Abstract
Chemokines adopt a conserved tertiary structure stabilized by two disulfide bridges and direct the migration of leukocytes. Lymphotactin (Ltn) is a unique chemokine in that it contains only one disulfide and exhibits large-scale structural heterogeneity. Under physiological solution conditions (37°C, 150 mM NaCl) Ltn is in equilibrium between the canonical chemokine fold (Ltn10) and a distinct 4-stranded β-sheet (Ltn40). Consequently, it has not been possible to address the biological significance of each structural species independently. To stabilize the Ltn10 structure independent of specific solution conditions, Ltn variants containing a second disulfide bridge were designed. Placement of the new cysteines was based on a sequence alignment of Ltn with either the first (Ltn-CC1) or third disulfide (Ltn-CC3) in the CC chemokine, HCC-2. NMR data demonstrate that both CC1 and CC3 retain the Ltn10 chemokine structure and no longer exhibit structural rearrangement. The ability of each mutant to activate the Ltn receptor, XCR1, has been tested using an intracellular Ca2+ flux assay. These data support the conclusion that the chemokine fold of Ltn10 is responsible for receptor activation. We also examined the role of amino- and carboxyl-terminal residues in Ltn-mediated receptor activation. In contrast to previous reports, we find that the 25 residues comprising the novel C-terminal extension do not participate in receptor activation, while the native N-terminus is absolutely required for Ltn function.
Chemokines are small, secreted proteins that signal leukocytes to migrate during an immune response. Binding of chemokines to G-protein coupled receptors (GPCRs)1 stimulates cell chemotaxis, while diffusion and glycosaminoglycan (GAG) binding are believed to establish the chemokine gradient that migrating cells follow. Approximately 50 chemokines have been identified and are classified into four subfamilies (CXC, CC, CX3C, and C) based on the spacing of two cysteine residues near the N-terminus, each of which contributes to a pair of conserved disulfide bonds (1).
Lymphotactin (Ltn), the only example of a C-type chemokine, lacks the first and third cysteine residues found in all other chemokines and contains a unique extended C-terminal sequence that is conserved across species (2). We have previously shown that under physiological conditions (37 °C, 150 mM NaCl), Ltn exhibits reversible conformational heterogeneity, converting between two distinct structural species. The relative population of the two conformations is highly dependent on the temperature and ionic strength of the solution (3). At 10°C in the presence of 200 mM NaCl, Ltn is a monomer that adopts the conserved chemokine fold (Ltn10) (4). However, at 40 °C in the absence of salt it switches to a dimeric 4-stranded β-sheet structure (Ltn40) completely distinct from other chemokines. Rearrangement to the Ltn40 structure involves unfolding of the canonical chemokine structure and assembly of a completely different secondary structure hydrogen bonding network (3).
The receptor for Ltn, XCR1, has been cloned (5) and in vitro assays for XCR1 activation have been described (5, 6). While other chemokines display a strict requirement for specific residues at the N-terminus for receptor activation, similar studies on Ltn yielded ambiguous results (2, 7–9). In contrast, multiple reports describe an absolute functional requirement for the C-terminal extension of Ltn (8, 10). Thus, it seems likely that Ltn binds and activates XCR1 in a novel manner relative to other chemokines.
Our overall objective is to correlate the dual structures of lymphotactin with functional chemokine activities like glycosaminoglycan binding and receptor activation. Although we can shift the conformational equilibrium such that either the Ltn10 or Ltn40 species predominates, it is technically challenging to conduct either in vitro or in vivo assays under such conditions. Moreover, the alternative conformation may not be completely absent, even at extremes of temperature and ionic strength. Variants of Ltn restricted to only one conformation would therefore be useful for examining the biological activity of each structural species.
To this end, we constructed two Ltn variants that incorporate a second disulfide bond designed to prevent structural interconversion. Each protein retains the canonical chemokine structure observed for wild-type Ltn at 10 °C in 200 mM NaCl, and neither protein exhibits the conformational heterogeneity displayed by wild-type Ltn, even at elevated temperatures. The 3D structure of one mutant has been determined by NMR. Functional studies indicate that the Ltn10 conformation is competent for receptor activation but requires a native N-terminus, while the novel C-terminal extension is entirely dispensable, contrary to previous reports. By restricting Ltn to a single conformation, we can begin to define the biological function of each structural species and the in vivo relevance of their unique interconversion.
EXPERIMENTAL PROCEDURES
Protein expression, purification and mutagenesis
Recombinant human lymphotactin was expressed using a modified pQE30 vector (Qiagen), purified by metal-affinity chromatography, and cleaved with TEV protease to remove the hexahistidine affinity tag as previously described (11). Purity, identity, and molecular weight were verified by matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) and NMR. To introduce cysteine substitutions or create amino-terminal variants Ltn(G−1VGSE), Ltn(G1GSE) and Ltn(A1GSE), site-directed mutagenesis was performed using pairs of complementary primers and the QuikChange kit (Stratagene). To truncate Ltn at specific residues, Ltn was PCR amplified from the pQE30 vector using a 5’ primer containing a StyI restriction site and a 3’ primer containing a HindIII restriction site at the appropriate truncation position. PCR products were then cut with StyI and HindIII, gel purified, and ligated back into the pQE30 vector. All expression vectors were verified by DNA sequencing. Purified Ltn was frozen, lyophilized, and stored at −20°C.
Generation of native Ltn(1–93) by cyanogen bromide cleavage
To accommodate proteolytic removal of the hexahistidine affinity tag, the Ltn expression construct described above lacks the N-terminal Val residue and the resulting protein therefore corresponds to Ltn(2–93). To generate an Ltn protein that retains the initial valine, codons for the tripeptide Gly-Met-Val were inserted between the TEV protease cleavage site and the Ltn sequence, and the amino acid substitutions M63V and M72A were introduced into the same expression vector. Recombinant Ltn was then expressed and purified as described resulting in Ltn containing the N-terminal sequence G−2M−1V1GSE. To remove the G-M dipeptide, lyophilized protein was resuspended in either 80% TFA or 6 M guanidine/0.1 N HCl followed by an overnight treatment at room temperature with cyanogen bromide (CNBr). Complete cleavage of the G-M dipeptide was verified by MALDI-MS analysis. Residual CNBr and solvent components were removed from the protein sample by lyophilization followed by reversed-phase HPLC as described by Peterson et al (11).
NMR spectroscopy
NMR experiments were performed on a Bruker DRX 600 equipped with a 1H/15N/13C Cyroprobe© or a conventional 1H/15N/13C probe equipped with three axis gradients. NMR samples contained 90% H20, 10% D20, and 0.02% NaN3, with 20 mM sodium phosphate at pH 6.0. Temperature and NaCl concentration were varied as specified in the results. Complete 1H, 15N, and 13C resonance assignments for CC3 (20 mM sodium phosphate, pH 6.0) were obtained at 25°C using the following experiments: 15N-1H HSQC (12), 3D SE HNCA (13, 14), 3D SE HNCO (13, 15), 3D SE HN(CO)CA (13), 3D SE HNCACB, 3D SE HNCACO, 3D SE HCCONH, 3D 15N SE TOCSY-HSQC (16), 3D SE C(CO)NH (17), 3D HCCH TOCSY (18), and 2D 13C constant time HSQC (19). Heteronuclear 15N-1H NOE values were determined from an interleaved pair of two-dimensional gradient sensitivity-enhanced correlation spectra of [U-15N]-CC3 Ltn acquired with and without a 5 s proton saturation period (20). NMR data were processed with NMRPipe (21), and XEASY (22) was used for resonance assignments and analysis of NOE spectra.
Structure Calculation and Analysis
A total of 1234 unique, non-trivial NOE distance constraints were obtained from 3D 15N-edited NOESY-HQSC (23), 13C(aromatic)-edited NOESY-HSQC, and 13C(aliphatic)-edited NOESY-HSQC (18) spectra (τmix = 80 ms). A total of 82 backbone and dihedral angle constraints (for residues 11–66) were generated from secondary shifts of the 1Hα, 13Cα, 13Cβ, 13C’, and 15N nuclei shifts using the program TALOS (24). Structures were generated in an automated manner using the NOEASSIGN module of the torsion angle dynamics program CYANA 2.1 (25), which produced an ensemble with high precision and low residual constraint violations that required minimal manual refinements. The 20 CYANA conformers with the lowest target function were subjected to a molecular dynamics protocol in explicit solvent (26) using XPLOR-NIH (27). Completely disordered residues 76–93 were not included in water refinement calculations, thus the ensemble of CC3 NMR structures deposited in the PDB contains only residues 1–75.
In vitro Calcium flux assay for XCR1 activation
HEK 293 cells stably expressing XCR1 (6) were maintained in DMEM media containing 10% heat-inactivated FBS, 2 mM glutamine, 1% penicillin-streptomycin, and 500 µg/mL G418. Cells were plated at an initial density of 5 × 04 cells/mL in T-75 flasks from frozen stocks. At confluency, cells were harvested using enzyme-free cell dissociation solution (Specialty Media), resuspended in culture media and allowed to rest for 2 hours under incubator conditions. After incubation, cells were centrifuged and washed twice in EBSSH buffer (26 mM HEPES, 125 mM NaCl, 5.6 mM glucose, 5 mM KCl, 2 mM CaCl, 1 mM MgSO4, 1 mM NaH2PO4, 0.2% BSA, pH 7.4) and resuspended to 4 × 106 cells/mL in 10 mL EBSSH containing 2.5 mM probenecid and 50 µg Fluo-3-AM (Molecular Probes). After loading for 1 hour at room temperature, cells were washed twice with EBSSH containing probenecid and aliquoted at a density of 1.5 × 106 cells/mL into eppendorf tubes. Immediately before assay, aliquots were spun down and resuspended in probenecid free EBSSH buffer at a density of 106 cells/mL and transferred to a 3 mL fluorescent cuvette. Lymphotactin-induced calcium flux was monitored at 30°C using a PTI spectrofluorometer with Peltier temperature control at 505 nm excitation and 525 nm emission wavelengths. After fluorescence signal returned to baseline, carbachol (1mM) was added to serve as an internal standard. Data for dose response curves are reported as the ratio of peak height for chemokine sample versus peak height of carbachol control as shown in equation 1:
| (1) |
RESULTS
Ltn undergoes a reversible conformational change
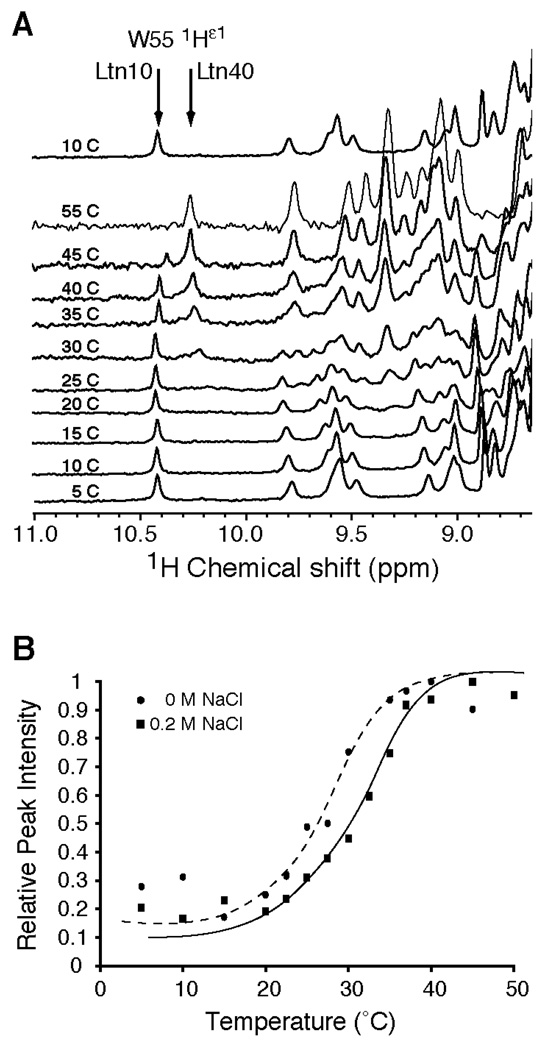
In addition to containing a single disulfide bond and extended C-terminus, Ltn is a unique chemokine that exhibits structural heterogeneity at physiological solution conditions (150 mM NaCl, 37 °C) (3). Rearrangement from the canonical chemokine structure exhibited by Ltn at 10°C (Ltn10) to a novel structure (Ltn40) can be directly observed in the 1D-1H NMR spectra by monitoring the chemical shift of the indole 1Hε1 resonance of W55 as a function of temperature (Figure 1A). In Ltn10, W55 is part of the C-terminal α-helix buried within the hydrophobic core, corresponding to a chemical shift of 10.4 ppm. Unfolding of the helix accompanies the structural rearrangement to Ltn40, shifting the resonance of 1Hε1 to 10.2 ppm as W55 becomes solvent exposed. These signal intensities reflect the abundance of each species in solution. The relative 1Hε1 peak height at 10.2 ppm (Ltn40) as a function of temperature and salt concentration is shown in Figure 1B. In the presence of 200 mM NaCl the midpoint transition temperature is higher than in the absence of salt, demonstrating the ionic stabilization of the Ltn10 structure as previously observed by intrinsic tryptophan fluorescence (3). Three other aspects are apparent from the 1D 1H NMR temperature series. First, the conformational rearrangement between Ltn10 and Ltn40 is a two–step process, with no evidence of stable intermediates. Secondly, the Ltn10 to Ltn40 rearrangement is freely reversible and is not affected by repeated temperature titrations. Third, and most importantly, at conditions where most biological assays are performed (25 to 37°C), there would exist a significant concentration of both structural species, as the equilibrium constant of the Ltn40 versus Ltn10 species is approximately 1 at 30°C in the presence of salt.
Figure 1.
Ltn exhibits a reversible structural rearrangement between two distinct structural species. A. 1D-1H NMR spectra of wild-type Ltn (200 µM in 20 mM sodium phosphate, 200mM NaCl, pH 6.0) were acquired at various temperatures. During the Ltn10 to Ltn40 transition, Trp-55 moves from a buried position in the hydrophobic core of Ltn10 to a solvent exposed, unstructured region of Ltn40, which reflected in the chemical shift value for the Nε-1H from 10.2 ppm to 10.4 ppm for the Ltn10 and Ltn40 structures, respectively. The fractional population of Ltn10 and Ltn40 species can be assessed from the intensity of the 1H-Nε peak at 10.2 versus 10.4 ppm. To demonstrate the reversibility of the conformational rearrangement, a spectrum reacquired at 10 °C following the temperature titration is illustrated after the 55 °C spectrum. B. A plot of 1H-Nε peak intensity at 10.4 ppm versus temperature was conducted using wild-type Ltn in the absence (closed circles and dashed trendline) or presence (closed squares and solid trendline) of 200mM NaCl. Peak intensities have been normalized to the maximal intensity obtained after conversion to Ltn40 species. Trendlines were visually fit to the data to represent the shift in relative peak intensity after inclusion of salt to the protein sample.
Generation of Ltn10-restricted Ltn proteins, CC1 and CC3
Ltn is a unique chemokine that contains only one of two conserved disulfide bonds. It is the first disulfide, located between the N-terminal region and 30s loop (loop connecting β-strands 1 and 2) that is specifically missing in Ltn. To test the hypothesis that addition of a second disulfide bridge would stabilize the Ltn10 conformation, we designed two Ltn mutants with cysteine substitutions at positions corresponding to two different disulfide bonds in HCC-2 (Figure 2A). HCC-2 contains three disulfide bonds, two of which correspond to the typical pair found in all CC chemokines, while the unique third disulfide tethers the N-loop to the C-terminal helix (28). Based on a sequence alignment between Ltn and HCC-2 (sequence identity 36%), the CC1 sequence was engineered to include a single residue substitution in the N-terminus, T10C, and an insertion that places an Ala-Cys dipeptide between Ltn residues G32 and S33, located within the 30s loop. It was our hypothesis that introduction of this missing disulfide back into Ltn (Ltn-CC1) may restrict Ltn to the Ltn10 conformation. However, we also considered the possibility that Ltn purposefully lacks this first disulfide as interactions or movements within the 30s-loop are critical for biological function. Therefore, we also wished to introduce the additional disulfide at a site that did not restrict the mobility of the 30s loop, yet was also incompatible with the Ltn40 structure. This was accomplished by introducing a disulfide, modeled after the third disulfide in HCC-2, between the 310 helix and α-helix (Ltn-CC3).
Figure 2.
Introduction of a second disulfide bond in Ltn stabilizes the Ltn10 structure. A. Placement of a second disulfide bond was based on alignment of Ltn with the CC chemokine, HCC-2. Construction of Ltn-CC1 (T10C/iG32ACS33) is based on alignment of the first disulfide in HCC-2, while the position of cysteine substitutions in Ltn-CC3 (V21C/V59C) are based on the unusual third disulfide present in HCC-2. B. 1H-15N HSQC spectra acquired for CC1 and CC3 at 40°C are compared with those taken for wild-type Ltn at 10, 37, and 40°C. Spectra for wild-type protein at 10 and 37°C were acquired in 20mM sodium phosphate, pH 6.0 containing 200 mM NaCl. Spectra of all other protein samples were acquired using 20mM sodium phosphate, pH 6.0 in the absence of salt. HSQC spectra of CC1 and CC3 Ltn resemble the patterns observed for Ltn10. Neither mutant displays evidence for a mixture of the two structural species as observed for the wild-type protein at 37°C. The 1HN peak for Val56 is boxed in the WT spectra acquired at 10°C and the CC1 and CC3 mutants. Owing to ring-current effects of W55, the proton chemical shift for Val56 is significantly upfield of other backbone amide protons, and is indicative of the conserved chemokine fold. C. Amide chemical shifts for the CC1 and CC3 mutants are plotted against the corresponding values for wild-type Ltn at 10 and 40 °C. The excellent correlation of the Ltn10 chemical shifts versus the disulfide-stabilized mutants as compared to the poor correlation with the Ltn40 chemical shifts, indicate that CC1 and CC3 adopt the chemokine-like fold. Outliers in each plot are labeled and correspond to residues at or adjacent to the substituted position in the amino acid sequence.
The second variant, CC3, was designed by aligning the NMR structures of Ltn10 (4) and HCC-2 (28) to identify the Ltn residues nearest to the third and sixth cysteine residues of HCC-2. By inspecting the overlaid structures, it was clear that V59 in Ltn occupies the position filled by one of the HCC-2 cysteines, and that the other cysteine residue corresponded to either P20 or V21 in Ltn. Because P20 is highly conserved in the chemokine family (including HCC-2) and might play an important structural role, we introduced V59C and V21C substitutions into Ltn to create the CC3 variant.
Both Ltn disulfide variants were isotopically labeled with 15N, purified and evaluated by 2D NMR to determine if the proteins exhibited characteristics of the Ltn10 fold (Figure 2B). At both 10 and 40 °C, each protein displayed an HSQC spectrum similar to that of Ltn10, including two characteristic upfield shifted resonances. In the chemokine fold, a number of hydrophobic interactions involving a conserved aromatic residue position the helix with respect to the β-sheet. Ring-current effects from W55 produce unusual 1HN chemical shifts for L19 and V56 at 5.32 and 5.91 ppm in Ltn10, and very similar values were observed for the same residues in CC1 and CC3. Because similarities between HSQC spectra are not always readily apparent, we compared chemical shift values of CC1 and CC3 with the corresponding values for Ltn10 and Ltn40 (Figure 2C). Strong correlations are observed for CC1 and CC3 with the Ltn10 chemical shifts but not with the Ltn40 assignments, indicating that both Ltn disulfide variants adopt the Ltn10 conformation.
We also compared 1D 1H spectra of CC1 and CC3 at various temperatures (Figure 2D) with wild-type Ltn. In contrast to wild-type Ltn (Figure 1A), the W55 1Hε1 signal for CC3 did not shift from 10.4 ppm to 10.2 ppm with increasing temperature (Figure 2D). Even in the absence of salt (a condition which destabilizes the Ltn10 structure in wild-type Ltn), we found no evidence of conformational heterogeneity or Ltn40 species formation as temperature was increased. Similar results were obtained for CC1 (data not shown). Thus, CC1 and CC3 maintain the Ltn10 conformation and do not interconvert with the Ltn40 species, even under conditions that normally favor Ltn40.
CC3 adopts the Ltn10 chemokine-like fold
We solved the structure of CC3 by NMR spectroscopy at 25°C. The final ensemble of 20 conformers (PDB entry 2HDM) is shown in Figure 3A, and a summary of the experimental restraints and structural statistics is provided in Table 1. Backbone r.m.s.d. (Figure 3B) and 15N-1H heteronuclear NOE (Figure 3C) values confirm that residues 1–10 and 69–93 are dynamically disordered in solution, as observed with the Ltn10 structure (4). As expected, CC3 adopts the chemokine fold and aligns closely with the Ltn10 NMR structure determined using wild-type protein at 10 °C in 200 mM NaCl (Figure 3D). The Cα r.m.s.d. between the Ltn10 and CC3 structures is 1.55 Å (residues 15–64). The additional disulfide in CC3 links the 310 loop (C21) and the C-terminal α-helix (C59), preventing CC3 from rearranging to the Ltn40 structure. The presence of a disulfide bond between C21 and C59 is supported by NOEs between the 1Hβ protons of C21 and 1Hα of V59.
Figure 3.
Solution structure for Ltn-CC3, demonstrates Ltn-CC3 adopts the Ltn10 fold. A. Ensemble of 20 conformers for CC3 (residues 9–75) is shown in Cα trace and rotated 180°about the y-axis. Disulfides are highlighted in yellow. B. Backbone Cα r.m.s.d. values and C. 1H-15N heteronuclear NOEs are plotted for each residue of CC3. D. Overlay of wild-type Ltn10 (cyan) and CC3 (magenta) structures (residues 9–68), rotated 90°about the y-axis. The r.s.m.d. difference for Cα trace is 1.55 Å between Ltn10 and CC3 for residues 15–64.
Table 1.
Structural statistics for Ltn CC3
| Experimental constraints | |
| Non-redundant distance constraints | |
| Long | 330 |
| Medium [1<(i−j)≤5] | 153 |
| Sequential [(i−j)=1] | 333 |
| Intraresidue [i=1] | 418 |
| Total | 1234 |
| Dihedral angle constraints (ϕandψ) | 82 |
| Average atomic R.M.S.D. to the mean structure (Å) | |
| Residues 13–66 | |
| Backbone (N, Cα, C’) | 0.52 ± 0.07 |
| Heavy atoms | 1.05 ± 0.11 |
| Deviations from idealized covalent geometrya | |
| Bond length RMSD (Å) | 0.016 |
| Torsion angle RMSD (°) | 1.2 |
| WHATCHECK quality indicators | |
| Z-score | −2.42 ± 0.27 |
| RMS Z-score | |
| Bond lengths | 0.82 ± 0.02 |
| Bond angles | 0.67 ± 0.03 |
| Bumps | 0 ± 0 |
| Lennard-Jones Energy (kJ/mol)b | −1343 ± 63 |
| Constraint violations | |
| NOE distance violations > 0.5 Åc | 0 ± 0 |
| NOE distance RMSD (Å) | 0.020 ± 0.002 |
| Torsion angle violations >5°d | 0 ± 0 |
| Torsion angle RMSD (°) | 0.657 ± 0.110 |
| Ramachandran statistics (% of residues 1–75) | |
| Most favored | 85.30 ± 4.10 |
| Additionally favored | 12.06 ± 3.70 |
| Generously allowed | 1.33 ± 1.64 |
| Disallowed | 1.32 ± 1.15 |
Final X-PLOR force constants were 250 (bonds), 250 (angles), 300 (impropers), 100 (chirality) and 100 (omega), 50 (NOE constraints), and 200 (torsion angle constraints).
Nonbonded energy was calculated in XPLOR-NIH.
The largest NOE violation in the ensemble of structures was 0.34 Å.
The largest torsion angle violation in the ensemble of structures was 4.7°.
The native Ltn N-terminus is required for XCR1 activation
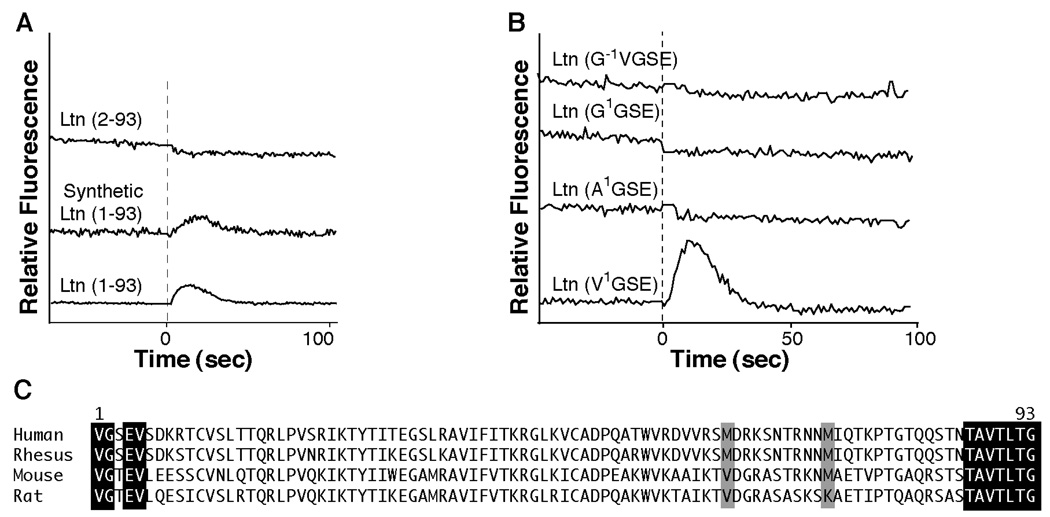
To assess biological activity of the Ltn10-restricted proteins, we monitored intracellular Ca2+ levels in HEK293 cells expressing the XCR1 receptor. Based on previous report (7) that a Ltn protein lacking the amino terminal valine residue is biologically active, our expression system was designed to yield residues 2–93 of the mature Ltn sequence (G2SE- rather than V1GSE-), after cleavage of the His8-affinity tag by TEV protease (11). However, this Ltn(2–93) protein failed to induce a Ca2+ flux response, while Ltn(1–93) from a commercial source and recombinant Ltn(1–93) used in our previous structural studies (4) both induced significant Ca2+-flux responses (Figure 4A). Therefore, before testing Ltn-CC1 and Ltn-CC3 for their ability to activate XCR1, we wished to confirm the importance of a native N-terminus to XCR1 agonist activity, producing a series of Ltn variants in which the N-terminal valine was substituted by alanine (A1GSE-) or glycine (G1GSE-), or prepended by an additional glycine (G−1V1GSE-). None of these alternative N-terminal variants gave a positive Ca2+ flux response as compared with the V1GSE-Ltn(1–93) controls (Figure 4B). Based on these results, Ltn displays a strict requirement for a native N-terminus for receptor activation.
Figure 4.
Ltn requires a native N-terminus for activation of XCR1. A. Comparison of Ca2+ flux response using Ltn(1–93) obtained from a commercial source (R&D Systems) or factor Xa recombinant Ltn versus His8-TEV recombinant Ltn(2–93). Addition of Ltn is indicated with dashed line. All protein concentrations were 100 nM. B. Ca2+ flux response of recombinant Ltn proteins containing either a +1 N-terminal sequence (G−1V1GSE-Ltn) or a substitution of the N-terminal valine with either a glycine or alanine are compared with native N-terminal, Ltn(1–93) obtained from cyanogen bromide cleavage of recombinant protein. All protein concentrations are 500 nM. C. Alignment of Ltn protein sequences from various species indicates that the N-terminus exhibits more sequence variability compared to the C-terminus of Ltn orthologs. To accommodate CnBr treatment, methionine residues at position 63 and 72 were substituted based on sequence alignments with other Ltn proteins. Neither methionine is absolutely conserved, therefore, an M63V and M72A double mutant was generated.
For high-level production of Ltn proteins with a native N-terminus, we developed a modified system employing cyanogen bromide cleavage. Since CNBr specifically cleaves the amide bond after methionine residues, we modified our expression system to include a methionine residue between the TEV protease cleavage site and the Ltn N-terminal valine. Human Ltn contains two internal methionine residues at position 63 and 72, neither of which is strictly conserved among mammalian orthologs (Figure 4C). Methionine 63 is located in the α-helix but does not make significant structural contacts and was replaced with valine, while M72 in the unstructured C-terminal tail was substituted by alanine. After purification and digestion with TEV protease, the residual G-M dipeptide was removed by CNBr treatment to yield the native V1GSE N-terminus. The Ltn(M63V/M72A) double mutant behaved identically to the wild-type Ltn(1–93) protein as determined by 15N-1H HSQC and Ca2+-flux measurements, suggesting that the methionine substitutions induce no significant structural or functional changes.
The chemokine fold of Ltn10 is responsible for XCR1 activation
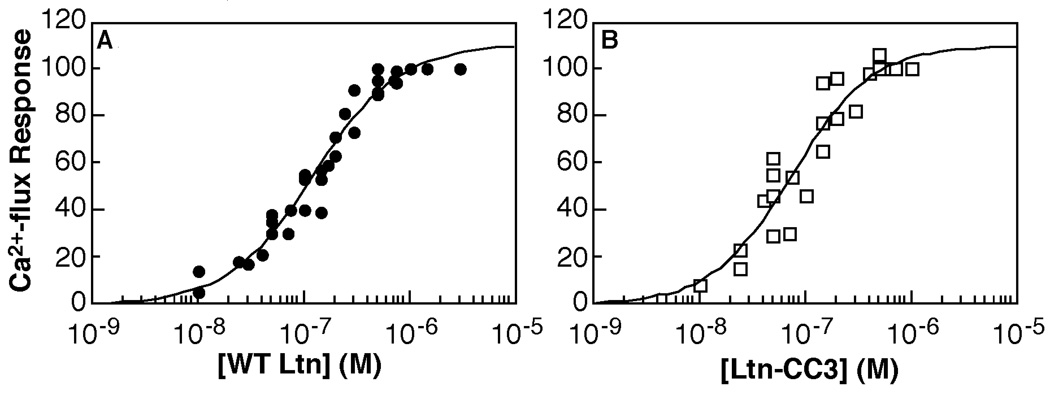
To test the competence of the CC1 and CC3 variants for receptor activation as compared with wild-type Ltn, the same cysteine substitutions were introduced into the Ltn(M63V/M72A) background. The Ltn(M63V/M72A)-CC1 and Ltn(M63V/M72A)-CC3 proteins were purified using the CNBr cleavage protocol. Both proteins induced Ca2+-flux responses similar to that obtained for Ltn(M63V/M72A) protein, which is reflected in the EC50 values and Hill coefficients determined for Ltn(M63V/M72A)-CC3 (75.8 ± 17.4 nM; Hill slope = 1.1 ± 0.3) and Ltn(M63V/M72A) (128.1 ± 17.0 nM; Hill slope = 1.1 ± 0.1) (Figure 5).
Figure 5.
Ltn-CC3 is an XCR1 agonist. CC3 (panel B) activates XCR1 with a similar EC50 value (75.8 ± 17.4 nM) as WT (panel A) (EC50 = 128.1 ± 17.0 nM) by in vitro Ca2+ flux assay. Both proteins consist of residues 1–93 and contain the M63V/M72A substitutions. Complete data from three independent data sets for wild-type and two independent data sets for CC3 were fit to determine EC50 values. Data were normalized to carbachol control as described in experimental procedures and plotted as a percent of maximum response.
The disordered Ltn C-terminus is not required for XCR1 activation
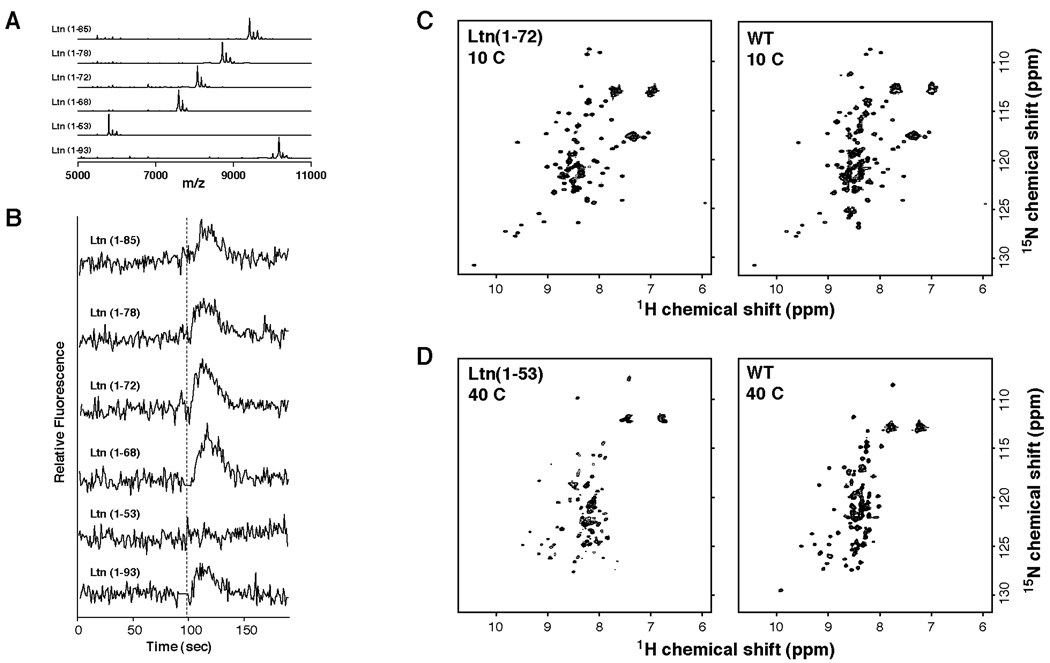
Previous reports ascribed a functional significance to the extended C-terminal Ltn sequence (10), but our structural studies on Ltn10 and CC3 showed that residues 70–93 are dynamically disordered in free solution (4). Therefore, we investigated the role of specific C-terminal residues in activating XCR1 by truncating Ltn after residues 53, 68, 72, 78, and 85 and assayed for Ca2+-flux activity. All C-terminal truncations were constructed on the M63V/M72A background. Purified proteins were analyzed by MALDI-MS to confirm purity and molecular weight for each truncated protein (Table 2, Figure 6A). Aside from Ltn(1–53), all of the C-terminally truncated proteins produced a Ca2+ flux response comparable to wild-type Ltn (Figure 6B). Ltn(1–72), which includes residues of the α-helix, adopts the Ltn10 structure at 10°C in 200mM NaCl based on HSQC comparisons (Figure 6C). The only C-terminal truncation that failed to induce Ca2+-flux, Ltn(1–53), lacks the C-terminal α-helix (residues 54–68) and cannot form the Ltn10 structure based on 15N-1H HSQC analysis (data not shown). Strikingly, instead of unfolding completely it adopts the Ltn40 structure (Figure 6D).
Table 2.
Calculated and observed molecular weights of full-length and C-terminally truncation Ltn proteins.
| Protein | Expected MWa (Da) |
Observed MWb (Da) |
|---|---|---|
| Ltn(1–93) | 10179 | 10175 |
| Ltn(1–53) | 5794 | 5797 |
| Ltn(1–68) | 7565 | 7595 |
| Ltn(1–72) | 8051 | 8079 |
| Ltn(1–78) | 8718 | 8718 |
| Ltn(1–85) | 9421 | 9418 |
Expected molecular weights were calculated from primary sequence using the ProtParam tool on the ExPASy proteomics server (http://ca.expasy.org).
Observed molecular weights were determined from MALDI-MS analysis of purified proteins (Figure 6A).
Figure 6.
The unstructured dynamically disordered C-terminus of Ltn is not required for activation of XCR1. A. Recombinant Ltn proteins containing C-terminal truncations were constructed on the Ltn(M63V/M72A) background. Protein purity and expected molecular weights for each truncation mutant were verified by MALDI-MS. B. Recombinant Ltn proteins containing C-terminal truncations were tested for their ability to induce Ca2+ flux response. Addition of chemokine is indicated with dashed line. All protein concentrations were 200nM. Only Ltn(1–53), which can not adopt the Ltn10 structure did not activate the Ltn receptor. C. Comparison of 1H-15N HSQC spectra for wild-type Ltn and Ltn(1–72) proteins obtained at 10°C in 20mM sodium phosphate buffer, pH 6.0 containing 200 mM NaCl indicate that truncation of last 22 residues does not affect maintenance of Ltn10 structure. D. Comparison of 1H-15N HSQC spectra for wild-type Ltn and Ltn(1–53) proteins obtained at 40°C in 20mM sodium phosphate buffer, pH 6.0. While Ltn(1–53) does not obtain the chemokine fold, it does maintain some characteristics of the Ltn40 structure.
DISCUSSION
Roles of N-and C-terminal regions in receptor activation
Chemokines function through interactions with their cognate receptor by a proposed two-step mechanism; an initial binding event, involving the N-terminus of the GPCR and the chemokine N-loop, followed by activation of the receptor through binding of the chemokine N-terminus. Published reports disagree on the extent to which specific Ltn N-terminal residues are necessary for receptor activation (2, 7–9). In addition, truncation of the last 22 residues resulted in a Ltn protein that did not trigger either a Ca2+ flux response in XCR1-transfected cells (10), or a chemotactic response from human peripheral blood lymphocytes and mouse splenocytes (8). Only four of the first 10 residues of the Ltn N-terminus are conserved among the sequences from human, rhesus, rat and mouse, but the last 7 C-terminal residues are absolutely conserved (Figure 4C). Taken together, these observations prompted us to consider a variation in the two-step model for chemokine receptor activation to explain the functional requirement for the unusual C-terminal extension in Ltn. In this model, the conserved chemokine fold of Ltn10 may bind to an extracellular binding site on XCR1, but employ the conserved C-terminus for receptor activation in contrast to the N-terminus as observed in most other chemokines.
Our previous structural studies utilized Ltn samples generated with a bacterial expression system that preserved the native N-terminal sequence (V1GSE) and is functional in a Ca2+-flux assay for XCR1 activation, but suffered from non-specific proteolysis by the factor Xa enzyme, employed for removal of the affinity tag (3, 4). Based on previous reports that addition or removal of an amino acid residue at the N-terminus had no effect on Ltn activity (2, 7, 8), we developed an improved bacterial expression system that incorporates an N-terminal His8 tag and tobacco etch virus (TEV) protease cleavage site (11). The TEV protease recognition sequence consists of ENLYFQ/G (29), with cleavage occurring between the Q and G residues, and will tolerate certain amino acid substitutions, like serine or alanine, at the P1’ site (30). We substituted the P1’ amino acid with valine residue in order to produce native Ltn(1–93) but TEV digestion failed, and a cleavable Ltn(2–93) protein (corresponding to G2SE at the N-terminus) was inactive in Ca2+-flux assays at concentrations ranging from 0.1 – 1 µM. Ultimately, we reengineered the expression system to use TEV protease digestion prior to protein refolding, followed by CNBr cleavage to expose the native N-terminal sequence. This scheme required substitution of two internal methionine residues (M63V and M72A); these substitutions did not alter the structural and biochemical properties of Ltn. M63A/M72A-Ltn(1–93) produced using the cyanogen bromide protocol was indistinguishable by NMR spectroscopy and equally active in the Ca2+-flux assay as native Ltn(1–93).
We have examined the ability of four N-terminal variant Ltn proteins to activate XCR1. Only the full-length protein, Ltn(1–93) containing the native N-terminal sequence (V1GSE) was competent in eliciting a Ca2+ flux. Although we have not examined every possible amino acid substitution, our data suggests a strong preference for valine as the N-terminal residue. These results are in contrast to previous studies that have used either +1 or −1 N-terminal variants. For instance, Kelner et al. (2) observed a positive Ca2+ flux and chemotactic response in CD4+-depleted, CD8+-enriched thymocytes using recombinant mouse Ltn(2–93), while two independent studies have reported +1 N-terminal variants of recombinant Ltn were functional (8, 9).
Our results clarify the role of the Ltn N-terminus in XCR1 activation, and show that additions, substitutions, or deletions abrogate its XCR1 agonist activity, similar to other chemokines. It should be noted that other investigators have reported negative chemotaxis and Ca2+ flux results even with wild-type Ltn(1–93) proteins. These results have been attributed to clonal differences in lymphocytes obtained from primary cultures. For instance, Dorner et al. (7) observed differential response of CD4+ and CD8+ T-cells taken from blood donors when either chemically synthesized Ltn(1–93) or naturally purified protein preparations of Ltn were used in parallel. Hedrick et al. (8) examined several human NK clones for their ability to respond to synthetic Ltn(1–93) in a chemotaxis assay. While some clones were found to respond very well, others showed little or no response to same protein preparation. They speculated that the varying ability of human NK clones to respond to Ltn may reflect differences in receptor level expression level or the relative mobility of particular clones. Alternatively, since these clones are maintained in culture by constant restimulation, they postulated the differential response may depend on how recently each clone had been stimulated.
In contrast with previous reports (8, 10), data presented here indicate that the C-terminus of Ltn is not required for in vitro activation of XCR1. Except for Ltn(1–53), a series of C-terminally truncated Ltn variants all elicited a Ca2+-flux response. However, the structure of Ltn(1–53) is significantly altered, since residues of the C-terminal α-helix have been eliminated. Structural studies of Ltn10 indicate that the protein is disordered beyond residue 69 (4). Therefore, it is not unexpected that truncations did not significantly alter either the 15N-1H HSQC peak pattern or the ability of Ltn(1–72) to exhibit conformational exchange. In addition, the Ltn(1–68), Ltn(1–72), Ltn(1–78), and Ltn(1–85) truncations were also prepared on the CC3 background to generate Ltn10-restricted C-terminal truncations and each of these generated a Ca2+ flux. It is not clear why other studies did not observe a functional response using Ltn(1–72) protein, especially given that Marcaurelle et al. (10) also reported that truncation of the last 22 residues did not alter the 15N-1H HSQC spectrum. Although it is possible that interactions with receptor may induce secondary structure formation in C-terminal residues, they are not necessary for in vitro activation. It is interesting to note that Ltn from chicken does not contain the extended Cterminus, and the recombinant protein is biologically active (31). Without further studies, we cannot rule out that the C-terminus provides regulatory functions or a protein-protein interaction surface that may be critical for in vivo function.
Restriction of protein conformation by site-directed mutagenesis
Ltn is the only chemokine to exhibit a dramatic conformational duality involving two entirely distinct tertiary structures. We hypothesized that introduction of a second disulfide bridge analogous to those found in other chemokine structures would stabilize the Ltn10 conformation and prevent interconversion with the novel Ltn40 species. We introduced cysteine residues corresponding either to the first disulfide found in all other chemokines or to the third disulfide seen in HCC-2 and selectively stabilized the Ltn10 conformation. The additional disulfide prevents interconversion between the Ltn10 and Ltn40 species because the new covalent crosslink is incompatible with the β-sheet configuration of Ltn40. Therefore, the unique ability of Ltn to undergo structural rearrangement may be a simple consequence of the lack of a second conserved disulfide.
It is noteworthy that the Ltn(1–53) species failed to induce any Ca2+ flux response, and that this protein does not adopt the Ltn10 structure, but instead displays an 15N-1H HSQC spectrum very similar to Ltn40 (Figure 6D). These data suggest that the Ltn40 structure is unlikely to function as an XCR1 agonist. A remaining question is what, if any, biological function(s) can be ascribed to the Ltn40 structure. Chemokines function through dual interactions with their cognate receptor(s) and extracellular glycosaminoglycans. Thus it is possible that the Ltn40 structure provides an essential high-affinity GAG binding site for Ltn in vivo.
By eliminating the Ltn40 structure we demonstrated that the canonical chemokine fold adopted by the Ltn10 structure is sufficient for receptor activation in vitro and the resulting EC50 value for Ltn-CC3 (76 µM) is similar to that of the WT protein (128 µM). As shown in Figure 1B, the relative concentration of Ltn10 is ~70% of the total WT Ltn under conditions corresponding to the Ca2+ flux assay (25 °C, 150 mM NaCl). After correcting for the fraction of WT Ltn present as Ltn40, the EC50 values for Ltn-CC3 and Ltn10 are indistinguishable, consistent with the hypothesis that the Ltn10 species is solely responsible for XCR1 activation by Ltn.
Although the additional disulfide in CC3 is not located at the same position as the first disulfide bridge found in all other chemokines (connecting the N-terminus and the β1–β2, or '30s', loop), CC3 remained functional in the XCR1 activation assay. This engineered Ltn variant will be a useful tool for analysis of structure-activity relationships. For instance, in comparison with other chemokines, Ltn exhibits a more flexible 30s loop. Because the 30s loop is unaltered in CC3, we will be able to determine whether those residues participate in XCR1 binding or activation. Future studies will focus on the use of these conformationally-restricted mutants to examine if a reversible structural rearrangement is a prerequisite to Ltn function in vivo, and what specific elements of Ltn10 structure are responsible for receptor binding and/or activation.
ACKNOWLEDGEMENTS
Cyanogen bromide cleavage, DNA sequencing, and MALDI-MS services were provided by the Protein and Nucleic Acid Facility (Medical College of Wisconsin). HEK293/XCR1 cells were a generous gift of Schering-Plough. We thank Dr. Christoph Seibert (Rockefeller University) for assistance with calcium flux assays.
This work was supported by NIH grants AI45843 and AI063325.
Footnotes
The abbreviations used are: GCPR, G-coupled protein receptor; GAG, glycosaminoglycan; Ltn, lymphotactin; MALDI-MS, matrix-assisted laser desorption ionization mass spectrometry; CNBr, cyanogen bromide; RANTES, regulated on activation normal T cell expressed and secreted; HCC-2, human chemokine 2; NK cells, natural killer cells.
REFERENCES
- 1.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 2.Kelner G, Kennedy J, Bacon K, Kleyensteuber S, Largaespada D, Jenkins N, Copeland N, Bazan J, Moore K, Schall T, Zlotnik A. Lymphotactin: a cytokine that represents a new class of chemokine. Science. 1994;266:1395–1399. doi: 10.1126/science.7973732. [DOI] [PubMed] [Google Scholar]
- 3.Kuloglu ES, McCaslin DR, Markley JL, Volkman BF. Structural rearrangement of human lymphotactin, a C chemokine, under physiological solution conditions. J Biol Chem. 2002;277:17863–17870. doi: 10.1074/jbc.M200402200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuloglu ES, McCaslin DR, Kitabwalla M, Pauza CD, Markley JL, Volkman BF. Monomeric Solution Structure of the Prototypical 'C' Chemokine Lymphotactin. Biochemistry. 2001;40:12486–12496. doi: 10.1021/bi011106p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida T, Imai T, Kakizaki M, Nishimura M, Takagi S, Yoshie O. Identification of single C motif-1/lymphotactin receptor XCR1. J Biol Chem. 1998;273:16551–16554. doi: 10.1074/jbc.273.26.16551. [DOI] [PubMed] [Google Scholar]
- 6.Shan L, Qiao X, Oldham E, Catron D, Kaminski H, Lundell D, Zlotnik A, Gustafson E, Hedrick JA. Identification of viral macrophage inflammatory protein (vMIP)-II as a ligand for GPR5/XCR1. Biochem Biophys Res Commun. 2000;268:938–941. doi: 10.1006/bbrc.2000.2235. [DOI] [PubMed] [Google Scholar]
- 7.Dorner B, Müller S, Entschladen F, Schröder JM, Franke P, Kraft R, Friedl P, Clark-Lewis I, Kroczek RA. Purification, structural analysis, and function of natural ATAC, a cytokine secreted by CD8+ cells. J. Biol. Chem. 1997;272:8817–8823. doi: 10.1074/jbc.272.13.8817. [DOI] [PubMed] [Google Scholar]
- 8.Hedrick JA, Saylor V, Figueroa D, Mizoue L, Xu Y, Menon S, Abrams J, Handel T, Zlotnik A. Lymphotactin is produced by NK cells and attracts both NK cells and T cells in vivo. J Immunol. 1997;158:1533–1540. [PubMed] [Google Scholar]
- 9.Kennedy J, Kelner G, Kleyensteuber S, Schall T, Weiss M, Yssel H, Schneider P, Cocks B, Bacon K, Zlotnik A. Molecular cloning and functional characterization of human lymphotactin. Journal of Immunology. 1995;155:203–209. [PubMed] [Google Scholar]
- 10.Marcaurelle LA, Mizoue LS, Wilken J, Oldham L, Kent SB, Handel TM, Bertozzi CR. Chemical synthesis of lymphotactin: a glycosylated chemokine with a C-terminal mucin-like domain. Chemistry. 2001;7:1129–1132. doi: 10.1002/1521-3765(20010302)7:5<1129::aid-chem1129>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 11.Peterson FC, Elgin ES, Nelson TJ, Zhang F, Hoeger TJ, Linhardt RJ, Volkman BF. Identification and characterization of a glycosaminoglycan recognition element of the C chemokine lymphotactin. J Biol Chem. 2004;279:12598–12604. doi: 10.1074/jbc.M311633200. [DOI] [PubMed] [Google Scholar]
- 12.Mori S, Abeygunawardana C, Johnson MO, van Zijl PCM. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. Ser. B. 1995;105:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 13.Grzesiek S, Bax A. Inproved 3D Triple-Resonance NMR Techniques Applied to a 31 kDa Protein. J. Magn. Reson. 1992;96:432–440. [Google Scholar]
- 14.Kay LE, Xu GY, Yamazaki T. Enhanced-Sensitivity Triple-Resonance Spectroscopy with Minimal H2O Saturation. J. Magn. Reson. Ser. A. 1994;109:129–133. [Google Scholar]
- 15.Muhandiram DR, Kay LE. Gradient-Enhanced Triple-esonance Three-Dimensional NMR Experiments with Improved Sensitivity. J. Magn. Reson. Ser. B. 1994;103:203–216. [Google Scholar]
- 16.Zhang O, Kay LE, Olivier JP, Forman-Kay JD. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 17.Grzesiek S, Anglister J, Bax A. Correlation of Backbone Amide and Aliphatic Side-Chain Resonances in 13C/15N-Enriched Proteins by Isotropic Mixing of 13C Magnetization. J. Magn. Reson. Ser. B. 1993;101:114–119. [Google Scholar]
- 18.Kay LE, Xu G-Y, Singer AU, Muhandiram DR, Forman-Kay JD. A Gradient-Enhanced HCCH-TOCSY Experiment for Recording Sidechain 1H and 13C Correlations in H2O Samples of Proteins. J. Magn. Reson. Ser. B. 1993;101:333–337. [Google Scholar]
- 19.Santoro J, King GC. A constant-Time 2D overbodenhausen experiment for inverse correlation of isotopically enriched species. J. Magn. Reson. 1992;97:202–207. [Google Scholar]
- 20.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and a phosphopeptide-complexed src homology 2 domain studeid by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 21.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 22.Bartels C, Xia T-H, Billeter M, Güntert P, Wüthrich K. The Program XEASY for Computer-Supported NMR Spectral Analysis of Biological Macromolecules. J. Biomol. NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 23.Talluri S, Wagner G. An optimized 3D NOESY-HSQC. J Magn Reson B. 1996;112:200–205. doi: 10.1006/jmrb.1996.0132. [DOI] [PubMed] [Google Scholar]
- 24.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 25.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 26.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 27.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 28.Sticht H, Escher SE, Schweimer K, Forssmann WG, Rosch P, Adermann K. Solution structure of the human CC chemokine 2: A monomeric representative of the CC chemokine subtype. Biochemistry. 1999;38:5995–6002. doi: 10.1021/bi990065i. [DOI] [PubMed] [Google Scholar]
- 29.Dougherty WG, Cary SM, Parks TD. Molecular genetic analysis of a plant virus polyprotein cleavage site: a model. Virology. 1989;171:356–364. doi: 10.1016/0042-6822(89)90603-x. [DOI] [PubMed] [Google Scholar]
- 30.Kapust RB, Tozser J, Copeland TD, Waugh DS. The P1' specificity of tobacco etch virus protease. Biochem Biophys Res Commun. 2002;294:949–955. doi: 10.1016/S0006-291X(02)00574-0. [DOI] [PubMed] [Google Scholar]
- 31.Rossi D, Sanchez-Garcia J, McCormack WT, Bazan JF, Zlotnik A. Identification of a chicken "C" chemokine related to lymphotactin. J Leukoc Biol. 1999;65:87–93. doi: 10.1002/jlb.65.1.87. [DOI] [PubMed] [Google Scholar]