Abstract
The ability to repair DNA damage is likely to play an important role in the survival of facultative intracellular parasites because they are exposed to high levels of reactive oxygen species and nitrogen intermediates inside phagocytes. Correcting oxidative damage in purines and pyrimidines is the primary function of the enzymes formamidopyrimidine (faPy)–DNA glycosylase (Fpg) and endonuclease VIII (Nei) of the base excision repair pathway, respectively. Four gene homologs, belonging to the fpg/nei family, have been identified in Mycobacterium tuberculosis H37Rv. The recombinant protein encoded by M. tuberculosis Rv2924c, termed Mtb-Fpg1, was overexpressed, purified and biochemically characterized. The enzyme removed faPy and 5-hydroxycytosine lesions, as well as 8-oxo-7,8-dihydroguanine (8oxoG) opposite to C, T and G. Mtb-Fpg1 thus exhibited substrate specificities typical for Fpg enzymes. Although Mtb-fpg1 showed nearly complete nucleotide sequence conservation in 32 M. tuberculosis isolates, the region upstream of Mtb-fpg1 in these strains contained tandem repeat motifs of variable length. A relationship between repeat length and Mtb-fpg1 expression level was demonstrated in M. tuberculosis strains, indicating that an increased length of the tandem repeats positively influenced the expression levels of Mtb-fpg1. This is the first example of such a tandem repeat region of variable length being linked to the expression level of a bacterial gene.
Keywords: Mycobacterium tuberculosis, DNA repair, formamidopyrimidine–DNA glycosylase, tandem repeats
Introduction
Mycobacterium tuberculosis is a predominant cause of tuberculosis infections worldwide, with the highest incidence found in developing countries. This pathogen is a member of the M. tuberculosis complex (MTC), which contains the genetically highly conserved species Mycobacterium bovis, Mycobacterium africanum, Mycobacterium canettii, M. bovis BCG, Mycobacterium caprae, Mycobacterium pinnipedii and Mycobacterium microti. As a facultative intracellular pathogen, M. tuberculosis survives and replicates inside human macrophages. Accordingly, it resides in a hostile environment where reactive oxygen and nitrogen radicals can induce deleterious effects, including DNA damage. Consequences of DNA damage comprise single- and double-strand breaks, abasic (apurinic/apyrimidinic, AP) sites and base damages that can have cytotoxic or mutagenic effects (Bjelland & Seeberg, 2003). Thus, in order to survive, intracellular bacteria must be particularly capable of repairing oxidative and nitrosative DNA lesions.
In Escherichia coli, subtle base alterations in DNA of endogenous origin are primarily repaired by the base excision repair (BER) pathway (Seeberg et al., 1995). The repair of oxidized bases is initiated by the activity of a DNA glycosylase belonging to the formamidopyrimidine (faPy)–DNA glycosylase (Fpg)/endonuclease VIII (Nei) family. Escherichia coli Fpg (Fpg Ec), encoded by the mutM gene, primarily catalyses excision of 8oxoG and other oxidatively damaged purines from DNA, while the principal substrates of E. coli Nei (Nei Ec) are oxidized pyrimidines. Although Fpg and Nei have different substrate specificities, they share common N- and C-terminal domains and a similar enzymatic mode of action with three types of activities: hydrolysis of the N-glycosidic bond with transient formation of an AP site (DNA glycosylase activity) and cleavage of the sugar–phosphate backbone 3′ to the abasic site (β-elimination) before the 5′-cleavage (δ-elimination) (Melamede et al., 1994; Bhagwat & Gerlt, 1996; Zharkov et al., 2003). Consecutive activities of these three functions remove the lesion from duplex DNA, leaving a single-nucleotide gap in the damaged strand flanked by phosphate residues.
While there is an abundance of studies addressing oxidative damage repair mechanisms in E. coli, reports on these aspects in mycobacteria are scarce. Based on the genome sequences of M. tuberculosis strains, no less than four homologs of the fpg/nei E. coli genes have been identified (Mizrahi & Andersen, 1998). Recently, two of these genes from M. tuberculosis, encoding Mtu-Nei2 and Mtu-Fpg2, were cloned and characterized (Sidorenko et al., 2008). Another study on an Fpg (MutM)-deficient Mycobacterium smegmatis revealed a remarkable increase in the accumulation of A to G (T to C) mutations, in contrast to Fpg-deficient E. coli, where C to A mutations predominate (Jain et al., 2007).
The genome of M. tuberculosis contains several kinds of repetitive DNA sequences, including insertion sequences, major polymorphic tandem repeats, polymorphic GC-rich repetitive sequences, direct repeats, variable number of tandem repeats (VNTR) and Mycobacterium interspersed repetitive units (MIRU) (van Soolingen et al., 1993; Supply et al., 1997; Smittipat & Palittapongarnpim, 2000). The VNTRs often differ in copy number between isolates (Frothingham & Meeker-O'Connell, 1998). Structures consisting of 40–100-bp repetitive sequences, termed MIRUs, have been found scattered in 41 locations in the M. tuberculosis H37Rv chromosome; 12 of these were polymorphic in MIRU copy numbers between isolates (Supply et al., 1997, 2000; Magdalena et al., 1998). The tandem repeats, predominantly MIRU-VNTR, are commonly used in genotyping of M. tuberculosis isolates for routine epidemiological discrimination (Supply et al., 2006), and one of these, previously termed exact tandem repeat (ETR)-F and VNTR3239, has been detected upstream of Rv2924c (Frothingham & Meeker-O'Connell, 1998; Smittipat & Palittapongarnpim, 2000). However, the exact function of these repeats in M. tuberculosis is not well understood. Proposed possible functions for MIRU-VNTR include regulation of gene expression, differential translation of genes within a polycistronic operon, or some may serve as structural components for chromosome organization (Supply et al., 1997).
In this study, the ORF of the main fpg-homolog Rv2924c was cloned, and its gene product, termed Mtb-Fpg1, was biochemically characterized. Recombinant Mtb-Fpg1 was assessed with regard to its enzymatic activity toward a panel of DNA substrates containing single lesions. The DNA sequences of Mtb-fpg1 homologs and their upstream regions in M. tuberculosis and other mycobacterial species were compared and led to the characterization of the MIRU-VNTR tandem repeat region upstream of Mtb-fpg1. This tandem repeat region was characterized in a panel of M. tuberculosis isolates and other mycobacterial species, and the impact of their length on Mtb-fpg1 gene expression was assessed.
Materials and methods
Mycobacterial genome sequences and bioinformatics tools
The genome sequences of M. tuberculosis H37Rv (http://genolist.pasteur.fr/TubercuList/) (Cole et al., 1998), F11 (http://www.broad.mit.edu/), and CDC1551 (Accession no. NC_002755) (Fleischmann et al., 2002), M. bovis AF2122/97 (Garnier et al., 2003) (http://genolist.pasteur.fr/BoviList), Mycobacterium leprae TN (Cole et al., 2001) (http://genolist.pasteur.fr/Leproma), Mycobacterium avium ssp. paratuberculosis K10 (Accession no. NC_002944), M. smegmatis mc2155 (Accession no. NC_008596), M. avium 104 (http://tigrblast.tigr.org/), Mycobacterium ulcerans Agy99 (http://genolist.pasteur.fr/BuruList/index.html), Mycobacterium sp. JLS (http://www.jgi.doe.gov/), KMS (http://www.jgi.doe.gov/), MCS (http://www.jgi.doe.gov/), Mycobacterium vanbaalenii PYR-1 (http://www.jgi.doe.gov/) and Mycobacterium marinum (Accession no. NC_010612), and the incomplete genomes of M. tuberculosis 210 (http://tigrblast.tigr.org/ufmg), C (accession no. NZ AAKR00000000), M. microti OV254 (http://www.sanger.ac.uk/Projects/M_microti/) and Mycobacterium flavescens (accession no. NZ AAPA00000000) were searched for the presence of fpg/nei homologs and, when feasible, repetitive sequences between the rnc and fpg genes. The different fpg/nei orthologs and single-repeat motifs within each Mycobacterium genome were identified by blast searches (http://genolist.pasteur.fr). Tandem repeats were identified using tandemrepeatsfinder (http://tandem.bu.edu/trf/trf.html) (Benson, 1999). The parameters were set to 1, 4, 4, 80, 10, 100 and 500, aiming to find tandem repeats containing near-identical repeats of some length. Overlapping tandem repeat regions were joined and regarded as one region. Operons were predicted using VIMSS Operon Prediction (http://www.microbesonline.org/) (Price et al., 2005). The VIMSS database was also used to identify fpg/nei orthologs in bacterial species other than mycobacteria.
Media, bacterial strains and DNA manipulations
Bacterial strains and plasmids used in this study are listed in Supporting Information, Table S1. The M. tuberculosis collection included a wide variety of genotypes, such as the Bejing and Haarlem genotypes, which were previously shown to have mutations in putative mutator genes (Rad et al., 2003). Some of the strains were previously characterized in detail by multiple genetic markers (Kremer et al., 1999, 2005; Viana-Niero et al., 2001). DNA isolation, PCR amplification and cloning were performed according to standard techniques (Sambrook & Russell, 2001). DNA sequencing was conducted using an Applied Biosystems 3730 Genetic Analyser System (Applied Biosystems, Foster City, CA), ABI BigDye Terminator v. 3.1 DNA sequencing kit (Applied Biosystems, Foster City, CA) and the primers listed in Table S2. The M. tuberculosis H37Rv Rv2924c (fpg) gene was amplified by PCR and cloned into the expression vector pET22b (Novagen, Madison, WI) encoding a C-terminal His-tag. Escherichia coli ER2566 (New England Biolabs, Beverly, MA) and BK3004 (Alseth et al., 1999) were transformed with the pET22b plasmids by standard methods (Sambrook & Russell, 2001). For the gene expression studies, the M. tuberculosis strains were selected so as to be representative for the VNTR3239 tandem repeat types (TRT) 2, 3, 4 and 6. Mycobacterium tuberculosis strain 29593, representing the VNTR3239 TRT 6, had IS6110 restriction fragment length polymorphism (RFLP) genotype 01403120 (similar to strain NLA009700438).
Mycobacterium tuberculosis Fpg purification
Escherichia coli strain BK3004 (fpg−) was used for overexpressing M. tuberculosis Mtb-Fpg1 from the plasmid pET22b-Rv2924c. BK3004 was grown in Luria–Bertani broth with ampicillin (100 μg mL−1) at 37 °C while shaking until the OD600 nm was 1.0, when 1 mM isopropyl-d-thiogalactopyranoside was added and the cells were incubated with shaking for 3 h. Cells were harvested and washed in lysis buffer (300 mM NaCl, 25 mM Tris and 10 mM imidazole, pH 8.0) before subjecting them to mechanical lysis in a French Pressure Cell (SLM Aminco, Spectronic Instruments Inc., Rochester, NY). The cleared lysate was loaded onto an Ni-NTA column (Qiagen, Hilden, Germany), washed with 300 mM NaCl, 25 mM Tris, pH 7.5, and eluted with 100, 150 and 200 mM imidazole. The purified protein was dialyzed against 300 mM NaCl and 25 mM Tris, pH 8.0, overnight.
Assay for enzymatic cleavage of DNA substrates
Duplex DNA substrates containing a single 8oxoG opposite of C, A, G, T, 5-hydroxycytosine (5OHC): G, 5-hydroxyuracil (5OHU): G, DHU: G, U: A or U: G base pair were generated by 32P-5′ end-labeling of oligonucleotides using T4 polynucleotide kinase (New England Biolabs) as described previously (Eide et al., 1996). The oligonucleotide sequences of the DNA substrates used are listed in Table S2. DNA glycosylase reactions were performed by mixing purified protein as indicated, with 10–50 fmol DNA substrate in a total volume of 15 μL. The enzyme activities were assayed in a reaction buffer containing 70 mM 3-(N-morpholino) propane sulfonic acid, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol and 5% glycerol, and incubated at 37 °C for 1 h. Fpg Ec (New England Biolabs) was included as the positive control. Products of the reactions were separated by 20% denaturing polyacrylamide gel electrophoresis (PAGE) and visualized by phosphorimaging.
Assessment of alkylbase and faPy–DNA glycosylase activities
Alkylated DNA and met-faPy substrates were prepared as described previously, using N-[H3]-methyl-N′-nitro sourea (1.5 Ci mmol−1) and calf thymus DNA (6000 d.p.m. μg−1 DNA) (Riazuddin & Lindahl, 1978) or poly(dG–dC) (12 000 d.p.m. μg−1) (Boiteux et al., 1984), respectively. Removal of bases was measured in a total reaction volume of 50 μL containing 7 μg DNA substrate and enzyme as indicated.
Molecular strain typing
The recommended international standard protocol for IS6110-based RFLP typing was performed (Kamerbeek et al., 1997). The IS6110-based RFLP patterns were analyzed using bionumerics software (Applied Maths, Sint-Martems-Latem, Belgium). Similarities between RFLP patterns were calculated using the Dice coefficient, and the dendrogram was prepared with the unweighted pair group method using arithmetic averages algorithm.
RNA isolation
Mycobacterium tuberculosis strains were grown in Middlebrook 7H9 medium with ADC and 0.05% Tween 80 at 37 °C while shaking until OD600 nm was 0.5. For the quantitative reverse transcriptase (RT)-PCR, bacterial cells were collected in tubes containing RNA-later, harvested by centrifugation and resuspended in Trizol (Invitrogen, Carlsbad, CA). Cell suspensions were transferred to FastRNA BLUE Tubes (BIO 101 Inc., La Jolla, CA) and processed in a MagNA Lyser (Roche Applied Science, Penzberg, Germany). Chloroform was then added to the cell lysates and mixed well. The tubes were centrifuged and supernatants were transferred to microcentrifuge tubes containing an equal volume of ethanol. After mixing, the solutions were applied to RNeasy spin columns (Qiagen) and treated according to the manufacturer's protocol. Total RNA was eluted with RNAse-free water and DNA was removed with TURBO DNA-free™ (Ambion, Hurtingdon, UK). The amount of total RNA was quantified using NanoDrop (ND-1000, NanoDrop Technologies, Wilmington, DE). All RNA isolations were performed, under cold conditions when required, on at least three independent cultures for each strain studied.
Quantitative RT-PCR
For quantitative real-time RT-PCR, M. tuberculosis strains representing TRT 2, NLA000100560 (2-1) and NLA009801353 (2-2); TRT 3, NLA009802122 (3-2) and H37Rv (3-3); TRT 4, NLA009700438 (4); and TRT 6, 29593 (6) were grown, and RNA was isolated as described above. cDNA was synthesized from isolated total RNA using high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol.
Primers for Mtb-fpg1, nei2 and the housekeeping gene sigA were designed to obtain a product of similar size and the same optimal annealing temperature. The Power SYBR® Green PCR master mix (Applied Biosystems, Warrington, UK) and StepOnePlus™ instrument (Applied Biosystems, Foster City, CA) were used in the two-step RT-PCR. The quantitative PCR reaction was performed in quadruplicates on cDNAs with specific primers under the following conditions: 10 min hold at 95 °C and 40 cycles of 95 °C for 15 s and 60 °C for 60 s, and then melting curve analysis was performed at 95 °C for 15 s, 60 °C for 60 s and 95 °C for 15 s. Negative controls, consisting of no-template (water) and RNA in the reaction mixtures, were run with all reactions to test for DNA contamination. The experiments were repeated at least six times. PCR products were also run on agarose gels to confirm the formation of a single product of the desired size. For the relative quantification of mRNA, the comparative Ct method (ΔΔCt) was used to calculate the relative expression levels in each strain, normalized to sigA (endogenous control) and then given as fold change relative to Mtb-fpg1 and nei2 gene expression of M. tuberculosis H37Rv (calibrator). The two-sample t-test was performed to check for statistically significant differences in Mtb-fpg1 and nei2 expression level between M. tuberculosis strains.
Results
Mycobacterial genes with homology to E. coli fpg and nei
The fpg/nei orthologs of M. tuberculosis H37Rv, M. bovis AF2122/9, M. leprae TN, M. a. paratuberculosis K10, M. avium 104 and M. smegmatis mc2155 were identified and presented in Table 1. Four putative ORFs with homology to the fpg/nei family are present in the genome sequence of M. tuberculosis H37Rv: Rv0944 (Mtu-fpg2), Rv2464c (Mtu-nei2), Rv2924c (Mtb-fpg1) and Rv3297 (putative nei) (Cole et al., 1998; Sidorenko et al., 2008). The amino acid sequences exhibited significant similarities to characteristic motifs and conserved residues of the bacterial Fpg/Nei family, including a DNA glycosylase catalytic domain and a helix–two-turns–helix DNA-binding motif (Fig. S1). The catalytic residues at the N-terminus were conserved in Rv2464c, Mtb-Fpg1 and Rv3297, while Rv0944 encoded a truncated protein lacking the N-terminal part. Highly conserved proteins with 100% amino acid identity to all these four DNA glycosylases are also present in the M. bovis genome sequences (Table 1). In contrast, only a single complete fpg ortholog of Mtb-fpg1 is present in the M. leprae genome (80% identity and 88% similarity at the deduced amino acid level). Mycobacterium leprae orthologs to Rv0944 and Rv2464c are, however, present as pseudogenes, which are abundant in the M. leprae genome (Cole et al., 2001). The M. smegmatis genome also harbors orthologs to the four fpg/nei genes with amino acid similarities ranging from 75% to 82%. In M. smegmatis, however, the Rv0944c ortholog is complete. The M. avium and M. a. paratuberculosis genome sequences contain all four fpg/nei orthologs exhibiting amino acid similarities of 85% to 90%, including a full-length Rv0944 ortholog. Surprisingly, a fifth fpg/nei homolog whose predicted gene product contains all the conserved domains of enzymes belonging to the Fpg/Nei family was identified in the genome of M. a. ssp. paratuberculosis K10.
Table 1.
Fpg/Nei DNA glycosylase orthologs present in the order Actinomycetales, including mycobacterial species
| Fpg/Nei orthologs |
|||||
|---|---|---|---|---|---|
| Species | Putative Fpg | Putative Fpg | Putative Nei | Putative Nei | Putative Fpg/Nei? |
| M. leprae TN | ML1658 | – | – | – | – |
| M. tuberculosis CDC1551 | MT2994 | MT0970 | MT2539 | MT3396 | – |
| M. tuberculosis H37Rv | Rv2924c* | Rv0944 | Rv2464c | Rv3297 | – |
| M. bovis AF2122/97 | Mb2949c | Mb0969 | Mb2491C | Mb3325 | – |
| M. marinum | MM1783 | MM4559 | MM3812 | MM1237 | – |
| M. ulcerans | MUL2031 | MUL3737 | MUL2650 | MUL4418 | – |
| M. avium 104 | MAV3782 | MAV3149 | MAV1066 | MAV1708 | – |
| M. avium ssp. paratuberculosis K10 | MAP2994c | MAP0889 | MAP2284c | MAP3416 | MAP1328c |
| M. smegmatis mc2 155 | SMEG2419 | SMEG4683 | SMEG5545 | SMEG1756 | – |
| Streptomyces avermitilis MA-4680 | SAV2664 | SAV7289 | SAV2501 | SAV5427 | – |
| Streptomyces coelicolor A3(2) | SCO0945 | SCO5573 | SCO2626 | SCO5760 | – |
| Corynebacterium efficiens YS-314 | CE1975 | CE2834 | CE0922 | – | – |
| Corynebacterium glutamicum | NCg10813 | NCg11993 | NCg12898 | – | – |
| Corynebacterium diphtheriae | DIP1543 | DIP0829 | DIP2304 | – | – |
| Leifsonia xyli ssp. xyli str. CTCB07 | Lxx00140 | Lxx09800 | Lxx20840 | Lxx08780 | – |
| Nocardia farcinica | Nfa13280 | Nfa41830 | NFA50200 | NFA9770 | – |
| Propionibacterium acnes KPA171202 | PPA1451 | PPA1623 | – | – | – |
| Tropheryma whippeli TW08/27 | TW374 | – | – | – | – |
| Tropheryma whippeli str. Twist | TW396 | – | – | – | – |
| Bifidobacterium longum NCC2705 | – | – | – | – | – |
| E. coli† | MutM | – | Nei | – | – |
Now termed Mtb-Fpg1.
Included as a reference strain.
–, none.
Biochemical characterization of M. tuberculosis Mtb-Fpg1
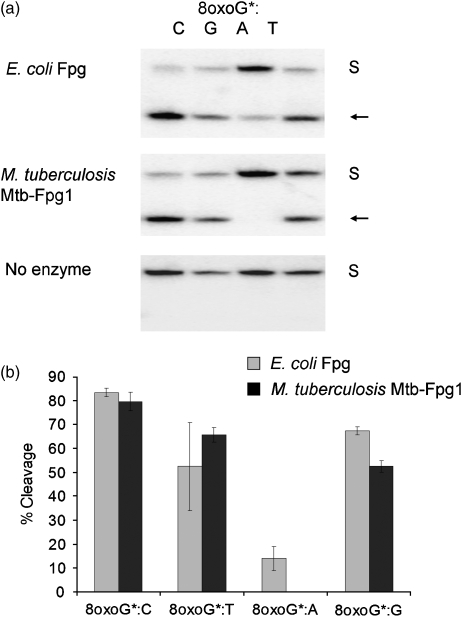
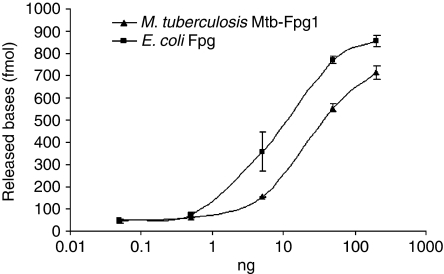
The Mtb-fpg1-encoded protein was purified to homogeneity and found to migrate at c. 32 kDa in sodium dodecyl sulfate-PAGE, corresponding to the molecular weight predicted from the genome sequence (31.95 kDa) (data not shown). The ability of recombinant M. tuberculosis Mtb-Fpg1 to remove an 8oxoG lesion was compared with that of Fpg Ec. Mtb-Fpg1 was able to remove 8oxoG when paired with C, G and T, while no activity was detected against 8oxoG:A (Fig. 1). In addition to DNA glycosylase activity, recombinant Mtb-Fpg1 protein also possessed strand cleavage activity (Fig. 1a). Mtb-Fpg1 has not been tested previously for activity toward substrates other than 8oxoG. Another typical substrate for Fpg protein is faPy residues. The M. tuberculosis Mtb-Fpg1 protein was tested for the ability to remove met-faPy lesions in DNA, and a similar capacity for Mtb-Fpg1 to excise met-faPy residues was observed as for Fpg Ec (Fig. 2). The recombinant Mtb-Fpg1 protein was further assessed for its ability to remove oxidized pyrimidines, 5OHC, 5OHU and diHU, in addition to uracil, alkylated bases and adenine opposite 8oxoG. Some cleavage activity was detected toward 5OHC, whereas no activity was present toward the other substrates (Table S3). In conclusion, these results demonstrate that the protein encoded by M. tuberculosis Mtb-fpg1 is an Fpg DNA glycosylase.
Fig. 1.
DNA glycosylase activity of Mycobacterium tuberculosis Mtb-Fpg1. (a) An aliquot of 50 ng of purified Mtb-Fpg1 or Fpg Ec was incubated with 10–50 fmol of a 24-bp duplex oligodeoxyribonucleotide containing a single 8oxoG residue opposite G, C, A or T. Base excision and strand cleavage were analyzed by 20% PAGE and phosphorimaging. The arrow indicates the cleaved DNA substrate. *P32-labeled strand. S, substrate. (b) Quantification of strand cleavage activity. The results represent the averages of three independent experiments and error bars indicate the SE of the mean.
Fig. 2.
FaPy–DNA glycosylase activity of Mycobacterium tuberculosis Mtb-Fpg1. Removal of met-faPy from [3H]-methyl-faPy-poly(dG–dC) DNA by increasing amounts of purified Mtb-Fpg1 (triangles) and Fpg Ec (squares). Results represent the averages of three independent experiments and error bars indicate SE of the mean.
Characterization of the Mtb-fpg1 and rnc intergenic region in mycobacterial species with MIRU-VNTR repeats
The sequences of the Mtb-fpg1 gene and flanking regions were compared in a panel of mycobacterial species for which the complete genome sequences were available. The rnc, Rv2927c and Rv2926c genes were present in all the mycobacterial species examined, and their location and deduced amino acid sequences were highly conserved (data not shown). However, the intergenic region between rnc and Mtb-fpg1 revealed significant differences in the number of base pairs. The corresponding intergenic region in M. tuberculosis (strains H37Rv, CDC1551, 210, C and F11), M. bovis and M. microti contained 358, 303 and 279 bp (Table 2), respectively, and these regions consisted of a complex organization of multiple direct repeats. In contrast, in the non-MTC members, the corresponding intergenic region contained only a few base pairs from the stop codon of rnc to the start codon of the Mtb-fpg1 orthologs (Table 2).
Table 2.
Mycobacterial species with MIRU-VNTR repeats
| Mycobacterium species | Intergenic region rnc-Mtb-fpg1 (bp) |
|---|---|
| M. tuberculosis H37Rv* | 358 |
| M. tuberculosis CDC1551* | 358 |
| M. tuberculosis 210† | 358 |
| M. tuberculosis C† | 358 |
| M. tuberculosis F11* | 358 |
| M. bovis AF2122/97* | 303 |
| M. microti OV254† | 279 |
| M. leprae TN* | 54 |
| M. a. paratuberculosis K10* | 20 |
| M. avium 104* | 20 |
| M. smegmatis mc2 155* | 6 |
| M. sp. JLS* | 6 |
| M. sp. KMS* | 6 |
| M. sp. MCS* | 6 |
| M. vanbaalenii PYR-1* | 6 |
| M. flavescens PYR-GCK†‡ | 6 |
| M. marinum (M strain)* | 3 |
| M. ulcerans Agy99* | 1 |
Completely sequenced genome.
Incompletely sequenced genome.
Sequence homology to Mycobacterium sp. JLS, Mycobacterium sp. KMS and Mycobacterium sp. MCS, as well as Mycobacterium vanbaalenii indicate no more than 6 bp from the stop codon of rnc to the start codon of fpg.
Species belonging to the Mycobacterium tuberculosis complex (above the line) exhibit long intergenic regions between rnc and Mtb-fpg1 genes, contrasting those in non-M. tuberculosis complex species (below the line). The length of the intergenic region is a marker for the presence of repeats.
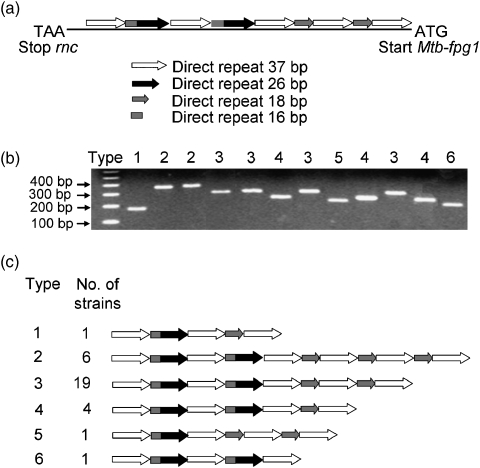
The intergenic region between Mtb-fpg1 and rnc in M. tuberculosis H37Rv described above consisted of a complex structure of exact, direct repeats (Fig. 3a). This structure contained five identical copies of a 37-bp repeat, where the last two repeats were preceded by a smaller 18-bp repeat. The second and third repeats were both preceded by a 26-bp repeat element and a 16-bp element that was identical to the 5′ end of the 18-bp repeat (Fig. 3a). The attenuated M. tuberculosis H37Ra is predicted to have an ORF encoding a 63 amino acid long hypothetical protein, MRA_2951, within this intergenic region (Zheng et al., 2008). Even though the identical DNA sequence is found in other completed M. tuberculosis genome sequences, this ORF is not annotated in those. Furthermore, the complete genome sequence of M. tuberculosis H37Ra is not yet validated, weakening the significance of the putative ORF MRA_2951. To assess whether this repeat organization was representative for that in other M. tuberculosis isolates, the corresponding regions upstream of Mtb-fpg1, in addition to the Mtb-fpg1 genes themselves, from 32 clinical isolates were subjected to PCR and DNA sequence analysis. The results showed that the Mtb-fpg1 gene was almost completely conserved, and only a single point mutation, inducing an amino acid change in a region flanking an active domain, was found in a M. tuberculosis isolate (NLA000301029). In contrast, a total of six different classes of repeat organization patterns upstream of Mtb-fpg1 were observed among these isolates (Fig. 3b). The most predominant variant, termed type 3, was identical to the repeat structure found in M. tuberculosis H37Rv, while the type 4 was the variant also found in M. bovis AF2122/97. The structures of the various repeats are shown in Fig. 3c.
Fig. 3.
The Mycobacterium tuberculosis Mtb-fpg1 region contains variable numbers of repeat units. (a) Organization of the repetitive elements in M. tuberculosis H37Rv. The start of the repetitive structure is located 40 bp after the stop codon of rnc and ends at the start codon of Mtb-fpg1. Arrows indicate length of the repeats. In direct repeat 18 bp, 16 out of 18 consecutive base pairs are identical to direct repeat 16 bp. (b) Variation in the rnc-Mtb-fpg1 intergenic region in M. tuberculosis isolates monitored by PCR amplification. Number indicates PCR product size/repeat type according to (c). (c) The organization of the different types of M. tuberculosis Mtb-fpg1 tandem repeat regions found in 32 M. tuberculosis strains. Arrows as in (a).
Correlation of M. tuberculosis strain typing and repeat occurrence
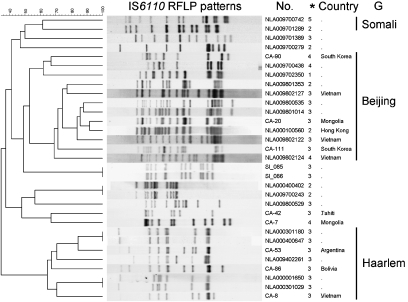
IS6110 RFLP typing of 31 selected M. tuberculosis strains, originating from various parts of the world, showed a high degree of DNA polymorphism among the isolates (Fig. 4). Eight RFLP patterns were clustered into four groups of two identical patterns each. These isolates represented microepidemics of tuberculosis in the Netherlands. In order to investigate possible differences between nonclustered, endogenous reactivation cases (from which no transmission had occurred) and clustered cases, isolates representing each of the two identical patterns were also included in the collection (Table S1). However, no clear correlation was observed between the rnc-Mtb-fpg1 TRT and IS6110 RFLP type, genotype or level of success among the strains. In isolates of the Somali and Beijing genotype, various rnc-Mtb-fpg1 TRTs were observed. For instance, the Beijing strains collectively exhibited TRTs designated 1, 2, 3 and 4, although TRT 3 was generally most abundant. In contrast, for the isolates belonging to other IS6110 RFLP clusters and Haarlem genotype, identical rnc-Mtb-fpg1 TRTs, were observed within each of these groups, indicating that these repeats evolve more slowly than IS6110, or that other repeat variants are selected against.
Fig. 4.
IS6110 RFLP patterns of the Mycobacterium tuberculosis strains. IS6110 RFLP patterns of the M. tuberculosis strains used in this study. *rnc-Mtb-fpg1 repeat type; G, genotype.
The relationship between tandem repeat length and Mtb-fpg1 expression levels
The four ORFs, Rv2927c (hypothetical protein), Rv2926c (hypothetical protein), rnc and Mtb-fpg1, were located on the same strand, and they were predicted to be part of the same operon in M. a. paratuberculosis and M. leprae using VIMSS Operon Prediction. This algorithm might, however, not be ideal for mycobacterial operon prediction. In M. tuberculosis species, due to the long distance between rnc and Mtb-fpg1, only Rv2927c, Rv2926c and rnc were predicted to be in the same operon.
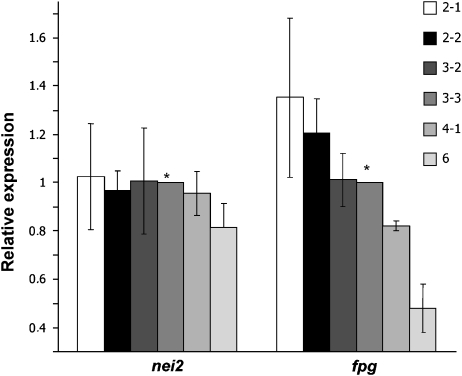
The transcription levels of M. tuberculosis Mtb-fpg1 was assessed using quantitative real time RT-PCR in representative M. tuberculosis strains containing different TRTs (Fig. 5). The transcription level of Mtb-fpg1 mRNA was compared with the transcription level of nei2 mRNA. While the transcription of nei2 mRNA level was more or less the same in the strains examined (P>0.05), the level of Mtb-fpg1 transcription decreased with decreasing intergenic repeat length (Fig. 5). An increase in Mtb-fpg1 expression level was observed in strains NLA000100560 (2-1) (P<0.05) and NLA009801353 (2-2) (P<0.05), which contain the longest repeat, as compared with H37Rv, while strain NLA009802122 (3-2) (P>0.05) exhibited an Mtb-fpg1 expression level equal to H37Rv (3-3). Strain NLA009700438 (4) (P<0.05) showed lower Mtb-fpg1 expression level than H37Rv. Strain 29593 (6) (P<0.05) had the lowest expression level of Mtb-fpg1. Even though the fold changes in the Mtb-fpg1 expression levels were modest among the strains tested, the differences between the changes observed were statistically significant. In general, the longer tandem repeat regions within a strain in our experiments were correlated with higher mRNA levels of Mtb-fpg1. Taken together, the results indicate that the length of the tandem repeat units influences Mtb-Fpg1 expression levels.
Fig. 5.
Expression analysis of Mycobacterium tuberculosis Mtb-fpg1. Analysis of the relative expression of Mtb-fpg1 and nei2 (Rv2464c) mRNA in M. tuberculosis strains containing different TRTs upstream of the Mtb-fpg1. Data are expressed as fold change relative to M. tuberculosis H37Rv Mtb-fpg1 or nei2 expression when normalized to sigA. Data represent the average of six independent experiments. Error bars indicate SE of the mean. TRT as in Fig. 3c. Strains: 2-1, NLA0001000560; 2-2, NLA009801353; 3-2, NLA009802122; 3-3, H37Rv; 4, NLA 009700438; 6, 29593. *Calibrator.
Discussion
The M. tuberculosis genome sequence has revealed the presence of putative gene homologs encoding proteins involved in BER, nucleotide excision repair, recombinational repair and SOS-induced DNA damage response (Cole et al., 1998, 2001; Mizrahi & Andersen, 1998). However, no M. tuberculosis gene homologs encoding mismatch repair components have been recognized so far. Notably, only a few M. tuberculosis DNA repair components have been characterized to date. However, the lifestyle and importance of this pathogen, in particular its ability to replicate or stay dormant and survive within human macrophages, warrant the elucidation of the role of DNA repair in M. tuberculosis. Here, we report the characterization of M. tuberculosis Mtb-Fpg1, a DNA glycosylase shown to be involved in the defence against oxidative DNA damage. In addition, the repeat sequence motif immediately upstream of Mtb-fpg1 was characterized, and genome sequence analyses demonstrated that this particular sequence motif was unique to species belonging to MTC. Gene expression analysis was used to assess whether there was a link between the tandem repeat length and Mtb-fpg1 expression levels, and a potential correlation was observed. This finding suggests that the rnc-Mtb-fpg1 tandem repeat units may be adaptive and could potentially affect the regulation of M. tuberculosis oxidative DNA damage repair.
Multiple genes encoding DNA glycosylases belonging to the Fpg/Nei family are present in MTC and the M. avium complex (Table 1), suggesting that the repair of oxidative DNA lesions is well developed in these mycobacteria. Alternatively, these components might exert different functions in genome maintenance. The highly human-adapted M. leprae genome, which has undergone major reductive evolution, has retained a single conserved fpg gene, indicating that the Fpg family of enzymes plays a central role in this pathogen. The presence of multiple fpg/nei homologs in other mycobacteria, on the other hand, suggests a need for versatile mechanisms to remove oxidized bases, and may indicate that the various fpg gene copies might be differentially expressed under variable conditions. Despite the limited data available on gene expression profiles of Fpg/Nei DNA glycosylases, it has been shown previously that M. tuberculosis Rv3297 (Nei) is induced by DNA damage by a RecA-independent pathway (Rand et al., 2003). None of the four M. tuberculosis putative Fpg/Nei homologs have been shown to be essential for optimal growth (Sassetti et al., 2003). Moreover, a study of M. tuberculosis transposon mutants did not exhibit reduced virulence of H37Rv Mtb-fpg1 mutants in severe combined immunodeficiency mice (McAdam et al., 2002). However, none of these studies were conducted with the entire mycobacterial Fpg/Nei family inactivated, and, thus, the significance of mycobacterial Fpg/Nei family components in fitness for survival and pathogenesis remains unknown. From both prokaryotic and eukaryotic organisms, it is known that there are multiple back-up systems among DNA glycosylases, to the extent that a single null mutant might not exhibit a clear phenotype.
Mtb-Fpg1 has been annotated as an Fpg homolog due to the presence of conserved domains and residues in the deduced amino acid sequence (Cole et al., 1998). We have demonstrated that purified Mtb-Fpg1 possesses both DNA glycosylase and strand-nicking activities on representative Fpg substrates. Our results corroborate with recent characterization of M. tuberculosis and M. smegmatis Fpg/Nei orthologs (Table S4). Recently, Jain et al. (2007) showed that an M. smegmatis mutant lacking the Mtb-Fpg1 ortholog is devoid of 8oxoG repair in cell-free extracts and that this mutant is more sensitive to H2O2 than the wild type. Both 8oxoG repair and survival can be restored by complementing the mutant with a plasmid containing M. tuberculosis Mtb-fpg1 (Jain et al., 2007). Mtb-Fpg1 is thus a distinct part of the defence system against oxidative DNA damage in M. tuberculosis.
The MTC genomes are genetically highly conserved and variation seems to be more frequently caused by insertions and deletions rather than by base substitutions (Zheng et al., 2008). Furthermore, a large amount of variation between and within the genomes of Mycobacterium species is due to the presence of repetitive sequence elements (Cole et al., 1998; Supply et al., 2000). Here, we show that the Mtb-fpg1 gene itself was almost completely conserved among M. tuberculosis isolates, while polymorphisms in the tandem repeat sequence immediately upstream of Mtb-fpg1 were identified. Organized repetitive sequence elements showing polymorphisms consisting of different numbers of tandem repeats are quite common in the M. tuberculosis genome and have been referred to as VNTRs, MIRUs or ETRs (Frothingham & Meeker-O'Connell, 1998; Smittipat & Palittapongarnpim, 2000; Supply et al., 2000; Spurgiesz et al., 2003). The tandem repeat units in the rnc-Mtb-fpg1 intergenic region characterized here have previously been designated as ETR-F and VNTR3239 and have been used in M. tuberculosis strain typing (Frothingham & Meeker-O'Connell, 1998; Smittipat & Palittapongarnpim, 2000; Supply et al., 2000; Spurgiesz et al., 2003). This VNTR3239 is more complex than other VNTRs and is composed of 55-bp (37-bp+18-bp) and 79-bp (37-bp+26-bp+16-bp) repeat units of tandem repeats (Fig. 3a), while other VNTRs of M. tuberculosis are usually simple repeats of a single kind of sequence (Smittipat & Palittapongarnpim, 2000; Smittipat et al., 2005). It has been shown that this locus exhibited variation in Beijing strains (Spurgiesz et al., 2003), corroborating with our results. These repeats may be predicted to contribute to mycobacterial genome dynamics. Potentially, different tandem repeat constellations might affect DNA folding and consequent affinity, binding and interactions of transcription factors in a diversified manner, thereby affecting gene expression. Alternatively, the tandem repeats themselves might act as an enhancer by containing a variable number of binding sites for a regulator, as is the case, for example, in the regulation of serotonin transporter gene expression in brain disorders (Haddley et al., 2008). Our data showed that the Mtb-fpg1 gene was indeed differentially expressed in M. tuberculosis strains with long and short tandem repeat regions under standard cultivation condition. Further studies on the influence of tandem repeats on the expression of this and other genes will extend the knowledge about the function of these repeats in M. tuberculosis genome dynamics. Finally, the multiplicity of mycobacterial Fpg/Nei homologs and their relative role(s) in oxidative DNA damage repair awaits clarification, in order to elucidate the individual and collective physiological contributions of these homologs in intracellular survival and virulence under oxidative stress.
Acknowledgments
This work was supported by grants from the Research Council of Norway. We thank M. Bjørås and S.A. Frye for constructive discussions, O.H. Ambur for critical reading of the manuscript, and L. Hestvik for technical assistance.
Authors' contribution
I.O. and S.V.B. contributed equally to this work.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Predicted domains and motifs in Fpg/Nei homologs.
Table S1. Bacterial strains and plasmids included in the study.
Table S2. DNA sequences of oligonucleotides employed in the study.
Table S3. Substrate specificity of Mycobacterium tuberculosis Mtb-Fpg1.
Table S4. Overview of characterization of mycobacterial Fpg/Nei orthologs to date.
Please note: Wiley-Blackwell is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Alseth I, Eide L, Pirovano M, Rognes T, Seeberg E, Bjørås M. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol Cell Biol. 1999;19:3779–3787. doi: 10.1128/mcb.19.5.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat M, Gerlt JA. 3′- and 5′-strand cleavage reactions catalyzed by the Fpg protein from Escherichia coli occur via successive beta- and delta-elimination mechanisms, respectively. Biochemistry. 1996;35:659–665. doi: 10.1021/bi9522662. [DOI] [PubMed] [Google Scholar]
- Bjelland S, Seeberg E. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res. 2003;531:37–80. doi: 10.1016/j.mrfmmm.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Boiteux S, Huisman O, Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO J. 1984;3:2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Eide L, Bjørås M, Pirovano M, Alseth I, Berdal KG, Seeberg E. Base excision of oxidative purine and pyrimidine DNA damage in Saccharomyces cerevisiae by a DNA glycosylase with sequence similarity to endonuclease III from Escherichia coli. P Natl Acad Sci USA. 1996;93:10735–10740. doi: 10.1073/pnas.93.20.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Alland D, Eisen JA, et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frothingham R, Meeker-O'Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- Garnier T, Eiglmeier K, Camus JC, et al. The complete genome sequence of Mycobacterium bovis. P Natl Acad Sci USA. 2003;100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddley K, Vasiliou AS, Ali FR, Paredes UM, Bubb VJ, Quinn JP. Molecular genetics of monoamine transporters: relevance to brain disorders. Neurochem Res. 2008;33:652–667. doi: 10.1007/s11064-007-9521-8. [DOI] [PubMed] [Google Scholar]
- Jain R, Kumar P, Varshney U. A distinct role of formamidopyrimidine DNA glycosylase (MutM) in down-regulation of accumulation of G, C mutations and protection against oxidative stress in mycobacteria. DNA Repair. 2007;6:1774–1785. doi: 10.1016/j.dnarep.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Kamerbeek J, Schouls L, Kolk A, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer K, van Soolingen D, Frothingham R, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol. 1999;37:2607–2618. doi: 10.1128/jcm.37.8.2607-2618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer K, Au BK, Yip PC, Skuce R, Supply P, Kam KM, van Soolingen D. Use of variable-number tandem-repeat typing to differentiate Mycobacterium tuberculosis Beijing family isolates from Hong Kong and comparison with IS6110 restriction fragment length polymorphism typing and spoligotyping. J Clin Microbiol. 2005;43:314–320. doi: 10.1128/JCM.43.1.314-320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdalena J, Vachee A, Supply P, Locht C. Identification of a new DNA region specific for members of Mycobacterium tuberculosis complex. J Clin Microbiol. 1998;36:937–943. doi: 10.1128/jcm.36.4.937-943.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam RA, Quan S, Smith DA, et al. Characterization of a Mycobacterium tuberculosis H37Rv transposon library reveals insertions in 351 ORFs and mutants with altered virulence. Microbiology. 2002;148:2975–2986. doi: 10.1099/00221287-148-10-2975. [DOI] [PubMed] [Google Scholar]
- Melamede RJ, Hatahet Z, Kow YW, Ide H, Wallace SS. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- Mizrahi V, Andersen SJ. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol Microbiol. 1998;29:1331–1339. doi: 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- Price MN, Huang KH, Alm EJ, Arkin AP. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 2005;33:880–892. doi: 10.1093/nar/gki232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rad ME, Bifani P, Martin C, et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg Infect Dis. 2003;9:838–845. doi: 10.3201/eid0907.020803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand L, Hinds J, Springer B, Sander P, Buxton RS, Davis EO. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol Microbiol. 2003;50:1031–1042. doi: 10.1046/j.1365-2958.2003.03765.x. [DOI] [PubMed] [Google Scholar]
- Riazuddin S, Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978;17:2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory press; 2001. [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Seeberg E, Eide L, Bjørås M. The base excision repair pathway. Trends Biochem Sci. 1995;20:391–397. doi: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- Sidorenko VS, Rot MA, Filipenko ML, Nevinsky GA, Zharkov DO. Novel DNA glycosylases from Mycobacterium tuberculosis. Biochemistry. 2008;73:442–450. doi: 10.1134/s0006297908040093. [DOI] [PubMed] [Google Scholar]
- Smittipat N, Palittapongarnpim P. Identification of possible loci of variable number of tandem repeats in Mycobacterium tuberculosis. Tubercle Lung Dis. 2000;80:69–74. doi: 10.1054/tuld.2000.0236. [DOI] [PubMed] [Google Scholar]
- Smittipat N, Billamas P, Palittapongarnpim M, et al. Polymorphism of variable-number tandem repeats at multiple loci in Mycobacterium tuberculosis. J Clin Microbiol. 2005;43:5034–5043. doi: 10.1128/JCM.43.10.5034-5043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurgiesz RS, Quitugua TN, Smith KL, Schupp J, Palmer EG, Cox RA, Keim P. Molecular typing of Mycobacterium tuberculosis by using nine novel variable-number tandem repeats across the Beijing family and low-copy-number IS6110 isolates. J Clin Microbiol. 2003;41:4224–4230. doi: 10.1128/JCM.41.9.4224-4230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol. 1997;26:991–1003. doi: 10.1046/j.1365-2958.1997.6361999.x. [DOI] [PubMed] [Google Scholar]
- Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- Supply P, Allix C, Lesjean S, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol. 2006;44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Soolingen D, de Haas PE, Hermans PW, Groenen PM, van Embden JD. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viana-Niero C, Gutierrez C, Sola C, Filliol I, Boulahbal F, Vincent V, Rastogi N. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J Clin Microbiol. 2001;39:57–65. doi: 10.1128/JCM.39.1.57-65.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zharkov DO, Ishchenko AA, Douglas KT, Nevinsky GA. Recognition of damaged DNA by Escherichia coli Fpg protein: insights from structural and kinetic data. Mutat Res. 2003;531:141–156. doi: 10.1016/j.mrfmmm.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Zheng H, Lu L, Wang B, et al. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One. 2008;3:e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.