Abstract
Purpose
Marked reactive stroma formation, designated as Grade 3 reactive stroma, is associated with poor outcome in clinically localized prostate cancer. In order to understand the biological processes and signaling mechanisms underlying the formation of such reactive stroma, we carried out microarray gene expression analysis of laser captured reactive stroma and matched normal stroma.
Experimental Design
Seventeen cases of reactive stroma grade 3 cancer were used to laser capture tumor and normal stroma. Expression analysis was carried out using Agiilent 44K arrays. Upregulation of selected genes was confirmed by quantitative RT-PCR. Expression data was analyzed to identify significantly up and down regulated genes and Gene Ontology analysis was used to define pathways altered in reactive stroma.
Results
A total of 544 unique genes were significantly higher in the reactive stroma and 606 unique genes were lower. Gene Ontology analysis revealed significant alterations in a number of novel processes in prostate cancer reactive stroma including neurogenesis, axonogenesis and the DNA damage/repair pathways as well as evidence of increases in stem cells in prostate cancer reactive stroma.
Conclusions
Formation of reactive stroma in prostate cancer is a dynamic process characterized by significant alterations in growth factor and signal transduction pathways and formation of new structures including nerves and axons.
Keywords: prostate cancer, microenvironment, neurogenesis, expression microarrays, pathology
INTRODUCTION
Prostate cancer (PCa) remains the most common malignancy affecting men and the third-leading cause of cancer-related death of men in the United States. It has been appreciated for many years that the tumor microenvironment plays an important role in the initiation and progression of prostate and other cancers (1–3). The tissues surrounding the cancer cells in PCa are distinct from the normal mesenchymal tissues of the prostate and consist of a mixture of fibroblasts, myofibroblasts, endothelial cells, immune cells, other cells and altered extracellular matrix. We have previously shown an increase in PCa stroma of cells with a myofibroblastic phenotype (4) that synthesize collagen I (4) and expressed tenascin, which is shown to be involved in modulation of cell growth and tumorigenesis. Using in vitro models, we demonstrated that TGF-β1 enhances the transformation of prostatic fibroblasts into the myofibroblastic phenotype (4). Other key processes previously identified in PCa stroma include angiogenesis (5) as well as infiltration of immune cells (3).
We recently examined the clinical implications of the histological variability of reactive stroma in prostate cancer (6, 7). While some prostate cancers have little histological alteration of the surrounding stroma compared to normal stroma in benign prostate, a subset of cancers reveals an obvious alteration in the surrounding stroma. Of note, we have shown men with tumors having the most profound histological alterations of reactive stroma, which is termed Grade 3 reactive stroma, had reduced biochemical recurrence-free survival in both tissue microarray (6) and biopsy studies (7). This predictive ability was independent of Gleason score and other pathologic parameters.
To understand the mechanisms by which reactive stroma can influence tumor behavior, we examined global changes in gene expression in Grade 3 reactive stroma relative to paired benign prostatic stroma using expression microarray analysis on laser captured RNAs from these two tissue types. By focusing on Grade 3 reactive stroma, which is associated with prostate cancer progression, we hope to indentify key changes in prostate reactive stroma that are associated with aggressive prostate cancer. Our analysis has revealed significant alterations in a number of novel pathways not previously identified in PCa reactive stroma including neurogenesis, axonogenesis and the DNA damage/repair pathways as well as evidence of a possible increase in stem cells in PCa reactive stroma.
MATERIALS and METHODS
Laser capture microdissection of stromal tissues
Laser capture microdissection (LCM) was performed using HistoGene LCM Frozen Section Staining Kit (Molecular Devices, Sunnyvale, CA). Briefly 10 um-thick frozen sections were cut from tissue cores of normal and cancerous area of fresh, unfixed prostatectomy specimen. All cancer tissues were from cases with Grade 3 reactive stroma as identified on paraffin-embedded whole mount sections and frozen sections were confirmed to have Grade 3 reactive stroma by a pathologist with experience in stromal grading (GA) prior to laser capture. Reactive stroma is less eosinophilic than normal stroma due to loss of smooth muscle and there is deposition of collagen fibrils and extracellular matrix. Reactive stroma has a higher cellularity than normal stroma, due to increased fibroblasts and myofibroblasts. The cells, which now resemble fibroblasts more than muscle cells, are characteristically disordered and show irregular length and thickness. A characteristic of reactive stroma is that these cells are arranged in an irregular fascicular pattern in a delicate fibrillary background. Grade 3 reactive stroma has at least 51% reactive stroma in the tumor section as described previously (7). Frozen sections were placed on non-charged glass slides and stored at −80° C before use. Slides were stained and dehydrated according the manufacturer protocol. LCM microdissection was performed using a Pixcell II LCM microscope (Molecular Devices), using a 15 um laser spot size, 0.75–0.90 ms laser pulse, and 45 mV beam power. Approximately 4000–8000 laser pulses of stroma between the glands in normal and prostate cancer tissues were captured on 2–3 caps (1500–2500 spots).
Amplification and expression microarray analysis
Total RNA was extracted from LCM-captured stroma using PicoPure RNA Isolation Kit (Molecular Devices) according to the manufacturer’s protocol. It was then amplified using RiboAmp RNA Amplification Kit (Molecular Devices). Two rounds of in vitro amplification were performed according to the manufacturer’s protocol. The quantity of amplified RNA was measured by spectrophotometer at OD 260 nm. Different starting amounts of control RNA were used to confirm linear RNA amplification. The cDNA reverse transcription and fluorescent labeling reactions were carried out using Invitrogen SuperScript Plus Direct cDNA Labeling System with Alexa Fluor aha-dUTPs (Invitrogen, Carlsbad, CA). Briefly 2.5 ug of aRNA and Universal Human Reference RNA (Stratagene La Jolla, CA) were labeled in reverse transcription with Alexa647 and Alexa555 aha-dUTPs respectively. After labeling, 2.5 ug of each aRNA was mixed with 2.5 ug of reference RNA and purified with SuperScript Plus Direct cDNA Labeling System purification module according to manufacturer’s instructions. Eluted sample was mixed with 10xBlocking Solution and 2xHiRPM buffer (Agilent Technologies, Palo Alto, CA), incubated for 2 min at 95–98° C and hybridized on 4x44K Whole Human Genome Oligo Microarray chip (Agilent Technologies, Santa Clara, CA) using SureHyb DNA Microarray Hybridization Chambers in DNA Microarray hybridization oven (Agilent Technologies) at 10rpm, 65°C for 20 hrs. After hybridization, slides were washed in Gene Expression Wash Buffer 1 and 2 for 1 min and than dried by centrifugation at 2000 rpm for 2 min. Microarrays were scanned with a dynamic autofocus microarray scanner (Agilent Microarray Scanner- G2565BA, Agilent Technologies,) using Agilent-provided parameters (Red and Green PMT were each set at 100%, and scan resolution was set to 5 um). The Feature Extraction Software v9.1.3.1 (Agilent Technologies) was used to extract and analyze the signals. To validate the amplification process amplified and unamplified RNAs were labeled and hybridized to an Agilent 21K oligonucleoide array.
Statistical analysis microarray expression data
Expression arrays were processed and loess-normalized using BioConductor (8). Array data has been deposited in the public Gene Expression Omnibus (GEO) data base, accession GSE11682. Arrays were processed in batches at different times (with an equal number of cancer and normal samples in each batch); to correct for batch effects, the estimated expression values within each batch were transformed to standard deviations from the mean (the results when using this method being very similar to what was observed using the alternative “Combat” algorithm (9)). Two-sided t-tests determined significant differences in mean gene mRNA levels between groups. False Discovery Rate (FDR) was estimated both by the method of Storey and Tibshirani (10) and by 100 permutations of the profile group labels. Expression values were visualized as color maps using the Cluster (11) and Java TreeView (12) software. Gene Ontology (GO) annotation terms were searched within gene sets essentially as described previously (13). Protein interaction network analysis was carried out essentially as described previously (14). The entire set of protein-protein interactions catalogued in the Human Protein Reference Database1 was obtained in September 2007. Interactions involving the same gene product interacting with itself were excluded from the analysis. Graphical visualization of the interaction network was generated using the Cytoscape software (15).
Quantitative RT-PCR
Real-time PCR was carried out in sealed 96-well microtiter plates (BioRad, Hercules, CA) with Absolute Blue QPCR SYBR Green Fluorescein Mix (Thermo Fisher Scientific Inc, Rochester, NY) and Bio-Rad I-cycler QPCR Machine (Bio-Rad,). All 17 tumor and normal stromal RNAs were analyzed in triplicate. Primers used are summarized in Supplementary Table S1. The PCR conditions were 20 sec denaturation, 30 sec annealing at the primer’s optimal temperature and 30 sec extension at 72° C for 40 cycles. cDNA from Universal Human Reference RNA (Stratagene) which was serially diluted from 1:5 to 1:10000 was used to generate standard curves. The relative copy number of transcript for each gene was normalized by transcript level of the housekeeping gene (hprt1) in each sample. Each PCR experiment was carried out in triplicate. Student t-test was performed to validate statistically significant differences. Data with P<0.05 in normal vs. reactive stroma comparison were considered significant.
Immunohistochemistry
Immunohistochemistry was performed on a small tissue microarrays with 10 reactive stroma grade 3 cancers and matched normal tissues essentially as described previously (16). For ERK1 immunohistochemistry antigen retrieval was carried out with 0.1 M Tris, pH 9.0 for 25 min with steam heat. Anti-ERK1 antibody (Cat# 32537, Abcam, Cambridge, MA) was used at 1:500 dilution for 30 minutes at room temperature. For anti-GSK-3β immunohistochemistry, antigen retrieval was performed in10 mM citrate buffer, pH 6.0, for 20 minutes in steam heat. Anti-GSK-3β antibody (Cat# 9332, Cell Signaling Technology, Beverly, MA), was used at 1:60 dilution overnight at 4°C. Antigen retrieval for c-KIT immunohistochemistry was performed in Reveal Decloaker Buffer (Biocare Medical, Concord, CA) in a pressure cooker for 10 min. Anti-c-Kit antibody (Cat #A4502, Dako, Carpinteira, CA), was used at 1:100 dilution for 30 min at room temperature.
Analysis of breast stroma microarray expression data
We obtained the expression profile dataset from Finak et al (17) of breast cancer stroma and normal stroma (GEO Accession GSE9014, dye swap profiles not used); for duplicate profiles, log-transformed ratios were averaged. Log ratios of each profile were scale normalized across the ~45K transcript probes to median value of zero. Sample E1527 appeared an obvious outlier, and so was removed from the analysis. Mapping between the prostate and breast stroma profile datasets was done using the Agilent transcript probe identifier. For heat map representation of the breast stroma data, the expression values for each gene were centered on the mean centroid between the normal and cancer groups. For each sample, the mean centroid was first defined without the given sample; the genes in the left out profile were then centered on this mean centroid (in this way, when using the average values to classify cancer from normal in Figure 4, the labeling of the classified sample itself played no role in the prediction).
Figure 4. Expression patterns in breast cancer reactive stroma for genes over-expressed in prostate cancer reactive stroma.

Referring to the profiling dataset from Finak et al.(15) of normal breast stroma (n=6) and breast cancer reactive stroma (n=52), the 776 transcript probes represented in that dataset and showing over-expression in Grade 3 prostate cancer reactive stroma (Figure 2A) were examined. Top panel, gene expression heat map (genes ordered from high to low expression in reactive stroma). Bottom panel, average expression of the (mean centroid-centered) genes for the corresponding sample profiles. Unique genes (those with a common name) showing over-expression in both prostate and breast reactive stroma in indicated region (P<0.01 each dataset) are listed.
RESULTS
Laser capture microdissection and expression array analysis of Grade 3 reactive stroma in prostate cancer
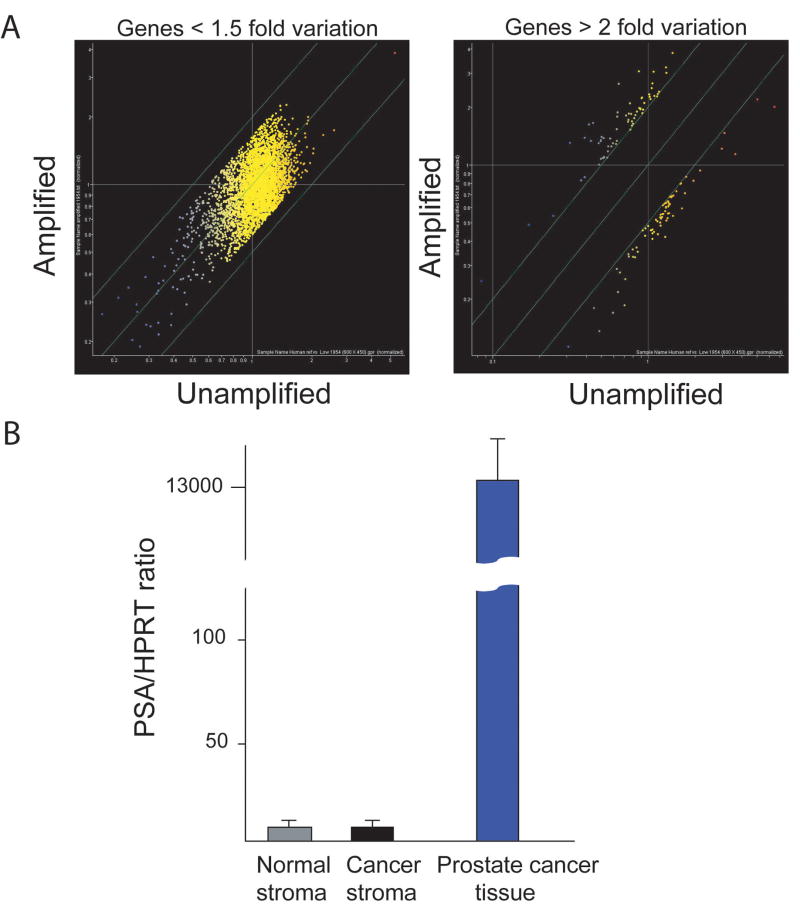
In order to evaluate global gene expression changes in Grade 3 reactive stroma versus normal peripheral zone stroma we performed laser capture microdissection and expression array analysis of stroma from 17 Grade 3 reactive stroma prostate cancer tissues and matched normal peripheral zone tissues. Captured RNAs were amplified and hybridized to Agilent 60- mer oligonucleotide 4x44K Whole Human Genome expression microarrays, which represent approximately 41000 unique genes and transcripts. To confirm that our amplification protocol is not introducing significant bias we amplified a prostate cancer RNA and then labeled and hybridized the RNA to a 21K oligonucleotide array and compared the expression results to unamplified RNA (Figure 1A). After normalization and filtering for the signals with low intensity, 88.7% of the genes showed no more than 1.5 fold difference. In another way of comparison, only 3.04% (140/4,602) of the genes changed their expression levels by 2-fold or more after amplification similar to the results of other groups (18) Of note, none of the 140 genes showed a reversal of relative expression i.e. increased to decreased. In all cases the difference was based on fold change in the same direction i.e. 3-fold Vs 6 fold. Thus, while there may be some variation introduced by amplification, this is unlikely to significantly impact our overall analysis.
Figure 1. Validation of RNA amplification and laser capture.
(A) Genes with no greater than 1.5-fold variation between amplified and unamplified RNA; 88.7% of genes met this criteria (left); genes with greater than 2-fold variation between amplified and unamplified RNAs (right), which constitute only 3% of the total.
(B) Quantitative RT-PCR of PSA expression in laser captured normal and tumor stroma and prostate cancer tissue. PSA content of normal and tumor stroma was not significantly different and both were 1600-fold less than undissected prostate cancer tissue. Mean +/− of triplicate determinations is shown. PSA copy number was normalized to HPRT copy number relative to the Universal Reference RNA.
To assess the purity of the captured stromal RNAs we carried out quantitative RT-PCR for both PSA and keratin-18, two epithelial markers expressed in both normal epithelium and prostate cancer, indicating some epithelial contamination of the stroma was present. Both were barely detectable. We were concerned that the intimate admixture of cancer epithelial cells and stroma might lead to increased contamination with epithelial cells in the cancer stroma samples when compared to normal stroma. However, there was no significant difference in the mean PSA and keratin-18 mRNA levels between cancer stroma and normal stroma RNAs (p<0.96 PSA and p<0.26 keratin 18, t-test). When compared to prostate cancer tissue, expression of PSA was more than 1600-fold higher in the tissue compared to the stroma samples (Figure 1B), indicating that there was minimal epithelial contamination of the laser captured stromal tissues.
We next analyzed the expression of ~19075 genes giving detectable signals in the 17 Grade 3 stroma RNAs and matched normal peripheral zone stromal samples using expression microarrays (Figure 2A). A total of 1675 probes representing 1141 unique genes were differentially expressed at p<0.01; 544 unique genes were higher in the reactive stroma, and 606 unique genes were lower (9 genes in the 544-set were represented in the 606-set by different probes). The 1675 significant probes significantly exceed the number of probes that would be expected to be differentially expressed by chance alone at this p-value (~450 probes by probability calculation, 382 by permutation testing). Thus there are significant differences in gene expression between Grade 3 reactive stroma and normal prostate stroma. One obvious histological and immunohistochemical characteristic of Grade 3 reactive stroma is the decreased amounts of differentiated smooth muscle in reactive stroma (see Supplementary Fig 1). Consistent with this, we noted that tropomyosins (multiple probes of each) and caveolin-1 (multiple probes), which are characteristically expressed in differentiated smooth muscle, were all significantly downregulated in Grade 3 reactive stroma.
Figure 2. A gene transcription signature of Grade 3 reactive stroma in prostate cancer.

(A) Heat map representation of 1675 gene probes (representing 1141 unique named genes) differentially expressed (P<0.01) in reactive prostate stroma compared to normal stroma (rows: genes, columns: profiled samples, yellow: high expression). (B) Quantitative RT-PCR validation of selected genes from the list of 1141 (P<0.05 for each, t-test). Mean +/− standard deviation of triplicate determinations is shown. HPRT was used for copy number normalization.
Validation of expression microarray results by quantitative RT-PCR
To validate the results of our expression microarrays we next carried out quantitative RT-PCR analysis of the RNAs used in the expression microarray studies. We analyzed a total of 7 genes that were upregulated in cancer stroma (FOXA1, TGFBR2, DVL1, ERBB2, GSK3B, SEMA4F and GLI2) and two that were downregulated (CAV1 and TPM1). As shown in Figure 2B, in all cases we were able to confirm statistically significant up or downregulation (p<0.05, t-test), as predicted from the microarrays, validating the accuracy of our microarray expression data.
Gene Ontology analysis of expression microarray data
To develop an objective analysis of key processes altered in the Grade 3 reactive stroma we carried out Gene Ontology (GO) analysis of the 1141 genes that were significantly altered. Analysis of the 544 unique genes high in reactive stroma revealed a total 66 GO annotation terms that were enriched (p<0.01, one-sided Fisher’s exact test) in this gene list in comparison to all the genes on the array and of these 46 were represented by at least 10 genes on the array. All 2309 GO terms representing at least 10 genes on the array were considered, and a number of these GO terms could conceivably have been nominally significant by multiple testing. Nevertheless, simulation testing using 1000 random gene sets yielded on average only about one-fourth the number of enriched GO terms (p<0.01) that were observed with the actual gene set (Supplementary Figure 2) indicating that the genes are involved in specific processes and not random. To further increase the stringency, only GO terms that were at least 2-fold enriched relative to all genes were considered, leaving 33 GO terms. These are shown in Table 1, grouped functionally, and component genes are shown in Supplementary Table S2. Some of these gene class associations as represented by GO terms are as would be predicted, while others are unexpected. The largest and most interesting is the 8 neural related GO terms. These primarily relate to neurogenesis, axonogenesis, synaptogenesis and neural biogenesis. This large cluster of GO terms indicates that active neurogenesis and axonogenesis is occurring in Grade 3 reactive stroma. While angiogenesis is a well known biological process in tumors, the role of axonogenesis and neurogenesis in cancer stroma is relatively unexplored. Another surprising cluster was a set of 4 GO terms related to DNA damage checkpoint response. This finding implies that some factor in the overall tumor microenvironment, perhaps increased reactive oxygen species, is leading to DNA damage in tumor stromal cells. Genomic lesions have been reported in tumor stroma (19), which is consistent with increased DNA damage in the tumor microenvironment. Other enriched terms, such as development and morphogenesis (5 terms), metabolic processes (6 terms), membrane and ion transport (4 terms) and positive regulation of RNA polymerase II (1 term) would be expected in a metabolically activated tissue compartment undergoing rapid remodeling and formation of new structures such as nerves and blood vessels. Kinase activity was also well represented (3 terms), consistent with the regulatory role of kinases in cell activation. Two apoptosis terms are also identified indicating active cell death, perhaps as a result of DNA damage or as part of a morphogenic process, is increased in reactive stroma.
Table 1.
Significantly enriched Gene Ontology terms (p<0.01) for genes upregulated (p<0.01) in reactive stroma
| Category/Term | P-value | Fold |
|---|---|---|
| NEUROGENESIS | ||
| dendrite development | 0.0003 | 11.7 |
| neurite development | 0.0024 | 10.5 |
| neural crest cell migration | 0.0081 | 7 |
| axonogenesis | 0.0008 | 5.4 |
| synapse organization and biogenesis | 0.0023 | 5.3 |
| neurite morphogenesis | 0.0013 | 4.9 |
| cell soma | 0.0088 | 4.8 |
| synaptic transmission | 0.0096 | 2.2 |
| DNA DAMAGE AND REPAIR | ||
| DNA damage checkpoint | 0.0068 | 5.2 |
| DNA damage response, signal transduction | 0.0015 | 4.8 |
| DNA integrity checkpoint | 0.0100 | 4.7 |
| response to DNA damage stimulus | 0.0058 | 2 |
| MORPHOGENESIS AND DEVELOPMENT | ||
| cell part morphogenesis | 0.0013 | 4.9 |
| cell projection morphogenesis | 0.0013 | 4.9 |
| cell projection organization and biogenesis | 0.0021 | 4.5 |
| epidermis development | 0.0025 | 3.4 |
| cellular structure morphogenesis | 0.0015 | 3.1 |
| METABOLIC PROCESSES | ||
| cellular polysaccharide catabolic process | 0.0053 | 8.1 |
| polysaccharide catabolic process | 0.0053 | 8.1 |
| oxidoreductase activity, acting on CH-OH group of donors | 0.0098 | 2.7 |
| monocarboxylic acid metabolic process | 0.0009 | 2.5 |
| fatty acid metabolic process | 0.0045 | 2.5 |
| regulation of hydrolase activity | 0.0063 | 2.4 |
| KINASES ACTIVITY | ||
| calmodulin regulated protein kinase activity | 0.0098 | 6.6 |
| protein kinase cascade | 0.0035 | 2.6 |
| protein serine/threonine kinase activity | 0.0016 | 2 |
| MEMBRANE AND ION TRANSPORT | ||
| chloride transport | 0.0080 | 4 |
| porter activity | 0.0089 | 2.2 |
| electrochemical potential-driven transporter activity | 0.0092 | 2.2 |
| carrier activity | 0.0053 | 2 |
| TRANSCRIPTION REGULATION | ||
| positive regulation of transcription from RNA pol II promoter | 0.0051 | 2.6 |
| APOPTOSIS | ||
| DNA fragmentation during apoptosis | 0.0066 | 7.5 |
| cell structure disassembly during apoptosis | 0.0018 | 7.4 |
A similar analysis revealed 33 GO terms enriched with at least 10 genes on the array enriched at least 2-fold and p<0.01 for the genes decreased in reactive stroma (Table 2 and Supplementary Table S3). Terms related to muscle cells (3 terms) and cytoskeleton structure (4 terms) were prominent, consistent with displacement of smooth muscle by reactive stroma as noted above. Also of interest is a group of 6 GO terms related to oxygen transport and superoxide metabolism. Two neurogenesis related terms were also identified, indicating that neurogenesis is a regulated process within the reactive stroma, as would be expected in a non-transformed tissue compartment.
Table 2.
Significantly upregulated (p<0.01) Gene Ontology terms for genes downregulated in reactive stroma
| Category /Term | P-value | Fold |
|---|---|---|
| OXYGEN TRANSPORT | ||
| hemoglobin complex | 0.0044 | 8.6 |
| superoxide metabolic process | 0.0003 | 8.3 |
| gas transport | 0.0089 | 6.8 |
| oxygen transporter activity | 0.0089 | 6.8 |
| oxygen transport | 0.0089 | 6.8 |
| oxygen binding | 0.0067 | 4.1 |
| CYTOSKELETON | ||
| actin filament bundle | 0.0014 | 7.9 |
| stress fiber | 0.0014 | 7.9 |
| perinuclear region of cytoplasm | 0.0012 | 4.2 |
| cytoskeleton | 0.0002 | 2 |
| MUSCLE | ||
| structural constituent of muscle | 0.0002 | 5.5 |
| regulation of heart contraction | 0.0103 | 3.8 |
| contractile fiber part | 0.0092 | 3.3 |
| ION CHANNEL ACTIVITY | ||
| channel inhibitor activity | 0.0044 | 8.6 |
| ion channel inhibitor activity | 0.0044 | 8.6 |
| PROTEIN FOLDING | ||
| unfolded protein response | 0.0044 | 8.6 |
| response to protein stimulus | 0.0019 | 3.9 |
| response to unfolded protein | 0.0019 | 3.9 |
| DEVELOPMENTAL PROCESSES | ||
| ureteric bud development | 0.0006 | 9.7 |
| positive regulation of developmental process | 0.0078 | 3.4 |
| regulation of developmental process | 0.0043 | 2.2 |
| NEUROGENESIS | ||
| positive regulation of axonogenesis | 0.0057 | 7.9 |
| positive regulation of neurogenesis | 0.0089 | 6.8 |
| IMMUNE SYSTEM | ||
| leukocyte activation | 0.0049 | 2.5 |
| cell activation | 0.0057 | 2.3 |
| BEHAVIOR | ||
| learning | 0.0017 | 7.4 |
| locomotory behavior | 0.0055 | 2.2 |
| CELL MOTILITY | ||
| cell motility | 0.0010 | 2.1 |
| CELL MEMBRANE | ||
| lipid raft | 0.0099 | 4.7 |
| OTHERS | ||
| histone binding | 0.0001 | 11.2 |
| ER-nuclear signaling pathway | 0.0089 | 6.8 |
| Golgi trans face | 0.0039 | 6 |
| copper ion binding | 0.0021 | 3.9 |
Protein interaction networks involving reactive stroma genes
We queried the Human Protein Reference Database (HPRD) catalogue of over 35,000 known physical protein-protein interactions (20), in order to determine how the reactive stroma-associated genes relate to each other in terms of interactions between their associated proteins. In the set of 1141 genes differentially expressed in reactive compared to normal stroma, 292 HPRD interactions were represented (both members being included in the 1141 gene set). Randomly selected gene sets yielded 124 interactions on average (SD = 23, Figure 3A), indicating that many of the 292 interactions are actually involved within the reactive stroma. The 292 interactions were visualized as a graphical network (Figure 3B). A view of the central node is shown in Figure 3C. Of note is the prominence of signal transduction molecules in this network (STAT1, STATIP1, LCK, MA2K11P1, PTPRF, MAPK3, PRKCD, PRKCA, HRAS, PLA2G4A etc) This finding indicates that there are profound changes in cellular signaling in reactive stroma that may play a key role in the reactive stroma phenotype and may be targets for signal transduction inhibitors used for treatment of prostate cancer.
Figure 3. Interaction network analysis of reactive stroma genes.
(A) Number of known human protein-protein interactions (from HPRD) involving the 1141 unique genes differentially expressed in reactive prostate stroma, as compared to that expected from a random gene set of 1141 (with +/− 1 SD, based on 1000 simulations).
(B) Network representation of the entire set of 292 HPRD protein-protein interactions involving the 1141 named genes differentially expressed in reactive prostate stroma.
(C) Subnetwork of HPRD protein-protein interactions from part (B). Nodes represent genes: yellow nodes, genes over-expressed in reactive stroma; blue nodes, genes under-expressed in reactive stroma. A line between two nodes signifies that the corresponding protein products of the genes can physically interact (according to the literature). Colored edges (other than gray) represent a common GO term annotation shared by both of the connected genes. Active hubs, or genes connected to a significant number (P < 0.01, one-sided Fisher’s exact test) of the other genes in the network, appear enlarged over the other nodes.
Upregulated growth factor pathways in reactive stroma
To identify potential modulators of the profound changes seen in the reactive stroma we identified growth factors, growth factor receptors or growth factor receptor modulators that were upregulated in reactive stroma (p<0.01). These are shown in Table 3. Several major growth factor families are represented. Of note are the number of these proteins or protein families that have been linked to stem cell maintenance including the Notch1, Kit and the Wnt pathways. Other proteins such as ErbB2, FGF19, fibronectin leucine repeat transmembrane protein-1 (FLRT1), endothelin receptor B (EDNRB) and glial cell derived neurotrophic factor (GDNF) have been shown to be linked to neurogenesis, angiogenesis and matrix remodeling. Some of these factors may also act as paracrine growth factors for prostate cancer cells. Wnt10b can bind Frizzled receptors which are known to be expressed by prostate cancer cells (21). The GDNF receptor RET is also known to be expressed in prostate cancer cells (22). Of course, Wnts and endothelins expressed by prostate cancer cells can also potential act on the stromal cells as well. The endothelin 1 and TGF-β signaling pathway stimulation are important in upregulation of collagen production and matrix remodeling (23). FGF19 specifically interacts with FGFR-4, which is expressed in prostate cancer cells (24), but it requires the Klotho co-receptor to do so (25). We have found that Klotho is not expressed in prostate cancer cells and FGF19 is a poor mitogen for these cells (unpublished data) so FGF19 is probably not acting as a paracrine factor for prostate cancer cells but may be important in neurogenesis. FLRTs have been shown to be up-regulated by FGF signaling and interact with FGFR-1, and are expressed at sites of FGF signaling in neural structures (26). Finally the role of the TGF-β family is well established in prostate cancer stromal-epithelial interactions (4). Of note, it has been shown that increased expression of TGFBR2 in breast cancer stroma is associated with adverse outcome (27).
Table 3.
Genes encoding growth factors, growth factor receptors, extracellular matrix proteins and matrix interacting proteins significantly upregulated in reactive stroma (p<0.01)
| Growth factor families | Name | Symbol | P-value | Fold |
|---|---|---|---|---|
| EGF | v-erb-b2 erythroblastic leukemia viral oncogene homolog 2 | ERBB2 | 0.0046 | 1.7 |
| FGF | Fibroblast growth factor 19 | FGF19 | 0.0041 | 1.7 |
| Fibronctin leucine rich transmembrane protein 1 | FLRT1 | 0.0022 | 1.8 | |
| NOTCH | Notch homolog 1, translocation-associated | NOTCH1 | 0.0078 | 1.7 |
| WNT | wingless-type MMTV integration site family, member 10B | WNT10b | 0.0023 | 1.8 |
| TGF-beta | transforming growth factor, beta receptor II | TGFBR2 | 0.0087 | 1.7 |
| KIT | v-kit feline sarcoma oncogene homolog | KIT | 0.0067 | 1.7 |
| ENDOTHELIN | endothelin receptor type B | EDNRB | 0.0058 | 1.7 |
| NEUROTROPHIN | glial cell derived neurotrophic factor | GDNF | 0.0032 | 1.8 |
| Extracellular matrix | Cartilage oliomeric matrix protein | COMP | 0.0013 | 1.8 |
| Extracellular phosphoglycoprotein with ASARM motif | MEPE | 0.0034 | 1.8 | |
| Laminin alpha chain 2, merosin | LAMA2 | 0.0077 | 1.7 | |
| Fibrinogen alpha chain | FGA | 0.005 | 1.8 | |
| Matrix interacting proteins | Syndecan 4 | SDC4 | 0.000003 | 2.3 |
| Low density lipoprotein-related protein 1 | LRP1 | 0.0086 | 1.7 | |
| Collagen Type 1 and thrombospondin receptor | CD36 | 0.0001 | 2.1 | |
| Thrombospondin 4 | THBS4 | 0.0001 | 2.1 |
Alterations of reactive stroma matrix composition and proteins interacting with matrix
In addition to changes in cellular components we also noted changes in cell matrix and matrix interacting proteins in Grade 3 reactive stroma. These are shown in Table 3. COMP is expressed in cartilage but can also be expressed in other sites and is known to be upregulated by TGF-β in prostatic stromal cells (28). It is known to be expressed by vascular smooth muscle cells and plays important role in adhesion and migration (29). MEPE has been linked to phosphate metabolism and plays a role in bone resorption and formation (30). MEPE is a member of the small integrin binding ligand N-linked glycoprotein family (SIBLING) gene family and are expressed in various cancers (31). Some of these proteins bind to and modulate matrix metalloprotease (MMP) activity, but this has not been demonstrated for MEPE to date. LAMA2 (laminin alpha 2 subunit) is a component of both laminin 2 and laminin 4 which are expressed in peripheral nerve and neuromuscular junction respectively (32). Laminin alpha 2 is a component of capillary vessels in the central nervous system (32) and interestingly, upregulated expression of laminin alpha 2 may indicate changes in capillary basal membrane matrix favoring metastases in neuroendocrine carcinomas (33). Other proteins identified are well known cellular proteins which interact with the extracellular matrix such as CD36 (thrombospondin receptor, collagen type 1 receptor). Low-density lipoprotein-related protein 1 (LRP1) is a known receptor internalization factor enhancing TGF-β signaling, regulating extracellular matrix proteins such as fibronectin, MMP-9, and plasma membrane proteins like uPAR and integrins (34, 35). It is a catabolic receptor for ECM structural proteins and for proteins that bind to ECM. Syndecan4 is involved in focal adhesion, cell spreading, integrin signaling and stress fiber formation (36). Thrombospondin 4 is involved in neurite outgrowth as well many other processes (37).
Gene expression changes in reactive stroma are associated with increased protein expression
To determine if increased mRNA levels in reactive stroma were associated with increased protein levels we analyzed protein expression levels of ERK1, c-KIT and GSK3β in normal and reactive stroma by immunohistochemistry using a small tissue microarray with 10 cancers with Grade 3 reactive stroma and matched benign tissue. Typical results are shown in Supplementary Fig 3. In all cases increased protein expression was seen in the cancer stromal cells when compared to normal stroma.
Comparison of gene expression in prostate and breast cancer reactive stroma
Finak et al (17) have recently reported the results of their gene expression microarray analysis of laser captured breast cancer stroma from 53 primary breast cancers using methodology quite similar to ours. To identify similarities and differences between Grade 3 reactive stroma from PCa and breast cancer reactive stroma, we compared their expression data to our data set. A total of 75 transcript probes, representing 63 unique genes, were significantly upregulated (p<0.01, t-test) in both Grade 3 PCa and breast cancer reactive stroma when compared to their respective normal stromas. This overlap had a borderline statistical significance (p<0.02, one-sided Fisher’s exact test). The expression patterns of the 776 probes upregulated in Grade 3 reactive PCa stroma in the breast cancer stroma and normal breast stroma are shown in Fig 4 (along with the average expression for the entire gene set). Overall, the Grade 3 PCa upregulated stromal genes were increased in breast cancer stroma compared to normal breast stroma but many individual genes were decreased. Furthermore, the overlap between the downregulated genes was not statistically significant (p=0.30). Thus, there were some similarities between PCa stroma and breast cancer stroma, but there were differences as well. A Gene Ontology analysis of the commonly upregulated genes found seven of the 9 top enriched GO terms (p<0.001) being related directly or indirectly to DNA damage response and two being related to caspase regulation (see Supplementary Table S4).
DISCUSSION
During PCa progression stromal reaction occurs resulting in development of a reactive stroma microenvironment that is different from normal prostate stroma. Here we have analyzed expression changes taking place in Grade 3 reactive stroma, which has been shown to be associated with prostate cancer progression. A total of 1141 genes were differentially expressed in reactive stroma (at p<0.01) which was significantly higher than would be expected by chance alone and indicates that there are profound changes in gene expression associated with formation of reactive stroma in PCa. This is presumably due to both changes in the types of cells present and changes in gene expression of individual cells. Gene Ontology analysis reveals significant functional clustering of these genes. Of particular interest is a significant increase in the level of expression for genes responsible for neurogenesis, axonogenesis, synaptogenesis and neural biogenesis indicating that active neurogenesis and axonogenesis is occurring in Grade 3 reactive stroma. We observed an increased expression of several genes involved in neural development such as GDNF, NOTCH1, FGF19, GLI2, and CDK5. GDNF is neurotrophic stem cell factor, promoting survival and differentiation of neurons and preventing an apoptosis of motor neurons induced by axotomy. In some neural processes GDNF can increase CDK5 activity (38, 39). CDK5 is well known kinase involved in neuronal migration and guidance. Another up-regulated neural development factor GLI2, is a well known zinc-finger protein of the GLI family. It is a transcriptional mediator of Shh signaling, involved in patterning of dorsal-ventral axis of the spinal cord. It has been shown that TGF-β induces expression of GLI2 in many type of cells including fibroblasts and LNCaP prostate cancer cells (40). We have demonstrated previously significant biological interactions of nerves and cancer cells in PCa (41, 42) and our findings indicate that not only do cancer cells interact with pre-existing nerves but they may actually induce formation of nerves in vivo. Recent data from our group has demonstrated the occurrence of axonogenesis and neurogenesis in prostate cancer by quantitative analysis nerves and neurons in prostate cancer and normal controls (43). Of note, our in vitro studies have identified semaphorin 4F as a key factor in prostate cancer induced neurogenesis and our current data shows that semaphorin 4F is significantly upregulated in prostate cancer reactive stroma in vivo. Other molecules identified in analysis may also play important roles the complex process of axonogenesis and neurogenesis.
Another novel observation is the prominence of the DNA damage signaling pathway in reactive stroma. Our results show up-regulated expression of MRE11A, a member of Mre11-Nbs1- Rad50 complex responsible for DNA double strand break repair (44). At the same time we found an increased expression of 9-1-1 complex members HUS1 and RAD17, which are both involved in cellular response to many types of DNA damage and participate in DNA replication fork stabilization, G2/M arrest and DNA repair (45). The reason for the DNA damage response observed in reactive stroma is not clear but may be due to reactive oxygen species generated within the stroma, perhaps by inflammatory cells or by the cancer cells (46).
A potential important finding is the enrichment for terms related to somatic stem cells. There has been increasing evidence in recent years that somatic stem cells may play a role in generation of reactive stroma. This term was not identified in our GO analysis since only a few genes have been identified that are critical in stem cell biology and thus 10 genes were not present on our array. Among the up-regulated factors we identified was BMI-1, which is well known as a gene that is expressed in stem cells from various tissues and plays an important role in their maintenance (47). NOTCH1 is also up-regulated in reactive stroma and plays an important role stem cell self renewal (48). In addition both C-KIT and components of the Wnt pathway are also upregulated and have been shown to be involved in stem cell proliferation (49, 50). A recent report by Mishra et al (51) has shown that human mesenchymal stem cells can be induced in vitro by tumor conditioned media to differentiate into myofibroblast cells, typically observed in tumor-associated reactive stroma. It is of particular interest that gene expression analysis of mesenchymal stem cells treated with tumor cell conditioned medium showed elevated expression of several neurogenesis/axonogenesis related genes that we have shown to be upregulated in reactive stroma in vivo, including CDK5 and Semaphorin 4F, which were linked to the axon guidance pathway by these authors. These in vitro studies imply that mesenchymal stem cells may give rise to neural cells as well as myofibroblastic cells in prostate cancer reactive stroma. As discussed by Mishra et al (51), the origin of mesenchymal stem cells in tumor reactive is not completely established but at least a subset are derived from circulating marrow cells.
To date there have only been limited studies of gene expression in prostatic stroma and even fewer of reactive stroma in PCa. To our knowledge, the only direct comparison of normal and reactive stroma in PCa is the study by Richardson et al (52) of 5 cases using laser captured normal and tumor associated stroma. A total of 44 genes were consistently upregulated in all 5 cases in the tumor stroma. While their analytical approach was quite different than ours we still noted overlap of their 44-gene set and our 544 upregulated gene including FOXA1, COMP, STEAP1, and CPNE4. This group also found increases of FOLH1 (PSMA). This gene is expressed in PCa tumor vasculature (53) and we also found this gene to be increased although it was slightly outside our statistical cut-off (p=0.011). It should be noted that their study used unselected PCa cases while we focused on grade 3 reactive stroma. We are currently assessing gene expression changes in Grade 1 and 2 reactive stroma to identify the key components in the Grade 3 stroma that may be enhancing PCa progression.
Our comparison of breast cancer stroma to PCa Grade 3 reactive stroma reveals a partial overlap of gene expression signatures in these two tissues. It is not surprising that there are significant differences in the reactive stroma between these two cancers given the very significant differences in the biology of these cancers and the fact that the normal prostate stroma is predominantly smooth muscle while breast stroma is predominantly fibrous and adipose tissue. However, the finding that the DNA damage pathway is also upregulated in breast cancer stroma supports our finding that this pathway is increased in PCa stroma. As noted above, mutations of p53 have been identified in breast cancer stroma (19), consistent with the occurrence of DNA damage. In particular, increased expression of HUS1 and RAD17 was seen in stroma of both cancer types. Interestingly, both FOXA1 and COMP were increased in breast cancer stroma and in PCa stroma, both in our dataset and that of Richardson et al (52), supporting their role in reactive stroma biology. Thus, there are significant similarities, as well as differences, in reactive stroma from these two cancer sites. Of note, we did not find a statistically significant overlap of our gene set with the “wound healing” gene expression signature derived from serum stimulated fibroblasts as defined by Chang et al (54) that has been shown to have prognostic significance for a number of cancers, including breast cancer, when non-microdissected tissues are analyzed (data not shown). This is not completely surprising since many of the genes that are components of this signature are expressed predominantly in cancer epithelial cell when analyzed by immunohistochemistry or in situ hybridization (54). Further studies in various common cancer types may identify common and tissue type specific pathways activated and repressed in cancer stroma.
In summary, our studies indicate that formation of PCa reactive stroma is a complex dynamic process involving a number of growth factor pathways and characterized by activation of multiple processes including neurogenesis and axonogenesis. Further studies are needed to identify the key individual biomarkers of progression in Grade 3 reactive stroma. In addition, we have identified potential therapeutic targets in PCa stroma and key signal transduction pathways that may be useful in treatment of patients with aggressive PCa.
Acknowledgments
The technical assistance of Deanna Killen and Shantu Dixit is gratefully acknowledged.
Grant Support: This work was supported by grants from National Cancer Institute to the Tumor Microenvironment Network (1U54CA126568, DR), the P30 Cancer Center support grant (P30 CA125123) and the Baylor Prostate Cancer SPORE (P50CA058204, MI); the Diana Helis Henry Medical Research Foundation (MI, GA, DR); the Dept of Veterans Affairs Merit Review program (MI) and by the use of the facilities of the Michael E. DeBakey VAMC.
Footnotes
STATEMENT OF CLINICAL RELEVANCE
The tumor microenvironment can influence cancer progression in a variety of malignancies. In prostate cancer, a marked stromal response is associated with poor outcome independent of Gleason score and other pathological parameters. In this study we use microarray gene expression analysis to define key processes and signaling pathways associated with the formation of prostate cancer reactive stroma. Future studies may identify individual biomarkers within the reactive stroma that are associated with prostate cancer progression. Furthermore, we have identified a number of growth factor and signal transduction pathways that are potential drug targets in reactive stroma.
References
- 1.Tuxhorn JA, Ayala GE, Rowley DR. Reactive stroma in prostate cancer progression. J Urol. 2001;166:2472–83. [PubMed] [Google Scholar]
- 2.Chung LW, Baseman A, Assikis V, Zhau HE. Molecular insights into prostate cancer progression: the missing link of tumor microenvironment. J Urol. 2005;173:10–20. doi: 10.1097/01.ju.0000141582.15218.10. [DOI] [PubMed] [Google Scholar]
- 3.Alberti C. Prostate cancer progression and surrounding microenvironment. Int J Biol Markers. 2006;21:88–95. doi: 10.1177/172460080602100204. [DOI] [PubMed] [Google Scholar]
- 4.Tuxhorn JA, Ayala GE, Smith MJ, Smith VC, Dang TD, Rowley DR. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Cancer Res. 2002;8:2912–23. [PubMed] [Google Scholar]
- 5.Tuxhorn JA, McAlhany SJ, Dang TD, Ayala GE, Rowley DR. Stromal cells promote angiogenesis and growth of human prostate tumors in a differential reactive stroma (DRS) xenograft model. Cancer Res. 2002;62:3298–307. [PubMed] [Google Scholar]
- 6.Ayala G, Tuxhorn JA, Wheeler TM, et al. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res. 2003;9:4792–801. [PubMed] [Google Scholar]
- 7.Yanagisawa N, Rowley D, Kadmon D, Miles BJ, Wheeler TM, Ayala GE. Stromogenic prostatic carcinoma pattern (carcinomas with reactive stromal grade 3) in needle biopsies predicts biochemical recurrence-free survival in patients after radical prostatectomy. Hum Pathol. 2008;39:282–91. doi: 10.1016/j.humpath.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 8.Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer Science+Business Media, Inc; 2005. [Google Scholar]
- 9.Johnson W, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using Empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 10.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 13.Creighton C, Kuick R, Misek DE, et al. Profiling of pathway-specific changes in gene expression following growth of human cancer cell lines transplanted into mice. Genome Biol. 2003;4:R46. doi: 10.1186/gb-2003-4-7-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creighton CJ, Bromberg-White JL, Misek DE, et al. Analysis of tumor-host interactions by gene expression profiling of lung adenocarcinoma xenografts identifies genes involved in tumor formation. Mol Cancer Res. 2005;3:119–29. doi: 10.1158/1541-7786.MCR-04-0189. [DOI] [PubMed] [Google Scholar]
- 15.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Y, Wang J, Li R, Ayala G, Ittmann M, Liu M. GGAP2/PIKE-a directly activates both the Akt and nuclear factor-kappaB pathways and promotes prostate cancer progression. Cancer Res. 2009;69:819–27. doi: 10.1158/0008-5472.CAN-08-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finak G, Bertos N, Pepin F, Sadekova S, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B, Xu F, Baba Y. An evaluation of linear RNA amplification in cDNA microarray gene expression analysis. Mol Genet Metab. 2006;87:71–9. doi: 10.1016/j.ymgme.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Patocs A, Zhang L, Xu Y, et al. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N Engl J Med. 2007;357:2543–51. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- 20.Peri S, Navarro J, Amanchy R, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–71. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joesting MS, Perrin S, Elenbaas B, et al. Identification of SFRP1 as a candidate mediator of stromal-to-epithelial signaling in prostate cancer. Cancer Res. 2005;65:10423–30. doi: 10.1158/0008-5472.CAN-05-0824. [DOI] [PubMed] [Google Scholar]
- 22.Dawson DM, Lawrence EG, MacLennan GT, et al. Altered expression of RET proto-oncogene product in prostatic intraepithelial neoplasia and prostate cancer. J Natl Cancer Inst. 1998;90:519–23. doi: 10.1093/jnci/90.7.519. [DOI] [PubMed] [Google Scholar]
- 23.Horstmeyer A, Licht C, Scherr G, Eckes B, Krieg T. Signalling and regulation of collagen I synthesis by ET-1 and TGF-beta1. FEBS J. 2005;272:6297–309. doi: 10.1111/j.1742-4658.2005.05016.x. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Stockton DW, Ittmann M. The fibroblast growth factor receptor-4 Arg388 allele is associated with prostate cancer initiation and progression. Clin Cancer Res. 2004;10:6169–78. doi: 10.1158/1078-0432.CCR-04-0408. [DOI] [PubMed] [Google Scholar]
- 25.Kurosu H, Choi M, Ogawa Y, et al. Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem. 2007;282:26687–95. doi: 10.1074/jbc.M704165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haines BP, Wheldon LM, Summerbell D, Heath JK, Rigby PW. Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Dev Biol. 2006;297:14–25. doi: 10.1016/j.ydbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Barlow J, Yandell D, Weaver D, Casey T, Plaut K. Higher stromal expression of transforming growth factor-beta type II receptors is associated with poorer prognosis breast tumors. Breast Cancer Res Treat. 2003;79:149–59. doi: 10.1023/a:1023918026437. [DOI] [PubMed] [Google Scholar]
- 28.Untergasser G, Gander R, Lilg C, Lepperdinger G, Plas E, Berger P. Profiling molecular targets of TGF-beta1 in prostate fibroblast-to-myofibroblast transdifferentiation. Mech Ageing Dev. 2005;126:59–69. doi: 10.1016/j.mad.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Riessen R, Fenchel M, Chen H, Axel DI, Karsch KR, Lawler J. Cartilage oligomeric matrix protein (thrombospondin-5) is expressed by human vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2001;21:47–54. doi: 10.1161/01.atv.21.1.47. [DOI] [PubMed] [Google Scholar]
- 30.White KE, Larsson TE, Econs MJ. The roles of specific genes implicated as circulating factors involved in normal and disordered phosphate homeostasis: frizzled related protein-4, matrix extracellular phosphoglycoprotein, and fibroblast growth factor 23. Endocr Rev. 2006;27:221–41. doi: 10.1210/er.2005-0019. [DOI] [PubMed] [Google Scholar]
- 31.Fisher LW, Jain A, Tayback M, Fedarko NS. Small integrin binding ligand N-linked glycoprotein gene family expression in different cancers. Clin Cancer Res. 2004;10:8501–11. doi: 10.1158/1078-0432.CCR-04-1072. [DOI] [PubMed] [Google Scholar]
- 32.Vitolo D, Ciocci L, Cicerone E, et al. Laminin alpha2 chain (merosin M chain) distribution and VEGF, FGF(2), and TGFbeta1 gene expression in angiogenesis of supraglottic, lung, and breast carcinomas. J Pathol. 2001;195:197–208. doi: 10.1002/path.938. [DOI] [PubMed] [Google Scholar]
- 33.Vitolo D, Ciocci L, Deriu G, et al. Laminin alpha2 chain-positive vessels and epidermal growth factor in lung neuroendocrine carcinoma: a model of a novel cooperative role of laminin-2 and epidermal growth factor in vessel neoplastic invasion and metastasis. Am J Pathol. 2006;168:991–1003. doi: 10.2353/ajpath.2006.041310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabello-Verrugio C, Brandan E. A novel modulatory mechanism of transforming growth factor-beta signaling through decorin and LRP-1. J Biol Chem. 2007;282:18842–50. doi: 10.1074/jbc.M700243200. [DOI] [PubMed] [Google Scholar]
- 35.Gaultier A, Salicioni AM, Arandjelovic S, Gonias SL. Regulation of the composition of the extracellular matrix by low density lipoprotein receptor-related protein-1: activities based on regulation of mRNA expression. J Biol Chem. 2006;281:7332–40. doi: 10.1074/jbc.M511857200. [DOI] [PubMed] [Google Scholar]
- 36.Rapraeger AC. Molecular interactions of syndecans during development. Semin Cell Dev Biol. 2001;12:107–16. doi: 10.1006/scdb.2000.0239. [DOI] [PubMed] [Google Scholar]
- 37.Arber S, Caroni P. Thrombospondin-4, an extracellular matrix protein expressed in the developing and adult nervous system promotes neurite outgrowth. J Cell Biol. 1995;131:1083–94. doi: 10.1083/jcb.131.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Airaksinen MS, Saarma M. The GDNF family: signalling, biological functions and therapeutic value. Nat Rev Neurosci. 2002;3:383–94. doi: 10.1038/nrn812. [DOI] [PubMed] [Google Scholar]
- 39.Ledda F, Paratcha G, Ibanez CF. Target-derived GFRalpha1 as an attractive guidance signal for developing sensory and sympathetic axons via activation of Cdk5. Neuron. 2002;36:387–401. doi: 10.1016/s0896-6273(02)01002-4. [DOI] [PubMed] [Google Scholar]
- 40.Dennler S, Andre J, Alexaki I, et al. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67:6981–6. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- 41.Ayala GE, Wheeler TM, Shine HD, et al. In vitro dorsal root ganglia and human prostate cell line interaction: redefining perineural invasion in prostate cancer. Prostate. 2001;49:213–23. doi: 10.1002/pros.1137. [DOI] [PubMed] [Google Scholar]
- 42.Ayala GE, Dai H, Tahir SA, et al. Stromal antiapoptotic paracrine loop in perineural invasion of prostatic carcinoma. Cancer Res. 2006;66:5159–64. doi: 10.1158/0008-5472.CAN-05-1847. [DOI] [PubMed] [Google Scholar]
- 43.Ayala GE, Dai H, Powell M, Li R, Ding Y, Wheeler TM, Shine D, Kadmon D, Miles BJ, Ittmann MM, Rowley D. Cancer related axonogenesis and neurogenesis in prostate cancer. Clin Cancer Res. 2008 doi: 10.1158/1078-0432.CCR-08-1164. In Press. [DOI] [PubMed] [Google Scholar]
- 44.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–58. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 45.Parrilla-Castellar ER, Arlander SJ, Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 2004;3:1009–14. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 46.Kumar B, Koul S, Khandrika L, Meacham RB, Koul HK. Oxidative stress is inherent in prostate cancer cells and is required for aggressive phenotype. Cancer Res. 2008;68:1777–85. doi: 10.1158/0008-5472.CAN-07-5259. [DOI] [PubMed] [Google Scholar]
- 47.Glinsky GV. “Stemness” genomics law governs clinical behavior of human cancer: implications for decision making in disease management. J Clin Oncol. 2008;26:2846–53. doi: 10.1200/JCO.2008.17.0266. [DOI] [PubMed] [Google Scholar]
- 48.Fox V, Gokhale PJ, Walsh JR, Matin M, Jones M, Andrews PW. Cell-cell signaling through NOTCH regulates human embryonic stem cell proliferation. Stem Cells. 2008;26:715–23. doi: 10.1634/stemcells.2007-0368. [DOI] [PubMed] [Google Scholar]
- 49.Bashamboo A, Taylor AH, Samuel K, Panthier JJ, Whetton AD, Forrester LM. The survival of differentiating embryonic stem cells is dependent on the SCF-KIT pathway. J Cell Sci. 2006;119:3039–46. doi: 10.1242/jcs.03038. [DOI] [PubMed] [Google Scholar]
- 50.Nemeth MJ, Topol L, Anderson SM, Yang Y, Bodine DM. Wnt5a inhibits canonical Wnt signaling in hematopoietic stem cells and enhances repopulation. Proc Natl Acad Sci U S A. 2007;104:15436–41. doi: 10.1073/pnas.0704747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mishra PJ, Humeniuk R, Medina DJ, et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68:4331–9. doi: 10.1158/0008-5472.CAN-08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richardson AM, Woodson K, Wang Y, et al. Global expression analysis of prostate cancer-associated stroma and epithelia. Diagn Mol Pathol. 2007;16:189–97. doi: 10.1097/PDM.0b013e3180de20ac. [DOI] [PubMed] [Google Scholar]
- 53.Chang SS, Reuter VE, Heston WD, Bander NH, Grauer LS, Gaudin PB. Five different anti-prostate-specific membrane antigen (PSMA) antibodies confirm PSMA expression in tumor-associated neovasculature. Cancer Res. 1999;59:3192–8. [PubMed] [Google Scholar]
- 54.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi J, van de Rijn M, Botstein D, Brown P. PLoS Biol. 2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]