Abstract
The three human LIG genes encode polypeptides that catalyze phosphodiester bond formation during DNA replication, recombination and repair. While numerous studies have identified protein partners of the human DNA ligases (hLigs), there has been little characterization of the catalytic properties of these enzymes. In this study, we developed and optimized a fluorescence-based DNA ligation assay to characterize the activities of purified hLigs. Although hLigI joins DNA nicks, it has no detectable activity on linear duplex DNA substrates with short, cohesive single-strand ends. By contrast, hLigIIIβ and the hLigIIIα/XRCC1 and hLigIV/XRCC4 complexes are active on both nicked and linear duplex DNA substrates. Surprisingly, hLigIV/XRCC4, which is a key component of the major non-homologous end joining (NHEJ) pathway, is significantly less active than hLigIII on a linear duplex DNA substrate. Notably, hLigIV/XRCC4 molecules only catalyze a single ligation event in the absence or presence of ATP. The failure to catalyze subsequent ligation events reflects a defect in the enzyme-adenylation step of the next ligation reaction and suggests that, unless there is an in vivo mechanism to reactivate DNA ligase IV/XRCC4 following phosphodiester bond formation, the cellular NHEJ capacity will be determined by the number of adenylated DNA ligaseIV/XRCC4 molecules.
1. Introduction
DNA ligases catalyze a common step in DNA replication, genetic recombination and DNA repair. Consequently, defects in DNA ligation can cause cell lethality, increased genomic instability and hypersensitivity to DNA damage [1]. The DNA ligation reaction has three distinct catalytic steps and involves covalent reaction intermediates. In eukaryotes, DNA ligases react with ATP to form a covalent enzyme-adenylate intermediate. This first step occurs independently of DNA whereas the subsequent two steps involve interactions between the DNA ligase and its DNA substrate. In the second step, the AMP moiety is transferred to the 5’ phosphate terminus at a strand break to generate a covalent DNA adenylate intermediate. Finally, the non-adenylated DNA ligase catalyzes phosphodiester bond formation in a reaction that involves nucleophilic attack by the -OH group at the 3’ terminus on the activated 5’ DNA adenylate and the release of AMP [1].
There are three human genes that encode DNA ligases, LIG1, LIG3 and LIG4 [1]. Unlike the LIG1 and LIG4 genes which encode a single DNA ligase polypeptide, the LIG3 gene encodes nuclear and mitochondrial versions of DNA ligase IIIα by alternative translation initiation and a germ cell-specific version, DNA ligase IIIβ, by alternative splicing [2,3]. Mutations in the LIG1 and LIG4 genes have been identified in humans and have been linked to cancer predisposition and immunodeficiency [4-6]. Although no examples of human LIG3 mutations have been observed, mutations in the genes encoding two proteins that associate with DNA ligase IIIα have been linked with neurodegenerative diseases, suggesting that DNA ligase IIIα-dependent repair pathways are critical in terminally differentiated neuronal cells [7,8].
The human DNA ligase polypeptides contain a conserved catalytic domain that is flanked by different sequences that target these enzymes to various DNA metabolic pathways [1]. Both nuclear DNA ligase IIIα and DNA ligase IV have a partner protein, XRCC1 and XRCC4 respectively, that is necessary for the stability and activity of the DNA ligase in vivo [9,10]. A large number of other proteins that interact either directly with the DNA ligase polypeptide or indirectly via their partner protein have been identified, and the functional consequences of their interactions characterized [1]. Despite the fact that DNA ligases I and III are predominantly involved in joining DNA nicks whereas DNA ligase IV completes the repair of DNA double strand breaks in the major non-homologous end joining pathway [1,11,12], relatively few studies have addressed the catalytic activities and substrate specificities of the human DNA ligases.
To date, most biochemical studies on human DNA ligases have utilized a radiolabeled DNA substrate to measure DNA joining. In these assays, the conversion of the radioactively labeled oligonucleotide or polynucleotide substrate into a labeled, higher molecular weight product by ligation is detected after separation of the substrate and products by gel electrophoresis. A limitation of gel-based ligation assays is that they are not suitable for carrying out large numbers of reactions. Recently, we developed a fluorescence-based assay that we used to screen and identify inhibitors of the human DNA ligases [13-15]. Here, we first describe the optimization and validation of this assay and demonstrate its suitability for kinetic analyses of DNA ligases. Subsequently, we used this technique to characterize the substrate specificity and catalytic properties of the DNA ligases encoded by the human LIG genes.
2. Materials and Methods
2.1 Oligonucleotide substrates
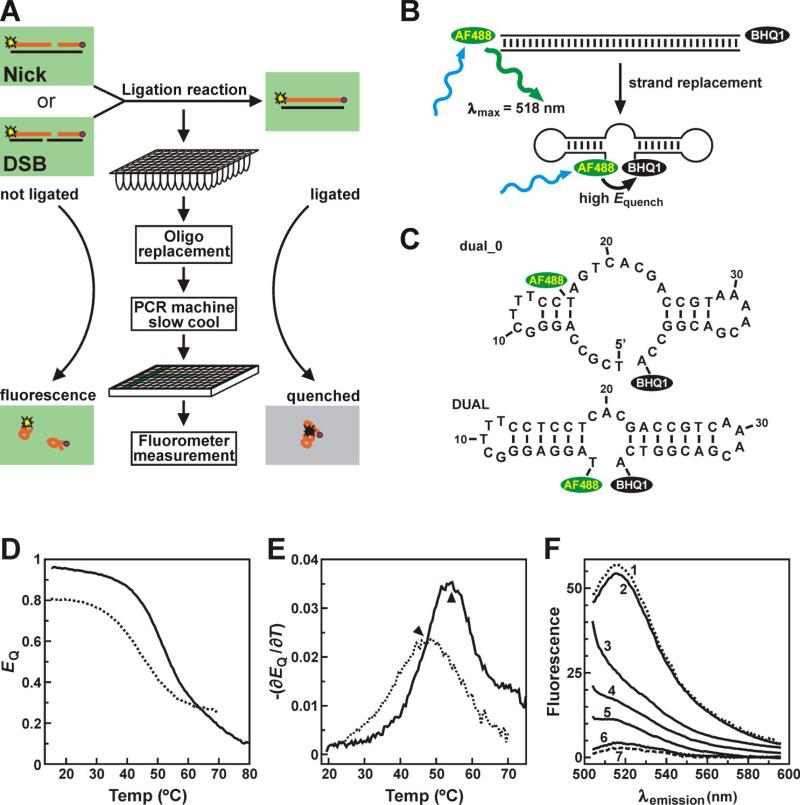
The oligonucleotides listed in Table I were purchased from Integrated DNA Technologies. To generate the nicked DNA substrate, oligonucleotide *U, containing a 5’-terminal AlexaFluor488 (AF488) dye, and D°, containing a 3’-terminal Black Hole Quencher-1 (BHQ1) group, were annealed to the complementary 40-mer oligonucleotide, T. This yielded a duplex DNA with a single nick in the AF488+BHQ1-labeled strand, positioning the fluorophore and dark quencher moieties 40 nucleotides apart (Fig. 1A). The double strand break (DSB) DNA substrate was generated by annealing the AF488-labeled upstream primer (*U), BHQ1-labeled downstream primer (D°), upstream template (UT), and downstream template (DT) primers, yielding linear duplexes with complementary 4 nucleotide single strand overhangs (Fig. 1A). Oligonucleotide UD encompasses the sequences of U and D and was used as an unlabeled competitor in DNA joining assays (described below). DUAL is a 5’-AF488 + 3’-BHQ1-labeled single-stranded oligonucleotide corresponding to completely ligated up- (*U) and down- (D°) stream primers. Primer dual_0 represents the 5’-AF488 + 3’-BHQ1-labeled strand of a ligation product in an un-optimized fluorescence-based ligase assay that we described previously [15].
Table I.
DNA oligonucleotides used in this study.
| Name | Sequence (5′→3′)a |
|---|---|
| dual_0 | TCG CCA GGG CTT TCC (AF-T)AG TCA CGA CCG TAA AAC GAC GGC CA-Q |
| DUAL | AF-TAG GAG GGC TTT CCT CCT CAC GAC CGT CAA ACG ACG GTC A-Q |
| UD | TAG GAG GGC TTT CCT CCT CAC GAC CGT CAA ACG ACG GTC A |
| *U | AF-TAG GAG GGC TTT CCT CCT CAC GAC C |
| D° | GTC AAA CGA CGG TCA-Q |
| T | CTG ACC GTC GTT TGA CGG TCG TGA GGA GGA AAG CCC TCC TA |
| UT | CTG ACC GTC GTT TGA CGG TCG |
| DT | TGA GGA GGA AAG CCC TCC TA |
“AF” and “Q” indicate the positions of the Alexa Fluor 488 and Black Hole Quencher-1 moieties, respectively, linked to the 5′- or 3′-termini of applicable DNA substrates. For probe dual_0, the Alexa Fluor 488 dye was conjugated to an internal T residue.
Fig. 1.
(A) A schematic of the fluorescence-based DNA joining assay starting with either nicked (Nick) or double strand break (DSB) substrates. Labeled substrate strands (orange lines) are covalently linked to the fluorophore AF488 (yellow star) or the dark quencher BHQ1 (purple circle). Details of the ligation reaction are described under “Materials and Methods”. (B) Following ligation, double stranded DNA products (top) are denatured and re-annealed in the presence of an unlabeled version of the labeled product strand. This displaces the labeled strand and permits juxtaposition of the AF488 and BHQ1 moities by intramolecular base pairing (bottom), thus quenching fluorescence emission from AF488. (C) The most stable folded DNA structures for the pre-optimized (dual_0) and optimized (DUAL) labeled strand products, calculated using mFold software [18]. (D) Thermal denaturation analyses of the dual_0 (dotted line) and DUAL (solid line) DNA oligonucleotides were performed as described under “Materials and Methods”. (E) The partial derivative of EQ (ΔT = 4°C) with respect to temperature plotted as a function of T for each model DNA in (D). The local maxima (arrowheads) indicate the characteristic melting temperatures (Tm) for structural transitions within each DNA. (F) Fluorescence emission spectra (λex = 488 nm) of the DUAL DNA oligonucleotide annealed to a complementary template strand in the presence of 5 mM Mg2+ (1, dotted line) or as a single stranded DNA in 10 mM TrisHCl [pH 8.0] without Mg2+ or K+ (2). Enhanced quenching of AF488 emission at room temperature as Mg2+ concentration was increased to 0.5 mM (3), 1 mM (4), 2 mM (5), and 5 mM (6), or in an alternative annealing buffer containing 50 mM K+ (7, dashed line).
2.2 Proteins
Recombinant full length hLig I, a fragment of hLigI containing the adenylation and OB-fold domains, and hLigIIIβ were purified from E. coli as described [15,16]. The hLigIV/XRCC4 complex was expressed in and purified from Sf9 cells as previously described [15]. An independent hLigIV/XRCC4 sample isolated from insect cells was purchased from Trevigen. An E. coli expression plasmid that co-expresses hLigIV and XRCC4 was a gift from Drs. Ellenberger and Tsai [17]. After overexpression in the E. coli Rosetta(DE3)pLysS strain (Novagen), the hLigIV/XRCC4 complex was purified by sequential P11 phosphocellulose, nickel agarose, Resource Q, and Sephacryl 200 gel filtration chromatography steps. The purification of the human DNA ligase IIIα/XRCCI complex from baculovirus-infected insect cells will be described elsewhere.
2.3 Fluorescence characterization of DNA substrates
Steady-state fluorescence of AF488-labeled DNA oligonucleotides was measured in 1 cm × 1 cm quartz cuvettes using a Cary Eclipse Spectrofluorometer (Varian) with 10 nm excitation and emission slits. Excitation was at 488 nm. In cases where a single wavelength is monitored (eg. thermal denaturation, ligase kinetics), fluorescence emission was measured at 518 nm. DNA ligation reactions link upstream AF488-labeled DNA oligonucleotides to downstream BHQ1-labeled strands. To measure the thermodynamic stability of folded DNA products generated by these reactions, thermal denaturation assays were performed using 20 nM DNA in 10 mM Tris-HCl [pH 8.0] containing 50 mM KCl. Cuvettes were equilibrated at 12°C for 10 minutes before initiating a 1°C/minute temperature gradient, recording AF488 fluorescence at 0.5°C intervals using the Cary Eclipse fluorometer equipped with a Peltier temperature controller and in-cell temperature probe. Measurement of parallel samples containing upstream AF488-labeled probe lacking the quencher moiety permitted the calculation of the quenching efficiency (EQ) at each measured temperature using the equation EQ = 1 – (FAF-Q/FAF), where FAF-Q and FAF are the blank-corrected fluorescence of the dual (AF488+BHQ1) and singly (AF488)-labeled DNA oligonucleotides, respectively. The apparent melting temperature (Tm) for each folded DNA substrate was then determined as the extremum of the derivative of fluorescence with respect to temperature.
2.4 Fluorescence-based DNA joining assays
Purified human DNA ligases were incubated with the fluorescent nicked or DSB DNA substrates (2−100 pmol) in ligation buffer (60 mM Tris-HCl [pH 8.0] containing 10 mM MgCl2, 5 mM DTT, 1 mM ATP, 50 μg/ml BSA and 50 mM NaCl for hLigI or 150 mM NaCl for hLigIII and hLigIV) at 25°C in a total volume of 20 μl. Following incubation, reactions were diluted to 200 μl with a 20-fold molar excess of the unlabeled competing oligonucleotide (UD) in annealing buffer (10 mM Tris-HCl [pH 8.0] containing 50 mM KCl, 1 mM EDTA and 5 mM MgCl2) and heated to 95°C for 5 minutes in a Thermocycler (Bio-Rad) before transferring products to 384- or 96-well plates. After cooling to room temperature at a rate of 2°C/minute, fluorescence was measured by fitting the Cary Eclipse fluorometer with a multi-plate reader (Fig. 1A) using the optical settings described above. The amount of substrate ligated as a percentage of input is equivalent to the quenching efficiency, calculated as EQ = (F-Fmin)/(Fmax-Fmin), where Fmax is the fluorescence of products from reactions lacking ligase (ie: 0% ligation) and Fmin is the emission from a completely ligated DNA substrate.
2.5 Analyses of enzyme kinetics
To measure ligase kinetics under multiple turnover conditions, purified human DNA ligases (1 pmol) were incubated with varying amounts of DNA substrate (varying from 2−100 pmol) for 5 min as described above. Initial velocity (V) was calculated as the appearance of ligated product over time and was measured across a range of substrate concentrations ([S]). Maximum velocity (Vmax) and the Michaelis-Menten constant for substrate (Km) were resolved by non-linear regression of V versus [S] plots using equation 1 and PRISM v3.03 software (GraphPad).
| (Eq. 1) |
Values of kcat were determined by dividing Vmax by the total enzyme concentration utilized in the experiments. To assess single turnover kinetics, purified pre-adenylated human DNA ligases (1−100 pmol) were incubated with DNA substrate (100 pmol) in ligation reactions lacking ATP for 5 min.
2.6 Ligase adenylation
Purified human DNA ligases (10 pmol) were incubated with Ni2+-affinity beads or Resource Q beads (10 μl of each) as appropriate at 4°C for 30 min. The beads were washed three times with buffer A (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1 mM EDTA and 1 mM DTT), and then incubated in AMP buffer (60 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM DTT and 50 μg/ml BSA) containing either nicked DNA (100 pmol) or pyrophosphate (PPi, 10 mM) for 15 min at 25°C. After washing three times with buffer A, the beads were incubated in AMP buffer containing [α32P]ATP (10 pmol) for 15 min at 25°C. DNA ligases were eluted from the nickel beads and Resource Q beads with 1 M imidazole and 1 M NaCl, respectively. After separation by SDS-PAGE, labeled, adenylated DNA ligases were visualized and quantitated by PhosphorImager analysis. To directly quantitate the number of labeled adenylated DNA ligase molecules, the labeled bands were cut out and the radioactivity determined by liquid scintillation counting.
3. Results
3.1 Optimization and validation of the fluorescence-based ligation assay
Previously we described a fluorescence based-ligation assay suitable for high throughput screening of chemical libraries [15]. In this assay, phosphodiester bond formation in the context of nicked DNA duplexes or overlapping double strand breaks results in the physical linkage of an oligonucleotide containing the fluorophore Alexa Fluor 488 (AF488) to an oligonucleotide containing the fluorescence quencher Black Hole Quencher-1 (BHQ1) (Fig. 1A). Since the distance between the fluorescent donor and quencher within the DNA duplex product precludes significant quenching (Fig. 1B, top), the fluorescent DNA molecules are denatured by heating and then slowly cooled in the presence of a 20-fold excess of an oligonucleotide competitor identical in sequence to the labeled strand but lacking the fluorophore or quencher (Fig. 1A). This strand replacement step releases both ligated and unligated fluorescent oligonucleotides as single stranded DNA, permitting structural condensation of labeled strands by intramolecular base pairing (Fig. 1B, bottom).
In our original assay, the labeled strand produced in the ligase reaction corresponds to the probe dual_0 (Fig. 1C, top). While this ligation product sufficed for the identification of enzyme inhibitors by high throughput screening [13,14], its use in quantitative characterization of enzyme activities was complicated by two concerns. First, the limited structural potential of the dual_0 product provides little restraint on the relative positioning of the AF488 dye and the BHQ1 quencher, thus precluding high efficiency quenching by either collisional or through-space interactions. Second, the dual_0 probe presents significant heterogeneity in folding potential, particularly near the ambient temperatures required for high throughput fluorescence measurements. This combination of factors limits the dynamic range of the assay and introduces the risk of significant temperature-dependent effects on quenching efficiency near ambient temperatures. As such, the first objective of this study was to optimize the dynamic range and robustness of this in vitro DNA ligase assay, to permit rigorous kinetic characterization of DNA ligase activities.
We reasoned that both the quenching efficiency and thermal stability of the labeled strand product could be improved by increasing the number of intramolecular base pair contacts and limiting the potential for alternatively folded conformations. These considerations led to the design of ligation substrates containing oligonucleotides *U and D° (Table I), which yield the product DUAL following ligation and strand replacement (Fig. 1C, bottom). The product DUAL is predicted to form extensive intramolecular base pairs within two separate stem-loop structures that stably juxtapose the AF488 and BHQ1 moieties [18]. Furthermore, independent localization of base pair contacts within the upstream (*U) and downstream (D°) labeled strand domains minimizes the risk of non-covalent linkage between substrate oligonucleotides following strand replacement in reactions with minimal ligase activity.
To confirm that AF488 fluorescence was effectively quenched by BHQ1 in the context of folded single-stranded DNA products and to assess the relative stability of folded DNA structures adopted by the dual_0 and DUAL probes, we first performed thermal denaturation analyses on these oligonucleotides. At low temperatures, both dual_0 and DUAL adopted condensed conformations, denoted by high quenching efficiencies (Fig. 1D). At low temperatures, fluorescence from the DUAL probe was quenched more efficiently (EQ = 0.95) relative to the dual_0 probe (EQ = 0.80), indicating that the AF488 dye was maintained in closer proximity to the BHQ1 quencher in the DUAL substrate than was possible in the context of the dual_0 product. Quenching of the AF488 moiety was dependent on DNA folding, since quenching was released as temperature increased. The enhanced intramolecular base pair potential of DUAL relative to dual_0 was also reflected in higher apparent melting temperature (Tm) of the former. As a consequence of the enhanced folded stability of DUAL, the quenching efficiency of this oligonucleotide was less sensitive to temperature in the ambient temperature range required for high throughput measurement (Fig. 1E; compare values of ∂EQ/∂T between 20−30°C). Since nucleic acid folding is stabilized by cations [19,20], we also characterized the ionic requirements for maximal quenching efficiency by the DUAL oligonucleotide at room temperature. Fluorescence from the AF488 dye was profoundly diminished in a dose-dependent fashion as Mg2+ was added to the solution (Fig. 1F, lines 2−6), with >95% quenching observed in 5 mM Mg2+. Addition of a monovalent cation (K+) to 50 mM (Fig. 1F, line 7) yielded a quenching efficiency similar to that observed in the thermal denaturation analyses. Finally, fluorescence from the DUAL probe was completely unquenched when hybridized to a complementary DNA oligonucleotide (Fig. 1F, line 1). This further confirmed that AF488 quenching depends on formation of an intramolecularly folded DNA product, thus necessitating the strand replacement step of the ligase assay (Fig. 1B).
To ensure that fluorescence quenching would solely reflect the progress of the ligation reaction, the steady state fluorescence of the AF488-labeled oligonucleotide was extensively characterized in the presence of other DNA molecules to mimic conditions that occur during the fluorescence based-ligation assay (Fig. 2). Together, these experiments yielded three important findings. First, AF488 fluorescence exhibited no quenching if the BHQ1 moiety was absent (panels 1−3, 7−9, 12, 13) or if AF488 and BHQ1 were not covalently linked (panels 4−6). The latter observation indicates that no quenching occurs for unligated substrates. Second, if AF488 and BHQ1 are covalently linked, fluorescence quenching is only observed when the labeled DNA strand was capable of intramolecular folding (cf. panels 11 versus 10, 15 versus 14 and 16). Finally, the presence of other reaction components (template strands, unlabeled competitor strands, etc.) did not directly impact fluorescence from unligated AF488-labeled substrates (cf. panels 2, 3 versus 1) or (AF488+BHQ1)-labeled ligation products (cf. panel 16 versus 14). These experiments demonstrate that AF488 emission is quenched solely following ligation of labeled upstream (*U) and downstream (D°) primers and labeled strand displacement, and indicate that this fluorescence-based ligase assay is a robust yet sensitive in vitro platform for quantitative characterization of DNA ligase activities.
Fig. 2.
Validation of ligase assay output showing that emission from AF488-labeled DNA products is quenched solely when covalently linked to BHQ1 and following displacement of the labeled strand. AF488-labeled upstream primer (*U) or (AF488+BHQ1)-labeled product (DUAL) oligonucleotides were assembled in various complexes to mimic possible ligation reaction outcomes (left panels). Reactions were slowly cooled in annealing buffer to facilitate intra- and intermolecular base pair formation where appropriate as described under “Materials and Methods”. The AF488-labeled upstream primer was tested alone (1), annealed to a complementary template as dsDNA (2), or replaced by excess competing unlabeled DNA (UD) to restore the labeled strand to a ssDNA format (3). The AF488-labeled upstream primer was incubated with the BHQ1-labeled downstream primer (D°) alone (4), hybridized to a common template (T) strand (5), or displaced by the unlabeled UD competitor (6). A similar set of reactions substituted an unlabeled downstream primer (D) for the BHQ1-labeled version (7−9). Subsequent reactions included AF488-labeled upstream primers ligated to BHQ1-labeled (10, 11) or unlabeled (12, 13) downstream primers, either as intact duplexes (10, 12) or released as ssDNA following strand replacement (11, 13). Finally, the (AF488+BHQ1)-labeled DUAL oligonucleotide which mimics completely ligated up- and downstream primers was incubated alone (14), hybridized to an unlabeled complementary strand (15), or displaced by the unlabeled DNA competitor (16). Reactions containing 10 pmol of the DNA substrate were incubated for 2 h at 25 °C in the absence or presence of 1 pmol of hLigI. For each assembled reaction, fluorescence emission was measured at 518 nm (λex = 488 nM). Average fluorescence values are listed as the mean ± standard deviation of three independent replicates (right), relative to the AF488-labeled upstream primer alone. “Q” is the efficiency of AF488 quenching for each reaction expressed as a percentage. “Oligos” indicates the oligonucleotide combinations employed in each reaction (Table I) and “AF488 strand” identifies whether the AF488 moiety is contained within a single-stranded (ss) or double-stranded (ds) DNA context.
3.2 Substrate specificity and catalytic efficiency of human DNA ligases on nicked and DSB DNA substrates
Three experiments were performed to ensure that the fluorescence-based DNA joining assay could faithfully track the progression of ligase reactions. First, fluorescence quenching was enhanced in reactions containing the AF488-labeled nicked DNA substrate as the reaction was enhanced in reactions containing the AF488-labeled nicked DNA substrate as the reaction was pushed to completion by increasing the concentration of DNA ligase, using hLigIIIβ as a model (Fig. 3A). Second, time-dependent accumulation of ligated DNA products was observed following addition of hLigI to AF488-labeled nicked substrates (Fig. 3B). From these reactions, initial reaction velocities (V) were calculated across a range of substrate concentrations ([S]), permitting resolution of Km and kcat as described in Materials and Methods (Fig. 3C). Finally, using gel-based assays we compared the ligation kinetics of the fluorescent nicked substrate with a comparable substrate lacking the AF488 dye and BHQ1 quencher moieties. These experiments indicated that ligase kinetics measured using the fluorescence-based assay were indistinguishable from those monitored using the gel-based system for both nicked (Fig. 3C) and DSB substrates (data not shown). Furthermore, these experiments confirmed that adding the fluorescent dye/quencher pair to DNA substrates does not affect their recognition by DNA ligases or ligation efficiency.
Fig. 3.
Resolution of ligase kinetic parameters using the fluorescence-based ligation assay. (A) A typical experiment showing the conce substrate. (B) Representative time courses of ligation reactions using hLigI programmed with indicated concentrations of nicked substrates. (C) Substrate-velocity plots of hLigI activity on nicked DNA substrates (5 − 100 pmol; 0.25 − 5μM) using the fluorescence-based (●) and gel-based (○) ligation assays.
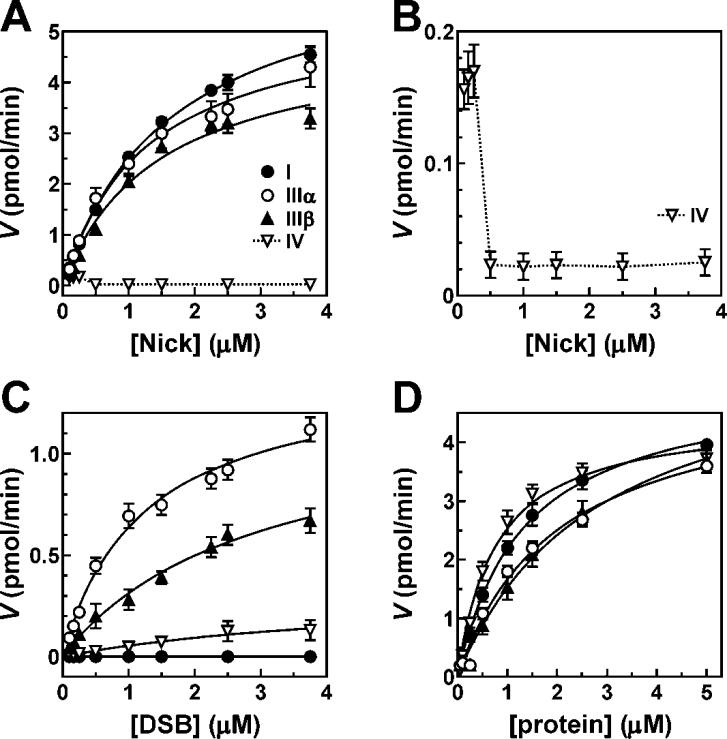
There is compelling evidence that the DNA ligases encoded by the three human LIG genes are directed to different DNA transactions by specific protein-protein interactions [1]. Since hLigI and hLigIII are primarily involved in pathways that are completed by nick ligation whereas hLigIV is a key factor in the major pathway that repairs DNA double strand breaks [1,4,11,12], we measured the catalytic properties of purified recombinant versions of DNA ligases I, III and IV on nicked and double strand break DNA substrates using the fluorescence-based assay. In reactions with the nicked DNA substrate, the activities of purified hLigI, hLigIIIα/XRCCI and hLigIIIβ were similar when comparing initial reaction velocity as a function of substrate concentration (Fig. 4A). The calculated kinetic parameters, kcat and Km for these enzymes are shown in Table II. Although hLigIV/XRCC4 is able to join nicked DNA, the amount of ligated product did not increase with increasing amounts of nicked DNA substrate and never exceeded the amount of hLigIV/XRCC4 added in reactions at either 25°C (Fig. 4B and Supplementary Table I) or 37°C (data not shown). Notably, when hLigIV/XRCC4 is significantly more abundant than the nicked substrate, the levels of ligation can reach more than 95% but there is no detectable increase in the initial reaction velocity (data not shown). Even under conditions where hLigIV/XRCC4 is active, the estimated kcat based on the observable Vmax is only 0.2 min−1, which is about 50-fold lower than hLigI in the same reaction. Similar results were obtained with commercially available hLigIV/XRCC4 purified from insect cells and with the functionally homologous Dnl4/Lif1 complex from Sacccharomyces cerevisiae (data not shown).
Fig. 4.
Multiple and single turnover kinetics of human DNA ligases. (A) Substrate-velocity plots from multiple turnover ligation reactions of hLigI ((I;●), hLigIIIα/XRCC1 (IIIα;○), hLigIIIβ (IIIβ;▲) and hLigIV (IV;▽) on nicked DNA substrates using the fluorescence-based ligation assay. (B) Substrate concentration dependence of hLigIV activity on the nicked DNA substrate under multiple turnover conditions. (C) Substrate-velocity plots from multiple turnover ligation reactions on DSB substrates. Symbol designations are as indicated in (A). (D) Single turnover ligation reactions of pre-adenylated human DNA ligases on nicked substrates in the absence of additional ATP. Symbols are as shown in (A). All enzyme activity results represent the average of at least three independent experiments with exhibited standard deviations.
Table II.
Kinetics of DNA ligation by hLigI, IIIα, IIIβ, and IV on nicked and DSB substrates. Enzyme activity parameters were obtained from reactions containing 1 pmol of each ligase in a 20 μl reaction.
| ligase | substrate | Km (μM) | kcat (min−1) | relative kcat | kcat/Km (M−1·s−1) |
|---|---|---|---|---|---|
| I | nick | 1.6 ± 0.2 | 6.6 ± 0.2 | 100 | 69,000 |
| I | dsb | 0 ± 0.0 | 0 ± 0.0 | 0 | n/a |
| IIIα | nick | 1.3 ± 0.2 | 5.5 ± 0.3 | 83 | 71,000 |
| IIIα | dsb | 1.2 ± 0.2 | 1.4 ± 0.1 | 21 | 20,000 |
| IIIβ | nick | 1.4 ± 0.3 | 4.9 ± 0.4 | 74 | 58,000 |
| IIIβ | dsb | 2.9 ± 0.5 | 1.2 ± 0.1 | 18 | 7,000 |
| IV | nick | n/a | 0.2 ± 0.1 | 3 | n/a |
| IV | dsb | n/a | 0.3 ± 0.1 | 5 | n/a |
In assays measuring the joining of linear DSB substrates with 4-nucleotide complementary single strand ends, hLigI had no detectable activity whereas both hLigIIIα/XRCC1 and hLigIIIβ had robust intermolecular joining activity (Fig. 4C). Notably, the hLigIIIα/XRCC1 complex had a higher kcat and a lower Km than hLigIIIβ (Fig. 4C and Table II), resulting in kcat/Km ratio that is three times higher than that of hLigIIIβ. This suggests that the hLigIIIα/XRCC1 complex has both a higher affinity for the DNA substrate and higher recycling rate than hLigIIIβ. Surprisingly, even though hLigIV is a key participant in NHEJ [12], the major pathway for repairing DSBs in vivo, the apparent kcat for hLigIV/XRCC4 (0.3 min−1) is almost 5-fold lower than that of hLigIIIα/XRCC1 (1.4 min−1) (Table II). Unlike our observations with nicked DNA substrates, the reaction velocity of hLigIV/XRCC4 increased as a function of DSB substrate concentration (Fig. 4C). However, this may reflect increased annealing of the DNA substrate molecules at higher concentrations, since the amount of ligated product generated by hLigIV/XRCC4 was always less than the amount of hLigIV/XRCC4 present in the assay (Supplementary Table I), similar to its activity on nicked DNA.
3.3 hLigIV/XRCC4 activity is similar to that of other human DNA ligases under single turnover conditions
To determine whether the limitations on hLigIV/XRCC4 activity were an intrinsic property of this DNA ligase rather than a consequence of differences in protein purification procedures, we measured the kinetics of the human DNA ligases in single turnover reactions (Fig. 4D). In these assays, the DNA ligases were pre-adenylated prior to incubation with an excess of DNA substrate in the absence of ATP. Under these reaction conditions, the activity of hLigIV/XRCC4 was similar to that of the other human DNA ligases (Fig. 4D). Thus, the different behavior of hLigIV/XRCC4 is only evident in multiple turnover reactions.
3.4 Defect in readenylation of hLigIV/XRCC4 following phosphodiester bond formation
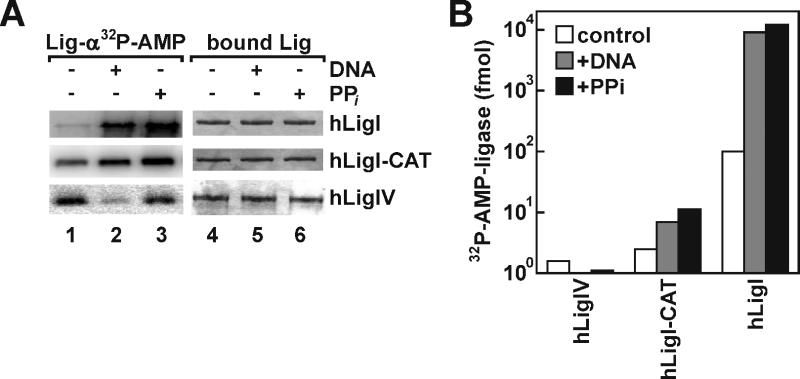
Based on the robust activity of hLIgIV/XRCC4 under single- but not multiple-turnover reaction conditions, we hypothesized that this enzyme is defective in the first step of the next catalytic cycle, formation of the enzyme-adenylate intermediate. Although purified DNA ligases contain a mixture of adenylated and non-adenylated molecules, the number of adenylated DNA ligase molecules can be estimated by performing ligation reactions with an excess of DNA substrate but in the absence of ATP. Using this approach, we determined that at least 99% of hLigI and hLigIV/XRCC4 molecules in the purified fractions were already adenylated. To confirm this, we incubated the purified DNA ligases with an excess of [α32P]ATP to label the non-adenylated molecules (Fig. 5A, lane 1). Under these conditions, we estimate that about 0.1 pmol (1%) of hLigI and 0.002 pmol (0.02%) of hLigIV/XRCC4 molecules were labeled by adenylation. In similar assays with the catalytic core of hLigI that is composed of the adenylation and OB-fold domains [16], about 0.002 pmol (0.02%) of the molecules were labeled by adenylation. The adenylate group can be removed either as a consequence of phosphodiester bond formation or by reversal of the first step of the ligation reaction. As expected, pre-incubation of hLigI with either nicked DNA in the absence of ATP or an excess of pyrophosphate, followed by incubation with [α32P]ATP resulted in about a 100-fold increase in the formation of labeled adenylated hLigI (Fig. 5A, cf. lanes 2 and 3 versus lane 1, and Fig. 5B). A similar pattern was observed in assays with the catalytic core of hLigI albeit with less of an increase in the formation of labeled adenylated intermediate. This may reflect inefficient release of AMP from the pre-adenylated protein by incubation with either nicked DNA in the absence of ATP or pyrophosphate. Alternatively, the subsequent re-adenylation reaction may be inefficient. In contrast, pre-incubation of hLigIV/XRCC4 with nicked DNA, and to some extent excess pyrophosphate, followed by incubation with [α32P]ATP actually decreased formation of labeled adenylated hLigIV (Fig. 5A, cf. lanes 2 and 3 versus lane 1, and Fig. 5B). Although it is possible that pyrophosphate fails to reverse the first step of the ligation reaction, the robust activity of hLigIV/XRCC4 in single turnover reactions indicates that transfer of AMP from hLigIV to the DNA occurs efficiently. Thus, we conclude that there is a defect in re-adenylation of hLigIV following phosphodiester bond formation that explains the abnormal behavior of hLigIV/XRCC4 in multiple turnover reactions.
Fig. 5.
Analyses of ligase adenylation. (A) Purified full length hLigI (hLigI), a fragment of hLigI containing the adenylation and OB-fold domains (hLigI-CAT) and hLigIV/XRCC4 (hLigIV) were immobilized on beads as described in Materials and Methods. The immobilized DNA ligases (10 pmol of each) were incubated with: lanes 1 and 4, 10 pmol of [α32P] ATP; lanes 2 and 5, 100 pmol nicked DNA followed by incubation with 10 pmol of [α32P] ATP; lanes 3 and 6, 10 mM pyrophosphate followed by incubation with10 pmol of [α32P] ATP as described in Materials and Methods. After elution from beads, proteins were separated by SDS-PAGE. Total proteins were detected by Coomassie blue staining (right panel) and labeled, adenylated proteins were detected and quantitated by PhosphorImager analysis (left panel). (B) Graphical representation of the ligase-adenylation assay. The horizontal axis numbers correspond to the lane designations in (A). The amount of labeled adenylated ligase was determined by liquid scintillation counting of the radioactive band.
4. Discussion
DNA strand breaks are common intermediates in DNA replication, recombination and almost all DNA repair pathways. The DNA ligases encoded by the three mammalian LIG genes have been purified from mammalian tissues and cells [21-23] and after overexpression in heterologous systems [16,17,24-27]. To date, the activities of the mammalian DNA ligases with different DNA substrates have only been compared in qualitative studies [22,24,28]. In addition, kinetic parameters have been determined for these enzymes in several studies but these are not directly comparable because of the use of different DNA substrates and/or reaction conditions [16,17,29]. Here we have quantitated and compared the activities of purified recombinant hLigI, hLigIIIα/XRCC1, hLigIIIβ and hLigIV/XRCC4 on DNA substrates that mimic DNA nicks and double strand breaks that are repaired in vivo. Notably, this is the first biochemical and kinetic characterization of the hLigIIIα/XRCC1 complex which is the active form of this enzyme involved in nuclear DNA repair events and the first systematic quantitative analysis of the catalytic properties of the three human DNA ligases under the same defined reaction conditions.
The DNA joining activities of the human DNA ligases were measured using a fluorescent-based ligation assay [15]. We initially developed this assay to screen for inhibitors of human DNA ligases in a high throughput format [13,14]. Here we have optimized and validated this assay for quantitative determination of kinetic parameters for the DNA ligases with both a nicked DNA substrate that measures intramolecular ligation and linear duplex DNA substrates with cohesive ends that measures intermolecular ligation. This assay method has several advantages over traditional gel-based assays. It is more versatile in terms of reaction conditions, gives more rapid results and is better suited for handling a large number of reactions.
The catalytic properties of hLigI and hLigIIIα/XRCC1 were similar for nick ligation, but unlike hLigIIIα/XRCC1 and hLigIIIβ, hLigI had no detectable intermolecular ligation activity. This is consistent with the results of gel-based assays comparing hLigI and hLigIIIβ and is compatible with the participation of hLigI in DNA replication and excision repair [30] and hLigIIIα/XRCC1 in excision repair and single strand break repair [11], all of which involve nick ligation. A unique feature of the DNA ligases encoded by the LIG3 gene is an N-terminal zinc finger that binds to nicks and gaps in DNA and resembles the pair of zinc fingers at the N-terminus of PARP-1 that constitute the DNA binding domain of this enzyme [26,31]. Although the DNA ligase III zinc finger is not required for nick ligation [26,31], it greatly stimulates intermolecular ligation [29,32] and presumably accounts for the high kcat values determined for intermolecular ligation by hLigIIIα/XRCC1 and hLigIIIβ. Although hLigIV/XRCC4 is a key component of the major DNA protein kinase-dependent NHEJ pathway [12], there is evidence that hLigIIIα/XRCC1 participates in a back-up alternative NHEJ pathway [33,34]. Interestingly, the steady state levels of [13] and, in chronic myelogenous leukemia cells expressing BCR-ABL, the hLigIIIα-dependent alternative NHEJ pathway is up-regulated and contributes to the error prone-repair of DNA double strand breaks in these cells [13,33]. Thus, the robust intermolecular ligation activity of hLigIIIα/XRCC1 is consistent with the participation of this complex in alternative NHEJ.
Under multiple turnover conditions, the behavior of hLigIV/XRCC4 was markedly different than that of hLigI, hLigIIIα/XRCC1 and hLigIIIβ. In accord with a published study [35], the number of ligated molecules generated by hLigIV/XRCC4 never exceeded the number of hLigIV/XRCC4 molecules in the reactions. Moreover, the initial reaction velocity either did not increase or increased only slightly with increasing amount of DNA substrate. In contrast, the activity of hLigIV/XRCC4 was similar to that of the other human DNA ligases in single turnover reactions. Thus, it appears that adenylated hLigIV/XRCC4 molecules catalyze phosphodiester bond formation but are then defective in undergoing the next catalytic cycle. This is consistent with a recent study showing that, under steady state conditions, the joining of nicked DNA by hLigIV/XRCC4 is biphasic with rapid initial phase followed by a subsequent slow phase [17]. In accord with published studies [17,36,37], the re-adenylation of hLigIV/XRCC4 was very inefficient. If each hLigIV/XRCC4 molecule is effectively restricted to a single ligation event in vivo then the DNA double strand break repair capacity of the major NHEJ pathway, at least in the initial phase, will be limited by the number of hLigIV/XRCC4 molecules present in the cell. Alternatively, there may be cellular factors that promote the re-adenylation of hLigIV/XRCC4. In support of this idea, it has been shown recently that the NHEJ factor XLF-Cernunnos promotes the re-adenylation of hLigIV/XRCC4 [37]. Since almost all the hLigIV molecules in the fraction purified from E. coli were adenylated, XLF-Cernunnos is not essential for adenylation of hLigIV but the efficiency of hLigIV adenylation may be significantly less than that of the other human DNA ligases.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health [GM47521, GM57479 and ES012512 to A.E.T., CA92584 to A.E.T. and M.-S.T, CA102428 to G.M.W.].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
X.C., A.E.T., and G.M.W. are co-inventors on a patent that covers the development and utility of the fluorescence-based DNA ligase assay.
References
- 1.Ellenberger T, Tomkinson AE. Eukaryotic DNA ligases: Structural and functional insights. Ann. Rev. Biochem. 2008;77:313–338. doi: 10.1146/annurev.biochem.77.061306.123941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakshmipathy U, Campbell C. The human DNA ligase III gene encodes nuclear and mitochondrial proteins. Mol. Cell. Biol. 1999;19:3869–3876. doi: 10.1128/mcb.19.5.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackey ZB, Ramos W, Levin DS, Walter CA, McCarrey JR, Tomkinson AE. An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination. Mol. Cell. Biol. 1997;17:989–998. doi: 10.1128/mcb.17.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T. Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents. Cell. 1992;69:495–503. doi: 10.1016/0092-8674(92)90450-q. [DOI] [PubMed] [Google Scholar]
- 5.Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett CF. Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol. 1999;9:699–702. doi: 10.1016/s0960-9822(99)80311-x. [DOI] [PubMed] [Google Scholar]
- 6.O'Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer SE, Seidel J. DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency. Mol. Cell. 2001;8:1175–1185. doi: 10.1016/s1097-2765(01)00408-7. [DOI] [PubMed] [Google Scholar]
- 7.Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 8.El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature. 2005;434:108–113. doi: 10.1038/nature03314. [DOI] [PubMed] [Google Scholar]
- 9.Bryans M, Valenzano MC, Stamato TD. Absence of DNA ligase IV protein in XR-1 cells: evidence for stabilization by XRCC4. Mutat. Res. 1999;433:53–58. doi: 10.1016/s0921-8777(98)00063-9. [DOI] [PubMed] [Google Scholar]
- 10.Caldecott KW, Tucker JD, Stanker LH, Thompson LH. Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Zhong S, Zhu X, Dziegielewska B, Ellenberger T, Wilson GM, MacKerrell AD, J., Tomkinson AE. Rational Design of Human DNA ligase Inhbitors that Target Cellular DNA Replication and Repair. Cancer Res. 2008;68:3169–3177. doi: 10.1158/0008-5472.CAN-07-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong S, Chen X, Zhu X, Dziegielewska B, Bachman KE, Ellenberger T, Ballin JD, Wilson GM, Tomkinson AE, MacKerell AD., Jr. Identification and validation of human DNA ligase inhibitors using computer-aided drug design. J. Med. Chem. 2008;51:4553–4562. doi: 10.1021/jm8001668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, Pascal J, Vijayakumar S, Wilson GM, Ellenberger T, Tomkinson AE. Human DNA ligases I, III, and IV-purification and new specific assays for these enzymes. Methods Enzymol. 2006;409:39–52. doi: 10.1016/S0076-6879(05)09003-8. [DOI] [PubMed] [Google Scholar]
- 16.Pascal JM, O'Brien PJ, Tomkinson AE, Ellenberger T. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004;432:473–478. doi: 10.1038/nature03082. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Lamarche BJ, Tsai MD. Human DNA ligase IV and the ligase IV/XRCC4 complex: analysis of nick ligation fidelity. Biochemistry. 2007;46:4962–4976. doi: 10.1021/bi0621516. [DOI] [PubMed] [Google Scholar]
- 18.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai Y, Greenfeld M, Travers KJ, Chu VB, Lipfert J, Doniach S, Herschlag D. Quantitative and comprehensive decomposition of the ion atmosphere around nucleic acids. J. Am. Chem. Soc. 2007;129:14981–14988. doi: 10.1021/ja075020g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heilman-Miller SL, Pan J, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme. II. Counterion-dependence of folding kinetics. J. Mol. Biol. 2001;309:57–68. doi: 10.1006/jmbi.2001.4660. [DOI] [PubMed] [Google Scholar]
- 21.Husain I, Tomkinson AE, Burkhart WA, Moyer MB, Ramos W, Mackey ZB, Besterman JM, Chen J. Purification and characterization of DNA ligase III from bovine testes. Homology with DNA ligase II and vaccinia DNA ligase. J. Biol. Chem. 1995;270:9683–9690. doi: 10.1074/jbc.270.16.9683. [DOI] [PubMed] [Google Scholar]
- 22.Robins P, Lindahl T. DNA ligase IV from HeLa cell nuclei. J. Biol. Chem. 1996;271:24257–24261. doi: 10.1074/jbc.271.39.24257. [DOI] [PubMed] [Google Scholar]
- 23.Tomkinson AE, Lasko DD, Daly G, Lindahl T. Mammalian DNA ligases, Catalytic domain and size of DNA ligase I. J. Biol. Chem. 1990;265:12611–12617. [PubMed] [Google Scholar]
- 24.Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J. Biol. Chem. 2000;275:26196–26205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- 25.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 26.Mackey ZB, Niedergang C, Murcia JM, Leppard J, Au K, Chen J, de Murcia G, Tomkinson AE. DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III. J. Biol. Chem. 1999;274:21679–21687. doi: 10.1074/jbc.274.31.21679. [DOI] [PubMed] [Google Scholar]
- 27.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomkinson AE, Roberts E, Daly G, Totty NF, Lindahl T. Three distinct DNA ligases in mammalian cells. J. Biol. Chem. 1991;266:21728–21735. [PubMed] [Google Scholar]
- 29.Cotner-Gohara E, Kim IK, Tomkinson AE, Ellenberger T. Two DNA-binding and Nick Recognition Modules in Human DNA Ligase III. J. Biol. Chem. 2008;283:10764–10772. doi: 10.1074/jbc.M708175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 2000;10:919–922. doi: 10.1016/s0960-9822(00)00619-9. [DOI] [PubMed] [Google Scholar]
- 31.Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell. Biol. 2003;23:5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor RM, Whitehouse CJ, Caldecott KW. The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining. Nucleic Acids Res. 2000;28:3558–3563. doi: 10.1093/nar/28.18.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sallmyr A, Tomkinson AE, Rassool F. Up-regulation of WRN and DNA ligase IIIa in Chromic myeloid leukemia: Consequences for the repair of DNA double strand breaks. Blood. 2008;112:1413–1423. doi: 10.1182/blood-2007-07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–4030. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 35.Lee KJ, Huang J, Takeda Y, Dynan WS. DNA ligase IV and XRCC4 form a stable mixed tetramer that functions synergistically with other repair factors in a cell-free end-joining system. J. Biol. Chem. 2000;275:34787–34796. doi: 10.1074/jbc.M004011200. [DOI] [PubMed] [Google Scholar]
- 36.Riballo E, Doherty AJ, Dai Y, Stiff T, Oettinger MA, Jeggo PA, Kysela B. Cellular and biochemical impact of a mutation in DNA ligase IV conferring clinical radiosensitivity. J. Biol. Chem. 2001;276:31124–31132. doi: 10.1074/jbc.M103866200. [DOI] [PubMed] [Google Scholar]
- 37.Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res. 2009;37:482–492. doi: 10.1093/nar/gkn957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.