Abstract
The increasing availability of data related to genes, proteins and their modulation by small molecules, paralleled by the emergence of simulation tools in systems biology, has provided a vast amount of biological information. However, there is a critical need to develop cheminformatics tools that can integrate chemical knowledge with these biological databases, with the goal of creating systems chemical biology.
Hailed as a departure from the “reductionist approach”, where investigators dedicate their efforts to the study of a single gene or protein, systems biology is generally regarded as the “comprehensive approach”. Large networks describing the regulation of entire genomes, metabolic or signal transduction pathways are analyzed in their totality at different levels of biological organization1. Systems biology, which blends theory, computational modeling, and high-throughput experimentation2, has already led to advances in the understanding of cell signaling3, developmental biology4, cell physiology5, and metabolic networks6. However, despite these advances in biological insight, what is currently lacking from these approaches is any holistic understanding of how small molecules affect biological systems. The introduction of cheminformatics tools that can be seamlessly integrated with currently available bio- and cheminformatic databases and biological network simulations and software will be required to move toward a systematic understanding of the way small molecules impact biological systems – a field which we call “systems chemical biology.”
Although some systems biology approaches have been applied to targets in lead7 and drug discovery8,9 within the pharmaceutical industry, this type of general integration of chemistry and systems biology has not yet been seen in academics. However, a tremendous opportunity now exists because academic biomolecular screening efforts, particularly the Molecular Libraries Screening Centers Network (MLSCN) being funded by the NIH Roadmap via the Molecular Libraries Initiative (MLI)10 have created a wealth of systematic data describing the biological effects of small molecules. The new data that is being generated is particularly valuable because it extends information beyond the relatively limited number of biological and screening data available from industry to the entire array of macromolecules and macromolecular networks that are being experimentally evaluated in academic laboratories. Via the MLI, the effects of hundreds of thousands of small molecules on biological systems of varied complexity, ranging from screens with purified targets to simultaneous multi-target (multiplex) screens, phenotypic screens, and even whole organism assays, are being investigated. Central to the MLI is public access, and bioassay data from MLSCN screens are being deposited in PubChem, a freely accessible database. This unprecedented effort has created an opportunity to integrate chemical data with the vast biologically relevant data being deposited in publicly available databases (see Box 1). However, this plethora of small molecule data has yet to reach the fields of computational and systems biology.
Box 1: Resources for Systems Chemical Biology (*)
Genes
Entrez Gene: http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene
Proteins
SwissProt: http://expasy.org/sprot/
Structures of biological macromolecules
PDB: http://www.rcsb.org/pdb/home/home.do
Structral Genomics Consortium: http://www.sgc.utoronto.ca/
Pathways
KEGG: http://www.genome.jp/kegg/
MetaCyc: http://metacyc.org/
BioCarta: http://www.biocarta.com/genes/index.asp
Reactome: http://www.reactome.org/
Receptors
GPCRdb: http://www.gpcr.org/7tm/
NHRs: http://www.nursa.org/
Ion Channels: http://www.iuphar-db.org/iuphar-ic/index.html
Biochemical pathway reaction kinetic
SABIORK: http://sabio.villa-bosch.de/SABIORK/
BRENDA: http://www.brenda.uni-koeln.de/
Small Molecules
PubChem: http://pubchem.ncbi.nlm.nih.gov/
Network Simulators
Xyce: http://www.cs.sandia.gov/Xyce/
BioNetGen: http://cellsignaling.lanl.gov/bionetgen/index.shtml
Annotated Biological Model
Uncertainty analysis
DAKOTA: http://www.cs.sandia.gov/DAKOTA/
Cheminformatics Tools
Open Eye software: http://www.eyesopen.com/
(*) This non-exhaustive list illustrates sources of data used in this commentary
To make the most of this new data describing the modulation of genes and proteins via diverse libraries of small molecules, there is a critical unmet need to develop a “chemistry-smart” systems biology interface. Cheminformatics, a discipline that emerged only a decade ago11, focuses, in part, on methods for retrieving and analyzing information from chemical databases. As such, it is advances in cheminformatics that will be needed to develop the tools necessary to capitalize on the assay data in PubChem. Cheminformatics has already become an integral part of the drug discovery decision-making process12 and is currently the main resource for computer-based studies of chemistry-modulated biological systems13. Cheminformatics is also increasingly being applied to in silico profiling of small molecule bioactivities for arrays of targets11,14,15. However, the goal of bring chemical cognizance to systems biology will provide new challenges to the field. While, as Leroy Hood describes, the accumulation of genomics and proteomics databases can “transform how we think about biology and medicine16”, new chemical databases have the potential to transform the thinking of both chemists and biologists and to pave the way toward the chemical biology vision of rationally modulating all proteins via small molecules17. This Commentary pursues two goals: First, we discuss the potential of systems chemical biology to analyze and integrate large-scale chemical and biological data. Second, we illustrate the power of a hypothetical systems chemical biology interface, based on seamless integration of current cheminformatic predictive tools18,19 with the array of mathematical and bioinformatics models already available in computational and systems biology.
Thinking big about small molecule data
For the past two decades, innovative technologies enabling rapid synthesis and high throughput screening of large chemical libraries have been adopted by the industrial sector. This resulted in a massive increase of compounds routinely screened against new targets and pathways, and their associated data. Such technologies, by contrast, were rarely available to the academic research community and the resulting data was largely proprietary.. Within the past decade, though, high-throughput screening has become increasingly common in academics and the resulting data is typically published and often made available in relevant databases. Most significantly, since the creation of the MLSCN in 2005 through May 25, 2007, 256 different MLSCN bioassays, encompassing over 140,000 chemicals, have been deposited in PubChem. Within this data, 29,558 compounds have been categorized as “active” in at least one MLSCN bioassay, and 65,118 compounds have been annotated as “inactive” in at least 55 bioassays. In addition to this new public repository of more “traditional” small molecule screening data,, there are at least twenty databases that characterize different types of biological activities for small molecules20, with many of them capturing quite complex data. These more complex datasets include cases where multiple measures of biological endpoints are captured simultaneously for a compound library (chemical biology), where the endpoint is measured in the form of gene or protein expression profiles across an array of genes (chemical genomics), or where diverse compounds are tested against a complex assay where multiple mechanisms could lead to the measured response (phenotypic or in vivo screens – chemical genetics). This information can be currently found in many different databases (see Box 2), and although several initiatives are focused on development and standardization of the reporting of small molecule biological effects, for instance in PubChem, GPCR binding21, NCI, FDA, NIEHS, and EPA. analyzing the complex data contained in even one of these databases remains a significant challenge.
Box 2: Standard Biological Endpoint Data Sources (*)
Small Molecules
PubChem: http://pubchem.ncbi.nlm.nih.gov/
Metabolites
Drugs and Clinical Candidates
NLM’s Dailymed: http://dailymed.nlm.nih.gov/
Toxicology Data
NIEHS: http://ntp.niehs.nih.gov/ntpweb/
EPA DSS-Tox: http://www.epa.gov/ncct/dsstox/index.html
(*) Non-exhaustive list
Modeling both traditional high-throughput screening data and more complex datasets, especially in chemical genomics, is likely to require advances in data mining, clustering, and visualization 22 techniques, that are suited for large multi-dimensional datasets. The availability of this new data presents an important challenge to computer scientists, because through the development of new cheminformatic tools, there will be the possibility of understanding the complex relationship between chemical structures and their effects in living systems and the hope of being able not only to model, but accurately predict the effects of chemicals in biological assays.
Integrating currently available bioinformatic tools
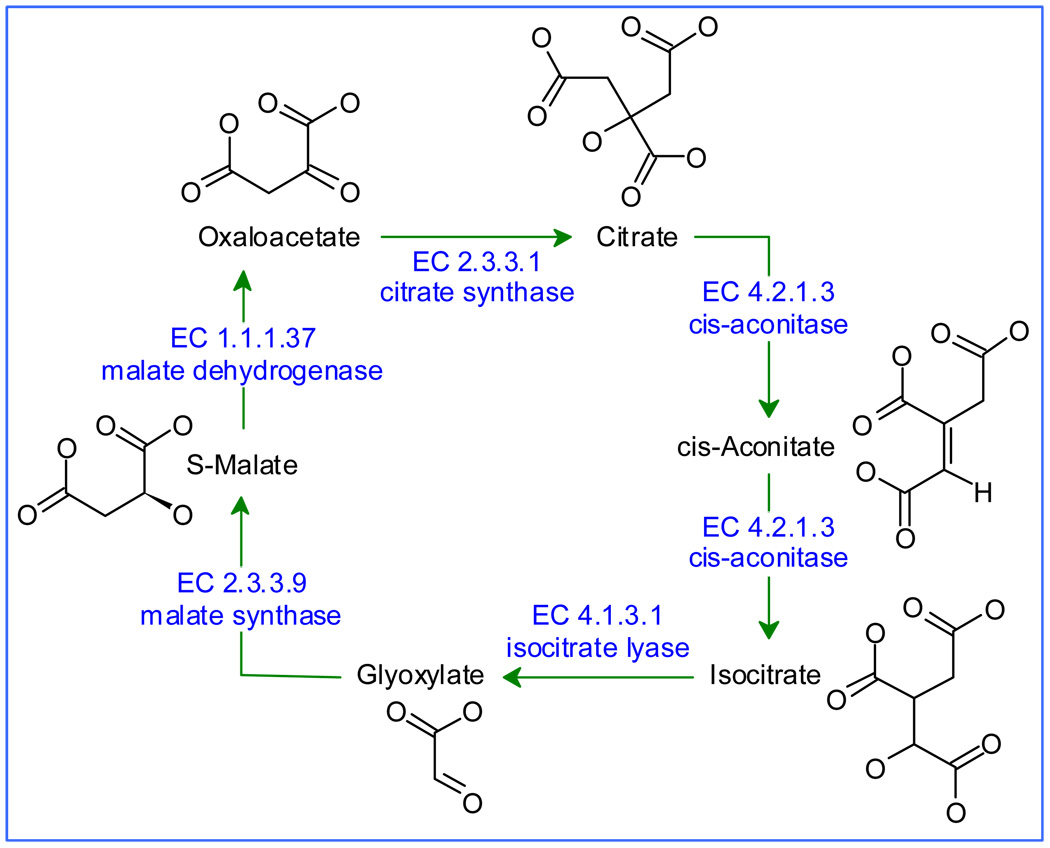
Relative to cheminformatic tools, bioinformatic tools are readily available and have been widely adopted by the biological community. For instance, KEGG, the Kyoto Encyclopedia of Genes and Genomes, aims to provide a “complete computer representation of the cell, the organism and the biosphere which will enable computational prediction of higher-level complexity of cellular processes and organism behaviors from genomic and molecular information” (http://www.genome.jp/kegg/). KEGG offers an effective summary of a vast array of metabolic and signal transduction data (Fig. 1).. In these databases, all objects are clickable, and lead to additional information related to reactions and pathways (all objects), and to gene and protein data, including links to other on-line databases like SwissProt and PubChem.
Figure 1.
A simplified representation of the glyoxylate pathway, extracted from KEGG. Object information, such as chemical structures, is one click away in KEGG, but have been added here for clarity.
However, there are also a lot of computational biology tools and biochemical data that are not currently integretated with KEGG, but would be useful for gaining an increased understanding of biological pathways. For instance, BioXyce is a biological network modeling tool 23 that can simulate the behavior of enzymes within pathways based on reasonable kinetic and microenvironment parameters 24. If tools like BioXyce could be integrated with KEGG, it would become possible to simulate metabolic pathways such as the one illustrated in Fig. 1. Such simulations could gain further accuracy by an interface that could incorporate reaction kinetics data such as KM, the Michaelis-Menten constant, and kcat, the turnover rate, which are both available from BRENDA25, and by optimizing the input parameters with DAKOTA, an uncertainty analysis tool (Box 1). However, even if all the state of the art computational biology and bioinformatic tools were fully integrated, the analysis would not take chemical knowledge into account. Currently, the introduction of a perturbing ligand on any systems biology network is simulated by a break in the electrical circuit, and does not take into account any specifics related to the small molecule per se, resulting in the loss of valuable information. This lack of chemistry awareness can only be addressed by developing new cheminformatics tools.
Cheminformatics meets bioinformatics
Why are cheminformatic tools for data mining not available to the same extent as bioinformatic tools? On the one hand, it is a surprising given all the commonalities between bioinformatics and cheminformatics. Cheminformatics and bioinformatics share common goals: In cheminformatics, one searches for similar compounds, while in bioinformatics one seeks homologous sequences; in cheminformatics one predicts activities and properties of small molecules, whereas in bioinformatics one predicts functions and properties of macromolecules; in cheminformatics one computes binding affinities between chemicals and proteins, and in bioinformatics one predicts the possibility of two biomolecules to interact. Bioinformatics techniques use Enzyme Commission (EC) numbers to predict the metabolites a given sequence can catalyze26–28, and structure- and sequence-based methods to locate homologous ligand binding sites29. Cheminformatics techniques such as virtual screening seek to identify novel compounds for a given target19, 30 , can classify metabolic 31 and organic 32 reactions, and predict the EC number given a metabolic reaction 33. Beyond common goals, cheminformatics and bioinformatics also share common computational techniques: clustering and machine learning based on regression, support vector machine (SVM), neural network, Bayesian networks, hidden Markov models, and decision trees, for example.
Yet, despite these common goals and techniques, a separation between the two cultures exists, as knowledge exchange and know-how transfers between these two fields have been quite limited. The reasons may relate to different scientific cultures (e.g., background in molecular biology vs. chemistry), different applications and funding sources, and different objects being studied, namely chemical compounds and biological sequences. Last but not least, high quality bioinformatics databases and resources have been developed and made available at no cost to the scientific community, whereas cheminformatics databases and resources have typically been made available for fee.
Towards systems chemical biology
Moving towards systems chemical biology, it will be essential to interface network simulators such as BioXyce, a “traditional” systems biology approach, with an interface that enables the scientist to apply bioinformatics and cheminformatics tools. Particularly important will be the development of cheminformatic approaches to take maximum advantage of the large-scale small molecule data that is now available. Ideally, these tools would permit chemical structures to be specifically described, visualized and modeled within system biology networks, would allow comparisons of the bioactivity properties of chemicals across all the available databases, and would enable relevant predictions of the effects of small molecules on biologic processes. Such chemical tools should be able to recognize structures from sketches, from linear notations such as SMILES34, and from common name input. Cheminformatics tools could then become the engine for predicting the influence of putative perturbing ligands on systems biology networks.
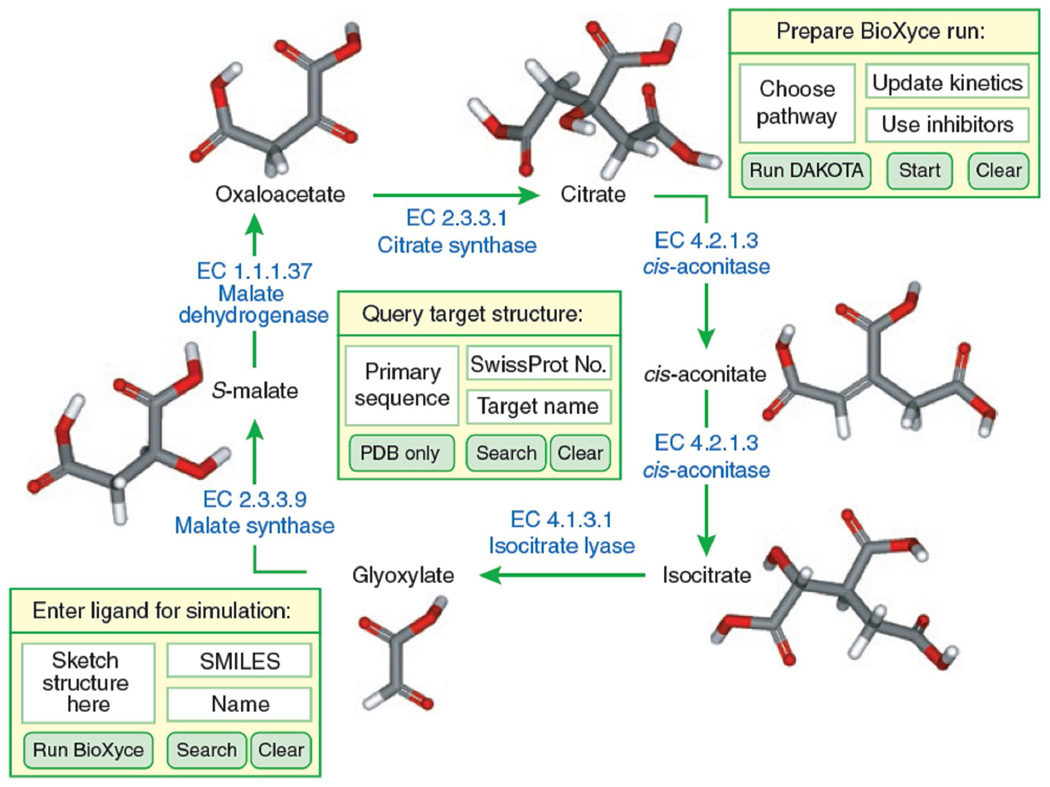
An example of a potential systems chemical biology approach based on the glyoxylate pathway is illustrated in Fig. 2. In an integrated interface, biochemical networks, target function and the effects of small molecules could all be simulated. In this scenario, one could begin by developing a BioXyce network that captures appropriate reaction and metabolite data to simulate the glyoxylate pathway, in both the forward and reverse direction. Using BRENDA, this generic pathway can be further adapted to compare the flux of the pathway in specific organisms, e.g., Mycobacterium tuberculosis vs. human. These pathways could then be optimized with DAKOTA under uncertainty conditions. After generating a realistic model of the network, the interactions of small molecules could be predicted using a combination of bioinformatic and cheminformatic tools.. As an example, in cases where a protein structures were not known, bioinformatic tools could be used to generate homology models in preparation for virtual screening.. To query the impact of any novel small molecule on the enzyme cycle, a switch to cheminformatics would permit ligand-based or structure-based virtual screening. By feeding predicted Ki values back into BioXyce at user-defined concentrations, it becomes possible to simulate the influence of small molecules across biological pathways.
Figure 2.
Conceptual representation of the systems chemical biology approach, applied to the glyoxylate pathway. Input boxes illustrate various levels of simulation.
The development of an integrated systems chemical biology interface could dramatically alter our way of thinking about complex biological networks, and unlock the true potential of in silico chemical biology studies of cellular and organism functions. By gaining access to the “known” as well as the “predictive” aspects of small molecule-biological nework interactions, scientists could be guided to understand, for example, the potential therapeutic impact of a small-molecule blockade of a critical step in a pathway. This may, ultimately, allow an understanding of why some, but not all, proteins within a pathway make good drug targets, and encourage an early focus on those targets that are the most likely to be clinically useful. We are concerned about the recent move by NIH to cancel its only funding opportunity for cheminformatics (http://grants.nih.gov/grants/guide/notice-files/NOT-RM-07-010.html), an X02 that was designed to foster cheminformatic tool developments to analyze PubChem and related public data (http://grants.nih.gov/grants/guide/pa-files/PAR-07-353.html). Funding centers that create small molecules and generate bioassay data on a massively unprecedented scale is commendable and within the MLI vision. Not funding the development of tools to analyze and mine the associated data defeats its purpose.
Acknowledgments
This work was supported in parts by the National Institutes of Health grant U54 MH074425-01 (TIO), by the National Institutes of Health planning grant P20-HG003898 (AT). Sandia is operated by Sandia Corporation, a Lockheed Martin Company, for the United States Department of Energy under Contract DE-AC04-94AL85000 (JLF, MDR). The authors express their gratitude to Mr. Tharun K. Allu and Dr. Dan C. Fara (UNM) for the PubChem bioassay data analysis.
References
- 1.Voit E, Neves AR, Santos H. Proc. Natl. Acad. Sci. USA. 2006;103:9452–9457. doi: 10.1073/pnas.0603337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kell DB. FEBS J. 2006;273:873–894. doi: 10.1111/j.1742-4658.2006.05136.x. [DOI] [PubMed] [Google Scholar]
- 3.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. BioSystems. 2006;83:136–151. doi: 10.1016/j.biosystems.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 4.Ochi H, Westerfield M. Develop. Growth Differ. 2007;49:1–11. doi: 10.1111/j.1440-169X.2007.00905.x. [DOI] [PubMed] [Google Scholar]
- 5.Brandman O, Ferrell JE, Li R, Meyer T. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 7.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Nat. Biotechnol. 2007;25:197–206. doi: 10.1038/nbt1284. [DOI] [PubMed] [Google Scholar]
- 8.Morphy R, Rankovic Z. Drug Discov. Today. 2007;12:156–160. doi: 10.1016/j.drudis.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Loging W, Harland L, Williams-Jones B. Nat. Rev. Drug Discov. 2007;6:220–230. doi: 10.1038/nrd2265. [DOI] [PubMed] [Google Scholar]
- 10.Austin CP, Brady LS, Insel TR, Collins FS. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 11.Brown F. Curr. Opin. Drug Discov. Devel. 2005;8:298–302. [PubMed] [Google Scholar]
- 12.Olsson T, Oprea TI. Curr. Opin. Drug Discov. Devel. 2001;4:308–313. [PubMed] [Google Scholar]
- 13.Willett P. Aslib Proc. 2007 in press. [Google Scholar]
- 14.Paolini GV, Shapland RHB, van Hoorn WP, Mason JS, Hopkins AL. Nat. Biotechnol. 2006;24:805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 15.Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Proc. Natl. Acad. Sci. USA. 2005;102:261–266. doi: 10.1073/pnas.0407790101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hood L. 2001 Interview available at http://www.technologyreview.com/Biotech/12575/ [Google Scholar]
- 17.Schreiber SL. Nat. Chem. Biol. 2005;1:64–66. doi: 10.1038/nchembio0705-64. [DOI] [PubMed] [Google Scholar]
- 18.Tropsha A. Predictive QSAR Modeling. In: Triggle D, Taylor J, editors. Comprehensive Medicinal Chemistry II. vol 4. Elsevier; 2006. pp. 113–126. [Google Scholar]
- 19.Fara DC, Oprea TI, Prossnitz ER, Bologa CG, Edwards BS, Sklar LA. Drug Discov Today Technol. 2006;3:377–385. doi: 10.1016/j.ddtec.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oprea TI, Tropsha A. Drug Discov Today Technol. 2006;3:357–365. doi: 10.1016/j.ddtec.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth BL, Kroeze WK. Curr. Pharm. Des. 2006;12:1785–1795. doi: 10.2174/138161206776873680. [DOI] [PubMed] [Google Scholar]
- 22.Burchiel SW, Thompson TA, Lauer FT, Oprea TI. Toxicol. Applied Pharmacol. 2007;221:203–214. doi: 10.1016/j.taap.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin S, Davidson G, May E, Faulon J-L, Werner-Washburne M. Proc. IEEE Comput. Syst. Bioinform. Conf; 2004. pp. 566–569. [Google Scholar]
- 24.Schiek RL, May EE. Proc. IEEE Comput. Syst. Bioinform. Conf; 2003. pp. 620–622. [Google Scholar]
- 25.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borgwardt KM, Ong CS, Schoenauer S, Vishwanathan SVN, Smola AJ, Kriegel HP. Bioinformatics. 2005;21 (Suppl. 1):i47–i56. doi: 10.1093/bioinformatics/bti1007. [DOI] [PubMed] [Google Scholar]
- 27.Cai CZ, Han LY, Ji ZL, Chen X, Chen YZ. Nucleic Acids Res. 2003;31:3692–3697. doi: 10.1093/nar/gkg600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunik V, Solan Z, Edelman S, Ruppin E, Horn D. Proc. IEEE Comput. Syst. Bioinform. Conf; 2005. pp. 80–85. [DOI] [PubMed] [Google Scholar]
- 29.Kalinina OV, Novichkov PS, Mironov AA, Gelfand MS, Rakhmaninova AB. Nucleic Acids Res. 2004;32:W424–W428. doi: 10.1093/nar/gkh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oprea TI, Matter H. Curr. Opin. Chem. Biol. 2004;8:349–358. doi: 10.1016/j.cbpa.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Latino DARS, Aires-de-Sousa J. Angew. Chem. Int. Ed. 2006;45:2066–2069. doi: 10.1002/anie.200503833. [DOI] [PubMed] [Google Scholar]
- 32.Gasteiger J. J. Comput.-Aided Mol. Design. 2007;21:33–52. doi: 10.1007/s10822-006-9097-4. [DOI] [PubMed] [Google Scholar]
- 33.Kotera M, Okuno Y, Hattori M, Goto S, Kanehisa M. J. Am. Chem. Soc. 2004;126:16487–16498. doi: 10.1021/ja0466457. [DOI] [PubMed] [Google Scholar]
- 34.Weininger D. J. Chem. Inf. Comput. Sci. 1988;28:31–36. doi: 10.1021/ci950169+. [DOI] [PubMed] [Google Scholar]