Abstract
Eukaryotic cells store neutral lipids in cytoplasmic lipid droplets1,2 enclosed in a monolayer of phospholipids and associated proteins3,4. These dynamic organelles5 serve as the principal reservoirs for storing cellular energy and for the building blocks for membrane lipids. Excessive lipid accumulation in cells is a central feature of obesity, diabetes and atherosclerosis, yet remarkably little is known about lipid-droplet cell biology. Here we show, by means of a genome-wide RNA interference (RNAi) screen in Drosophila S2 cells that about 1.5% of all genes function in lipid-droplet formation and regulation. The phenotypes of the gene knockdowns sorted into five distinct phenotypic classes. Genes encoding enzymes of phospholipid biosynthesis proved to be determinants of lipid-droplet size and number, suggesting that the phospholipid composition of the monolayer profoundly affects droplet morphology and lipid utilization. A subset of the Arf1–COPI vesicular transport proteins also regulated droplet morphology and lipid utilization, thereby identifying a previously unrecognized function for this machinery. These phenotypes are conserved in mammalian cells, suggesting that insights from these studies are likely to be central to our understanding of human diseases involving excessive lipid storage.
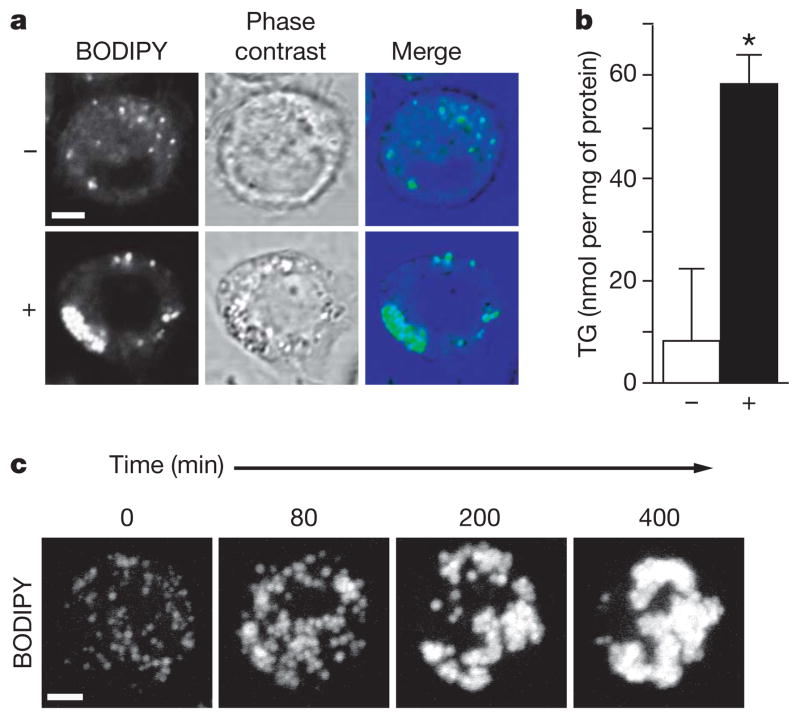
We studied lipid-droplet formation in Drosophila Schneider 2 (S2) cells, a proven system for functional genomic studies with efficient gene inactivation by RNAi6. We induced lipid-droplet formation by incubation with 1 mM oleate for 24 h. Staining with 4,4-difluoro-1,3,5,7,8-pentamethyl-4-bora-3a,4a-diaza-s-indacene (BODIPY493/503) showed that droplet size, number and overall volume were increased (Fig. 1a); cellular triacylglycerol content increased sevenfold (Fig. 1b). We confirmed that BODIPY-stained fluorescent signals corresponded to lipid droplets with a red fluorescent protein mCherry7 fused with lipid storage droplet-1 (LSD1), which localizes exclusively to the surface of lipid droplets3 (not shown).
Figure 1. Oleate increases the formation of lipid droplets in Drosophila S2 cells.
a, S2 cells incubated for 24 h without (upper) or with (lower) 1 mM oleate. Staining with BODIPY, phase-contrast image, and overlay are shown. b, Oleate-loaded cells have an increased triacylglycerol (TG) content. Cells were incubated as in a and their TG contents were measured. Results are means and s.d. for three experiments; P <0.05. c, Lipid-droplet formation occurs in steps. Cells treated as in a were stained with BODIPY. Single cells were followed by four-dimensional confocal time-lapse microscopy. Representative maximum projections of three-dimensional stacks at the indicated times are shown. Scale bars, 3 μm.
Imaging this process by time-lapse microscopy of BODIPY-labelled cells after oleate addition (Supplementary Movie 1) showed that droplet formation occurred in steps (Fig. 1c). First, increased numbers of small droplets formed in dispersed locations throughout the cell. Next, droplets increased in size and finally aggregated into one or several large clusters, resembling grapes. Electron microscopy confirmed the tight clustering of the droplets, which were often near the nucleus (Supplementary Fig. 3).
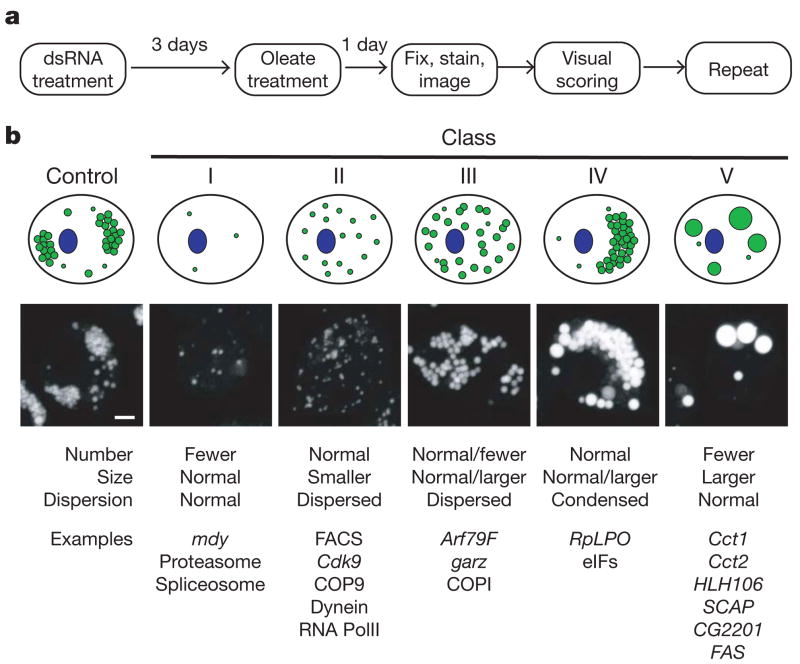
To unravel the molecular mechanisms governing this progression of changes during lipid-droplet formation, we performed a genome-wide RNAi screen in S2 cells (Fig. 2a). Images were acquired and examined by two independent observers, who scored them for alterations in droplet number, size and dispersion. The same data were analysed computationally (Supplementary Methods). From visual screening, both observers identified 847 candidate genes with altered lipid-droplet morphology. To verify these genes and to minimize the misidentification of genes from off-target effects of RNAi treatments8,9, RNAi experiments for these genes were repeated with a second, distinct set of double-stranded (ds)RNAs10. Visual analyses identified 132 genes whose knockdown consistently and repeatedly altered lipid-droplet morphology (Supplementary Table 1) and an additional 48 genes for which knockdown phenotypes were scored in two of three rounds (Supplementary Table 2). Computational analysis confirmed 86 of these 180 genes and added 47 genes with altered lipid-droplet morphology (Supplementary Table 3). Thus, we identified 227 genes (about 1.5% of the genome) that we conclude, with high confidence, affect lipid-droplet morphology. However, the high stringency of our criteria may have caused us to miss some genes involved in lipid-droplet morphology.
Figure 2. Genome-wide screen identified genes regulating the formation of lipid droplets.
a, Outline for strategy to screen for genes involved in lipid-droplet biogenesis. See the text for details. b, Genes involved in the screen for lipid-droplet biogenesis fall into distinct phenotypic classes. The 132 most striking phenotypes were classified according to lipid-droplet number, size and dispersion. From this classification, five major classes emerged; a graphic representation (top), an example image (middle) and some gene examples (bottom) are shown. Scale bar, 3 μm.
The 132 genes with striking phenotypes were categorized into five distinct phenotypic classes (Fig. 2b and Supplementary Table 1), which were validated for selected knockdowns by electron microscopy (Supplementary Fig. 3). Class I genes showed reduced numbers of droplets and included midway (encoding a diacylglycerol acyl-transferase), subunits of the proteasome and the spliceosome, and several uncharacterized open reading frames. Class II genes gave smaller, more dispersed, droplets and included subunits of the COP9–signallosome complex, dynein, and RNA polymerase II subunits. Class III genes showed more dispersed droplets of slightly larger size and were members of the Arf1–COPI vesicular transport machinery. Class IV genes yielded highly condensed clusters of droplets and included members of the translational machinery. Class V genes contained one or a few very large droplets and included an orthologue of sterol regulatory element binding-protein (SREBP), a master transcriptional regulator of lipid metabolism, and SREBP cleavage activating protein (SCAP)11. In Drosophila, the SREBP pathway is sensitive to and regulates phospholipid biosynthesis12. This class also included Cct1 and Cct2, which encode isoforms of phosphocholine cytidylyltransferase, the enzyme that catalyses the rate-limiting step in phosphatidylcholine synthesis13, and CG2201, which is predicted to have choline kinase activity that phosphorylates and activates choline14. Thus, most class V genes were linked directly or indirectly to phospholipid biosynthesis.
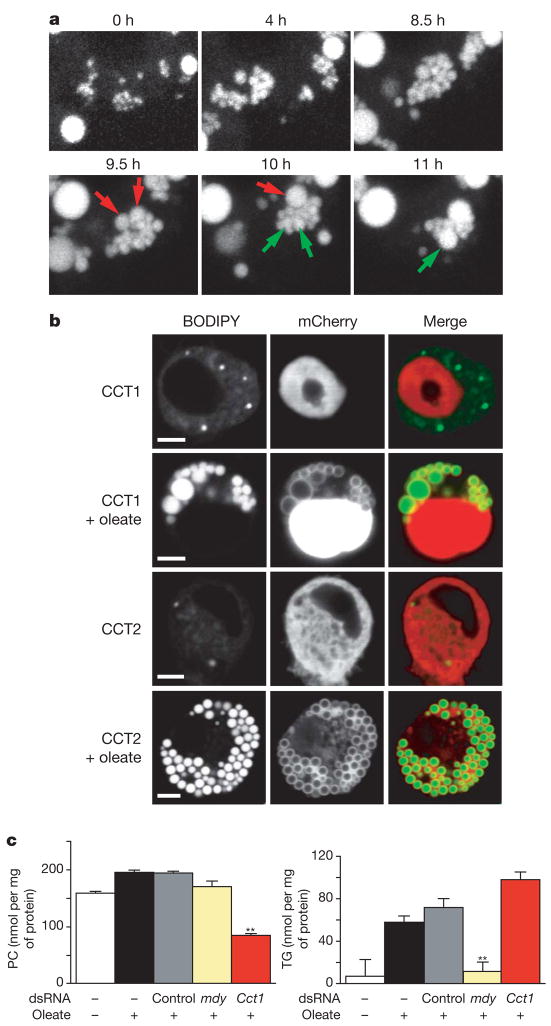
To further explore how phospholipid metabolism regulates lipid-droplet formation, we characterized the Cct1 and Cct2 knockdowns. Larger droplets in Cct knockdowns could arise from a failure to form new droplets, forcing newly synthesized neutral lipids into a few large droplets, or from the fusion of independently formed droplets. To distinguish between these possibilities, we observed the dynamics of lipid-droplet formation by time-lapse microscopy (Supplementary Movie 2) and found evidence that the droplets fuse (Fig. 3a and Supplementary Fig. 2).
Figure 3. Phosphatidylcholine content regulates the size and abundance of lipid droplets.
a, Lipid-droplet formation induced by oleate in Cct1 knockdown cells (Fig. 1a). Single cells were followed by time-lapse confocal microscopy. Representative projections revealed that droplets first proliferate normally (upper) and then fuse (lower). Examples of fusion are indicated (red and green arrows). b, CCT enzymes localize to the surface of droplets after induction with oleate. Cct1 and Cct2 were transiently expressed in S2 cells as amino-terminal mCherry-tagged fusion proteins and were stained and imaged before or after induction. BODIPY staining, mCherry fluorescence, and merge are shown. Scale bar, 3 μm. c, Cct1 knockdown cells have less phosphatidylcholine (PC) and more triacylglycerol (TG). S2 cells were treated with dsRNAs as indicated, loaded with oleate (as in Fig. 1a) and lysed. PC (upper) and TG (lower) levels in the extract were measured. Results are means and s.d. for three independent experiments. **, P <0.01 versus control RNAi.
We then examined where CCT proteins act. In untreated cells, mCherry-tagged Cct1 localized exclusively to the nucleus (Fig. 3b), similar to mammalian cytidylyltransferase-α (CT-α)13,15. After treatment with oleate, a significant portion of Cct1 localized to the lipid-droplet surface. By contrast, similarly tagged Cct2 localized to the cytoplasm but was also concentrated on droplet surfaces after treatment with oleate. This marked translocation of CCT enzymes to the droplet surface may serve to provide adequate phosphatidylcholine to the phospholipid monolayers of growing lipid droplets. If so, the ratio of surface phospholipids to core neutral lipids may regulate lipid-droplet morphology: when phospholipids are limiting (as in Cct1 or Cct2 knockdowns), fusion is induced to decrease the surface-to-volume ratio of droplets. In fact, the content of phosphatidylcholine in cells with Cct1 knockdown was decreased by about 60% (Fig. 3c, upper panel), and the triacylglycerol content was increased by about 40% (Fig. 3c, lower panel). The increase in triacylglycerol may reflect compensatory channelling of diacylglycerol into neutral lipids. A decreased phosphatidylcholine content would increase the relative amount of phosphatidylethanolamine in the droplet monolayer (as observed in flies lacking Cct1 (ref. 16)), which itself may directly promote droplet fusion17. Our results suggest a model in which phosphatidylcholine availability is a crucial regulator of lipid-droplet size and number.
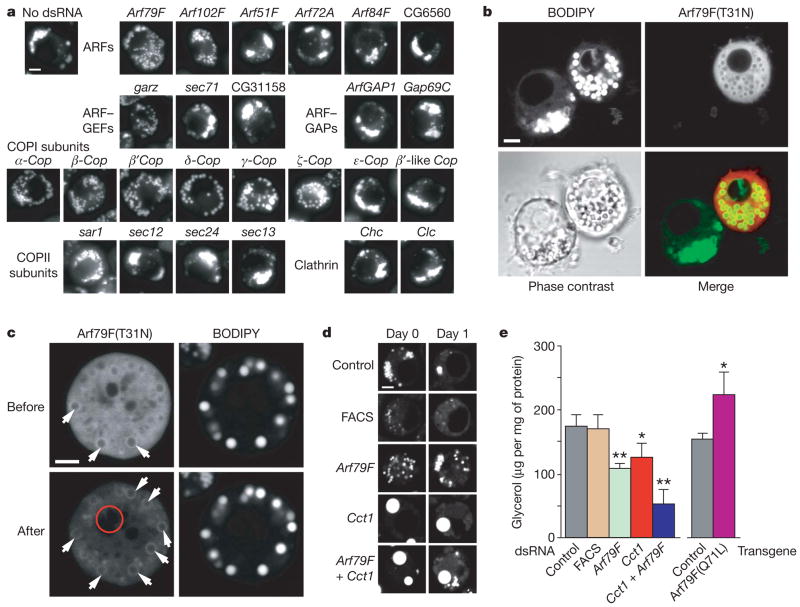
We also investigated class III genes, whose knockdowns showed slightly larger and more dispersed droplets. All class III genes were members of the Arf1–COPI machinery, including Arf79F, encoding an Arf1 family member, a gene encoding guanine nucleoside exchange factor (GEF), garz, and genes encoding components of the COPI coat. Similar effects were obtained by incubating cells with brefeldin A, a specific inhibitor of Arf1 exchange factors (Supplementary Fig. 5), and by expressing a dominant-negative version of Arf79F, encoding the T31N mutant, analogously to dominant-negative mutants for Ras or Ran18 (Fig. 4b). To test the specificity of this phenotype, we separately repeated RNAi knockdowns with dsRNAs for Drosophila genes encoding six ARF proteins, three GEFs, two GTPase-activating proteins, and all COPI subunits. We also tested other coat proteins, such as clathrin subunits and components of the COPII coat (Fig. 4a). Only Arf79f, garz and six of eight members of the COPI coat (α-, β-, β′-, δ-, γ- and ζ-Cop) exhibited the class III phenotype, indicating that our screen identifies a highly specific subset of vesicular transport components. Arf102F knockdown gave a partial phenotype. This function of the Arf1–COPI machinery in lipid-droplet formation seems to have been evolutionarily conserved; similar phenotypes were found in yeast and human cells (Supplementary Fig. 4).
Figure 4. ArfI–COPI complex members function in the formation of lipid droplets.
a, Knockdowns of Arf79F, the GEF gene gartenzwerg (garz) and specific subunits of the COPI coat affected lipid-droplet formation similarly. Representative images are shown. For descriptions of controls see Supplementary Information. GAP, GTPase-activating protein. b, Arf79F(T31N) localizes to the droplet surface and causes a similar phenotype to Arf79F knockdown. Arf79F(T31N) expressed as a carboxy-terminal mCherry-tagged fusion protein in S2 cells was observed by confocal microscopy after loading with oleate and staining with BODIPY. A representative confocal midsection is shown for BODIPY (top left), mCherry fluorescence (top right) and a merge (bottom right). c, Arf79F(T31N)–mCherry localizes to the droplet surface. The photobleached region (6 min) is indicated by a red circle. Arrows indicate the association of Arf79F(T31N) with the surface of droplets. d, Arf79F, Cct1 and double knockdowns lead to decreased lipolysis. S2 cells were treated with dsRNAs for three days as indicated, loaded with 1 mM oleate for one day, and imaged by confocal microscopy after staining with BODIPY (day 0, left panels). Representative confocal midsections are shown. Oleate was removed from the medium and the cells were starved for one day in serum-free medium to induce lipolysis (day 1, right). Scale bars, 3 μm. FACS, gene encoding a long-chain-fatty-acid-CoA ligase (CG8732). e, Arf79F, Cct1 and double knockdowns lead to decreased glycerol release to the medium. A transgene encoding Arf79F(Q71L) leads to increased release of glycerol. Experiments were as in d, and the glycerol released was measured. Results are means and s.d. for three independent experiments. *, P <0.05, ** P <0.01 versus control RNAi (left) and versus control transgene (right).
We next sought to determine whether Arf79F acts directly on lipid droplets. ARF proteins exchange rapidly between active (GTP-bound) and inactive (GDP-bound) forms, making it difficult to localize only the active form. However, Arf79F(T31N) binds its exchange factor tightly, and the distribution of the exchange factor is predicted to reflect the localization of active ARF protein. Expressed Arf79F(T31N) appeared diffusely in the cytosol but was enriched at the droplet surface (Fig. 4c). Thus, Arf79F may act at the lipid-droplet surface where, as for other ARF proteins, it interacts with its GEF (presumably encoded by garz) and recruits COPI components. A recent in vitro study showed that Arf1 and several subunits of the COPI complex are recruited from the cytosol to purified lipid droplets in the presence of GTP-γS (ref. 4). Although the recruitment of Arf1 to lipid droplets was reported to activate phospholipase D (ref. 19), we found no effect of phospholipase D knockdown on lipid-droplet formation in Drosophila cells (not shown).
The well-established functions of class III genes in Arf1–COPI-mediated vesicular transport20,21 implicate this machinery in a similar budding mechanism at the surface of lipid droplets, possibly to promote the budding-off of droplets during lipid mobilization. Lipolysis is associated with the break-up of larger droplets into smaller ones, presumably to provide more surface area for lipases22. We examined the effect of the Arf79F knockdown on lipolysis by inducing lipid-droplet formation (Fig. 4d) and then inducing lipid mobilization by incubation with serum-free medium lacking oleate. After 24 h, control cells had few droplets. By contrast, many droplets remained when Arf79F was inactivated. In addition, much less glycerol, a product of lipolysis, was released by cells lacking the Arf1–COPI machinery (Fig. 4e). Supporting a function of the Arf1–COPI machinery in lipolysis, more glycerol was released by cells expressing a dominant-active form of Arf79F encoding Arf79F(Q71L). These data indicate that the Arf1–COPI machinery is required for efficient lipolysis. Our data agree with a report showing that lipolysis in murine adipocytes is accompanied by a brefeldin A-sensitive process that is required for the mobilization of cholesterol from storage pools in droplets23.
Increased droplet surface area during lipolysis would require more phospholipids in the surrounding monolayer. Because Cct1 knockdown limits phosphatidylcholine amounts, we tested its effect on lipolysis. As predicted, Cct1 knockdown markedly decreased the efficiency of lipolysis, as seen by lipid-droplet staining (Fig. 4d) and glycerol release (Fig. 4e). The effects of knockdowns of Arf79F and Cct1 on lipolysis were additive, suggesting that these genes function independently.
Arf1–COPI complexes mediate retrograde vesicular trafficking of membranes and proteins from the Golgi apparatus to the endoplasmic reticulum21 and are also involved in vesicular transport processes from the trans-Golgi network and endosomes21. Notably, the members of the Arf1–COPI complex we identified were recently found in a Drosophila screen for genes involved in protein secretion and Golgi organization24. Although the primary defect in class III knockdowns is as yet unknown, the phenotype on lipid-droplet formation is not likely to be an indirect consequence of inhibition of protein secretion. The effects are highly specific and are not observed with knockdowns of other proteins mediating secretory transport (endoplasmic reticulum translocation, COPII and clathrin). In addition, Arf79F is recruited to the lipid-droplet surface, as shown by us and others4, where it is presumably activated by the loading of GTP on its exchange factor.
Our study provides an initial systematic examination of the Drosophila genome to identify genes involved in lipid-droplet formation and utilization. Many genes that we identified sort to distinct classes of morphological changes, with each class containing functionally related proteins. These classes potentially link diverse processes, such as protein synthesis and degradation, the cell cycle, and organelle movement, with lipid-droplet biology. The variety of genes identified lends support to the emerging view of lipid droplets as dynamic organelles that are functionally connected to a variety of organelles and cellular processes, including the replication of intra-cellular pathogens such as Chlamydia trachomatis and hepatitis C25,26. Many components of these processes are likely to be highly conserved across species. Our studies in S2 cells may therefore be directly relevant to cellular lipid storage in general, holding the promise of identifying pathways and mechanisms central to human diseases involving excessive lipid storage and to the engineering of cellular lipid storage in organisms for the improved production of oils and biofuels.
METHODS SUMMARY
RNAi-mediated genomic screen
RNAi screening with University of California, San Francisco (UCSF), DmRNAi libraries versions 1 and 2 (15,683 genes) was as described10. Examination of selected genes demonstrated 80–90% mRNA knockdown (Supplementary Fig. 1); previous studies showed protein levels decreased by 80% (refs 27, 28). S2 cells (2.7 ×106 cells ml−1) were treated with 1 μg of dsRNA in 96-well plates for three days, and 1 mM oleic acid was added for 24 h. Cells were fixed for 1 h in 4% paraformaldehyde/PBS and stained for 1 h with BODIPY493/503 in PBS. Images were obtained with IC100 (Beckman) or ImageXpress Micro (Molecular Devices) automated microscopes and a 40×, 0.95 numerical aperture PlanApo dry objective lens (Nikon). Six image fields (400–600 cells per well) were acquired per RNAi experiment.
The primary visual screen was validated by treating S2 cells with an independent set of RNAs8–10. Details of the visual and computational screening procedures are given in Supplementary Information. For all gene knockdowns, the penetrance of the phenotypes was high, affecting more than 70–80% of the cells.
Primers used for the original screen can be found at http://rnai.ucsf.edu/dropletscreen, and primers for secondary validation can be found at http://mpibcms.biochem.mpg.de/en/rg/lipidrophe/absatz_01.html.
Protein localization
mCherry was from R. Tsien7. mCherry–Cct1, mCherry–Cct2 and Arf79F–mCherry expression vectors (actin promoter) were made with the Gateway system (Invitrogen). Arf79F(T31N)–mCherry and Arf79F(Q71L)–mCherry were generated by QuickChange II mutagenesis (Stratagene).
Lipid measurements
Cells were cultured in six-well plates (16 μg of dsRNAs per well) for three days; 1 mM oleate was then added for 24 h. Cells were lysed in 50 mM Tris-HCl pH 7.4, 0.25 M sucrose; samples (250 μg of protein) were assayed for lipids. Phosphatidylcholine was quantified with a colorimetric method29. Triacylglycerol content was measured from extracted lipids by thin-layer chromatography30.
Lipolysis studies
Cells were cultured with 1 mM oleate for 24 h. To stimulate lipolysis, oleate was removed and cells were cultured in serum-free medium for 24 h. Aliquots were analysed for glycerol (Sigma-Aldrich glycerol colorimetric assay) and protein (Bio-Rad DC protein assay) content.
Supplementary Material
Acknowledgments
We thank I. Vasenkova and R. De Breuil for help with dsRNA synthesis; K. Warner for help with yeast work; M. Schuldiner, E. Griffis, T. Fazzio, E. Herker, S. Stymne, B. Panning and M. Ott for reagents; D. B. Jones and G. Howard for assistance with manuscript preparation; G. Schoenhofer for web access for the database; members of the Farese, Vale and Walter laboratories for discussions; and D. Srivastava and S. Yamanaka for critical reading of the manuscript. This work was supported by a Freedom to Discover Award from Bristol–Myers Squibb and National Institutes of Health grant R21 DK078254-01 (to R.F.), a David and Mary Phillips postdoctoral fellowship award (to Y.G.), the Human Frontier Science Program Organization (T.C.W.), the Howard Hughes Medical Institute (P.W. and R.D.V.) and the J. David Gladstone Institutes.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Bartz R, et al. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Brown DA. Lipid droplets: proteins floating on a pool of fat. Curr Biol. 2001;11:R446–R449. doi: 10.1016/s0960-9822(01)00257-3. [DOI] [PubMed] [Google Scholar]
- 3.Miura S, et al. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- 4.Bartz R, et al. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 5.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nature Rev Mol Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 6.Ulvila J, et al. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- 7.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nature Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 8.Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- 9.Kulkarni MM, et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nature Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 10.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobrosotskaya IY, Seegmiller AC, Brown MS, Goldstein JL, Rawson RB. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science. 2002;296:879–883. doi: 10.1126/science.1071124. [DOI] [PubMed] [Google Scholar]
- 13.Kent C. Regulatory enzymes of phosphatidylcholine biosynthesis: a personal perspective. Biochim Biophys Acta. 2005;1733:53–66. doi: 10.1016/j.bbalip.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Morrison DK, Murakami MS, Cleghon V. Protein kinases and phosphatases in the Drosophila genome. J Cell Biol. 2000;150:57–62. doi: 10.1083/jcb.150.2.f57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornell RB, Northwood IC. Regulation of CTP:phosphocholine cytidylyltransferase by amphitropism and relocalization. Trends Biochem Sci. 2000;25:441–447. doi: 10.1016/s0968-0004(00)01625-x. [DOI] [PubMed] [Google Scholar]
- 16.Weber U, Eroglu C, Mlodzik M. Phospholipid membrane composition affects EGF receptor and Notch signaling through effects on endocytosis during Drosophila development. Dev Cell. 2003;5:559–570. doi: 10.1016/s1534-5807(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 17.Hafez IM, Cullis PR. Roles of lipid polymorphism in intracellular delivery. Adv Drug Deliv Rev. 2001;47:139–148. doi: 10.1016/s0169-409x(01)00103-x. [DOI] [PubMed] [Google Scholar]
- 18.Dascher C, Balch WE. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J Biol Chem. 1994;269:1437–1448. [PubMed] [Google Scholar]
- 19.Nakamura N, Banno Y, Tamiya-Koizumi K. Arf1-dependent PLD1 is localized to oleic acid-induced lipid droplets in NIH3T3 cells. Biochem Biophys Res Commun. 2005;335:117–123. doi: 10.1016/j.bbrc.2005.07.050. [DOI] [PubMed] [Google Scholar]
- 20.Spang A, Matsuoka K, Hamamoto S, Schekman R, Orci L. Coatomer, Arf1p, and nucleotide are required to bud coat protein complex I-coated vesicles from large synthetic liposomes. Proc Natl Acad Sci USA. 1998;95:11199–11204. doi: 10.1073/pnas.95.19.11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nature Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 22.Marcinkiewicz A, Gauthier D, Garcia A, Brasaemle DL. The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J Biol Chem. 2006;281:11901–11909. doi: 10.1074/jbc.M600171200. [DOI] [PubMed] [Google Scholar]
- 23.Verghese PB, Arrese EL, Soulages JL. Stimulation of lipolysis enhances the rate of cholesterol efflux to HDL in adipocytes. Mol Cell Biochem. 2007;302:241–248. doi: 10.1007/s11010-007-9447-0. [DOI] [PubMed] [Google Scholar]
- 24.Bard F, et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 2006;439:604–607. doi: 10.1038/nature04377. [DOI] [PubMed] [Google Scholar]
- 25.Kumar Y, Cocchiaro J, Valdivia RH. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr Biol. 2006;16:1646–1651. doi: 10.1016/j.cub.2006.06.060. [DOI] [PubMed] [Google Scholar]
- 26.Miyanari Y, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nature Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 27.Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hojjati MR, Jiang XC. Rapid, specific, and sensitive measurements of plasma sphingomyelin and phosphatidylcholine. J Lipid Res. 2006;47:673–676. doi: 10.1194/jlr.D500040-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Monetti M, et al. Dissociation of hepatic steatosis and insulin resistance in mice overexpressing DGAT in the liver. Cell Metab. 2007;6:69–78. doi: 10.1016/j.cmet.2007.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.