of 7-amino-4-methylcoumarin (amc) from the fluorogenic peptide substrate LLVY-amc (N-Suc-Leu-Leu-Val-Tyr-amc; Peptides International) (25). Epoxomicin, a potent and specific inhibitor of the proteasome, was included with some samples to identify specific proteasomal proteolytic activity (20 μm; Peptides International). The incubation conditions for release of the fluorogen (amc) were 37 C for 1 h, and fluorescence was measured in a fluorometer (Shimadzu Scientific Instruments) using a 380-nm excitation wavelength and a 460-nm emission wavelength. Free amc (Chemicon International) was used to prepare a standard curve, and all readings were calculated relative to the fluorescence intensity of the standard curve. The difference between the substrate cleavage activity levels in the presence and absence of epoxomicin was used to calculate the contribution of proteasome.
Caspase-3 activity
Cardiac muscle was pulverized under liquid nitrogen and homogenized in lysis buffer [100 mm HEPES, 10% sucrose, 0.1% Nonidet P-40, and antiprotease cocktail (1 tablet/10 ml; Roche) (pH 7.4)]. The activity of caspase-3 was measured using a fluorogenic substrate (Ac-DEVD-amc), with or without a caspase-3 inhibitor (Ac-DEVD-CHO) in the caspase assay buffer (CaspACE Assay System; Promega Corp., Madison, WI). Incubations were performed at 37 C for 1 h, and fluorescence was measured in a fluorometer (Shimadzu Scientific Instruments) with a 380-nm excitation wavelength and a 460-nm emission wavelength. The difference between the substrate cleavage activity in the presence and absence of caspase-3 inhibitor was used to calculate caspase-3 activity (14).
Statistical analysis
Results are presented as mean ± sem. Densitometric data for blots are expressed as a percentage of the control mean density after normalization to loading controls. To identify significant differences between two groups, comparisons were made using the Student’s t test. Differences with P values less than 0.05 were considered significant.
Results
Protein degradation is increased in cardiac muscle of diabetic mice
We studied nine pairs of mice [diabetic (STZ-treated) mice and pair-fed controls]. In addition, insulin was administered to six STZ-treated mice (Table 1). Heart weights of diabetic, control, and STZ plus insulin mice were 84.5 ± 6, 101 ± 4, and 100 ± 3 mg, respectively (P < 0.01; diabetes vs. control or STZ plus insulin group). The body weights of the diabetic mice (18.2 ± 1.8 g) were lower than either control (22.5 ± 2.2 g; P < 0.05) or STZ plus insulin (21.7 ± 3.5 g; P < 0.05) mice. The heart weight to body weight ratios were not statistically different in any group.
Table 1.
Physiological parameters
| Control (n = 9) | Diabetes (n = 9)a | Diabetes + Ins (n = 6) | |
|---|---|---|---|
| Blood glucose (mg/dl) | 133.8 ± 2.7 | 423.3 ± 11.6b | 169.7 ± 8.6 |
| Plasma insulin (ng/ml) | 1.12 ± 0.03 | 0.65 ± 1.1b | 1.23 ± 0.08 |
| Body weight (g) | 22.5 ± 2.2 | 18.2 ± 1.8c | 21.7 ± 3.5 |
| Heart weight (mg) | 101 ± 4 | 84.5 ± 6b | 100 ± 3 |
All data are presented as the mean ± se. Ins, Insulin.
Ten days after STZ treatment.
P < 0.01 vs. control.
P < 0.05 vs. control.
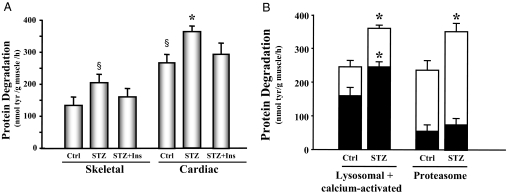
We measured total protein degradation rates in slices of the left ventricle and compared them with the rates measured in skeletal muscle. In control animals, protein degradation in cardiac muscle was 2-fold higher than in skeletal muscle (P < 0.01), indicating that cardiac muscle has a higher protein turnover rate compared with skeletal muscle (6). Compared with hearts of control mice, the rate of protein degradation was increased 31% in hearts of STZ-treated mice (Fig. 1A). Insulin replacement attenuated the increase in cardiac muscle protein degradation. When calcium-activated and lysosomal proteolysis was inhibited in cardiac tissue, there was a 30% decrease in the protein degradation in both control and diabetic mouse cardiac tissue. However, even with the calcium-activated and lysosomal proteolysis pathways inhibited, hearts from diabetic mice still exhibited increased protein degradation relative to the control mice. Thus, the increased protein degradation appears to result from stimulation of the ubiquitin proteasome system (Fig. 1B). To confirm this, epoxomicin, a specific proteasome inhibitor, was included in the incubation media. Under this condition, degradation due to the proteasome was 42% greater in the diabetic mouse heart than in the control mouse heart (Fig. 1B). To evaluate whether similar events occur in the right ventricle, we also measured protein degradation in this tissue. We found no significant difference in the rates of protein degradation between the right and left ventricles in either diabetic or control mice (data not shown). Thus, the accelerated protein degradation in cardiac muscle of diabetic mice is due to enhanced activity of a proteasome-dependent proteolytic system.
Figure 1.
Protein degradation is elevated in the cardiac muscle of STZ-treated diabetic mice. A, Protein degradation was measured as tyrosine (tyr) release from isolated skeletal and cardiac muscle from control (Ctrl), diabetic (STZ), or insulin-treated diabetic (STZ+Ins) mice. The protein degradation rates are depicted in the bar graph and represent the means ± se (n = 9 per group). §, P < 0.05 vs. skeletal muscle control; *, P < 0.05 vs. cardiac muscle control. B, Protein degradation was measured as tyrosine release from isolated cardiac muscle from control or diabetic (STZ) mice. The entire bar (white and black) represents total protein degradation. Black bars represent the remaining degradation activity after inhibition of the lysosomal plus calcium-activated proteolytic pathways (left) or proteasome proteolytic pathway (right). Therefore, the white portions of the bars only indicate the activities of the degradation pathways specified below the bars. Data are reported as the means ± se (n = 6 per group). *, P < 0.05 vs. control.
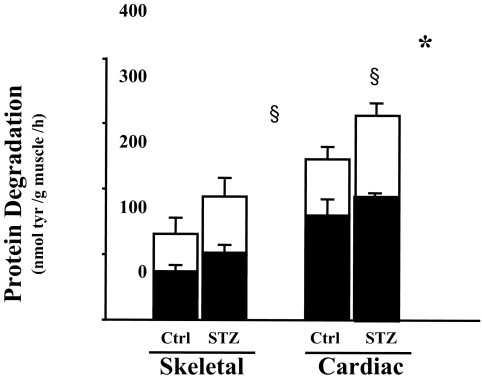
To evaluate this finding in more detail, we measured the degradation of soluble proteins in the heart muscle. Consistent with results from the measurement of total protein degradation in the cardiac muscle slice, degradation of soluble proteins was 1.8-fold higher in cardiac muscle when compared with skeletal muscle (Fig. 2). Evidence that the difference involved the ubiquitin-proteasome pathway was obtained by comparing protein degradation in cardiac extracts with or without addition of ATP. ATP is essential for the proteolytic activity of the ubiquitin proteasome system (21). In the absence of ATP, there was no significant difference between protein degradation rate in control vs. diabetic mice. In the presence of an ATP generation system, there was a greater increase in protein degradation in cardiac muscles from diabetic mice compared with those from control mice (Fig. 2). This indicated that the increased protein degradation in diabetic mouse heart is ATP dependent.
Figure 2.
Degradation of cardiac and skeletal muscle protein is ATP dependent. Protein degradation was measured by tyrosine (tyr) release from muscle extracts (soluble protein) of control (Ctrl) and diabetic (STZ) mice. The bar graphs show ATP-dependent protein degradation (white portion of bar) and ATP-independent degradation (black portion). Data are reported as the means ± se (n = 6 per group). §, P < 0.05 vs. skeletal muscle control; *, P < 0.05 vs. cardiac muscle control.
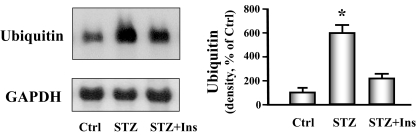
To provide additional evidence for activation of the ubiquitin proteasome system, we analyzed the level of ubiquitin mRNA in cardiac muscle of diabetic and control mice. There was a marked increase in ubiquitin mRNA in muscle of diabetic mice when compared with nondiabetic normal control mice. Insulin administration partially blocked the increase in ubiquitin mRNA (Fig. 3). We also found that there was a 2-fold increase in the chymotrypsin-like activity in the proteasome in heart muscle from diabetic mice compared with control mice. Again, insulin treatment partially blunted this response (Fig. 4). Together, these results suggest that diabetes activates the ubiquitin-proteasome system to degrade cardiac muscle protein.
Figure 3.
Ubiquitin mRNA is increased in cardiac muscle of diabetic mice. Ubiquitin mRNA was measured by Northern blot analysis of cardiac muscle of control (Ctrl), diabetic (STZ), or insulin-treated diabetic (STZ+Ins) mice. The bar graph of the band densities shows the quantity of ubiquitin mRNA expressed as the percentage of paired control after normalization with the density of the GAPDH band. Date are reported as the means ± se (n = 6 per group). *, P < 0.001 vs. control.
Figure 4.
Chymotrypsin-like activity of the proteasome is increased in cardiac muscle of diabetic mice. The chymotrypsin-like activity was measured using the fluorogenic substrate LLVY-amc, with or without the proteasome inhibitor epoxomicin (20 μm) in control (Ctrl), diabetic (STZ), or insulin-treated diabetic (STZ+Ins) mice. The difference between activity in the presence and absence of inhibitor was used to calculate proteasome activity. Data are reported as the means ± se (n = 6 per group). *, P < 0.05 vs. control.
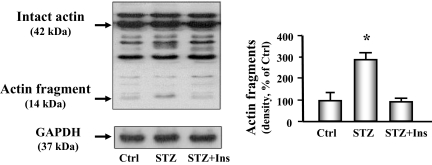
We next measured degradation of the nonsoluble, myofibrillar proteins by assessing actomyosin-actin cleavage (5,22). We found an increased level of the characteristic 14-kDa actin fragment that results from caspase-3-mediated proteolysis in the heart of diabetic mice (Fig. 5). Treating the diabetic mice with insulin significantly attenuated the increase in actin cleavage.
Figure 5.
Actin cleavage is increased in cardiac muscle of diabetic mice. Actin cleavage was measured in control (Ctrl), diabetic (STZ), or insulin-treated diabetic (STZ+Ins) mice. Western blot analysis (left) of heart muscle protein shows both intact actin (42 kDa) and the 14-kDa actin fragment cleavage product (indicated by arrows). GAPDH (bottom panel) was used as a loading control. The bar graph (right) shows the mean value of the 14-kDa fragment band intensity, expressed as a percentage of untreated control value after normalization by the density of the GAPDH bands. Insulin-deficient mice (middle bar) had substantially higher levels of the actin fragment compared with control mice or insulin-treated diabetic mice. Data are reported as the means ± se (n = 6 per group). *, P < 0.01 vs. control.
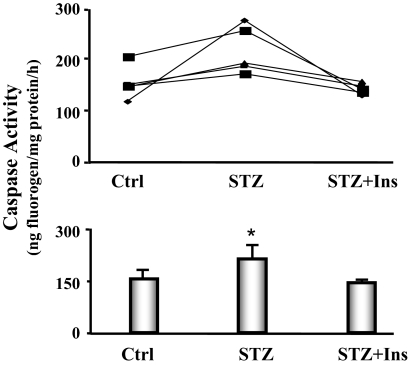
The activation of caspase-3 is an initial step that stimulates protein degradation by the ubiquitin proteasome system in skeletal muscle of diabetic mice. There was evidence for the increased proteolytic activity of caspase-3 because the level of the characteristic 14-kDa actin cleavage fragment was elevated in heart muscle of diabetic mice. To confirm this finding, we measured caspase-3 activity in cardiac muscles of pair-fed control, diabetic, and diabetic mice treated with insulin. The activity was increased 33% in diabetic heart vs. control heart, and insulin administration reversed this response (Fig. 6).
Figure 6.
Caspase-3 activity is increased in the cardiac muscle of diabetic mice. The activity of caspase-3 in cardiac muscle was measured using a fluorogenic substrate (Ac-DEVD-amc), in the presence or absence of the caspase-3 inhibitor (Ac-DEVD-CHO). Five feeding-matched triplets of mice were monitored. Each triplet contained one mouse with diabetes (STZ), one pair-fed, sham-injected control (Ctrl), and one pair-fed diabetic mouse receiving insulin treatment (STZ+Ins). Triplets are connected by a line (line graph). The bar graph shows the average caspase-3 activities. Data are reported as the means ± se (n = 5 per group). *, P < 0.05 vs. control.
We also evaluated the effect of insulinopenia on protein synthesis in isolated heart muscle slices. Protein synthesis was measured as the incorporation of l-[U-14C] phenylalanine (7). We found no significant differences in the rates of protein synthesis measured in heart slices from diabetic mice, control mice, and diabetic mice treated with insulin (data not shown).
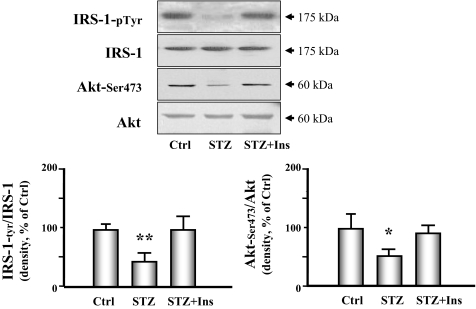
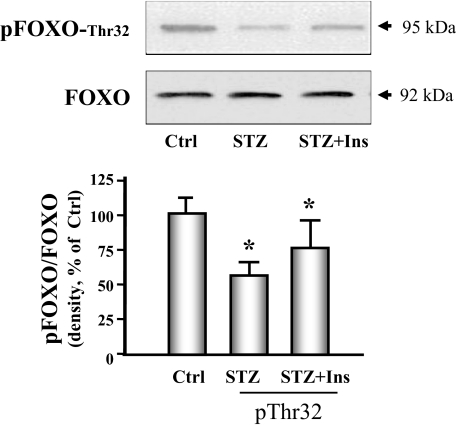
Finally, we examined whether signaling through the IRS-1/Akt pathway in the heart was altered by diabetes because signaling defects in this system have been linked to increased proteolysis in skeletal muscle (5). Diabetes caused a significant decrease in IRS-1 tyrosine phosphorylation and in the phosphorylation of Akt (Fig. 7). Insulin therapy improved both abnormalities. The forkhead O transcription factor (FOXO) is a substrate of Akt and activates the ubiquitin E3 ligase when it is dephosphorylated. We measured phosphorylation of FOXO and found that phosphorylation of threonine 32 was decreased in hearts of diabetic mice. Insulin partially restored FOXO phosphorylation to the level measured in control animals (Fig. 8).
Figure 7.
The IRS-1/Akt signaling pathway is down-regulated in cardiac muscle of STZ-treated diabetic mice. The components of the IRS-1/Akt signaling pathway were measured in cardiac muscle extracts in control (Ctrl), diabetic (STZ), or insulin-treated diabetic (STZ+Ins) mice. IRS-1, Akt, and p-Akt Ser473 were measured by Western analyses. IRS-1 phosphorylation was measured by immunoprecipitating with an antiphosphotyrosine antibody, followed by Western analysis for IRS-1. The bar graphs show the ratio of phosphorylated to total IRS-1 (left) or Akt (right) presented as a percent of control values. Data are reported as the means ± se (n = 6 per group). *, P < 0.05 or **, P < 0.01 vs. control.
Figure 8.
FOXO phosphorylation is decreased in cardiac muscle of diabetic mice. FOXO phosphorylation was measured by Western analyses using phosphospecific anti-pFOXO-Thr32 in control (Ctrl), diabetic (STZ), or insulin-treated diabetic (STZ+Ins) mice. The bar graph shows the ratio of phospho-FOXO to FOXO band densities reported as a percent of control values. Data are reported as the means ± se (n = 6 per group). *, P < 0.05 vs. control.
Discussion
Diabetes mellitus is an independent risk factor for the development of heart failure. This occurs from the observation that diabetes is associated with contractile dysfunction of the heart (26). Our results suggest that the decreased cardiac contractile activity in diabetes is due to the loss of cardiac muscle protein. Protein turnover is an important physiological process in the heart, considering that the entire complement of proteins in the heart is replaced on average every 30 d (27). Indeed, we found that diabetes caused a decrease in cardiac wet weight and that this was due to an increased degradation rate of cardiac muscle proteins. By measuring cardiac wet weight over dry weight, we were able to assess protein turnover with the same tissue sample.
Consistent with our results of decreased heart weights, others have reported that the heart atrophies in STZ-induced, type 1 diabetic animals. Singh et al. (28) and Liu et al. (29) reported a 25 and 20% decrease, respectively, in the heart weights in STZ mice compared with values from control mice. In our study the reduced heart weight in diabetes was due to an increased rate of degradation of total heart protein (Fig. 1A and Table 1). Interestingly, the rate of protein degradation was 1.8- to 2-fold higher in cardiac muscle than skeletal muscle, regardless of whether the animal was diabetic or a normal control (Figs. 1A and 2). Others have reported that individual cardiac muscle proteins turn over at rates 1.5- to 3-fold higher than skeletal muscle (29,30).
We evaluated the protein degradation process using several different techniques. We measured the release of tyrosine from isolated cardiac muscle slices and from soluble muscle proteins. Tyrosine release was used because muscle neither synthesizes nor degrades tyrosine, and its loss from the muscle reflects the net degradation of proteins (4). The release of tyrosine from incubated muscles has been used extensively with skeletal muscles isolated from small rodents, and the rate of protein degradation is linear with time (15). We studied small muscles to avoid potential limitations in the diffusion of oxygen, glucose, amino acids, etc. In addition, small muscles maintain levels of ATP and phosphocreatine, and show almost neutral protein balance during incubations for two or more hours (15). However, we recognize that results from incubation of cardiac muscle slices could differ because of the release of tyrosine from edges of the cardiac slice due to cut trauma. To minimize this possibility, the muscle slices were preincubated for 30 min before the experimental period was begun. This essentially washes out any tyrosine contributed by broken cells at the cutting interface. We confirmed our tissue slice data using an independent method of assessing protein degradation, namely by measuring the accumulation of the 14-kDa actin cleavage product from caspase-3 activity (22). An alternative approach would be to evaluate protein metabolism in isolated papillary muscles (31), but again, trauma could affect these measurements. In addition, it is possible that the rate of protein turnover in papillary muscles might be different from that in ventricular muscle. Surprisingly, we did not find significant differences in the measured rates of protein synthesis among diabetic hearts, control hearts, and the hearts of diabetic mice treated with insulin. This interesting result suggests that the large loss of cardiac mass in the diabetic animal is due predominantly to an increase in protein degradation.
By measuring protein degradation in the presence of different types of proteolytic inhibitors, we determined that the ubiquitin-proteasome system is the major proteolytic pathway degrading cardiac muscle in diabetic mice; inhibiting the proteasome blocked the protein degradation induced by insulin deficiency. Further evidence included the findings that: 1) ubiquitin mRNA was increased substantially in hearts of diabetic mice; and 2) the chymotrypsin-like activity in the proteasome was increased in hearts of diabetic mice. We also found that the lysosomal and calcium-activated proteolytic pathways comprised about 20–30% of the cardiac muscle degradation. This was the same in the diabetic heart as in the normal control heart. Consistent with our conclusions, others have reported that the ubiquitin-proteasome system is important for the degradation of key cardiac proteins such as myosin heavy chain (32) and cardiac troponin I (13). Liu et al. (29) have also reported that ubiquitin mRNA was increased in the hearts of diabetic rats.
The increase in protein degradation in cardiac muscle of diabetic animals is similar to the increases in skeletal muscle proteolysis in experimental animal and patients with sepsis (33), trauma (34), cancer (35), or uremia (36). Our data also show that activation of caspase-3 is associated with increased cardiac muscle protein degradation. Others have reported that caspase-3 is active in normal cardiac tissue and activated during ischemia-reoxygenation, suggesting a central role for this protease in regulating cardiac muscle protein metabolism (26). Our results indicate that the insulin signaling pathway, specifically the IRS-1/Akt pathway, is a key regulatory system in both cardiac and skeletal muscle.
In conclusion, the mechanisms by which diabetes mellitus confers an increased risk of heart diseases are multifactorial (2,3). Our results extend to the list of abnormalities by showing that diabetes activates proteolytic pathways in the heart that contribute to the loss of cardiac protein and, potentially, cardiac function. The latter is important because others have demonstrated that insulin deficiency depressed heart contractile function (28,37). This includes echocardiography experiments that reveal early abnormalities in diastolic function associated with diabetes in both animal models and humans (38). We conclude from this study that diabetes is characterized by increased cardiac muscle protein degradation. This degradation involves the breakdown of the myofibrillar protein actin as a result of activation of the ubiquitin-proteasome system. These responses could contribute to diabetic cardiomyopathy.
Footnotes
This study was supported in part by the University Research Committee of Emory University, a Norman S. Coplon Extramural Research Grant from Satellite Health, and National Institutes of Health (NIH) DK62796 (to X.H.W.), NIH DK062081 and AHA GIA0655280B (to J.D.K), and NIH HL70762 (to J.D.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online July 24, 2008
Abbreviations: amc, 7-Amino-4-methylcoumarin; DTT, dithiothreitol; E-64, trans-epoxysuccinyl-l-leucylamido-4-guanidino butane; FOXO, forkhead O transcription factor; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IRS-1, insulin receptor substrate-1; STZ, streptozotocin.
References
- Severson DL 2004 Diabetic cardiomyopathy: recent evidence from mouse models of type 1 and type 2 diabetes. Can J Physiol Pharmacol 82:813–823 [DOI] [PubMed] [Google Scholar]
- Stamler J, Vaccaro O, Neaton JD, Wentworth D 1993 Diabetes, other risk factors, and 12-yr cardiovascular mortality for men screened in the Multiple Risk Factor Intervention Trial. Diabetes Care 16:434–444 [DOI] [PubMed] [Google Scholar]
- Herrero P, Peterson LR, McGill JB, Matthew S, Lesniak D, Dence C, Gropler RJ 2006 Increased myocardial fatty acid metabolism in patients with type 1 diabetes mellitus. J Am Coll Cardiol 47:598–604 [DOI] [PubMed] [Google Scholar]
- Price SR, Bailey JL, Wang X, Jurkovitz C, England BK, Ding X, Phillips LS, Mitch WE 1996 Muscle wasting in insulinopenic rats results from activation of the ATP-dependent, ubiquitin-proteasome proteolytic pathway by a mechanism including gene transcription. J Clin Invest 98:1703–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu Z, Hu J, Du J, Mitch WE 2006 Insulin resistance accelerates muscle protein degradation: activation of the ubiquitin-proteasome pathway by defects in muscle cell signaling. Endocrinology 147:4160–4168 [DOI] [PubMed] [Google Scholar]
- Lee SW, Dai G, Hu Z, Wang X, Du J, Mitch WE 2004 Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J Am Soc Nephrol 15:1537–1545 [DOI] [PubMed] [Google Scholar]
- Hu Z, Lee IH, Wang X, Sheng H, Zhang L, Du J, Mitch WE 2007 PTEN expression contributes to the regulation of muscle protein degradation in diabetes. Diabetes 56:2449–2456 [DOI] [PubMed] [Google Scholar]
- Belke DD, Betuing S, Tuttle MJ, Graveleau C, Young ME, Pham M, Zhang D, Cooksey RC, McClain DA, Litwin SE, Taegtmeyer H, Severson D, Kahn CR, Abel ED 2002 Insulin signaling coordinately regulates cardiac size, metabolism, and contractile protein isoform expression. J Clin Invest 109:629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaim KE, Kochel PJ, Kira Y, Kobayashi K, Fossel ET, Jefferson LS, Morgan HE 1983 Insulin effects on protein synthesis are independent of glucose and energy metabolism. Am J Physiol 245:C133–C143 [DOI] [PubMed] [Google Scholar]
- Aikawa R, Nawano M, Gu Y, Katagiri H, Asano T, Zhu W, Nagai R, Komuro I 2000 Insulin prevents cardiomyocytes from oxidative stress-induced apoptosis through activation of PI3 kinase/Akt. Circulation 102:2873–2879 [DOI] [PubMed] [Google Scholar]
- Dickhout JG, Austin RC 2006 Proteasomal regulation of cardiac hypertrophy: is demolition necessary for building? Circulation 114:1796–1798 [DOI] [PubMed] [Google Scholar]
- Gomes AV, Zong C, Edmondson RD, Berhane BT, Wang GW, Le S, Young G, Zhang J, Vondriska TM, Whitelegge JP, Jones RC, Joshua IG, Thyparambil S, Pantaleon D, Qiao J, Loo J, Ping P 2005 The murine cardiac 26S proteasome: an organelle awaiting exploration. Ann NY Acad Sci 1047:197–207 [DOI] [PubMed] [Google Scholar]
- Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C 2004 Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc Natl Acad Sci USA 101:18135–18140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Hu J, Du J, Klein JD 2007 X-chromosome linked inhibitor of apoptosis protein inhibits muscle proteolysis in insulin-deficient mice. Gene Ther 14:711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracos VE, Goldberg AL 1986 Maintenance of normal length improves protein balance and energy status in isolated rat skeletal muscles. Am J Physiol 251(4 Pt 1):C588–C596 [DOI] [PubMed] [Google Scholar]
- Clark AS, Mitch WE 1983 Comparison of protein synthesis and degradation in incubated and perfused muscle. Biochem J 212:649–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitch WE, Bailey JL, Wang X, Jurkovitz C, Newby D, Price SR 1999 Evaluation of signals activating ubiquitin-proteasome proteolysis in a model of muscle wasting. Am J Physiol 276(5 Pt 1):C1132–C1138 [DOI] [PubMed] [Google Scholar]
- Bailey JL, Wang X, England BK, Price SR, Ding X, Mitch WE 1996 The acidosis of chronic renal failure activates muscle proteolysis in rats by augmenting transcription of genes encoding proteins of the ATP-dependent ubiquitin-proteasome pathway. J Clin Invest 97:1447–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM 1999 Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA 96:10403–10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida S, Towatari T, Kominami E, Katunuma N 1980 Inhibitions by E-64 derivatives of rat liver cathepsin B and cathepsin L in vitro and in vivo. J Biochem 88:1805–1811 [DOI] [PubMed] [Google Scholar]
- Fagan JM, Waxman L, Goldberg AL 1987 Skeletal muscle and liver contain a soluble ATP + ubiquitin-dependent proteolytic system. Biochem J 243:335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Wang X, Miereles C, Bailey JL, Debigare R, Zheng B, Price SR, Mitch WE 2004 Activation of caspase-3 is an initial step triggering accelerated muscle proteolysis in catabolic conditions. J Clin Invest 113:115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AS, Mitch WE 1983 Muscle protein turnover and glucose uptake in acutely uremic rats. Effects of insulin and the duration of renal insufficiency. J Clin Invest 72:836–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Du J, Hu Z, Walsh K, Wang XH 2007 Evidence for adipose-muscle cross talk: opposing regulation of muscle proteolysis by adiponectin and fatty acids. Endocrinology 148:5696–5705 [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL 2005 Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol 398:364–378 [DOI] [PubMed] [Google Scholar]
- Kannel WB, Hjortland M, Castelli WP 1974 Role of diabetes in congestive heart failure: the Framingham study. Am J Cardiol 34:29–34 [DOI] [PubMed] [Google Scholar]
- Razeghi P, Taegtmeyer H 2005 Cardiac remodeling: UPS lost in transit. Circ Res 97:964–966 [DOI] [PubMed] [Google Scholar]
- Singh J, Chonkar A, Bracken N, Adeghate E, Latt Z, Hussain M 2006 Effect of streptozotocin-induced type 1 diabetes mellitus on contraction, calcium transient, and cation contents in the isolated rat heart. Ann NY Acad Sci 1084:178–190 [DOI] [PubMed] [Google Scholar]
- Liu Z, Miers WR, Wei L, Barrett EJ 2000 The ubiquitin-proteasome proteolytic pathway in heart vs skeletal muscle: effects of acute diabetes. Biochem Biophys Res Commun 276:1255–1260 [DOI] [PubMed] [Google Scholar]
- McNulty PH, Young LH, Barrett EJ 1993 Response of rat heart and skeletal muscle protein in vivo to insulin and amino acid infusion. Am J Physiol 264(6 Pt 1):E958–E965 [DOI] [PubMed] [Google Scholar]
- Lesch M, Taegtmeyer H, Peterson MB, Vernick R 1976 Mechanism of the inhibition of myocardial protein synthesis during oxygen deprivation. Am J Physiol 230:120–126 [DOI] [PubMed] [Google Scholar]
- Eble DM, Spragia ML, Ferguson AG, Samarel AM 1999 Sarcomeric myosin heavy chain is degraded by the proteasome. Cell Tissue Res 296:541–548 [DOI] [PubMed] [Google Scholar]
- Tiao G, Hobler S, Wang JJ, Meyer TA, Luchette FA, Fischer JE, Hasselgren PO 1997 Sepsis is associated with increased mRNAs of the ubiquitin-proteasome proteolytic pathway in human skeletal muscle. J Clin Invest 99:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansoor O, Beaufrere B, Boirie Y, Ralliere C, Taillandier D, Aurousseau E, Schoeffler P, Arnal M, Attaix D 1996 Increased mRNA levels for components of the lysosomal, Ca2+-activated, and ATP-ubiquitin-dependent proteolytic pathways in skeletal muscle from head trauma patients. Proc Natl Acad Sci USA 93:2714–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A, Sun X, Fischer JE, Hasselgren PO 1999 The expression of genes in the ubiquitin-proteasome proteolytic pathway is increased in skeletal muscle from patients with cancer. Surgery 126:744–749 [PubMed] [Google Scholar]
- Bailey JL, Zheng B, Hu Z, Price SR, Mitch WE 2006 Chronic kidney disease causes defects in signaling through the insulin receptor substrate/phosphatidylinositol 3-kinase/Akt pathway: implications for muscle atrophy. J Am Soc Nephrol 17:1388–1394 [DOI] [PubMed] [Google Scholar]
- An D, Rodrigues B 2006 Role of changes in cardiac metabolism in development of diabetic cardiomyopathy. Am J Physiol Heart Circ Physiol 291:H1489–H1506 [DOI] [PubMed] [Google Scholar]
- Ahmed SS, Jaferi GA, Narang RM, Regan TJ 1975 Preclinical abnormality of left ventricular function in diabetes mellitus. Am Heart J 89:153–158 [DOI] [PubMed] [Google Scholar]