Abstract
Hfq is a highly conserved pleiotropically acting prokaryotic RNA-binding protein involved in the post-transcriptional regulation of many stress-responsive genes by small RNAs. In this study, we show that Hfq of the strictly human pathogen Neisseria meningitidis is involved in the regulation of expression of components involved in general metabolic pathways, iron metabolism and virulence. A meningococcal hfq deletion strain (H44/76Δhfq) is impaired in growth in nutrient-rich media and does not grow at all in nutrient-limiting medium. The growth defect was complemented by expression of hfq in trans. Using proteomics, the expression of 28 proteins was found to be significantly affected upon deletion of hfq. Of these, 20 proteins are involved in general metabolism, among them seven iron-responsive genes. Two proteins (PilE, TspA) are involved in adherence to human cells, a step crucial for the onset of disease. One of the differentially expressed proteins, GdhA, was identified as an essential virulence factor for establishment of sepsis in an animal model, studied earlier. These results show that in N. meningitidis Hfq is involved in the regulation of a variety of components contributing to the survival and establishment of meningococcal disease.
Keywords: Neisseria meningitidis, Hfq, proteomics, riboregulation
Introduction
A prokaryotic riboregulated network, in which small RNAs (sRNAs), in conjunction with specific proteins, regulate the translation and/or the decay of mRNAs has recently been discovered (Majdalani et al., 2005). These regulatory events commonly require the action of the Sm-like protein Hfq. Hfq is a strikingly conserved pleiotropically acting RNA-binding protein, facilitating base pairing between sRNA and mRNA, which in general may either decrease ribosome binding or unmask the RNAseE cleavage site, leading to mRNA decay, or may improve ribosome binding, leading to mRNA stability (Valentin-Hansen et al., 2004). Recent studies have shown that Hfq extensively impacts bacterial physiology, including control of virulence factors. Null mutants of hfq of a variety of pathogens are highly attenuated in animal models (Geissmann & Touati, 2004; Valentin-Hansen et al., 2004; Brennan & Link, 2007; Sittka et al., 2007).
The strictly human pathogen Neisseria meningitidis causes septicemia and meningitis, a life-threatening disease, especially in childhood, and is a serious public health problem worldwide (de Souza & Seguro, 2008). This pathogen possesses a variety of genes involved in the adaptation to the different environments encountered in the host, including iron depletion (Grifantini et al., 2003; Delany et al., 2004; Basler et al., 2006). All four available completely sequenced genomes of N. meningitidis contain a gene with significant homology to hfq (Parkhill et al., 2000; Tettelin et al., 2000; Bentley et al., 2007; Peng et al., 2008). Hfq of N. meningitidis was identified as an essential gene for the onset of septicemia using signature-tagged transposon-mutated meningococci in an infant rat model (Sun et al., 2000). This observation strongly suggests that Hfq and genes under control of Hfq in the meningococcus are of importance for establishing disease. To identify which genes of N. meningitidis are regulated by Hfq, we constructed an hfq knock-out strain and used a proteomic approach to identify proteins whose expression is under control of Hfq.
Materials and methods
Bacterial strains and culture conditions
Neisseria meningitidis strain H44/76, B: P1.7,16: F3-3: ST-32 (cc32), is closely related to the sequenced serogroup B strain MC58 and belongs to the same clonal complex (van der Ende et al., 1999). Meningococci were cultured in tryptic soy broth (TSB) (BD), GC broth or on GC plates (Difco) supplemented with 1% (v/v) Vitox (Oxoid) at 37 °C in a humidified atmosphere of 5% CO2 (van der Ende et al., 1999). If appropriate, plates or broth were supplemented with erythromycin (5 μg mL−1) and/or chloramphenicol (25 μg mL−1). Growth was monitored by measuring the OD600 nm of cultures at regular time intervals. Growth experiments were repeated five times.
Construction of an hfq knock-out mutant of N. meningitidis
An N. meningitidis H44/76 hfq knock-out mutant (H44/76Δhfq) was constructed using the PCR-ligation-PCR method (Ali & Steinkasserer, 1995; van der Ende et al., 1999). PCR products were generated with primer pairs ABHfq1/ABHfq2 and ABHfq3/ABHfq4, ligated and the ligation product was reamplified with the primer pair ABHfq1/ABHfq4. The resulting PCR product was cloned into pCR2.1 (Invitrogen). The EcoRI-digested erythromycin resistance cassette from pAErmC' (Zhou & Apicella, 1996) was introduced into the created unique MfeI restriction site, yielding plasmid pHfq10. H44/76Δhfq was generated by natural transformation of strain H44/76 with pHfq10 and selection for erythromycin resistance. Replacement of hfq (NMB0748) by the erythromycin cassette was confirmed by PCR with combinations of primers JP19-20, JP22, ABHfq1 and ABHfq4. Oligonucleotides are listed in Table 1.
Table 1.
Oligonucleotides used in this study
| Name | Sequence 5′–3′ | GenBank accession number | Location |
|---|---|---|---|
| ABHfq1 | CCGGCGGCATGGGCGCAT | AE002098.2* | 780118..780135 |
| ABHfq2 | TTTTAACTCCGTTATTATGATTGTG | AE002098.2 | 779576..779600 |
| ABHfq3 | ACAATTGAATCCGCACGAAGCATGA | AE002098.2 | 779281..779264 |
| ABHfq4 | CAGGTTTTCATGTCCGTCCA | AE002098.2 | 778947..778966 |
| ALHfq11 | GGGGCATATGACAGCTAAAGGACAA | AE002098.2 | 779570..779556 |
| ALHfq12 | CCCCTCATGATTCGGCAGGCTGCTGGAC | AE002098.2 | 779283..779300 |
| ALHfq13 | CAGAGAAGGCATGTGGAACA | AE002098.2 | 779891..779872 |
| ALHfq14 | TCAGGTTGAGTCTTTCGATCA | AE002098.2 | 779469..779449 |
| ALHfq15 | TGGGTGACGGAAGTGTTTCT | AE002098.2 | 779413..779432 |
| ALHfq16 | GATGTCGAATGCCCACACTT | AE002098.2 | 779037..779056 |
| NMB0747F | ATGTTATTGCAAAACATCCTTC | AE002098.2 | 779195..779174 |
| NMB0747R | TTATTTTTGACGCAGTTTTTCA | AE002098.2 | 778629..778650 |
| JP19 | TAAATACAAAACGCTCATTGGC | M17990.1† | 2086..2107 |
| JP20 | ACCTCTTTACTAATTCAAGGGT | M17990.1 | 1813..1833 |
| JP22 | AAATCGTCAATTCCTGCATGTT | M17990.1 | 2329..2350 |
Genomic localization according to
Tettelin et al. (2000), GenBank AE002098.2 and
Projan et al. (1987), GenBank M17990.1.
Complementation of H44/76Δhfq
To complement the hfq deletion, hfq from strain H44/76 was amplified with the primer pair ALHfq11/ALHfq12, containing NdeI and RcaI restriction sites, respectively. The resulting PCR product and shuttle vector pEN11-pldA (Bos et al., 2005) were digested with NdeI and RcaI, and the PCR product was cloned into NdeI–RcaI-predigested pEN11-pldA and transformed into Escherichia coli TOP10F' (Invitrogen). Chloramphenicol-resistant colonies were checked by colony PCR and sequencing, using universal M13 primers. Plasmid DNA of a clone containing a complete intact hfq-coding region gene (pEN11-hfq2) was isolated and used to transform H44/76Δhfq. The expression of hfq was induced by addition of IPTG to the culture medium to a final concentration of 1 mM. The DNA sequence of hfq of H44/76 was deposited into GenBank (FJ606876).
Reverse transcriptase (RT)-PCR
RNA was isolated using the Rneasy® Midi Kit (Qiagen). RT-PCR was performed using SuperScriptIII (Invitrogen). Primer pairs ALHfq13/ALHfq15, ALHfq13/ALHfq16 and ALHfq14/ALHfq16 were used to investigate whether NMB0747-NMB748-NMB749 is transcribed as a polycistronic operon and primer pair ALNMB0747F/ALNMB0747R to investigate the transcription of NMB0747 in H44/76Δhfq.
Cell fractionation
Meningococci were grown in broth until OD600 nm=0.6–0.8, harvested by centrifugation (10 min at 3000 g) and resuspended in 50 mM Tris-HCl (pH 7.8). Of the remaining culture medium, blebs were removed by centrifugation (100 000 g, 1 h, 4 °C). The supernatant thus obtained was used as a source of secreted proteins. Meningococcal cells were disrupted by sonication (Branson B15 Sonifier, 50 W, 10 min, 50% duty cycle, 4 °C), followed by centrifugation (3000 g, 10 min, 4 °C). The supernatant was centrifuged (28 000 g, 30 min, 4 °C) and pellets, containing the cell envelops (inner and outermembranes), were resuspended in 2 mM Tris-HCL (pH 6.8) containing 1% sodium lauroyl sarcosinate and incubated overnight at 4 °C to dissolve inner membranes. The outer membrane fraction was then obtained by centrifugation (100 000 g, 2 h, 4 °C) and dissolved overnight in 200 μL 2 mM Tris-HCl (pH 6.8) at 4 °C. All fractions were stored at −20 °C. Protein concentrations were determined by Protein Assay (Bio-Rad).
One-dimensional (1D) sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
Proteins were resolved by SDS-PAGE (Laemmli, 1970). Gels (12% or 25%) were stained with the PageBlue kit (Fermentas), washed in MilliQ water and stored in 1% acetic acid at 4 °C until the bands of interest were excised for further analysis.
2D gel electrophoresis and matrix-assisted laser deionization/ionization time of flight (MALDI-TOF) MS
Samples were dissolved in 2D sample buffer (7.7 M urea, 2.2 M thiourea, 30 mM Tris-HCl, pH 8.5, 4% CHAPS and a trace of bromophenol blue); 1.25 μL DeStreak solution (GE Healthcare) and 2.5 μL immobilized pH gradient (IPG) buffer, pH 4–7 (GE Healthcare) were added. First dimension IPG-strips (Immobiline DryStrip, pH 4–7, GE Healthcare) were applied on top of the sample solution, covered with oil and incubated overnight at room temperature (RT). Isoelectric focusing was performed on the Protean IEF cell (Bio-Rad) basically with gradually increasing voltage up to 3500 V according to the standard GE Healthcare protocol. After IEF, strips were incubated in 1 mL equilibration buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 2% SDS and a trace of bromophenol blue) with 10 mg dithiothreitol added for 20 min on a shaker at RT. Strips were transferred to 1 mL equilibration buffer with 25 mg iodoacetamide and again incubated for 20 min. The IPG strip was transferred from the tube to the top of a 12% SDS-PAGE gel and the gel was covered with a warm solution of 1% low-melting point agarose, 15% glycerol and a trace of bromophenol blue in Laemmli buffer. Electrophoresis was conducted at 200 V. Gels were stained with PageBlue, washed in MilliQ water and stored in 1% acetic acid at 4 °C until spots of interest were excised for further analysis. MALDI-TOF MS was carried out as described previously (Pannekoek et al., 2005). All included protein assignments are unambiguous. Monoisotopic peaks in the peptide mass fingerprint spectrum were searched against the complete nonredundant database of all organisms (MSDB at MASCOT). Only in case of a significant MOWSE score of a meningococcal protein as the top hit was the identification considered reliable (pIs and MWs were also not restricted in the search and were found to match). We only analyzed those proteins whose expression was reproducibly and markedly altered (‘on–off’ proteins) when comparing wt, H44/76Δhfq and pEN11-hfq2 complemented cells (in both 1D and 2D gels). This approach guarantees that the protein identified is indeed the one under Hfq control.
Results
Hfq is conserved among neisserial species and part of a polycistronic operon
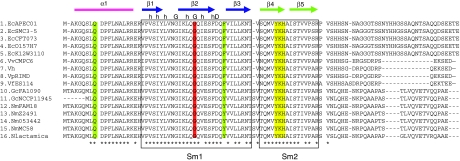
The complete genome sequences of four meningococcal, two strains of the closely related human pathogen Neisseria gonorrhoeae and one commensal neisserial species (Neisseria lactamica) are known (Parkhill et al., 2000; Tettelin et al., 2000; Bentley et al., 2007; Chung et al., 2008; Peng et al., 2008). The amino acid sequence of Hfq of the meningococcal strain used in this study (H44/76) is identical to the Hfq sequence of meningococcal strain FAM18 and 98% identical to the sequences of the other three meningococcal strains. In addition, the sequence of H44/76 is 98% identical to gonococcal Hfq and 95% identical to the Hfq sequence of N. lactamica. The H44/76 neisserial Hfq amino acid sequence shows 63% and 60% identities to Hfq proteins of E. coli and Vibrio spp., respectively (Fig. 1). Of importance, hardly any amino acid substitutions in the meningococcal Hfq sequence, compared with those of E. coli and Vibro spp., are observed in highly conserved regions of the protein shown to be important for functionality (Fig. 1) (Schumacher et al., 2002).
Fig. 1.
Amino acid sequence alignment of Hfq proteins of Escherichia coli serotypes, Vibrio species, Neisseria meningitidis, Neisseria gonorrhoeae strains and Neisseria lactamica. The secondary structural elements of Hfq protein are shown above the alignment with the N-terminal helix α1 in cyan. The Sm1 and Sm2 motif regions are boxed. The sole conserved residue Gly34 is indicated in red. Highly conserved hydrophobic residues within the Sm1 region are indicated by a lower case h, and the two highly conserved glycine and aspartic acid residues within the Sm1 motif are indicated by G and D, respectively. The absolutely conserved glutamine of helix α1 that is important for base recognition and the highly conserved tyrosine (or phenylalanine) are indicated in green. Within the Sm2 region, the ‘Hfq Sm2 motif YKH’ is colored yellow. Other conserved residues are indicated by an asterisk. Note the minimal sequence variation between Hfq of E. coli serotypes (lines 1–5), Vibrio vulnificus (line 6), Vibrio harveyi (line 7), Vibrio parahaemolyticus (line 8), Vibrio fischeri (line 9), N. gonorrhoeae strains (lines 10 and 11), N. meningitidis strains (lines 12-15) and N. lactamica (line 16) (adapted from Schumacher et al., 2002).
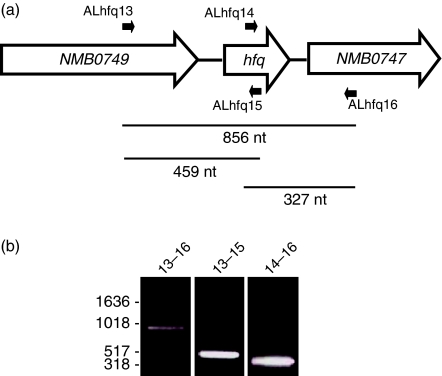
The genetic organization of the chromosomal locus of hfq among all, except one (N. gonorrhoeae FA1090), neisserial strains investigated is also conserved. In neisserial spp. the hfq gene is preceded by a gene encoding d-alanyl-d-alanine endopeptidase (NMB0749, pbp7). Eighty-four basepairs downstream of neisserial hfq, an ORF encoding a conserved hypothetical protein is found (NMB0747, Fig. 2a). To determine whether neisserial hfq was transcriptionally linked to either of the two flanking genes, RT-PCR analysis was performed, templated by total RNA from meningococcal strain H44/76 using primers annealing to pbp7 and the downstream gene. This RT-PCR yielded a product of c. 800 bp, indicating that hfq is transcriptionally linked to pbp7 and NMB0747 (Fig. 2b).
Fig. 2.
Transcriptional analysis of the Hfq operon. (a) Schematic representation of the Hfq polycistronic operon. Primers used in RT-PCR are indicated in black arrows. The size of the calculated RT-PCR products is indicated below the black lines. (b) RT-PCR results. Products obtained by RT-PCR were separated on agarose gel. Numbers on the left represent marker sizes; primer pairs used in the RT-PCR are indicated above the lanes. Reactions in which the addition of reverse transcriptase was omitted did not yield any products (not shown).
Hfq is required for optimal growth of N. meningitidis
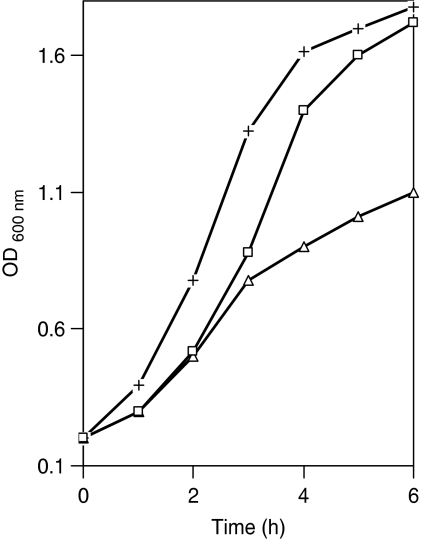
An H44/76Δhfq strain of N. meningitidis was created by complete gene replacement of hfq with an erythromycin resistance cassette. Upon examination of the growth characteristics of the H44/76Δhfq strain, it was observed that this strain formed very tiny colonies after overnight growth on rich solid media, compared with the wild-type (wt) strain. In addition, the strain did not grow in TSB, a relatively nutrient-poor broth, and exhibited a growth deficiency in GC broth, compared with the wt strain (Fig. 3). As the erythromycin resistance cassette used here does not contain a terminator, transcription of NMB0747 should be unaffected. This was confirmed by RT-PCR (not shown). In addition, the expression of hfq in trans in GC broth clearly restored growth (Fig. 3).
Fig. 3.
Growth kinetics of H44/76Δhfq and complementation of the growth defect by expression of neisserial hfq in trans. Growth of the strains at 37°C was followed by measuring the density of the cultures at intervals. +, H44/76 wt strain; ▵, H44/76Δhfq; □, H44/76Δhfq+pEN11-hfq2 (induced by IPTG). A representative experiment is shown.
Identification of proteins differentially expressed in H44/76 wt as compared with the H44/76Δhfq strain
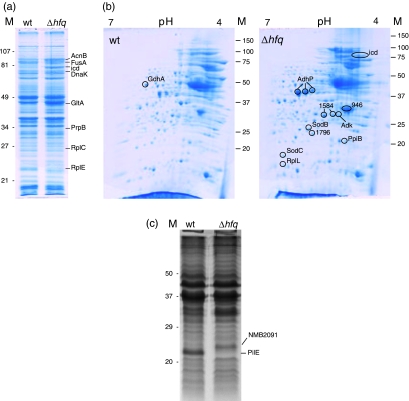
To identify genes whose expression is controlled by Hfq, protein patterns of the wt strain, the H44/76Δhfq strain and the complemented strain were compared by 1D and 2D gels. Only those proteins, whose differential regulation was confirmed in at least two independent experiments and whose expression was turned to wt strain levels in the complemented strain were considered to be truly differentially regulated. Proteomic analyses of patterns of whole-cell lysates, cytoplasm, cell envelops, outer membranes and growth medium (secreted proteins) showed that the expression of at least 28 proteins in N. meningitidis was affected by Hfq (Fig. 4). Of these 28 proteins, 23 were upregulated in the H44/76Δhfq strain while the other five proteins (PilE, RplE, GdhA, AtpA and AtpD) were downregulated in the absence of Hfq (Fig. 4, Table 2). The majority (n=19) of the differentially regulated proteins were identified in the whole-cell lysates and/or cytoplasmic fractions, eight proteins were identified in membrane fractions and one protein (isocitrate dehydrogenase: icd; NMB0920) was identified in both the whole-cell lysate as well as in the secreted protein fraction of the H44/76Δhfq strain (Fig. 4a and b, Table 2).
Fig. 4.
Proteins differentially regulated in H44/76Δhfq. (a) 1D analysis of protein patterns of a whole-cell lysate of wt and H44/76Δhfq cells. (b) 2D analysis of protein patterns of whole-cell lysates of wt (left panel) and H44/76Δhfq cells (right panel). (c) 1D analysis of cell envelops of wt and H44/76Δhfq cells. Differentially expressed proteins comparing wt and H44/76Δhfq cells are indicated. Whole-cell lysates were run on 12% gels, and cell envelops on 25% gels.
Table 2.
Differential regulated proteins in H44/76Δhfq
| Gene ID* | Name† | Location‡ | Reg−§ | Functions¶ | Functional class∥ | MW** | pI†† | Figure‡‡ |
|---|---|---|---|---|---|---|---|---|
| NMB0018 | PilE | CE | − | Type IV pilus assembly protein PilE | Surface structure | 18 072 | 9.6176 | 4c |
| NMB0138 | FusA | W | + | Elongation factor G | Protein translation and modification | 77 244 | 4.8184 | 4a |
| NMB0142 | RplC | W | + | 50S ribosomal protein L3 | Ribosomal protein synthesis and modification | 22 678 | 10.7606 | 4a |
| NMB0154 | RplE | W | − | 50S ribosomal protein L5 | Ribosomal protein synthesis and modification | 20 322 | 10.0609 | 4a |
| NMB0341 | TspA§§ | C | + | T-cell-stimulating protein A | Unknown | 92 488 | 4.0561 | NS |
| NMB0430 | PrpB | W, C | + | Putative carboxyphosphonoenol pyruvate phosphonomutase | Carbohydrate metabolism | 31 714 | 5.0352 | 4a |
| NMB0431 | PrpC | C | + | Methylcitrate synthase | Carbohydrate metabolism | 42 818 | 7.1147 | NS |
| NMB0546 | AdhP | W | + | Alcohol dehydrogenase (propanol preferring) | Fermentation | 36 547 | 5.8432 | 4b |
| NMB0554¶¶ | DnaK | W | + | Molecular chaperone | Environmental information processing | 68 791 | 4.5862 | 4a |
| NMB0589 | RplS | CE | + | 50S ribosomal protein L19 | Ribosomal protein synthesis and modification | 13 767 | 11.0643 | NS |
| NMB0634¶¶ | FbpA | C | + | Iron(III) ABC transporter/major ferric iron-binding protein | Transport/binding proteins | 35 827 | 10.1574 | NS |
| NMB0791¶¶ | PpiB | W | + | Peptidyl-prolyl cis–trans isomerase B (cyclophilin B) | Protein translation and modification | 18 852 | 4.8602 | 4b |
| NMB0823 | Adk | W | + | Adenylate kinase | Purine ribonucleotide biosynthesis | 23 190 | 4.7879 | 4b |
| NMB0884¶¶ | SodB | W, C | + | Superoxide dismutase, Fe–Mn family | Detoxification | 21 892 | 6.1483 | 4b |
| NMB0920 | icd | W, W, SP | + | Isocitrate dehydrogenase | Tricarboxylic acid cycle | 80 163 | 5.6646 | 4a, 4b |
| NMB0946 | – | W, OM | + | Peroxiredoxin 2 family protein | Unknown | 26 912 | 4.5382 | 4b |
| NMB0954 | GltA | W, C | + | Type II citrate synthase | Tricarboxylic acid cycle | 48 121 | 6.7886 | 4a |
| NMB1320¶¶ | RplI | W | + | 50S ribosomal protein L9 | Ribosomal protein synthesis and modification | 15 747 | 7.5349 | 4b |
| NMB1388 | Pgi-2 | C | + | Glucose-6-phosphate isomerase | Glycolysis | 62 084 | 6.5126 | NS |
| NMB1398 | SodC | W | + | Cu–Zn superoxide dismutase | Detoxification, periplasmic protein | 19 520 | 6.6244 | 4b |
| NMB1572¶¶ | AcnB | W, IM | + | Aconitate hydratase | Tricarboxylic acid cycle | 92 715 | 5.2810 | 4a |
| NMB1584 | – | W, C, C | + | 3-Hydroxyacid dehydrogenase | Amino acid metabolism | 30 378 | 5.1348 | 4b |
| NMB1710 | GdhA | W | − | Glutamate dehydrogenase | Amino acid biosynthesis | 48 490 | 6.1895 | 4b |
| NMB1796¶¶ | – | W, C | + | Putative oxidoreductase | Metabolism | 20 686 | 5.8148 | 4b |
| NMB1934 | AtpD | IM | − | ATP synthase F1, β chain | ATP-proton motive force | 50 391 | 4.7746 | NS |
| NMB1936 | AtpA | IM | − | ATP synthase F1, α chain | ATP-proton motive force | 55 291 | 5.2754 | NS |
| NMB2091 | – | CE | + | Putative hemolysin | Membranes, lipoproteins and porins | 21 745 | 10.2186 | 4c |
| NMB2129 | ArgG | IM | + | Argininosuccinate synthase | Amino acid biosynthesis | 49 664 | 4.9527 | NS |
Gene identification according to Tettelin et al. (2000).
Protein name according to Tettelin et al. (2000).
Fraction in which the protein was identified: W, whole cell lysate; C, cytoplasm; CE, cell envelop; IM, innermembrane; OM, outermembrane; SP, secreted protein fraction.
Up- or downregulation in hfq knockout.
Protein function according to KEGG (http://www.genome.jp/kegg).
Functional classification according to Parkhill et al. (2000).
Protein molecular weight according to JCVI-CMR (http://cmr.jcvi.org/cgi-bin/CMR/shared/Menu.cgi?menu=genome).
Isoelectric point according to JCVI-CMR (http://cmr.jcvi.org/cgi-bin/CMR/shared/Menu.cgi?menu=genome).
Figure in which spot and/or band is identified. NS, not shown.
Protein name according to Oldfield et al. (2007).
Iron-responsive gene (Grifantini et al., 2003; Delany et al., 2004; Basler et al., 2006).
Of the 28 proteins differentially expressed between the wt and the H44/76Δhfq strain, 12 are functionally involved in general metabolism and eight are involved in ribosomal protein synthesis/modification, amino acid biosynthesis, protein translation and modification. Two proteins involved in detoxification (SodB and SodC), one iron(III) ABC transporter (FbpA) and one molecular chaperone (DnaK) were also identified. In addition, two proteins with a largely unknown function (encoded by NMB2091 and NMB0946) were also differentially expressed in the H44/76Δhfq strain (Table 2). Of interest, in cell envelops of the H44/76Δhfq strain, PilE, the structural subunit of the Type IV pili (Tfp) and a well-characterized virulence factor involved in adherence to cells and cell motility of meningococci (Nassif et al., 1994; Oldfield et al., 2007), was no longer detectable (Fig. 4c). In addition, a protein designated T-cell-stimulating protein A (TspA), also implicated in adherence of meningococci to cells (Oldfield et al., 2007), was found to be upregulated in H44/76Δhfq.
Discussion
The Hfq protein is recognized as a major post-transcriptional regulator of bacterial gene expression participating as an RNA chaperone in numerous regulatory pathways (Valentin-Hansen et al., 2004). In this study, we explored the Hfq regulon of the strictly human pathogen N. meningitidis.
Loss of Hfq function in the meningococcus (coding capacity c. 2200 genes) (Parkhill et al., 2000) resulted in clear deregulation of c. 28 proteins (>1% of total coding capacity) as found by comparative proteomics. Using approaches comparable to ours, it was found that loss of Hfq function in Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa leads to deregulation of c. 2% and 5% of the total coding capacity, respectively (Sonnleitner et al., 2003; Sittka et al., 2007), and is thus comparable to what we found in meningococci.
The genetic organization of hfq of N. meningitidis, however, is different from that found in most other pathogens studied so far. For example, in E. coli, S. Typhimurium and Listeria monocytogenes, hfq is located downstream from miaA, which encodes a protein similar to tRNA isopentenylpyrophosphate transferase and upstream from hflx, encoding the putative GTP-binding protein (Christiansen et al., 2004; Sittka et al., 2007; Kulesus et al., 2008). In these pathogens, hfq is usually cotranscribed with miaA, and transcription is terminated directly after hfq In N. meningitidis hfq is located downstream from pbp7, encoding d-alanyl-d-alanine-endopeptidase (penicillin-binding protein 7) and upstream of a hypothetical gene encoding a predicted rRNA methylase. We demonstrated by RT-PCR that meningococcal hfq is part of a polycistronic operon and cotranscribed with both of the flanking genes, a situation similar to what is found in Moraxella catarrhalis. In this pathogen, hfq is also cotranscribed with both of the flanking genes (miaA and kpsF, the latter encoding a predicted arabinose-5-phosphate isomerase) (Sittka et al., 2007; Attia et al., 2008; Kulesus et al., 2008). Using an erythromycin cassette without a terminator, we avoided interfering with transcription of NMB0747, part of the polycistron and directly downstream of Hfq. The growth defect observed in meningococci upon deletion of hfq was substantially reversed upon expression of hfq in trans. This observation strongly suggests that Hfq plays an important role in the physiology of the meningococcus and that upon deletion metabolic processes are disturbed, resulting in a phenotype with impaired growth.
In some bacterial species, Hfq is also required for efficient translation of rpoS mRNA, encoding the general stress σ factor, and/or loss of Hfq results in activation of RpoE, the alternative σ factor mediating the response to envelope stress (Hengge-Aronis, 2002; Johansen et al., 2006; Sittka et al., 2007). Analyses of protein patterns on 1 and 2D gels showed that the expression of 28 genes of N. meningitidis is affected by Hfq, either directly or indirectly. The identified genes are most likely not under control of RpoS, because N. meningitidis does not possess an RpoS-like σ factor. Involvement of RpoE, however, cannot be ruled out, because the genome of N. meningitidis does contain a gene (NMB2144) encoding this alternative σ factor. The functionality of RpoE and its possible connection to the Hfq regulon in meningococci remains to be addressed.
Classification of the genes identified by comparative analyses shows that the majority of the encoded proteins belong to the functional classes of metabolism (n=12), ribosomal protein synthesis and modification, amino acid biosynthesis and protein translation and modification (n=8). Among these, the NADP-specific glutamate dehydrogenase (NADP-GDH, encoded by gdhA) was found to be downregulated in the absence of Hfq. This is an interesting observation, because gdhA has been shown to be an essential gene for systemic meningococcal infection in an infant rat model (Sun et al., 2000) and its expression levels are the highest in strains belonging to the hypervirulent lineages ET-5 (serogroup B) and IV-1 (serogroup A) (Pagliarulo et al., 2004). Downregulation of GdhA expression in the absence of Hfq suggests that Hfq might promote translation, for instance by stabilization of gdhA mRNA, and thus contribute to the virulence potential of N. meningitidis. Whether this occurs in conjunction with an sRNA remains to be addressed.
Of the 28 proteins identified, seven are encoded by genes whose expression is under control of iron and/or Fur (Grifantini et al., 2003; Delany et al., 2004; Basler et al., 2006). Recently, an iron- and Fur-regulated sRNA (NrrF) was identified in N. meningitidis (Mellin et al., 2007). This sRNA has been shown to repress expression of succinate dehydrogenase upon iron depletion and its interaction with the sdhCDAB transcript in vitro is enhanced in the presence of Hfq (Metruccio et al., 2009). Although none of the identified proteins belong to the succinate dehydrogenase complex, seven other iron-responsive proteins were differentially expressed in the H44/76Δhfq strain. This suggests that in N. meningitidis Hfq, possibly in conjunction with sRNAs, might also contribute to the fine-tuning of factors involved in iron homeostasis. One of the iron-responsive proteins, which is differentially expressed between wt and H44/76Δhfq, is superoxide dismutase (SodB). In E. coli, SodB is known to be regulated by RyhB, a Fur-regulated Hfq-dependent sRNA (Masse & Gottesman, 2002). Also, in other pathogens, SodB expression is controlled by Hfq and an sRNA (Wilderman et al., 2004; Mey et al., 2005). It is tempting to speculate that in meningococci, SodB expression is also regulated by Hfq in conjunction with an sRNA. This sRNA remains to be identified.
We detected two proteins (PilE and TspA) known to be involved in the adherence of meningococci to human cells. Tfp-mediated adherence of meningococci to human cells leads to clumps of bacteria associated with microvillus-like structures on the surface of the cells (Nassif et al., 1994; Wilderman et al., 2004). This is followed by contact-dependent downregulation of pili and intimate adhesion, mediated by adhesins including TspA (Oldfield et al., 2007). The absence of detectable PilE, the structural subunit of Tfp and the upregulation of TspA expression in H44/76Δhfq suggests a role of Hfq in the meningococcal adherence strategies. The mechanisms responsible for these observations remain to be elucidated, but riboregulatory processes have previously been shown to be involved in Tfp assembly and cell motility of other bacterial pathogens (Sonnleitner et al., 2003; Sittka et al., 2007; Dienst et al., 2008).
Acknowledgments
The authors thank Rolf Meijer for performing initial experiments, Julian Parkhill at the Wellcome Trust Sanger Institute for giving permission to make preliminary sequence data available and Karen S. Moore for critically reading the manuscript. This research was partly funded by the Sixth Framework Programme of the European Commission, Proposal/Contract no.: 512061 (Network of Excellence ‘European Virtual Institute for Functional Genomics of Bacterial Pathogens’, http://www.noe-epg.uni-wuerzburg.de).
References
- Ali SA, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. Biotechniques. 1995;18:746–750. [PubMed] [Google Scholar]
- Attia AS, Sedillo JL, Wang W, Liu W, Brautigam CA, Winkler W, Hansen EJ. Moraxella catarrhalis expresses an unusual Hfq protein. Infect Immun. 2008;76:2520–2530. doi: 10.1128/IAI.01652-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basler M, Linhartova I, Halada P, Novotna J, Bezouskova S, Osicka R, Weiser J, Vohradsky J, Sebo P. The iron-regulated transcriptome and proteome of Neisseria meningitidis serogroup C. Proteomics. 2006;6:6194–6206. doi: 10.1002/pmic.200600312. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Vernikos GS, Snyder LA, et al. Meningococcal genetic variation mechanisms viewed through comparative analysis of serogroup C strain FAM18. PLoS Genet. 2007;3:e23. doi: 10.1371/journal.pgen.0030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos MP, Tefsen B, Voet P, Weynants V, van Putten JP, Tommassen J. Function of neisserial outer membrane phospholipase a in autolysis and assessment of its vaccine potential. Infect Immun. 2005;73:2222–2231. doi: 10.1128/IAI.73.4.2222-2231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr Opin Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung GT, Yoo JS, Oh HB, Lee YS, Cha SH, Kim SJ, Yoo CK. Complete genome sequence of Neisseria gonorrhoeae NCCP11945. J Bacteriol. 2008;190:6035–6036. doi: 10.1128/JB.00566-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delany I, Rappuoli R, Scarlato V. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol Microbiol. 2004;52:1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- De Souza AL, Seguro AC. Two centuries of meningococcal infection: from Vieusseux to the cellular and molecular basis of disease. J Med Microbiol. 2008;57:1313–1321. doi: 10.1099/jmm.0.47599-0. [DOI] [PubMed] [Google Scholar]
- Dienst D, Duhring U, Mollenkopf HJ, Vogel J, Golecki J, Hess WR, Wilde A. The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology. 2008;154:3134–3143. doi: 10.1099/mic.0.2008/020222-0. [DOI] [PubMed] [Google Scholar]
- Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifantini R, Sebastian S, Frigimelica E, Draghi M, Bartolini E, Muzzi A, Rappuoli R, Grandi G, Genco CA. Identification of iron-activated and -repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. P Natl Acad Sci USA. 2003;100:9542–9547. doi: 10.1073/pnas.1033001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol R. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J Mol Biol. 2006;364:1–8. doi: 10.1016/j.jmb.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Kulesus RR, az-Perez K, Slechta ES, Eto DS, Mulvey MA. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect Immun. 2008;76:3019–3026. doi: 10.1128/IAI.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit Rev Biochem Mol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. P Natl Acad Sci USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA. A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J Bacteriol. 2007;189:3686–3694. doi: 10.1128/JB.01890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metruccio MM, Fantappie L, Serruto D, Muzzi A, Roncarati D, Donati C, Scarlato V, Delany I. The hfq-dependent small non-coding (s) RNA NrrF directly mediates Fur-dependent positive regulation of succinate dehydrogenase in Neisseria meningitidis. J Bacteriol. 2009;191:1330–1342. doi: 10.1128/JB.00849-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey AR, Craig SA, Payne SM. Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect Immun. 2005;73:5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif X, Beretti JL, Lowy J, Stenberg P, O'Gaora P, Pfeifer J, Normark S, So M. Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. P Natl Acad Sci USA. 1994;91:3769–3773. doi: 10.1073/pnas.91.9.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield NJ, Bland SJ, Taraktsoglou M, Dos Ramos FJ, Robinson K, Wooldridge KG, a'Aldeen DA. T-cell stimulating protein A (TspA) of Neisseria meningitidis is required for optimal adhesion to human cells. Cell Microbiol. 2007;9:463–478. doi: 10.1111/j.1462-5822.2006.00803.x. [DOI] [PubMed] [Google Scholar]
- Pagliarulo C, Salvatore P, De Vitis LR, et al. Regulation and differential expression of gdhA encoding NADP-specific glutamate dehydrogenase in Neisseria meningitidis clinical isolates. Mol Microbiol. 2004;51:1757–1772. doi: 10.1111/j.1365-2958.2003.03947.x. [DOI] [PubMed] [Google Scholar]
- Pannekoek Y, Heurgue-Hamard V, Langerak AA, Speijer D, Buckingham RH, van der Ende A. The N5-glutamine S-adenosyl-l-methionine-dependent methyltransferase PrmC/HemK in Chlamydia trachomatis methylates class 1 release factors. J Bacteriol. 2005;187:507–511. doi: 10.1128/JB.187.2.507-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Achtman M, James KD, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- Peng J, Yang L, Yang F, et al. Characterization of ST-4821 complex, a unique Neisseria meningitidis clone. Genomics. 2008;91:78–87. doi: 10.1016/j.ygeno.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Projan SJ, Monod M, Narayanan CS, Dubnau D. Replication properties of pIM13, a naturally occurring plasmid found in Bacillus subtilis, and of its close relative pE5, a plasmid native to Staphylococcus aureus. J Bacteriol. 1987;169:5131–5139. doi: 10.1128/jb.169.11.5131-5139.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner E, Hagens S, Rosenau F, Wilhelm S, Habel A, Jager KE, Blasi U. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb Pathog. 2003;35:217–228. doi: 10.1016/s0882-4010(03)00149-9. [DOI] [PubMed] [Google Scholar]
- Sun YH, Bakshi S, Chalmers R, Tang CM. Functional genomics of Neisseria meningitidis pathogenesis. Nat Med. 2000;6:1269–1273. doi: 10.1038/81380. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Saunders NJ, Heidelberg J, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Van der Ende A, Hopman CT, Dankert J. Deletion of porA by recombination between clusters of repetitive extragenic palindromic sequences in Neisseria meningitidis. Infect Immun. 1999;67:2928–2934. doi: 10.1128/iai.67.6.2928-2934.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. P Natl Acad Sc USA. 2004;101:9792–9797. doi: 10.1073/pnas.0403423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Apicella MA. Plasmid with erythromycin resitance and catechol 2,3-dioxigenase- or beta-galactosidase-encoding gene cassettes for use in Neisseria spp. Gene. 1996;171:133–134. doi: 10.1016/0378-1119(96)00103-5. [DOI] [PubMed] [Google Scholar]