Abstract
Optimized plasmid DNAs encoding the majority of SIVmac239 proteins and delivered by electroporation (EP) elicited strong immune responses in rhesus macaques. Vaccination decreased viremia in both the acute and chronic phases of infection after challenge with pathogenic SIVmac251. Two groups of macaques were vaccinated with DNA plasmids producing different antigen forms, “native” and “modified,” inducing distinct immune responses. Both groups showed significantly lower viremia during the acute phase of infection, whereas the group immunized with the native antigens showed better protection during the chronic phase (1.7 log decrease in virus load, P = 0.009). Both groups developed strong cellular and humoral responses against the DNA vaccine antigens, which included Gag, Pol, Env, Nef, and Tat. Vaccination induced both central memory and effector memory T cells that were maintained at the day of challenge, suggesting the potential for rapid mobilization upon virus challenge. The group receiving the native antigens developed higher and more durable anti-Env antibodies, including neutralizing antibodies at the day of challenge. These results demonstrate that DNA vaccination in the absence of any heterologous boost can provide protection from high viremia comparable to any other vaccine modalities tested in this macaque model.
Keywords: AIDS, DNA vaccine, macaque animal model, prophylactic vaccination
Genetic vaccination is a rapidly evolving technology with mostly unrealized potential. Some veterinary vaccines have recently emerged, and several human candidate DNA vaccines are in clinical trials (1). Low immunogenicity, due in part to poor delivery methods and to low DNA expression is a major obstacle for human DNA vaccination. Yet, genetic immunization continues to be attractive due to the potential power and flexibility of this approach, as shown in animal models. Here, we use the Indian rhesus macaque model to study protection by DNA vaccines against challenge with highly pathogenic simian immunodeficiency virus (SIVmac251). This model is considered a faithful representation of HIV infection of humans, recapitulating many aspects of pathogenesis and disease progression. DNA vaccination of macaques against SIV has shown promising results when optimized DNA expression vectors encoding multiple antigens were used (2). Very few macaque studies using DNA-only vaccination followed by challenge with high dose pathogenic SIVmac251 or SIVmac239 (2, 3) or other challenges (4, 5) have been performed, since it has been assumed that DNA alone is a weak immunogen in nonhuman primates. Studies using DNA as a prime and heterologous boosts with viruses or protein and SIVmac challenge have reported promising results in terms of durable protection from high viremia, including DNA/rNYVAC (6), DNA/recombinant adenovirus (Ad) (7–9), and DNA/recombinant HSV (10). Other studies have reported either no decrease in viremia or modest and transient decrease during primary infection only, which, in some cases, could still benefit the animals (11). Promising strategies not involving DNA include nonreplicating recombinant adenovirus rAd26/rAd5 (12), replicating recombinant adenovirus plus protein boost (13), or recombinant rhCMV (14). Despite improvements of DNA vaccines, human trials have indicated that the magnitude of immune responses after DNA vaccination remains low (1, 15–18) compared to levels reported in macaques. The ability to increase the magnitude and quality of the immune responses and to achieve protection in a strict macaque SIV challenge model may provide critical information to improve DNA vaccination efficacy in humans.
Recent studies by several groups including ours, have demonstrated that DNA in vivo electroporation (EP) enhances uptake and immunogenicity of SIV DNA vaccines (19–24). Here, we explored the prophylactic potential of DNA-only vaccination against SIVmac251, using optimized DNA expression vectors encoding the majority of SIV proteins delivered intramuscularly (IM) by the more efficient DNA EP method followed by high dose SIVmac251 intrarectal challenge. We show that DNA-only vaccination induces high immune responses and provides protection after challenge with SIVmac251 by lowering the levels of both acute and chronic viral loads. This level of protection is similar to other successful vaccine modalities applied to this model and highlight the potential of DNA-only based vaccines.
Results
We compared the efficacy of two polyvalent SIV DNA-only vaccines in Indian rhesus macaques. Two groups of macaques (n = 8) were vaccinated with DNA vectors producing the majority of SIVmac239 proteins. One group (Native) received DNA vectors expressing the “native” forms of SIV antigens Gag, Pol, Env, and the Nef-Tat-Vif (NTV) fusion protein, whereas the antigens delivered to the other group (Modified) were altered to change the trafficking of the proteins as described in Materials and Methods. The animals received four DNA immunizations by EP with a mixture of SIV plasmids together with a plasmid producing IL-12 as molecular adjuvant (Fig. 1A). Vaccinations EP1, EP2, and EP3 (weeks 0, 11, and 19) were performed with a low DNA dose (100 μg DNA/antigen), whereas EP4 (week 32) used a higher DNA dose (400 μg DNA/antigen).
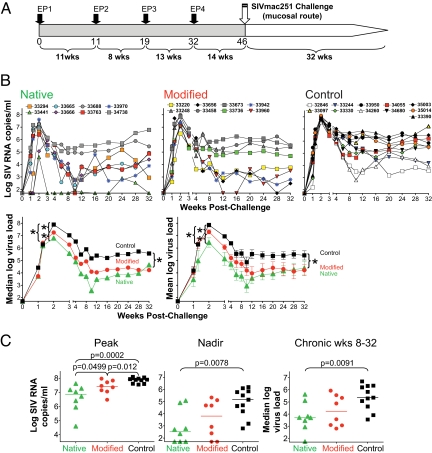
Fig. 1.
DNA vaccination and challenge. (A) Study design. Indian rhesus macaques were vaccinated with DNA at the indicated time points via IM route using EP, followed by intrarectal challenge with SIVmac251. (B) Sequential viral load determinations for individual animals in the two vaccinated groups and the control group are shown (log of SIV RNA copies/mL plasma). The bottom left panel shows the median log virus loads for the three groups. Asterisks indicate significant differences as shown in panel C. The bottom right panel shows the mean log virus load (using log-transformed values) with standard error bars for the three groups. Even though several means and standard error of the means (SEMs) are biased by assigning the threshold of detection of the assay to viral loads below it, this analysis resulted in signficant differences in peak, nadir, and chronic phase. (C) The log VL at peak, nadir, and chronic phase for each animal and the median for each group are shown.
The vaccinated animals and a group of 11 naive controls were challenged 14 weeks after the last vaccination by the intrarectal route using highly pathogenic SIVmac251 (Fig. 1B). Although all animals became infected, we found significant differences among the three groups. Vaccinated animals had substantially lower peak viremia compared to the controls. The median peak log viral loads of the vaccinees (Fig. 1C) were 6.9 log (Native) and 7.4 log (Modified), and both were lower than the controls (7.9 log). Thus, DNA vaccination resulted in a significant lower peak viremia of 1 log (P = 0.0002 control vs. the Native group) and 0.5 log (P = 0.012 control vs. the Modified group). The difference between the two vaccinated groups in the acute phase also reached statistical significance (P = 0.0499, two-tailed Wilcoxon rank sum test), suggesting that the combination of vectors expressing the native antigens was superior to the ones expressing the modified antigens, which also showed lower Env responses (see below, Figs. 2 and 3). Eleven macaques with three major histocompatibility complex (MHC) haplotypes (Mamu-A*01, B*08, and B*17) reported to affect viremia by some SIV stocks (25–28) were distributed over the three groups, as detailed in Table S1. To account for any possible effect on the intergroup comparisons, we adjusted the comparisons between the three groups using the presence of any protective haplotype as a stratification factor. The results were similar to the unadjusted tests above: P = 0.0001 for control vs. Native, P = 0.0059 for control vs. Modified, and P = 0.070 for Native vs. Modified (exact stratified Wilcoxon rank sum test), in agreement with the observation that the challenge stock used is not associated with MHC-linked spontaneous control of viremia [herein and (29)].
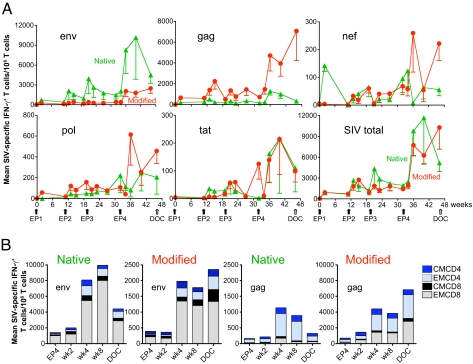
Fig. 2.
Development of SIV-specific cellular immune responses in immunized animals. (A) The mean and SEM of SIV-specific IFN-γ T cell responses to Env, Gag, Nef, Pol, and Tat, as well as the sum of all responses (SIV total) over the period of immunization are shown for the two groups of animals vaccinated with DNAs expressing the native (green) or modified (red) SIV antigens. Note the lower scale in vertical axis for the nef, pol, and tat panels. (B) The mean of Gag- and Env-specific Central (CM) and Effector (EM) memory responses after EP4 are shown for the two groups of animals. Note the lower scale in vertical axis of the two middle panels.
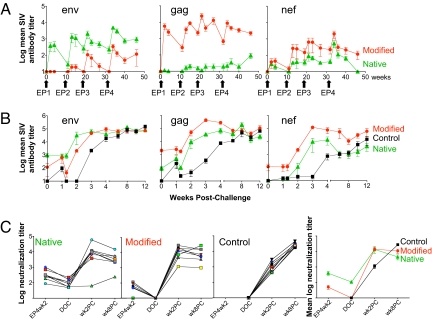
Fig. 3.
Development of SIV-specific humoral immune responses in immunized animals. (A) Mean and SEM of reciprocal endpoint titers to Env, Gag, and Nef are plotted over the vaccination period for the two groups of animals vaccinated with DNAs expressing the native antigens (green) or the modified antigens (red). (B) Mean and SEM of reciprocal endpoint titers after challenge for Env, Gag, and Nef for the vaccine and control groups. (C) Log of the SIVmac251-TCLA neutralizing antibody titers for all animals in the two vaccine groups and the control group, as well as the mean log neutralization titer per group before and after challenge.
Following peak viremia, several vaccinated animals rapidly decreased plasma virus load, and five animals (three in the Native and two in the Modified group) suppressed viremia below the threshold of the assay (50 copies/mL plasma, Fig. 1C, nadir). A total of 10 vaccinated animals and only one control showed dramatic decrease in viremia (<1,000 copies/mL plasma) early after the peak. These decreases in viremia were not sustained for most of the animals, although #33441 and #33970 in the Native group and #33656, #33942, #33960 in the Modified group continued to have undetectable to <1,000 copies of virus/mL plasma in most measurements during the chronic phase.
The difference in virus loads between vaccinees and controls persisted during the chronic phase (Fig. 1B). In the early phase of chronic infection (weeks 8–20), both the Native and Modified groups continued to show a statistically significant difference compared to the control (P = 0.0036 and 0.02, respectively, Wilcoxon rank sum test). Considering the entire chronic period (weeks 8–32, Fig. 1C), the median virus loads of the Native group was 3.7 log compared to 4.2 log for the Modified group and 5.4 log for the controls. Therefore, the Native group showed a 1.7 log difference in chronic viremia compared to the control (P = 0.0091; Wilcoxon rank sum test). This difference was not sustained for the Modified group, which continued to show a difference of 1.2 log compared to the control, but did not reach significance (P = 0.075). There was no statistical difference between the two vaccine groups for the entire chronic phase. The subset of macaques with reportedly protective MHC haplotypes did not have significantly lower levels of chronic infection (P = 0.26 for weeks 8–20, P = 0.27 for weeks 8–32, exact stratified Wilcoxon rank sum test). Nevertheless, we stratified the animals for the protective haplotypes and obtained results similar to those of the unstratified tests (for weeks 8–20, Native vs. controls, P = 0.0053, Modified vs. controls, P = 0.012; for weeks 8–32, P = 0.012 and P = 0.053, respectively). Thus, DNA-only vaccination achieved a significant reduction in both peak (1 log) and chronic (1.7 log) viremia. The changes in virus loads were also compared using mean values (Fig. 1B, right panel), which resulted in similar conclusions. By unbalanced analysis of variance (ANOVA), the comparison of the nadirs of the Native and Control groups had a significance level of P = 0.0042, and the comparison of the chronic viral loads of the same two groups had P = 0.0058. The difference in the untransformed peaks between the Native and control groups was highly significant (P < 0.0001), the difference between the Modified and control groups was significant (P = 0.0029), and the difference between the Native and Modified groups was not significant (P = 0.078). Thus, the analysis using median or mean values show significant differences in peak, nadir, and chronic phase between the vaccinee and the control group. These results demonstrate that optimized DNA vectors and more efficient DNA delivery were able to contain viremia for a long period after challenge. The Native group showed the best protection from high viremia during the acute and chronic phase. These results further suggest that the DNA vaccine delivered by EP was able to achieve protection similar to other methods of vaccination tested in this macaque model.
The development of cellular and humoral immune responses upon DNA EP was followed over time, and the results of the two different DNA vaccines were compared. After the first EP (EP1), SIV-specific IFN-γ T cell responses (primarily to Gag and Env) were detected in all vaccinated macaques, and subsequent vaccinations (EP2, EP3) led to further increases (Fig. 2A, mean values, and Fig. S1A, individual animals) with peak responses at 2 or 4 weeks post each vaccination. Low levels of Nef-, Pol-, and Tat-specific T cell responses were also detected in both groups. A great increase in immune responses was found upon EP4 (higher DNA dose), and these high levels were sustained for up to 14 weeks, indicating that the low dose DNA vaccine used during EP1-EP3 was suboptimal. Peak values after EP4 were ≈1.2% (range 0.3%–5.5%) of SIV-specific IFN-γ+ T cells in the blood (Fig. S1A). SIV-specific cells producing TNFα upon peptide stimulation were also generated (Fig. S2A). The range of peak TNFα responses after EP4 were 0.1% to 1.4% of blood T cells. Comparison of the two vaccines showed that the native and the modified antigens induced distinct cellular immune responses (Fig. 2 and Fig. S1A). The Native group showed a predominant response to Env, whereas the Modified group showed a predominant response to Gag. The native Env induced higher responses over the complete vaccination period from EP1week2 to EP4week14 (P = 0.011, repeated measures ANOVA). In addition to IFN-γ, the TNFα responses to native Env were also significantly higher after EP4 (EP4wk2–EP4wk14, P = 0.046 Wei-Johnson test) (Fig. S2A). The modified Gag antigen induced significantly higher IFN-γ responses throughout the vaccination period (P = 0.020, nonparametric Wei-Johnson test). Animals in the two groups also developed distinct responses to Pol and Nef. After EP4, higher Pol and Nef responses were found in the Modified group, which were significantly different from the Native group at the day of challenge (DOC) (Pol, P = 0.01; Nef, P = 0.018, Wilcoxon rank sum test, Fig. 2A, and Fig. S1A) indicating that, like Gag, the modified forms of Pol and Nef are more immunogenic. In addition to single-positive antigen-specific T cells, DNA vaccination also induced dual-positive (IFN-γ+ TNFα+) Gag- and Env-specific responses (Fig. S2B). SIV-specific cells producing IL-2 were not found, and this may be related to IL-12 DNA co-administration (30). Phenotypic analysis of the IFN-γ producing cells (Fig. 2B) revealed that Env (either native or modified) induced primarily a CD8+ T cell response, whereas Gag induced primarily a CD4+ T cell response. The use of the modified antigens decreased the difference between CD4+ and CD8+ responses, since the modified Gag resulted in more CD8+ and the modified Env in proportionally more CD4+ responses. Further analysis of the nature of the cellular immune responses after EP4 (EP4wk2 to DOC) showed that the modified Gag induced higher number of cells with central memory (CM) markers (P = 0.0008, Wei-Johnson) as well as higher effector memory (EM) responses (P = 0.0084, Fig. 2B and Fig. S1B). The responses to native Env revealed higher numbers in both CM and EM subsets than the modified Env (Fig. 2B). The increased CM cells were due to both CD4+ and CD8+ fractions, whereas the EM increase represented primarily CD8+ cells and to a lesser extend CD4+ cells (Fig. 2B). Evaluation of the functionality of the IFN-γ+ cells (Fig. S3) showed that the majority of Env- and Gag-specific CD4+ or CD8+ T cells are also producing Granzyme B, thus DNA-only vaccination induced functional cytotoxic effector T cells.
Analysis of the plasma antibody responses showed induction of Env, Gag, and Nef binding antibodies (Ab) (Fig. 3A, mean titers, and Fig. S4, individual animals). In general, the peak responses were achieved after EP4, reaching reciprocal titers of 103 to 105. The responses induced by the native Env and the modified Gag were statistically higher than the corresponding responses in the opposite groups throughout the vaccination period. At the DOC, the titers of the two groups remained significantly different (P = 0.0016 for Env and P = 0.0034 for the Gag exact Wilcoxon rank sum test). The modified Nef induced slightly higher Ab titers at DOC, but this did not reach significance (P = 0.078).
We also measured neutralizing antibody (Nab) against a highly neutralization-sensitive lab-adapted stock of SIVmac251 and against SIVmac239 at two time points during vaccination period (EP4wk2 and DOC) (Fig. 3C). Interestingly, Nab to SIVmac251 induced by the native Env at EP4 week 2 were significantly higher (P = 0.0006 by the exact Wilcoxon rank sum test) and persisted up to the DOC. In contrast, none of the animals in the Modified group had Nab at the DOC. No Nab to SIVmac239 were found. Thus, vaccination with DNA expressing the native Env led to the development of higher antibody titers and more durable Nab responses against SIVmac251 (Fig. 3).
Antibody analysis after challenge showed strong and rapid anamnestic responses to Gag, Env, and Nef (Fig. 3B, mean values, and Fig. S5, individual animals). Upon challenge, the Env antibody titers increased at a faster rate enabling an earlier plateau for the Native group. Conversely, the Gag responses increased faster and reached higher levels in the Modified group. SIV Nef binding antibodies were also higher in animals immunized with modified Nef. Nab were monitored at weeks 2 and 8 post-challenge (PC) (Fig. 3C). All vaccinees showed anamnestic responses, whereas no difference between the vaccine groups was noted. The vaccinated animals reached a plateau at week 2 PC, whereas the control group reached comparable levels of Nab by week 8.
There were significant differences in cellular immune responses between the vaccinated animals and controls during the first period after challenge (0–8 weeks, Fig. S1A, lower panel). Cellular immune responses in the control animals were detected PC at week 2 (five of eight animals) and week 8 (all eight animals). The frequency of IFN-γ producing SIV-specific T cells in controls was lower than in the vaccinated macaques. The median values of total IFN-γ cells (sum of six antigens) at week 8 PC were 10,981 in the Modified group, 3,853 in the Native group, and 659 in the control group. The differences have significance levels P = 0.028 for Modified vs. Native, P = 0.0002 for Modified vs. control, and P = 0.0030 for Native vs. control. The cellular immune responses detected after challenge were not uniformly induced in the vaccinated animals. SIV-specific IFN-γ cells increased in the majority of vaccinated animals during the 0–8 week period PC. In several animals, the responses remained at the same levels or decreased, and these animals tended not to benefit from vaccination. TNFα responses were also induced upon challenge, but remained lower than the prechallenge levels in 15 of 16 animals (Fig. S2A). An interesting observation was that the IFN-γ and TNFα responses to antigens other than Gag and Env increased dramatically in several animals.
Protection against SIV most likely depends on several factors, therefore, identification of immunological correlates is complicated by the distinct contribution of different responses to virus containment. As an example, one of the best protected animals, #33441, had an increase in SIV-specific IFN-γ+ T cells PC, but had the lowest Ab titers and no increase in Nab (Fig. 3C, and Figs. S1A and S5). A striking difference between the two vaccine groups is on the level of humoral immune responses at the DOC. We also compared the breadth of immune responses PC by measuring the number of peptide pools recognized by T cells of individual animals to produce IFN-γ. There was a strong negative association of the number of positive peptide pools with peak viral load (P = 0.0035, DOC to week 8 PC, Jonckheere-Terpstra test for trend in ordered categories corrected for multiple comparisons), indicating that the breadth of T cell response may also contribute to the protection. Interestingly, protection from high viremia correlated with the preservation of central memory CD4+ cells during the early phase of infection. For example, the levels of CD4+ CM cells at week 8 PC in 16 vaccinated animals showed a negative correlation with median VL (weeks 8–32, Spearman correlation coefficient 0.52, P = 0.039).
Discussion
Our study demonstrates that DNA vaccination as prime and boost strategy provides powerful protection against a high dose challenge with pathogenic SIVmac251. In this report, we used two sets of SIV expression plasmids delivered by in vivo EP to induce potent immune responses able to greatly reduce both acute (1 log) and chronic (1.7 log) viremia upon rectal high dose SIVmac 251 challenge. This vaccination strategy expands our previous DNA-only vaccine study, where immunization via the intramuscular route was able to decrease viremia by 1 log both in the acute and the chronic phase (P = 0.01) (2), and demonstrates that high levels of both cellular (range of 0.3% to 5.5% of peripheral T cells at peak) and humoral immune responses (103 to 105 Ab titers for different proteins) can be achieved by IM DNA EP, even with the low DNA doses used here (100 μg DNA plasmid/antigen and 400 μg in the last vaccination).
Studies by other investigators using the same animal model (Indian rhesus macaque challenged with SIVmac251 or SIVmac239) and IM DNA delivery, either reported no significant protection or only transient protection during the acute phase (3, 31). In fact, very few studies report the use of DNA-only vaccination and challenge with a high dose pathogenic SIVmac251/239, whereas the majority of reported data in macaques used in addition other vaccine modalities as boost. Some of these prime/boost studies reported no decrease in viremia, or decrease during the acute phase only (25, 32, 33), and few studies reported decrease in the chronic phase (7–10, 34). Here, we demonstrate that the DNA vaccine delivered by EP was able to achieve protection similar to other robust vaccination methods tested in this macaque model, including DNA prime/recombinant virus boost, rAd combinations, and replicating Ad with protein boost (6–13, 34). DNA vaccination showed clear evidence of protection in the majority of vaccinated animals (10 of 16, including six of eight in the Native and four of eight in the Modified group).
An additional objective of this challenge study was to compare immunogenicity of modified and native antigens included in the corresponding groups. We previously reported increased immunogenicity using mixtures of native and modified (fusion to MCP-3 and catenin) antigens in rhesus macaques (2). DNA vaccination studies in mice and macaques showed increased humoral and cellular immune responses by MCP3-Gag compared to the native Gag (2). LAMP-gag fusion produced strong cellular and humoral immunity in macaques (35, 36). Direct comparison in macaques using plasmids producing the native HIV-1 Gag versus LAMP-Gag showed similar levels of T cell responses but with significantly different phenotypes (induction of mainly CD4+ CM T cell responses by native Gag; induction of both CM and EM CD4+ and CD8+ T cell responses by the LAMP/gag chimera) (36). Thus, antigen modification can significantly alter immune responses.
Head-to-head comparisons in this study showed that the native Env and the modified Gag, Pol, and Nef-Tat-Vif fusion gave the highest immune responses. As a group, the animals vaccinated with the Native antigen vectors showed the best protection. A major difference between the two groups was the level of cellular and humoral Env responses, which were significantly higher in the Native group at the DOC, including Nab against a neutralization-sensitive stock of lab-adapted SIVmac251. Because the antibodies failed to neutralize a virus stock (SIVmac239) that more closely resembles the neutralization-sensitivity of the challenge virus, the contribution of Nabs in the protection described here remains uncertain. Although virus-specific T cell responses may have been more important than Nabs, we cannot rule-out other antibody mediated antiviral effector functions, such as ADCC and ADCVI (37). Since Gag is also an important target providing protection (12), it is critical to continue the evaluation of the modified Gag vectors, which showed dramatic increase in both cellular and humoral immune responses. Although fewer vaccinated animals in the Modified group (four of eight) benefited strongly compared to the Native group (six of eight), the four responders were solidly protected from high viremia during the chronic phase, which may be a reflection of the strong and broad cellular immune responses in this group. Requirements for protection are probably different during the acute and chronic phases of infection. Our results suggest that the higher Env antibody titers in the Native group may have provided an early advantage, but the strong cellular responses are also important for chronic virus containment. On the basis of this analysis, a vaccine containing the native Env and modified Gag, Pol, and Nef vectors may induce maximal desired immune responses. In conclusion, although no clear correlates of protection have emerged, our results show that DNA-only vaccination is an important vaccine modality able to provide protection in a strict macaque model and should be a high priority for further studies. DNA EP is feasible in humans and several protocols are presently being approved for clinical trials. Our results suggest methods to improve DNA vaccination by improving both the expression and delivery of optimized DNA vectors.
Materials and Methods
DNA Vectors.
Several plasmids encoding SIV antigens included in the vaccine mixes have been described, and include modified antigens fused to: MCP-3 chemokine, promoting secretion and attraction of antigen presenting cells; a catenin (CATE)-derived peptide for increased proteasomal degradation; and the lysosomal associated protein, LAMP1 for targeting the MHC II compartment. Plasmids encoding the native myristoylated p57gag (pCMVgagDX, 1S), a secreted p39gag MCP3-p39gag fusion (21S), or a rapidly degraded fusion of Gag to a catenin peptide (2S) (2, 38, 39); the Env expression plasmids producing the native protein (99S) or the MCP3-env (73S) fusion (2, 39) have been described. Optimized plasmids expressing a mutated, inactive Pol (88S) and a Nef-Tat-Vif (84S) fusion protein or fusions of these two antigens to LAMP, generating LAMP-Pol (103S) and LAMP-NTV (143S) (2) were selected after testing for immunogenicity in mice. The vaccine mixtures also contained the rhesus IL-12 plasmid (AG3) as a molecular adjuvant (40).
Immunization and Challenge.
Naive Indian rhesus macaques (Macaca mulatta) were housed and handled at the California National Primate Center, University of California, Davis, in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care International. Screening for 10 MHC class I alleles was performed by PCR (D. Watkins, Wisconsin Regional Primate Center) (Table S1). Each group had animals with the following “protective” haplotypes reported to affect viremia after challenge with certain SIVmac stocks (25–28): Control group (n = 11): Two Mamu-A*01, one Mamu-B*17, and one Mamu-A*01/B*17; Native group (n = 8): Two Mamu-A*01, one Mamu-B*08, and one Mamu-B*17; and Modified group (n = 8): Two Mamu-A*01 and one Mamu-B*17 animals. In vivo EP using the CELLECTRA adaptive constant-current electroporator (VGX Pharmaceuticals) was performed by injection of highly purified endotoxin-free DNA (Qiagen) IM (0.5 mL/injection) into the left and right thighs followed by EP as described (33). For EP1-EP3, a low DNA dose (100 μg plasmid/SIV antigen and 200 μg IL-12 plasmid, total of 600 μg in 1 mL water) was used at weeks 0, 11, and 19. At week 32, the animals received a fourth DNA dose (EP4) with a total of 2 mg DNA (400 μg each SIV antigen and 400 μg IL-12 plasmid in 1 mL water). The gag DNA for the Modified group contained an equimolar mixture of CATE-gag and MCP3-gag. All animals were challenged intrarectally with a 1:50 dilution of a pathogenic SIVmac251 virus stock used previously (2). The stock contains 5 × 103 TCID50/mL and 100 AID/mL by intrarectal titration. It was prepared by R. Desrosiers (Harvard Medical School, Boston) and became available through the National Institute of Allergy and Infectious Diseases (NIAID, contract N01-Al-60305). This stock has infected all naive animals at 1:10 and 1:50 dilutions in several studies. Analysis of the stock and of the transmitted viruses by single genome amplification (SGA) revealed that infected macaques harbored many different transmitted viruses of varied sequence, demonstrating that this is a high multiplicity stock.
Analysis of Antigen-Specific Immunity.
All analyses were performed using frozen cells. Viable cells were cultured at 1.5 × 106 PBMC/mL in complete RPMI in the presence or absence of pools of overlapping peptides (15 aa peptides overlapping by 11 aa) derived from SIVmac239 Gag, Env, Pol, Nef, Tat, and Vif. Vif responses were only monitored PC. Cells were cultured for 12 h with monensin (Golgi Stop; BD PharMingen) to inhibit cytokine secretion. Immunostaining and flow cytometric analysis was performed as described (38). To determine cytotoxic T cells, staining was performed using the following labeled antibodies: CD3-APCCy7, CD4-PerCPCy5.5 and CD8-AF405, IFN-γ-PECy7 (BD PharMingen), and Granzyme B-PE (Abcam).
Binding antibodies against SIV Env, Gag, and Nef were measured by ELISA using serial dilutions of the samples. The mean absorbance (A450) plus three standard deviations of non-immune rhesus macaque plasma was subtracted from the obtained values. Neutralizing antibodies against SIVmac251-TCLA (H9 cell-grown) and SIVmac239CS.23 (293T pseudovirus) were measured in M7-Luc cells and TZM-bl cells, respectively (41).
Viral Load Measurement.
SIV RNA copy numbers were determined by a real-time nucleic acid sequence-based isothermal amplification (NASBA) assay using SIVmac251-specific primers (42) with a threshold of detection of 50 copies/mL.
Supplementary Material
Acknowledgments.
We thank D.E. Weiss, J. Treece, E.-M. Lee, and I. Kalisz from the Advanced BioScience Laboratories, Inc., Kensington, MD, and K. Goncharova from Computer and Statistical Services, Frederick, MD, for assistance and T. Jones for editorial assistance. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Part of this work was supported by the grant RR-00169 from the National Center for Research Resources, National Institutes of Health, to the California National Primate Research Center.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902628106/DCSupplemental.
References
- 1.Kutzler MA, Weiner DB. DNA vaccines: Ready for prime time? Nat Rev Genet. 2008;9:776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosati M, et al. DNA vaccines expressing different forms of simian immunodeficiency virus antigens decrease viremia upon SIVmac251 challenge. J Virol. 2005;79:8480–8492. doi: 10.1128/JVI.79.13.8480-8492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lena P, et al. Co-immunization of rhesus macaques with plasmid vectors expressing IFN-gamma, GM-CSF, and SIV antigens enhances anti-viral humoral immunity but does not affect viremia after challenge with highly pathogenic virus. Vaccine. 2002;20(Suppl 4):A69–A79. doi: 10.1016/s0264-410x(02)00391-2. [DOI] [PubMed] [Google Scholar]
- 4.Haigwood NL, et al. Protection from pathogenic SIV challenge using multigenic DNA vaccines. Immunol Lett. 1999;66:183–188. doi: 10.1016/s0165-2478(98)00156-4. [DOI] [PubMed] [Google Scholar]
- 5.Fuller DH, et al. Induction of mucosal protection against primary, heterologous simian immunodeficiency virus by a DNA vaccine. J Virol. 2002;76:3309–3317. doi: 10.1128/JVI.76.7.3309-3317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hel Z, et al. Containment of simian immunodeficiency virus infection in vaccinated macaques: Correlation with the magnitude of virus-specific pre- and postchallenge CD4(+) and CD8(+) T cell responses. J Immunol. 2002;169:4778–4787. doi: 10.4049/jimmunol.169.9.4778. [DOI] [PubMed] [Google Scholar]
- 7.Casimiro DR, et al. Attenuation of simian immunodeficiency virus SIVmac239 infection by prophylactic immunization with DNA and recombinant adenoviral vaccine vectors expressing Gag. J Virol. 2005;79:15547–15555. doi: 10.1128/JVI.79.24.15547-15555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson NA, et al. Vaccine-induced cellular immune responses reduce plasma viral concentrations after repeated low-dose challenge with pathogenic simian immunodeficiency virus SIVmac239. J Virol. 2006;80:5875–5885. doi: 10.1128/JVI.00171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suh YS, et al. Reduction of viral loads by multigenic DNA priming and adenovirus boosting in the SIVmac-macaque model. Vaccine. 2006;24:1811–1820. doi: 10.1016/j.vaccine.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Kaur A, et al. Ability of herpes simplex virus vectors to boost immune responses to DNA vectors and to protect against challenge by simian immunodeficiency virus. Virology. 2007;357:199–214. doi: 10.1016/j.virol.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letvin NL, et al. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006;312:1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, et al. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature. 2008;457:87–91. doi: 10.1038/nature07469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Q, et al. Comparative evaluation of oral and intranasal priming with replication-competent adenovirus 5 host range mutant (Ad5hr)-simian immunodeficiency virus (SIV) recombinant vaccines on immunogenicity and protective efficacy against SIV(mac251) Vaccine. 2007;25:8021–8035. doi: 10.1016/j.vaccine.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 14.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tavel JA, et al. Safety and immunogenicity of a Gag-Pol candidate HIV-1 DNA vaccine administered by a needle-free device in HIV-1-seronegative subjects. J Acquir Immune Defic Syndr. 2007;44:601–605. doi: 10.1097/QAI.0b013e3180417cb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham BS, et al. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 DNA candidate vaccine. J Infect Dis. 2006;194:1650–1660. doi: 10.1086/509259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bodles-Brakhop AM, Heller R, Draghia-Akli R. Electroporation for the delivery of DNA-based vaccines and immunotherapeutics: Current clinical developments. Mol Ther. 2009;17:585–592. doi: 10.1038/mt.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu S. Immunogenicity of DNA vaccines in humans: It takes two to tango. Hum Vaccin. 2008;4:449–452. doi: 10.4161/hv.4.6.6179. [DOI] [PubMed] [Google Scholar]
- 19.Rosati M, et al. Increased immune responses in rhesus macaques by DNA vaccination combined with electroporation. Vaccine. 2008;26:5223–5229. doi: 10.1016/j.vaccine.2008.03.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luckay A, et al. Effect of plasmid DNA vaccine design and in vivo electroporation on the resulting vaccine-specific immune responses in rhesus macaques. J Virol. 2007;81:5257–5269. doi: 10.1128/JVI.00055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirao LA, et al. Combined effects of IL-12 and electroporation enhances the potency of DNA vaccination in macaques. Vaccine. 2008;26:3112–3120. doi: 10.1016/j.vaccine.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otten G, et al. Enhancement of DNA vaccine potency in rhesus macaques by electroporation. Vaccine. 2004;22:2489–2493. doi: 10.1016/j.vaccine.2003.11.073. [DOI] [PubMed] [Google Scholar]
- 23.Otten GR, et al. Potent immunogenicity of an HIV-1 gag-pol fusion DNA vaccine delivered by in vivo electroporation. Vaccine. 2006;24:4503–4509. doi: 10.1016/j.vaccine.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Zur Megede J, et al. A therapeutic SIV DNA vaccine elicits T-cell immune responses, but no sustained control of viremia in SIVmac239-infected rhesus Macaques. AIDS Res Hum Retroviruses. 2008;24:1103–1116. doi: 10.1089/aid.2008.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pal R, et al. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J Virol. 2002;76:292–302. doi: 10.1128/JVI.76.1.292-302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loffredo JT, et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mothe BR, et al. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2003;77:2736–2740. doi: 10.1128/JVI.77.4.2736-2740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yant LJ, et al. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patterson LJ, et al. Protection against mucosal simian immunodeficiency virus SIV(mac251) challenge by using replicating adenovirus-SIV multigene vaccine priming and subunit boosting. J Virol. 2004;78:2212–2221. doi: 10.1128/JVI.78.5.2212-2221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halwani R, et al. Therapeutic vaccination with simian immunodeficiency virus (SIV)-DNA+IL-12 or IL-15 induces distinct CD8 memory subsets in SIV-infected macaques. J Immunol. 2008;180:7969–7979. doi: 10.4049/jimmunol.180.12.7969. [DOI] [PubMed] [Google Scholar]
- 31.Allen TM, et al. Tat-vaccinated macaques do not control simian immunodeficiency virus SIVmac239 replication. J Virol. 2002;76:4108–4112. doi: 10.1128/JVI.76.8.4108-4112.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel TU, et al. Multispecific vaccine-induced mucosal cytotoxic T lymphocytes reduce acute-phase viral replication but fail in long-term control of simian immunodeficiency virus SIVmac239. J Virol. 2003;77:13348–13360. doi: 10.1128/JVI.77.24.13348-13360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horton H, et al. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J Virol. 2002;76:7187–7202. doi: 10.1128/JVI.76.14.7187-7202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hel Z, et al. Improved vaccine protection from simian AIDS by the addition of nonstructural simian immunodeficiency virus genes. J Immunol. 2006;176:85–96. doi: 10.4049/jimmunol.176.1.85. [DOI] [PubMed] [Google Scholar]
- 35.Chikhlikar P, et al. DNA encoding an HIV-1 Gag/human lysosome-associated membrane protein-1 chimera elicits a broad cellular and humoral immune response in Rhesus macaques. PLoS ONE. 2006;1:e135. doi: 10.1371/journal.pone.0000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valentin A, et al. Comparison of DNA vaccines producing HIV-1 gag and LAMP/gag chimera in rhesus macaques reveals antigen-specific T-cell responses with distinct phenotypes. Vaccine. 2009;27:4840–4849. doi: 10.1016/j.vaccine.2009.05.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forthal DN, et al. Rhesus macaque polyclonal and monoclonal antibodies inhibit simian immunodeficiency virus in the presence of human or autologous rhesus effector cells. J Virol. 2006;80:9217–9225. doi: 10.1128/JVI.02746-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosati M, et al. Increased immune responses in rhesus macaques by DNA vaccination combined with electroporation. Vaccine. 2008;26:5223–5229. doi: 10.1016/j.vaccine.2008.03.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Gegerfelt AS, et al. Long-lasting decrease in viremia in macaques chronically infected with simian immunodeficiency virus SIVmac251 after therapeutic DNA immunization. J Virol. 2007;81:1972–1979. doi: 10.1128/JVI.01990-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schadeck EB, et al. Plasmid encoded IL-12 functions as a DNA vaccine adjuvant and augments SIVgag-specific cell-mediated and humoral immune responses in Rhesus macaques. Vaccine. 2006;24:4677–4687. doi: 10.1016/j.vaccine.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 41.Montefiori D. Evaluating neutralizing antibodies against HIV, SIV and SHIV in a luciferase reporter gene assay. In: Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, Coico R, editors. Current Protocols in Immunology. New York, NY: John Wiley & Sons; 2004. pp. 12.11.11–12.11.15. [Google Scholar]
- 42.Romano JW, et al. Quantitative evaluation of simian immunodeficiency virus infection using NASBA technology. J Virol Methods. 2000;86:61–70. doi: 10.1016/s0166-0934(99)00184-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.