Abstract
Because of inherent differences between deceased donor (DD) and living donor (LD) liver grafts, we hypothesize that the molecular signatures will be unique, correlating with specific biologic pathways and clinical patterns.
Microarray profiles of 63 biopsies in 13 DD and 8 LD liver grafts done at serial time points (procurement, backbench, and post-reperfusion) were compared between groups using class comparisons, network and biological function analyses. Specific genes were validated by quantitative PCR and immunopathology. Clinical findings were also compared.
Following reperfusion, 579 genes in DD grafts and 1324 genes in LDs were differentially expressed (p<0.005). Many up-regulated LD genes were related to regeneration, biosynthesis and cell cycle, and a large number of down-regulated genes were linked to hepatic metabolism and energy pathways correlating with post-transplant clinical laboratory findings. There was significant up-regulation of inflammatory/immune genes in both DD and LD, each with a distinct pattern. Gene expression patterns of select genes associated with inflammation and regeneration in LD and DD grafts correlated with protein expression.
Unique patterns of early gene expression are seen in LD and DD liver grafts, correlating with protein expression and clinical results, demonstrating distinct inflammatory profiles and significant down-regulation of metabolic pathways in LD grafts.
Introduction
The use of liver grafts from living donors (LD) for adult recipients was developed to help overcome the disparity between the number of deceased donors (DD) and the growing waitlist. In adult-to-adult LD liver transplantation grafts are smaller than the standard liver volume, with surgical technique and a set of attributes that are distinctive from whole DD grafts and unique risk factors for graft failure (1). Cellular processes for rapid restoration of liver mass in these grafts may compete with the need to maintain systemic metabolic homeostasis and impact immediate graft function. By comparison, DD grafts experience a degree of hepatocellular damage from brain death, donor events, procurement, preservation, and reperfusion injury (2). Identifying these disparate responses to transplantation on a molecular level may provide clues to clinical interventions that can improve outcomes.
Rodent models of partial hepatectomy and transplant provide a large amount of information regarding molecular pathways of liver regeneration, inflammation and repair (3–6) and show that cellular proliferation causes a shift away from liver metabolic function (7–10). Several studies have used microarrays to investigate liver regeneration and injury in rodents (11–14), but few have utilized expression data from the human whole genome (15, 16).
Adult-to-adult LD liver transplantation provides a unique opportunity to explore the recovery of a human liver not subject to the confounding effects of brain death and organ recovery. The cellular and molecular differences between DD and LD grafts with regard to injury, repair and function are not known. We used whole genome-wide expression analysis to assess the immediate phase of liver graft recovery after transplantation comparing human adult-to adult right lobe LD and whole DD transplants. We hypothesized that molecular signatures of LD grafts will differ significantly from DD grafts, such that LD grafts will demonstrate early gene expression profiles linked to hepatic regeneration as well as unique pro-inflammatory profiles. Recognizing these distinctive differences at the molecular level provides an important basis for identifying potential targets for future studies and possible therapeutic intervention.
Methods and Materials
Patients and study design
All study protocols and consent procedures were approved by the Institutional Review Boards of the University of Pennsylvania, Virginia Commonwealth University, and the Medical Advisory Committee of Gift of Life Donor Program. Core biopsies were taken in the LD and from the DD before manipulation of the liver (PRE). A second biopsy was taken on the backbench following cold preservation (COLD). The final biopsy (POST) was taken after reperfusion following completion of the bile duct anastamosis. Cold ischemic time was measured from time of donor cross-clamp to removal from ice. Warm ischemic time was defined as time of removal of liver from ice to reperfusion. Clinical data was collected from donor and recipient charts and from the electronic transplant database at the University of Pennsylvania and Virginia Commonwealth University. This data included donor and recipient demographics, intra-operative details, and postoperative liver function.
Isolation of RNA
Biopsies were placed immediately in RNA Later (Ambion, Austin, TX) and frozen. Total RNA, extracted using Trizol (Invitrogen, Carlsbad, CA), was cleaned on RNEasy columns (Qiagen Inc, Valencia, CA). RNA integrity was confirmed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) (17, 18).
Preparation of labeled cRNA and hybridization
Five μg of mRNA was converted to double-stranded cDNA, then to cRNA using T7 RNA polymerase with biotinylated nucleotides (Ambion MessageAmp II Enhanced Biotin kit). 15 micrograms of fragmented cRNA was hybridized to each Affymetrix HG-U133Plus 2.0 GeneChip (Affymetrix, Santa Clara, CA). Chips were hybridized, washed and stained as described (20–22). Signal intensities were generated using the Affymetrix GeneChipR Operating System (GCOS).
Data analysis
CEL files were normalized with Robust Multichip Average (RMA Express 0.3 beta 1) (19). Class comparisons were done in BRB ArrayTools (http://linus.nci.nih.gov/BRB-ArrayTools.html). Two-class unpaired F-tests were performed using a random variance model. A False DiscoveryRate of <0.1 was chosen to identify differentially expressed genes at a user-defined p-value of 0.005.
Biological function analysis was done with Expression Analysis Systematic Explorer (EASE 2.0; http://david.niaid.nih.gov/david/ease.htm) and the Database for Annotation, Visualization and Integrated Discovery (DAVID; www.david.niaid.nih.gov/david). Functions were found using Gene Ontology (GO) (20). Pathway analysis and additional analysis of gene functions was done with Ingenuity Pathway Analysis (Ingenuity® Systems, Redwood City, CA). The resulting biological networks comprised up to 35 differentially expressed genes, called Focus genes.
RT-qPCR using TaqMan Low Density Arrays (TLDA)
TLDA microfluidic cards (384-well; Applied Biosystems, Foster City, CA) were used to validate differential gene expression of 63 genes, chosen for validation from the results of the class comparisons. Total RNA was extracted from the biopsies using TRIZOL (Invitrogen). Contaminating genomic DNA was removed using DNA-free (Ambion). For RT-qPCR, cDNA was transcribed from 2μg total RNA using the cDNA archive kit (Applied Biosystems). All transcript assays consisted of two unlabeled PCR primers and a FAM dye-labeled TaqMan® MGB probe, pre-spotted on the TLDA card. All amplifications were done in triplicate and threshold cycle (Ct) scores were averaged to calculate relative expression. The Ct scores were normalized against 18S rRNA controls by the RQ Manager 1.2 software (Applied Biosystems).
Immunopathology
Immunohistopathology was used to correlate gene expression with protein secretion. Paraffin-embedded PRE and POST liver sections from 4 LD and 4 DD grafts were stained for each of the chosen associated proteins. Sections were stained using an Envision immunoperoxidase kit (Dako) and antibodies to SOCS3 (Santa Cruz Biotech), IL1RL1 (Sigma), IL-8 (Epitomics) plus cell-lineage markers and isotype-matched controls.
Supplementary Data
The .CEL files for all the microarrays in this study are in the Gene Expression Omnibus (GEO): (www.ncbi.nlm.nih.gov/geo). Supplementary data tables for all the class comparisons are at: (www.scripps.edu/mem/eht/salomon/livertxdata/).
Results
Patient characteristics
A total of 21 patients, 13 DD recipients and 8 right lobe LD recipients had a full set of 3 serial liver biopsies (total 63 biopsies) and complete clinical information is presented in Table 1. Recipients of DD grafts had higher Model for End-stage Liver Disease (MELD) scores than LDs, reflecting the need for a higher MELD in order to be offered a DD graft. There were no significant differences in recipient/donor gender, age, or primary liver disease. There were more patients with hepatitis C (HCV) receiving DD (n=8) than LD grafts (n=2), p=NS. There were no biopsy complications, all grafts functioned well, and graft and patient survival was 100% at 12 months post-transplant. Cold ischemia times were shorter in LD recipients where the donor and recipient operations are tightly coordinated. Anhepatic and warm ischemia times were also shorter in the LD recipients due to the differences in surgical techniques. In most of the DDs, portal and arterial reperfusion occurred within minutes of each other because both anastomses were performed during the warm ischemic period. In LDs the arterial anastomosis often required microsurgical techniques and surgeon preference was to perform these after portal reperfusion to keep warm ischemic time to a minimum. There were no significant differences in total operative time or blood replacement.
Table 1.
Patient and donor demographics and characteristics of the recipient surgical procedure
| Deceased Donor Transplants (n=13) | Living Donor Transplants (n=8) | P-value | |
|---|---|---|---|
| Recipient Primary disease | |||
| - Hepatitis C | 5 | 1 | |
| - Hepatoma | - | 1 | |
| - Hepatitis C + Hepatoma | 3 | 1 | |
| - Post-alcoholic cirrhosis | 2 | - | |
| - Primary sclerosing cholangitis | - | 3 | |
| - Primary biliary cirrhosis | 1 | 1 | |
| - Acute liver failure | 1 | - | |
| - Other cholestatic disease | 1 | 1 | |
|
| |||
| Recipient: Male/Female | 10/3 | 3/5 | 0.09 |
|
| |||
| Recipient: Age (min-max, median) | 29 – 66 (49) | 37 – 63 (49) | 0.92 |
|
| |||
| Recipient MELD (min-max, median) | 18 – 37 (25.5) | 6 – 18 (13.8) | <0.001 |
|
| |||
| Liver Donor | |||
| - Gender (M/F) | 7/6 | 4/4 | 0.61 |
| - Age (min-max, median) | 19 – 74 (43) | 29 – 50 (40) | 0.70 |
|
| |||
| Donor Cause of death | |||
| - Cerebrovascular accident/intracerebral bleed | 7 | ||
| - Anoxia sec. to cardiac arrest | 3 | N/A | |
| - Traumatic brain injury | 3 | ||
|
| |||
| Transplant Surgery Details | |||
|
| |||
| Total operative time (min) | 298 ± 10 | 366 ± 39 | 0.22 |
|
| |||
| Hepatectomy phase (min) | 125 ± 9 | 169 ± 28 | 0.79 |
|
| |||
| Anhepatic phase (min) | 107 ± 8 | 79 ± 6 | 0.03 |
|
| |||
| Packed red blood cell concentrate (units) | 4.2 ± 0.7 | 2.3 ± 0.9 | 0.12 |
|
| |||
| Cold ischemic time (min) (Cross-clamp to removal from ice) | 282 ± 19 | 38 ± 7 | <0.001 |
|
| |||
| Warm ischemic time (min) (Removal from ice to portal reperfusion) | 59 ± 2 | 37 ± 3 | <0.001 |
|
| |||
| Time between base-line PRE biopsy and COLD biopsy at end CI (min) | 304 ± 12 | 272 ± 73 | 0.68 |
|
| |||
| Time between portal reperfusion and POST biopsy (min) | 53 ± 2 | 79 ± 13 | 0.10 |
|
| |||
| Length of stay in hospital days (min - max, median) | 7 – 21 (10) | 9 – 39 (15) | 0.31 |
Class comparisons between time points and graft types
We compared differential gene expression as a function of the PRE, COLD and POST stages of the liver transplant. The total number of genes differentially expressed between all the class comparisons is illustrated in Figure 1A. There were significant differences between the PRE biopsy in the LD and in the DD, illustrating the effect of donor status and brain death compared to a healthy LD. Interestingly, cold storage of a DD graft results in little change in gene transcription as compared to the PRE biopsy (only 10 genes differentially expressed between COLD and PRE). In contrast, resection of the LD graft prior to the COLD biopsy resulted in the differential expression of 180 genes, most likely due to the complex process of parenchymal dissection. While these comparisons are important and illustrate underlying differences between brain dead and living donors, for the purpose of focus in this manuscript we will not discuss these differences, but have included all comparisons in the Supplementary online tables. To address our underlying hypothesis, a detailed analysis was focused on the following 3 class comparisons: 1) DD POST vs. DD PRE, 2) LD POST vs. LD PRE and, 3) LD POST vs. DD POST. A large number of genes were differentially expressed in both graft types following reperfusion (POST) when compared to the PRE biopsies, but there were relatively few common genes expressed by both LD and DD grafts (Fig. 1B).
Figure 1.
A. Diagram of number of differentially expressed genes in each class comparison. The numbers of differentially expressed genes between groups are illustrated in the small boxes connecting the larger shaded boxes (at p-value 0.005).
B. Venn diagram of overlap of differentially expressed genes in LD and DD POST reperfusion, compared to PRE transplantation for each graft type.
Gene expression in DD grafts after reperfusion: DD POST vs. DD PRE
The first class comparison compared DD POST biopsies to donor baseline biopsies. The number of differentially expressed genes POST was 579 (375 up-regulated after reperfusion and 204 down-regulated). Table 2 shows 20 selected genes that are present in the over-represented biological processes based on GO classifications in the DD grafts after reperfusion (DD POST vs. DD PRE). The selection of these genes represents integration of two statistical metrics to rank genes for very high biological significance. First, our ANOVA-based tool for class comparison allows us to rank differentially expressed genes by their p values, a standard and useful procedure in organizing these large data sets. Second, Ingenuity Pathway Analysis of the differentially expressed genes provides a statistical metric to confirm the over-representation (i.e. significance) of any functional gene networks that are identified as opposed to simply being found by random chance in any large list of gene expression values. So the 20 genes selected represent the highest ranked differentially expressed genes by p values that are also identified as populating over-represented functional gene networks. There are other important genes and pathways that are beyond the space limitations and results for both differential gene expression and Ingenuity functional networks are available as Supplemental Data(www.scripps.edu/mem/eht/salomon/livertxdata/).
Table 2.
Differential gene expression in over-represented biological processes based on Gene Ontology classifications: 20 select genes from DD POST vs PRE class comparison and 20 select genes from LD POST vs PRE class comparison.
| Differential gene expression for DD grafts: POST reperfusion vs. PRE transplant | ||||||
|---|---|---|---|---|---|---|
| Gene symbol | Description | GO biological Process | Geom mean DD PRE | Geom mean DD POST | Fold change POST vs PRE | P-value |
| ATF3 | activating transcription factor 3 | (DNA dependent) transcription, regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 62.4 | 1503.1 | 23.81 | p < 1e-07 |
| JUN | v-jun sarcoma virus 17 oncogene homolog (avian) | (DNA dependent) transcription, regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 221.3 | 2018.7 | 9.09 | p < 1e-07 |
| TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 (A20) | (DNA dependent) transcription, regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 44.6 | 166.6 | 3.73 | p < 1e-07 |
| NFKBIZ | nuclear factor kappa B inhibitor, zeta (MAIL) | anti-apoptosis, (DNA-dependent) regulation of transcription, regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 176.7 | 805.1 | 4.57 | p < 1e-07 |
| CCL2 | chemokine (C-C motif) ligand 2 (MCP-1) | anti-apoptosis, inflammatory response, JAK-STAT cascade | 59.9 | 706.3 | 11.76 | p < 1e-07 |
| SOCS3 | suppressor of cytokine signaling 3 | anti-apoptosis, JAK-STAT cascade | 79.9 | 620.9 | 7.75 | 1.00E-07 |
| HSPA1B | heat shock 70kDa protein 1B | anti-apoptosis, negative regulation of biological and cellular process | 123.5 | 1161 | 9.43 | 3.00E-07 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | apoptosis, cell cycle arrest, negative regulation of cell proliferation | 260.7 | 707.1 | 2.71 | 4.00E-07 |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | inflammatory response, (DNA dependent) transcription, regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 111.5 | 1260.7 | 11.36 | 1.00E-07 |
| CCL4 | chemokine (C-C motif) ligand 4 (MIP1-beta) | inflammatory response, cell adhesion | 84.2 | 568.5 | 6.76 | p < 1e-07 |
| IL8 | interleukin 8 | inflammatory response, chemotaxis, cell cycle arrest | 40.2 | 243.7 | 6.06 | p < 1e-07 |
| IL1B | interleukin 1, beta | inflammatory response, chemotaxis, negative regulation of cell proliferation | 39.8 | 124.1 | 3.12 | 1.00E-07 |
| IL6 | interleukin 6 (interferon, beta 2) | inflammatory response, regulation of cell proliferation, negative regulation of metabolism | 23.4 | 33.2 | 1.42 | 6.22E-04 |
| ICAM1 | intercellular adhesion molecule 1 (CD54) | immune response, chemotaxis, cell adhesion | 45.1 | 195.3 | 4.33 | p < 1e-07 |
| IL10 | interleukin 10 | inflammatory response, anti-apoptosis, regulation of T cell proliferation | 12.9 | 20 | 1.55 | 2.35E-04 |
| DUSP5 | dual specificity phosphatase 5 | dephosphorylation, protein metabolism | 41.3 | 233.9 | 5.65 | p < 1e-07 |
| HBEGF | heparin-binding EGF-like growth factor | signal transduction | 46.5 | 165.7 | 3.56 | p < 1e-07 |
| GSTA4 | glutathione S-transferase A4 | response to stress, metabolism | 24 | 17.1 | −1.40 | 9.48E-04 |
| EIF4E2 | Eukaryotic translation initiation factor 4E member 2 | regulation of biosynthesis, protein metabolism | 90.9 | 64.7 | −1.40 | 1.96E-04 |
| ALB | albumin | apoptosis, negative regulation of cellular process | 915 | 115.2 | −7.94 | 3.00E-07 |
| Differential gene expression for LD grafts: POST reperfusion vs. PRE transplant | ||||||
| SOCS3 | suppressor of cytokine signaling 3 | regulation of cell growth, anti-apoptosis, JAK-STAT cascade, negative regulation of insulin receptor signaling pathway | 46.8 | 443.1 | 9.43 | 2.51E-04 |
| STAT3 | signal transducer and activator of transcription 3 | (DNA-dependent) regulation of transcription, signal transduction, JAK-STAT cascade | 531.6 | 1511.8 | 2.84 | 0.0018449 |
| NFKB1 | NF of kappa light polypeptide gene enhancer in B- cells 1 (p105) | (DNA-dependent) regulation of transcription, anti-apoptosis, inflammatory response, signal transduction | 102.5 | 159.6 | 1.56 | 0.0014409 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21) | apoptosis, cell cycle arrest, negative regulation of cell proliferation | 164.8 | 812.1 | 4.93 | 1.52E-05 |
| JUN | v-jun sarcoma virus 17 oncogene homolog (avian) | (DNA-dependent) regulation of transcription, regulation of progression through cell cycle, leading edge cell differentiation | 76.7 | 300.1 | 3.91 | 0.0015611 |
| ODC1 | ornithine decarboxylase 1 | polyamine biosynthetic process | 471.6 | 3665.4 | 7.75 | 0.0003283 |
| DHODH | dihydroorotate dehydrogenase | ‘de novo’ pyrimidine base biosynthetic process | 222 | 777.7 | 3.51 | 0.000586 |
| HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | mitosis | 34.2 | 50.2 | 1.47 | 0.0010275 |
| IL1RN | interleukin 1 receptor antagonist | inflammatory response, insulin secretion | 104.9 | 761.4 | 7.25 | 3.11E-05 |
| IL4R | interleukin 4 receptor | immune response, signal transduction | 194.2 | 458.1 | 2.36 | 0.0011693 |
| ILF2 | interleukin enhancer binding factor 2, 45kDa | (DNA-dependent) regulation of transcription, immune response | 190.1 | 423.9 | 2.23 | 0.0027839 |
| PFKFB2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 | fructose 2,6-bisphosphate metabolic process | 19.4 | 28 | 1.44 | 0.0027338 |
| CEBPA | CCAAT/enhancer binding protein (C/EBP), alpha | (DNA-dependent) regulation of transcription, generation of precursor metabolites and energy, cytokine and chemokine mediated signaling pathway | 620.5 | 140.1 | −4.43 | 0.0002454 |
| PPARA | peroxisome proliferative activated receptor, alpha | (DNA-dependent) regulation of transcription, lipid metabolic process, fatty acid transport, positive regulation of fatty acid beta-oxidation | 265.5 | 114.8 | −2.31 | 9.90E-06 |
| AMACR | alpha-methylacyl-CoA racemase | metabolic process | 316.6 | 70.3 | −4.50 | 8.00E-07 |
| GPD1 | glycerol-3-phosphate dehydrogenase 1 (soluble) | gluconeogenesis, carbohydrate metabolic process, glycerol-3- phosphate metabolic process | 166.3 | 82.7 | −2.01 | 2.95E-05 |
| ACOX2 | acyl-Coenzyme A oxidase 2, branched chain | bile acid metabolic process, electron transport, lipid metabolic process, fatty acid beta-oxidation | 1160.1 | 271.1 | −4.28 | 8.77E-05 |
| HNF4A | hepatocyte nuclear factor 4, alpha | lipid metabolic process, blood coagulation, (DNA-dependent) regulation of transcription | 448.5 | 262.7 | −1.71 | 0.0019787 |
| THRSP | thyroid hormone responsive (SPOT14 homolog, rat) | lipid metabolic process, regulation of transcription from RNA polymerase II promoter | 1033.3 | 118.9 | −8.69 | 0.0003317 |
| PCK2 | phosphoenolpyruvate carboxykinase 2 (PEPCK mitochondrial) | glucose metabolic process, gluconeogenesis | 1459.8 | 799.7 | −1.83 | 0.0009141 |
The DD grafts demonstrated a large number of up-regulated inflammatory genes after reperfusion, including cytokines, chemokines and other regulators of inflammation. Specific examples include the inflammatory chemokine IL-8, as well as CCL2 (MCP-1) and TNFAIP3/A20, described as hepatoprotective (21, 22), and ICAM1, involved in leukocyte recruitment (23). Other over-represented GO categories in deceased donors after reperfusion are transcription (DNA-dependent) and regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism, with ATF3, induced by stress, as one of the most up-regulated genes (23-fold increase).
Gene expression in LD grafts after reperfusion: LD POST vs. LD PRE
The second detailed analysis compared gene expression in the LD liver grafts after reperfusion (POST) to the PRE biopsy taken in the donor. After reperfusion in the LD the number of differentially expressed genes was 1324 (756 up-regulated and 568 down-regulated). Table 2 shows 20 selected genes that are present in the over-represented biological processes based on GO classifications in the LD grafts after reperfusion (LD POST vs. LD PRE).
The gene lists generated support our hypothesis that activation of molecular networks for regeneration is an immediate event in partial LD grafts, associated with activation of pro-inflammatory pathways. There was a marked increase in genes of the IL-6/STAT3 pathway (24), including SOCS3, a feedback mediator that attenuates pro-inflammatory signaling and regulates STAT3 after partial hepatectomy (25), hepatocytes growth factor (HGF), a known stimulator of liver regeneration (3), and NFκB1, involved in regeneration and hepatoprotection (26). Other up-regulated networks involve cell cycle progression and de-novo biosynthesis of polyamines and pyrimidines.
A very significant finding in the LD POST biopsies was that most of the down-regulated genes involve hepatic metabolic function: fatty acid metabolism (GPD1, AMCR), fatty acid oxidation and tissue oxidative metabolism (PPARA), lipogenesis (THRSP) and bile acid metabolism (ACOX2). Genes enhancing glycolysis (PFKFB2) were up-regulated, while a key gluconeogenesis enzyme (PCK2) is down-regulated. Down-regulation of specific hepatic metabolism genes (HNF4α and C/EPBα) matches those seen in several animal models and also includes PEPCK, glutathione S-transferase, and genes linked to steroid and hormone metabolism (14, 27).
Similar to the DD grafts, there was up-regulation of immune/inflammatory genes in LD POST grafts. However, they demonstrated a very different profile than in DD POST, with some down-regulated as well. There were only 17 overlapping immune/inflammatory genes common to both DD and LD (Table 3). For example, CCL2 (MCP-1) and CCL20 (MIP-3α) play a role in immune response and were up-regulated in both. LD POST grafts primarily demonstrated up-regulation of interleukin-associated receptor genes such as IL1R and IL4R, as well as genes associated with innate immunity, such lipopolysaccharide binding protein (LPB), while the DD POST grafts demonstrated genes associated with cytokines; interferon gamma, IL1β, IL6, IL8 and IL10. Some MHC class II genes were up-regulated in LD POST, a pattern not seen in the DD grafts.
Table 3.
Comparison of immune and inflammatory genes differentially expressed in LD and DD grafts in POST biopsies compared to PRE
| Genes differentially expressed in LD POST vs. LD PRE | Genes differentially expressed in DD POST vs. DD PRE | ||||||
|---|---|---|---|---|---|---|---|
| Gene symbol | Description | Fold change LD POST vs LD PRE |
GO category | Gene symbol | Description | Fold change DD POST vs DD PRE |
GO category |
| COMMON TO BOTH LD AND DD | COMMON TO BOTH LD AND DD | ||||||
|
| |||||||
| BCL3 | B-cell CLL/lymphoma 3 | 1.89 | immune response | BCL3 | B-cell CLL/lymphoma 3 | 2.75 | immune response |
| BCL6 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | 1.42 | inflammatory response | BCL6 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | 1.73 | inflammatory response |
| C5R1 | complement component 5 receptor 1 (C5a ligand) | 1.89 | immune response | C5R1 | complement component 5 receptor 1 (C5a ligand) | 2.48 | immune response |
| CALCA | calcitonin/calcitonin-related polypeptide, alpha | 5.99 | inflammatory response | CALCA | calcitonin/calcitonin-related polypeptide, alpha | 4.78 | inflammatory response |
| CCL2 | chemokine (C-C motif) ligand 2 (MCP-1) | 11.76 | inflammatory response | CCL2 | chemokine (C-C motif) ligand 2 (MCP-1) | 4.37 | inflammatory response |
| CCL4 | chemokine (C-C motif) ligand 4 (MIP-1-beta) | 6.76 | inflammatory response | CCL4 | chemokine (C-C motif) ligand 4 (MIP-1-beta) | 3.22 | inflammatory response |
| CCL18 | chemokine (C-C motif) ligand 18 | 1.47 | inflammatory response | CCL18 | chemokine (C-C motif) ligand 18 (pulmonary and | 1.46 | inflammatory response |
| CCL20 | chemokine (C-C motif) ligand 20 (MIP-3 alpha) | 5.81 | inflammatory response | CCL20 | chemokine (C-C motif) ligand 20 (MIP-3 alpha) | 5.49 | inflammatory response |
| IL1RL1 | interleukin 1 receptor-like 1 | 8.77 | immune response | IL1RL1 | interleukin 1 receptor-like 1 | 2.49 | immune response |
| IL1RN | interleukin 1 receptor antagonist | 7.25 | inflammatory response | IL1RN | interleukin 1 receptor antagonist | 4.88 | inflammatory response |
| NFKB2 | nuclear factor of kappa light polypeptide gene enhancer | 1.18 | immune response | NFKB2 | nuclear factor of kappa light polypeptide gene enhancer | 1.40 | immune response |
| RIPK2 | receptor-interacting serine-threonine kinase 2 | 1.67 | inflammatory response | RIPK2 | receptor-interacting serine-threonine kinase 2 | 1.62 | inflammatory response |
| THBS1 | thrombospondin 1 | 5.05 | inflammatory response | THBS1 | thrombospondin 1 | 4.67 | inflammatory response |
| ELF3 | E74-like factor 3 | 2.79 | inflammatory response | ELF3 | E74-like factor 3 | 1.94 | inflammatory response |
| FCN1 | ficolin (collagen/fibrinogen domain containing) 1 | 1.82 | immune response | FCN1 | ficolin (collagen/fibrinogen domain containing) 1 | 2.14 | immune response |
| FCAR | Fc fragment of IgA, receptor for | 1.42 | immune response | FCAR | Fc fragment of IgA, receptor for | 1.14 | immune response |
| ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 | 1.29 | immune response | ETS1 | v-ets erythroblastosis virus E26 oncogene homolog 1 | 1.25 | immune response |
|
| |||||||
| EXCLUSIVE TO LD | EXCLUSIVE TO DD | ||||||
|
| |||||||
| CRP | C-reactive protein, pentraxin-related | 8.06 | inflammatory response | FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | 11.36 | inflammatory response |
| SAA1 /// SAA2 | serum amyloid A1//A2 | 7.30 | inflammatory response | IL8 | interleukin 8 | 6.06 | inflammatory response |
| LBP | lipopolysaccharide binding protein | 3.16 | inflammatory response | SNF1LK | SNF1-like kinase | 5.59 | immune response |
| RAB27A | RAB27A, member RAS oncogene family | 3.07 | immune response | CCL3 | chemokine (C-C motif) ligand 3 (MIP-1-alpha) | 5.08 | inflammatory response |
| STAT3 | signal transducer and activator of transcription 3 | 2.84 | inflammatory response | NFIL3 | nuclear factor, interleukin 3 regulated | 3.75 | immune response |
| FPRL1 | formyl peptide receptor-like 1 | 2.50 | inflammatory response | PTGS2 | prostaglandin-endoperoxide synthase 2 | 3.58 | inflammatory response |
| IL4R | interleukin 4 receptor | 2.36 | immune response | IL1B | interleukin 1, beta | 3.12 | inflammatory response |
| ILF2 | interleukin enhancer binding factor 2, 45kDa | 2.23 | immune response | RGS1 | regulator of G-protein signalling 1 | 2.91 | immune response |
| CHUK | conserved helix-loop-helix ubiquitous kinase | 2.14 | immune response | ZFP36 | zinc finger protein 36, C3H type, homolog (mouse) | 2.91 | inflammatory response |
| DKFZP564J0863 | DKFZP564J0863 protein | 1.96 | immune response | IRF1 | interferon regulatory factor 1 | 2.75 | immune response |
| F11R | F11 receptor | 1.63 | inflammatory response | CD83 | CD83 antigen | 2.67 | immune response |
| IKBKG | inhibitor of kappa light polypeptide gene enhancer in B- | 1.63 | immune response | IRF4 | interferon regulatory factor 4 | 2.42 | immune response |
| NFKB1 | nuclear factor of kappa light polypeptide gene enhancer | 1.56 | inflammatory response | RGS1 | regulator of G-protein signalling 1 | 2.18 | immune response |
| HLA-DRB4 | major histocompatibility complex, class II, DR beta 4 | 1.24 | immune response | GBP1 | guanylate binding protein 1, interferon-inducible | 2.16 | immune response |
| LTA | lymphotoxin alpha (TNF superfamily, member 1) | 1.23 | immune response | GEM | GTP binding protein overexpressed in skeletal muscle | 2.15 | immune response |
| CD276 | CD276 antigen | 1.22 | immune response | CXCR4 | chemokine (C-X-C motif) receptor 4 | 2.01 | inflammatory response |
| NFE2L1 | nuclear factor (erythroid-derived 2)-like 1 | −1.36 | inflammatory response | ANXA1 | annexin A1 | 1.98 | inflammatory response |
| C2 | complement component 2 | −1.47 | inflammatory response | FPR1 | formyl peptide receptor 1 | 1.88 | inflammatory response |
| ATRN | attractin | −1.58 | inflammatory response | CXCL3 | chemokine (C-X-C motif) ligand 3 (CINC/MIP-2b) | 1.86 | inflammatory response |
| F11R | F11 receptor | −1.59 | inflammatory response | EBI2 | Epstein-Barr virus induced gene 2 | 1.83 | immune response |
| FCGRT | Fc fragment of IgG, receptor, transporter, alpha | −1.60 | immune response | CXCL1 | chemokine (C-X-C motif) ligand 1 | 1.74 | inflammatory response |
| DPP4 | dipeptidylpeptidase 4 (CD26) | −1.62 | immune response | CIAS1 | cold autoinflammatory syndrome 1 | 1.56 | inflammatory response |
| AQP9 | aquaporin 9 | −1.79 | immune response | IL10 | interleukin 10 | 1.55 | inflammatory response |
| PXMP2 | peroxisomal membrane protein 2, 22kDa | −1.99 | immune response | TREM1 | triggering receptor expressed on myeloid cells 1 | 1.54 | immune response |
| CNGA1 | cyclic nucleotide gated channel alpha 1 | −2.01 | immune response | CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | 1.52 | inflammatory response |
| CD4 | CD4 antigen (p55) | −2.02 | immune response | BMP2 | bone morphogenetic protein 2 | 1.44 | inflammatory response |
| TLR3 | toll-like receptor 3 | −2.04 | inflammatory response | IL6 | interleukin 6 (interferon, beta 2) | 1.42 | inflammatory response |
| CXCL12 | chemokine (C-X-C motif) ligand 12 | −2.20 | inflammatory response | SQSTM1 | Sequestosome 1 | 1.41 | immune response |
| EPHX2 | epoxide hydrolase 2, cytoplasmic | −2.26 | inflammatory response | FPRL1 | formyl peptide receptor-like 1///formyl peptide receptor | 1.38 | inflammatory response |
| FLJ20406 | hypothetical protein FLJ20406 | −2.73 | immune response | RELB | v-rel reticuloendotheliosis viral oncogene homolog B, | 1.35 | immune response |
| IFIT3 | interferon-induced protein with tetratricopeptide repeats | −2.75 | immune response | C1S | Complement component 1, s subcomponent | 1.28 | inflammatory response |
| KLKB1 | kallikrein B, plasma (Fletcher factor) 1 | −3.32 | inflammatory response | SLC11A1 | solute carrier family 11 (proton-coupled divalent metal | 1.27 | immune response |
| SCAP1 | src family associated phosphoprotein 1 | −3.42 | immune response | ICEBERG | ICEBERG caspase-1 inhibitor | 1.14 | inflammatory response |
| IFIT1 | interferon-induced protein with tetratricopeptide repeats | −5.54 | immune response | IFIH1 | Interferon induced with helicase C domain 1 | −1.10 | immune response |
| ZGPAT | zinc finger, CCCH-type with G patch domain | −6.00 | immune response | ADORA3 | adenosine A3 receptor | −1.62 | inflammatory response |
| TNFSF10 | tumor necrosis factor (ligand) superfamily, member 10 | −6.44 | immune response | ATRN | attractin | −1.75 | inflammatory response |
Differences between deceased donor and living donor grafts following reperfusion: LD POST vs. DD POST
The next analysis directly compared the expression profiles after reperfusion between LD and DD livers. 1506 genes were differentially expressed (726 up-regulated and 780 down-regulated, see online Supplementary Data). In this comparison, it was apparent that significant differences exist in expression of genes associated with biosynthesis, metabolism and energy pathways.
To better understand the functional significance of graft type on gene expression, we used an EASE analysis to assign p-values to each biological function rather than to individual genes (Table 4). The 12 functions that were increased in LDs are nearly all associated with cell proliferation and tissue regeneration. In contrast, all 17 down-regulated processes in LDs represent metabolic liver functions, consistent with our hypothesis that recovery of the partial LD graft requires redistribution of energy in favor of regeneration.
Table 4.
Biological theme analysis by EASE comparing LD grafts POST reperfusion vs. DD grafts POST reperfusion.
| Biological Functions Up-regulated in LD POST | EASE score (p-value) |
|---|---|
| RNA metabolism | 9.86E-05 |
| RNA processing | 2.28E-04 |
| nucleobase\, nucleoside\, nucleotide and nucleic acid metabolism | 2.69E-03 |
| antigen processing | 1.23E-02 |
| transcription from Pol II promoter | 1.61E-02 |
| rRNA processing | 2.15E-02 |
| protein amino acid phosphorylation | 2.16E-02 |
| rRNA metabolism | 3.35E-02 |
| nucleobase biosynthesis | 3.52E-02 |
| phosphorylation | 4.31E-02 |
| protein modification | 4.59E-02 |
| nucleobase metabolism | 4.62E-02 |
| Biological Functions Down-regulated in LD POST | |
| Lipid biosynthesis | 7.19E-03 |
| Alcohol metabolism | 9.34E-03 |
| Biosynthesis | 1.12E-02 |
| Carbohydrate metabolism | 1.38E-02 |
| Energy pathways | 1.61E-02 |
| Lipid metabolism | 1.72E-02 |
| Physiological process | 1.76E-02 |
| Main pathways of carbohydrate metabolism | 2.14E-02 |
| Macromolecule biosynthesis | 2.28E-02 |
| Photoreceptor maintenance | 2.44E-02 |
| Iron ion transport | 2.70E-02 |
| Xenobiotic metabolism | 2.46E-02 |
| Response to xenobiotic stimulus | 2.72E-02 |
| Metabolism | 3.23E-02 |
| Mismatch repair/Maintenance of fidelity during DNA replication | 3.47E-02 |
| Retinol metabolism | 3.47E-02 |
| Amino acid and derivative metabolism | 3.64E-02 |
We wished to determine if HCV status impacted on the 1506 differentially expressed genes identified in LD POST vs. DD POST. Only 23 (LD) and 7 (DD), respectively, were contained in the 1506 genes identified in earlier comparison of DD POST vs. LD POST. Thus, HCV status did not appear to influence the selection of genes identified in this study (complete analysis in Supplemental Data).
Biological networks analysis
Ingenuity Pathway Analysis organizes gene expression data into molecular networks using algorithms based on literature, known molecular functions and compendia of annotated databases. We utilized these networks to identify and graphically view biological mechanisms and inter-gene connectivity relevant to the datasets generated by our class comparisons and specific focus genes of interest. When all genes differentially expressed POST reperfusion in the LD to DD comparison were analyzed (N=1506), 778 genes were found to be linked to 43 networks with 10 or more Focus Genes per network. In 8 networks, the maximum of all 35 Focus Genes were present. In each network, there is a “centered” gene, which refers to the gene with the highest level of inter-gene connectivity. The 8 networks identified were: 1) cell growth and proliferation (centered around p53), 2) gene expression in cancer (centered around FOS), 3) connective tissue development (centered around JUN), 4) inflammation/immune responses (centered around NFκB), 5) lipid metabolism (centered around CEPBα/STAT3), 6) cell growth (centered around MAPK8), 7) cell cycle and DNA replication (centered around CDK4), and 8) cell death (centered around CAV1). These analyses further illustrated how cell growth and proliferation pathways are increased in LD grafts, and metabolism pathways are down-regulated. The complete sets of 8 networks are in the online Supplementary Data (Suppl. Figs 1–8). It is important to note that about 50% of the 1506 genes analyzed are not accounted for here. First, many genes in the human genome, even many with a known function, are not presently linked to discrete functional molecular networks in the literature and these account for the majority of these missing gene candidates. Second, we truncated our analysis for practical reasons at 43 different networks using a filter of >10 genes/network but another few hundred genes fall into smaller networks.
Validation of gene expression using TaqMan Low Density Arrays (TLDA)
Gene expression analysis requires independent validation of at least a subset of gene candidates. TLDA technology was used to validate 63 genes in triplicate plus controls. Validation genes were chosen from the results of the class comparisons: LD PRE vs. LD POST (28 genes), DD PRE vs. DD POST (22 genes), and LD POST vs. DD POST (18 genes) based on their biological interest. There was nearly complete concordance for gene expression between the microarrays and TLDA with the exception of 3 genes in the last class comparison (Suppl. Fig. 9). Because the same panel of genes was also measured for all samples profiled, we also validated expression of many genes in each sample that were not chosen for their biological interest or differential expression, with an 87% concordance for gene expression across all the samples studied. This amount of concordance of genes identified using Affymetrix arrays using a completely separate technology platform, and testing for many genes that were not differentially expressed in the group comparisons, is quite significant given the relatively high variability known for raw TaqMan PCR results and the fact that the fold-change differences for many of these comparisons were only 2-fold or less.
Relationship between gene expression and protein expression by immunopathology
To explore the relationship between our findings of early gene expression to protein expression, we identified several relevant cell proliferation and inflammatory genes that demonstrated significant expression differences in PRE and POST samples and used immunohistopathology to assess expression of their associated proteins in a representative sample of LD and DD biopsies. These genes were: SOCS3, a feedback regulator of regeneration; IL1RL1, a regulator of cell proliferation and member of the Toll superfamily; and IL-8, a proinflammatory CXC chemokine. We demonstrated protein expression patterns that matched gene expression. Immunoperoxidase comparison of biopsies from LD and DD showed comparable SOCS3 upregulation with gene expression in POST samples, which showed increased staining of hepatocytes, bile ducts and inflammatory cells post-reperfusion regardless of donor type (Fig. 2A). When comparing POST biopsies to PRE, SOCS3 demonstrated a 7.75 fold change in gene expression in DD and a 9.43 fold change in LD grafts (Table 2). IL1RL1 showed minimal baseline PRE staining but significant upregulation post-reperfusion primarily in LD tissues (Fig. 2B), correlating with an 8.77 fold change seen in the array data (Table 4). For other genes, such as IL-8, minimal or no expression was detected in the PRE or POST LD biopsies, whereas significant staining was detected post-reperfusion in the DD POST samples, correlating with a 6 fold increase seen in the array data (Fig. 2C, Table 2).
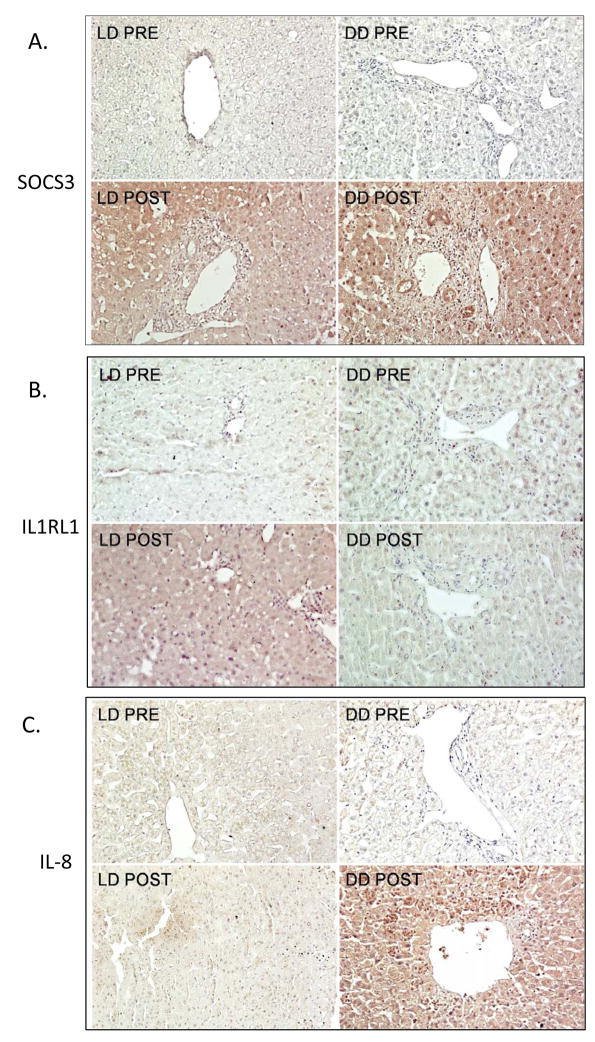
Figure 2.
Figure 2A–C. Immunoperoxidase analysis of gene expression in liver biopsies collected from the donor liver (PRE) and post-reperfusion (POST), showing representative data from 4 samples/group and comparing events using LD and DD liver tissues. Negligible staining for SOCS3, IL1RL1, or IL-8 was seen in the PRE biopsies. (Paraffin sections, hematoxylin counterstain, original magnifications ×250).
2A. Post-reperfusion, SOCS3 expression was upregulated in both LD and DD biopsies with staining of hepatocytes, bile ducts and inflammatory cells
2B. IL1RL1 demonstrates negligible expression in PRE LD biopsies but with upregulation by hepatocytes in post-reperfusion biopsies of LD tissues. There was minimal staining in DD livers, either PRE or POST.
2C. IL-8 was upregulated primarily in post-reperfusion biopsies from DD livers, with minimal LD expression. IL-8 staining was detected in hepatocytes and also in conjunction with infiltrating polymorphonuclear neutrophils.
Post-operative course and clinical correlation
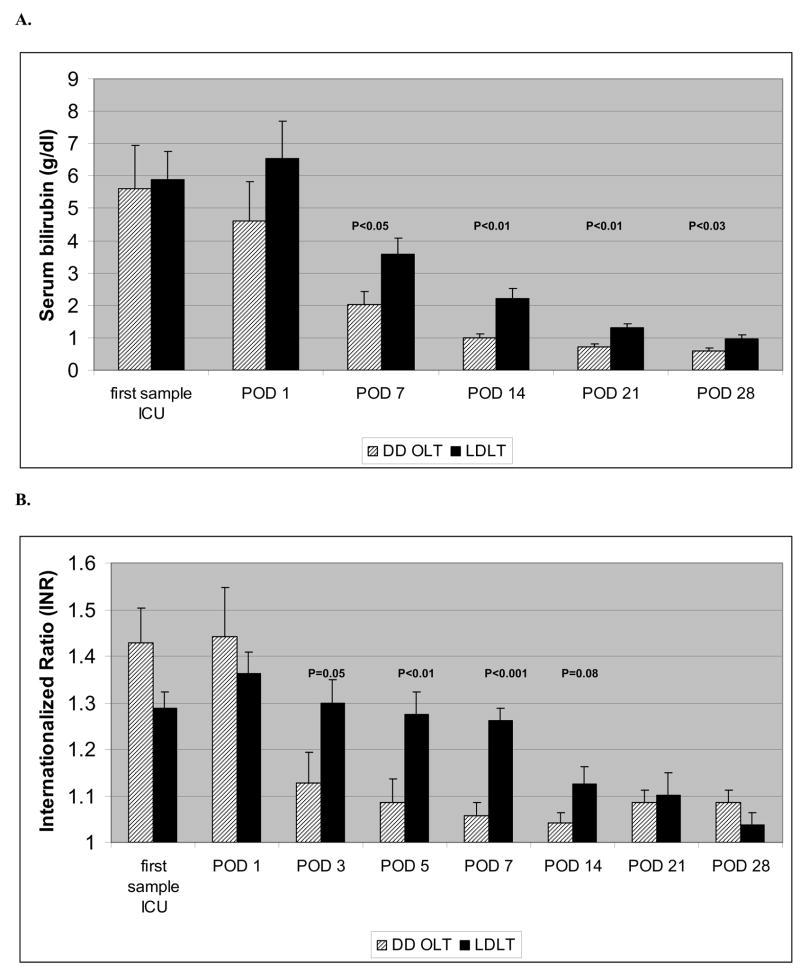
In order to determine if there may be clinical trends correlating with gene expression, we compared graft outcome, liver transaminases, bilirubin and coagulation factors in the first 4 weeks post-transplant in LD and DD recipients. All LD and DD recipients had good early allograft function with no graft loss within the first year post transplant, so these gene expression profiles were considered to correlate with good function. No significant differences were noted in transaminases levels (data not shown), however, bilirubin levels were elevated in LD compared to DD recipients, correlating to down-regulation of genes associated with bile synthesis in LD, significant from POD 7 to one month after transplantation (Figure 3A). In DD recipients, the INR was higher just after surgery, but improved quickly, whereas LD recipients demonstrated significantly higher INR in the first week after transplantation and slower return to normal (Figure 3B). Despite highly down-regulated albumin gene expression, no significant differences were seen for serum albumin, most likely due to albumin transfusions in both groups post-transplant (data not shown).
Figure 3.
A. Bilirubin serum levels in LD and DD liver transplant recipients in the perioperative period and the first month after transplant. POD = post-operative day.
B. Internationalized Ratios in recipients of a LD liver transplants (LDLT) and DD liver transplants (DD-OLT) in the first month after transplant.
Discussion
In attempts to reduce adult waitlist mortality, right lobe LD transplantation was introduced in the adult population, with comparable postoperative outcomes (1, 28). While it is agreed that both DD and LD livers can provide a suitable graft in the appropriate recipient, it must also be recognized that each is unique in surgical technique, the amount of damage incurred, the degree of metabolic demand and dysfunction, and the requirement for regeneration and recovery from injury. In DD liver transplantation, there is a loss of hepatocytes due to donor events and ischemic injury. In LD transplants, the liver parenchyma must be surgically divided, and a right lobe graft is required to regenerate a large amount of liver mass as well as recover from a short period of ischemia. Both graft types need to perform simultaneous basic and complex metabolic functions during this period of recovery. There is a necessary balance between the need for recovery and the need to maintain normal hepatic homeostasis post transplant. We propose that DD grafts and LD grafts respond to these needs with markedly different molecular mechanisms. The type of graft transplanted has a significant effect on this balance and each, as a consequence, has a unique and informative profile at the level of early gene expression.
Several animal studies have examined regeneration and gene expression following partial hepatectomy using various cDNA array technologies, confirming the importance of cell cycle progression genes, cytokines, and other novel immediate early genes (12–14, 29). There is also rodent experimental evidence illustrating the shift in metabolic function, energy balance, acute phase response and lipid metabolism in regenerating livers (9, 10) (30). In the transplant setting, microarray analysis of small-for-size rodent liver grafts demonstrated up-regulation of vasoconstrictive and adhesion molecule genes with increases in genes associated with inflammation and cell death, and down-regulation of genes related to energy metabolism (13, 31, 32). One clinical study demonstrated expression of acute stress genes in human liver grafts immediately after living donor transplantation, and another identified several genes in post-perfusion biopsies associated with early graft dysfunction (15, 33). While these studies identified specific gene expression profiles in certain classes of liver grafts, whole genome expression in serial samples was not assessed. A recent study utilizing whole genome oligonucleotide microarray analysis of 5 DD grafts identified over 700 genes differentially expressed after ischemia-reperfusion (16).
In our study, we were able to perform a time course analysis of the same human graft from procurement to cold storage to reperfusion with a careful comparison of LD to DD grafts at each stage. There are inherent differences between these two types of grafts due to multiple variables: donor status and quality, surgical technique at each stage, and transplanted liver mass. Each of these differences may have a significant impact on post-transplant recovery. Our analysis was directed toward the expression of genes that are known to be involved in metabolism, regeneration, and the proinflammatory response. The rationale to focus on these specific gene expression patterns is based on our working hypothesis that recovery is dependent on the graft’s ability to sustain metabolic activity and energy levels, initiate cell repair and cycle pathways, maintain liver synthetic function, and regulate the proinflammatory response.
There were significant differences noted in gene expression between LD and DD at all 3 time points, but the greatest disparities were noted after reperfusion. In the DD grafts, there was a large induction of genes, many inflammatory, and others associated with cell cycle regulation, consistent with findings in experimental studies (4, 34). A very different pattern was recognized in the LD graft recipients. There was differential expression of inflammatory, immune and cell stress genes in a pattern unique to the LD grafts, perhaps representing a mechanism of a small graft responding to the metabolic challenge created by size disparity. An interesting finding is the up-regulation of MHC class II genes observed in the LD grafts, consistent with rodent model data where regeneration of reduced-size allografts is accompanied by accelerated alloreactivity (35, 36). We also saw evidence for increased expression of genes associated with the innate immune system, which has been described to be important for initiation of liver regeneration (37). Weiss et al. recently noted a different pattern of inflammatory cytokines expression between DD and LD using RT-PCR (38). Our findings in the recipients of LD grafts also correlated well with previous results from Borozan et al, who described gene expression in LDs using a 19K-human microarray after reperfusion (15). Eleven of the 15 up-regulated genes and 7 of the 10 down-regulated genes, verified by RT-PCR in their study, were also found to be up-regulated in our gene list.
The LD POST biopsies also showed up-regulation of genes encoding purine, pyrimidine and structural protein synthesis, expected in a regenerating liver, while genes associated with higher metabolic liver functions such as bile acid metabolism and protein metabolism were markedly decreased. This shifting balance between metabolism and regeneration is further exemplified by the differential expression of genes encoding enzymes which play key roles in glycolysis and gluconeogenesis, correlating with previous observations in animal models describing down-regulation of glucose metabolism, lipid metabolism, bile secretion, and steroid and hormone metabolism (14, 39). We saw potential down-stream effect of this with higher bilirubin and INR in the LD recipients.
The biological network analysis provides important insights into the processes initiated in early phases of liver recovery and evidence supporting our initial hypothesis. While these networks are computed pathways and associations, they identify similarities between the 2 graft types, as well as significant disparities. In the LD grafts, network analysis showed an increased role of cellular proliferation, a marked increase in RNA biosynthesis and structural protein metabolism, and down-regulation of oxidative phosphorylation and global carbohydrate and lipid metabolism, none of which were seen in the DD grafts.
An important question for an analysis of functional molecular pathways is determining if genome-wide expression profiling studies reflect the actual molecular mechanisms driving states of health and disease. While it must always be presented in the correct context, these mechanistic connections drawn from unbiased genome-wide profiling are still of significant value. In the process of analyzing our genome-wide gene expression data we used tools such as EASE, DAVID, GO and Ingenuity to map highly significant differentially expressed genes to functional molecular pathways that were previously defined in the literature, resulting in mapping that is directly dependent on the value of the mechanistic data from individual genes and functional pathways. Therefore, the mechanistic insights drawn from genome-wide transcriptional analysis can also have significant value, similar to the gene-by-gene, pathway-by-pathway mechanistic data that it is based upon.
It is important to note that some of the differences described in gene and protein expression may be due to the contribution of variations in procurement and surgical technique, cold and warm ischemic times, and operative times, and not just due to the need for regeneration. These differences, however, emphasize the fact that these 2 types of transplants produce two very unique grafts. It is not possible to control many of these technical variables, but they clearly contribute to the different expression profiles and make the two types of grafts respond in a different way. In this limited sample size, we are not able to determine to what extent regeneration or ischemic injury plays in the differential gene expression compared to the other variables, but the patterns of gene expression seem to be illustrative of these 2 processes. Further work with more patients will be needed to determine the actual contribution of each element.
In summary, although this study was performed in a relatively small number of patients and is necessarily descriptive in nature, we have obtained detailed array data from serial biopsies of the same liver graft, which demonstrated a highly coordinated interplay of regenerative, metabolic and inflammatory pathways in the recovering human liver graft. The classical pattern of liver regeneration, known from rodent models, seems to be present in human partial liver transplantation, in association with a distinctive inflammatory profile. We were able to demonstrate correlation of relevant genes with protein expression, as well as clinical trends with serum levels in the patients. The down-regulation of genes associated with metabolic pathways in human LD grafts is unique and may be a key in understanding how the recipient/graft combination determines where efforts and energy should be placed. With further validation, this initial data may be used to explore and identify areas where intervention may be possible to support metabolic activity, decrease inflammation, or enhance regeneration.
Supplementary Material
Acknowledgments
Grant Support: KMO is supported by NIH grant RO1 DK07319202 and the Biesecker Center for Pediatric Liver Disease. We acknowledge the DNA Microarray Core of TSRI and funding for this project including NIH grant U19 AI063603 and the Molly Baber Research Fund. We also note the support of Applied Biosystems Incorporated (ABI) for a portion of the TLDA arrays. This research was also supported by The European Society for Organ Transplantation 2005 Research Grant and the Michael van Vloten Foundation by funding JdJ’s research fellowship. We would like to acknowledge the efforts of Dr. Kun Ming Chang in clinical data collection. We are also grateful to the Gift of Life Donor Program of Philadelphia, PA, and to Dr. Robert Fisher at Virginia Commonwealth University for providing additional clinical samples for our initial analysis.
Abbreviations
- DD
deceased donor
- LD
living donor
- PRE
pre-procurement biopsy
- COLD
cold backbench biopsy
- POST
post-reperfusion biopsy
- MELD
Model of Endstage Liver Disease
- HCV
hepatitis C virus
- GO
gene ontology
- TLDA
TaqMan Low Density Array
Footnotes
Financial disclosures: The authors have no conflict of interest and no financial disclosures
Transcript profiling: Gene expression data can be accessed via www.scripps.edu/mem/eht/salomon/livertxdata/
References
- 1.Olthoff KM, Merion RM, Ghobrial RM, Abecassis MM, Fair JH, Fisher RA, et al. Outcomes of 385 adult-to-adult living donor liver transplant recipients: a report from the A2ALL Consortium. Ann Surg. 2005;242(3):314–323. doi: 10.1097/01.sla.0000179646.37145.ef. discussion 323–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng S, Goodrich NP, Bragg-Gresham JL, Dykstra DM, Punch JD, DebRoy MA, et al. Characteristics associated with liver graft failure: the concept of a donor risk index. Am J Transplant. 2006;6(4):783–790. doi: 10.1111/j.1600-6143.2006.01242.x. [DOI] [PubMed] [Google Scholar]
- 3.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43(2 Suppl 1):S45–53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 4.Debonera F, Aldeguer X, Shen X, Gelman AE, Gao F, Que X, et al. Activation of interleukin-6/STAT3 and liver regeneration following transplantation. J Surg Res. 2001;96(2):289–295. doi: 10.1006/jsre.2001.6086. [DOI] [PubMed] [Google Scholar]
- 5.Debonera F, Wang G, Xie J, Que X, Gelman A, Leclair C, et al. Severe preservation injury induces Il-6/STAT3 activation with lack of cell cycle progression after partial liver graft transplantation. Am J Transplant. 2004;4(12):1964–1971. doi: 10.1111/j.1600-6143.2004.00626.x. [DOI] [PubMed] [Google Scholar]
- 6.Selzner N, Selzner M, Tian Y, Kadry Z, Clavien PA. Cold ischemia decreases liver regeneration after partial liver transplantation in the rat: A TNF-alpha/IL-6-dependent mechanism. Hepatology. 2002;36(4 Pt 1):812–818. doi: 10.1053/jhep.2002.35535. [DOI] [PubMed] [Google Scholar]
- 7.Arai M, Yokosuka O, Chiba T, Imazeki F, Kato M, Hashida J, et al. Gene expression profiling reveals the mechanism and pathophysiology of mouse liver regeneration. J Biol Chem. 2003;278(32):29813–29818. doi: 10.1074/jbc.M212648200. [DOI] [PubMed] [Google Scholar]
- 8.Haber BA, Chin S, Chuang E, Buikhuisen W, Naji A, Taub R. High levels of glucose-6-phosphatase gene and protein expression reflect an adaptive response in proliferating liver and diabetes. J Clin Invest. 1995;95(2):832–841. doi: 10.1172/JCI117733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue H, Ogawa W, Ozaki M, Haga S, Matsumoto M, Furukawa K, et al. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nat Med. 2004;10(2):168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 10.Leu JI, Crissey MA, Craig LE, Taub R. Impaired hepatocyte DNA synthetic response posthepatectomy in insulin-like growth factor binding protein 1-deficient mice with defects in C/EBP beta and mitogen-activated protein kinase/extracellular signal-regulated kinase regulation. Mol Cell Biol. 2003;23(4):1251–1259. doi: 10.1128/MCB.23.4.1251-1259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Liang X, Leu JI, Kovalovich K, Ciliberto G, Taub R. Global changes in interleukin-6-dependent gene expression patterns in mouse livers after partial hepatectomy. Hepatology. 2001;33(6):1377–1386. doi: 10.1053/jhep.2001.24431. [DOI] [PubMed] [Google Scholar]
- 12.Morita T, Togo S, Kubota T, Kamimukai N, Nishizuka I, Kobayashi T, et al. Mechanism of postoperative liver failure after excessive hepatectomy investigated using a cDNA microarray. J Hepatobiliary Pancreat Surg. 2002;9(3):352–359. doi: 10.1007/s005340200039. [DOI] [PubMed] [Google Scholar]
- 13.Nagano Y, Nagahori K, Yoshiro F, Hamaguchi Y, Ishikawa T, Ichikawa Y, et al. Gene expression profile analysis of regenerating liver after portal vein ligation in rats by a cDNA microarray system. Liver Int. 2004;24(3):253–258. doi: 10.1111/j.1478-3231.2004.0912.x. [DOI] [PubMed] [Google Scholar]
- 14.Fukuhara Y, Hirasawa A, Li XK, Kawasaki M, Fujino M, Funeshima N, et al. Gene expression profile in the regenerating rat liver after partial hepatectomy. J Hepatol. 2003;38(6):784–792. doi: 10.1016/s0168-8278(03)00077-1. [DOI] [PubMed] [Google Scholar]
- 15.Borozan I, Chen L, Sun J, Tannis LL, Guindi M, Rotstein OD, et al. Gene expression profiling of acute liver stress during living donor liver transplantation. Am J Transplant. 2006;6(4):806–824. doi: 10.1111/j.1600-6143.2006.01254.x. [DOI] [PubMed] [Google Scholar]
- 16.Conti A, Scala S, D’Agostino P, Alimenti E, Morelli D, Andria B, et al. Wide gene expression profiling of ischemia-reperfusion injury in human liver transplantation. Liver Transpl. 2007;13(1):99–113. doi: 10.1002/lt.20960. [DOI] [PubMed] [Google Scholar]
- 17.Flechner SM, Kurian SM, Head SR, Sharp SM, Whisenant TC, Zhang J, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004 doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurian SM, Flechner SM, Kaouk J, Modlin C, Goldfarb D, Cook DJ, et al. Laparoscopic donor nephrectomy gene expression profiling reveals upregulation of stress and ischemia associated genes compared to control kidneys. Transplantation. 2005;80(8):1067–1071. doi: 10.1097/01.tp.0000176485.85088.f7. [DOI] [PubMed] [Google Scholar]
- 19.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 20.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hokeness KL, Kuziel WA, Biron CA, Salazar-Mather TP. Monocyte chemoattractant protein-1 and CCR2 interactions are required for IFN-alpha/beta-induced inflammatory responses and antiviral defense in liver. J Immunol. 2005;174(3):1549–1556. doi: 10.4049/jimmunol.174.3.1549. [DOI] [PubMed] [Google Scholar]
- 22.Longo CR, Patel VI, Shrikhande GV, Scali ST, Csizmadia E, Daniel S, et al. A20 protects mice from lethal radical hepatectomy by promoting hepatocyte proliferation via a p21waf1-dependent mechanism. Hepatology. 2005;42(1):156–164. doi: 10.1002/hep.20741. [DOI] [PubMed] [Google Scholar]
- 23.Selzner N, Selzner M, Odermatt B, Tian Y, Van Rooijen N, Clavien PA. ICAM-1 triggers liver regeneration through leukocyte recruitment and Kupffer cell-dependent release of TNF-alpha/IL-6 in mice. Gastroenterology. 2003;124(3):692–700. doi: 10.1053/gast.2003.50098. [DOI] [PubMed] [Google Scholar]
- 24.Taub R. Hepatoprotection via the IL-6/Stat3 pathway. J Clin Invest. 2003;112(7):978– 980. doi: 10.1172/JCI19974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell JS, Prichard L, Schaper F, Schmitz J, Stephenson-Famy A, Rosenfeld ME, et al. Expression of suppressors of cytokine signaling during liver regeneration. J Clin Invest. 2001;107(10):1285–1292. doi: 10.1172/JCI11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuboki S, Okaya T, Schuster R, Blanchard J, Denenberg A, Wong HR, et al. Hepatocyte NF-kappaB activation is hepatoprotective during ischemia-reperfusion injury and is augmented by ischemic hypothermia. Am J Physiol Gastrointest Liver Physiol. 2007;292(1):G201–207. doi: 10.1152/ajpgi.00186.2006. [DOI] [PubMed] [Google Scholar]
- 27.White P, Brestelli JE, Kaestner KH, Greenbaum LE. Identification of transcriptional networks during liver regeneration. J Biol Chem. 2005;280(5):3715–3722. doi: 10.1074/jbc.M410844200. [DOI] [PubMed] [Google Scholar]
- 28.Berg CL, Gillespie BW, Merion RM, Brown RS, Jr, Abecassis MM, Trotter JF, et al. Improvement in survival associated with adult-to-adult living donor liver transplantation. Gastroenterology. 2007;133(6):1806–1813. doi: 10.1053/j.gastro.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Togo S, Makino H, Kobayashi T, Morita T, Shimizu T, Kubota T, et al. Mechanism of liver regeneration after partial hepatectomy using mouse cDNA microarray. J Hepatol. 2004;40(3):464–471. doi: 10.1016/j.jhep.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Strey CW, Winters MS, Markiewski MM, Lambris JD. Partial hepatectomy induced liver proteome changes in mice. Proteomics. 2004;4:1–8. doi: 10.1002/pmic.200400913. [DOI] [PubMed] [Google Scholar]
- 31.Man K, Lo CM, Lee TK, Li XL, Ng IO, Fan ST. Intragraft gene expression profiles by cDNA microarray in small-for-size liver grafts. Liver Transpl. 2003;9:425–432. doi: 10.1053/jlts.2003.50066. [DOI] [PubMed] [Google Scholar]
- 32.Zhong Z, Schwabe RF, Kai Y, He L, Yang L, Bunzendahl H, et al. Liver regeneration is suppressed in small-for-size liver grafts after transplantation: involvement of c-Jun N-terminal kinase, cyclin D1, and defective energy supply. Transplantation. 2006;82(2):241–250. doi: 10.1097/01.tp.0000228867.98158.d2. [DOI] [PubMed] [Google Scholar]
- 33.Berberat PO, Friess H, Schmied B, Kremer M, Gragert S, Flechtenmacher C, et al. Differentially expressed genes in postperfusion biopsies predict early graft dysfunction after liver transplantation. Transplantation. 2006;82(5):699–704. doi: 10.1097/01.tp.0000233377.14174.93. [DOI] [PubMed] [Google Scholar]
- 34.Que X, Debonera F, Xie J, Furth EE, Aldeguer X, Gelman AE, et al. Pattern of ischemia reperfusion injury in a mouse orthotopic liver transplant model. J Surg Res. 2004;116(2):262–268. doi: 10.1016/j.jss.2003.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Omura T, Nakagawa T, Randall HB, Lin Z, Huey M, Ascher NL, et al. Increased immune responses to regenerating partial liver grafts in the rat. J Surg Res. 1997;70(1):34–40. doi: 10.1006/jsre.1997.5115. [DOI] [PubMed] [Google Scholar]
- 36.Shiraishi M, Csete ME, Yasunaga C, Drazan KE, Jurim O, Cramer DV, et al. Regeneration-induced accelerated rejection in reduced-size liver grafts. Transplantation. 1994;57(3):336–340. doi: 10.1097/00007890-199402150-00004. [DOI] [PubMed] [Google Scholar]
- 37.Fausto N. Involvement of the innate immune system in liver regeneration and injury. J Hepatol. 2006;45(3):347–349. doi: 10.1016/j.jhep.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Weiss S, Kotsch K, Francuski M, Reutzel-Selke A, Mantouvalou L, Klemz R, et al. Brain death activates donor organs and is associated with a worse I/R injury after liver transplantation. Am J Transplant. 2007;7(6):1584–1593. doi: 10.1111/j.1600-6143.2007.01799.x. [DOI] [PubMed] [Google Scholar]
- 39.Rosa JL, Bartrons R, Tauler A. Gene expression of regulatory enzymes of glycolysis/gluconeogenesis in regenerating rat liver. Biochem J. 1992;287 (Pt 1):113–116. doi: 10.1042/bj2870113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.