Abstract
Mutations in the FBN1 gene cause Marfan syndrome (MFS) and a wide range of overlapping phenotypes. The severe end of the spectrum is represented by neonatal MFS, the vast majority of probands carrying a mutation within exons 24–32. We previously showed that a mutation in exons 24–32 is predictive of a severe cardiovascular phenotype even in non-neonatal cases, and that mutations leading to premature truncation codons are under-represented in this region. To describe patients carrying a mutation in this so-called ‘neonatal' region, we studied the clinical and molecular characteristics of 198 probands with a mutation in exons 24–32 from a series of 1013 probands with a FBN1 mutation (20%). When comparing patients with mutations leading to a premature termination codon (PTC) within exons 24–32 to patients with an in-frame mutation within the same region, a significantly higher probability of developing ectopia lentis and mitral insufficiency were found in the second group. Patients with a PTC within exons 24–32 rarely displayed a neonatal or severe MFS presentation. We also found a higher probability of neonatal presentations associated with exon 25 mutations, as well as a higher probability of cardiovascular manifestations. A high phenotypic heterogeneity could be described for recurrent mutations, ranging from neonatal to classical MFS phenotype. In conclusion, even if the exons 24–32 location appears as a major cause of the severity of the phenotype in patients with a mutation in this region, other factors such as the type of mutation or modifier genes might also be relevant.
Keywords: Neonatal Marfan syndrome, FBN1 mutations, exons 24–32, clinical and mutation-type analysis
Introduction
Marfan syndrome (MFS; MIM 154700) is a connective tissue disorder, with autosomal dominant inheritance and a prevalence of 1 in 5000–10 000 individuals.1 The cardinal features of MFS involve the ocular, cardiovascular and skeletal systems.2 Neonatal MFS is considered as the severe end of the MFS phenotype, and most cases are sporadic. Rare homozygote forms and a few compound heterozygote patients born to parents each displaying or not displaying a MFS phenotype, have been reported.3, 4
Although the known mutations of FBN1 are spread over the entire gene, the mutations causing neonatal MFS seem to cluster in a specific region from exons 24–32.5, 6, 7 Besides neonatal MFS, atypically severe phenotypes also cluster in exons 24–32.8 This region includes the central longest stretch of 12 cbEGF repeats, which is thought to form a rigid rod-like structure that may be important for microfibril assembly. Earlier, we showed that mutations in exons 24–32 were associated with a more severe phenotype than mutations located in other exons of the gene, including younger age at diagnosis of type I fibrillinopathy, higher probability of ectopia lentis, ascending aortic dilatation, aortic surgery, mitral valve abnormalities, scoliosis and shorter survival.9 The majority of these results was replicated even when neonatal cases were excluded, leading to the conclusion that exon 24–32 mutations define a high-risk group for cardiac manifestations, associated with severe prognosis at all ages.9 We also showed an under-representation of nonsense mutations and an over-representation of missense mutations in this region, when compared with other exons of the gene. Here, we focus on the clinical and molecular characterization of patients with a mutation in the so-called exons 24–32 ‘neonatal region‘, out of a series of 1013 probands with MFS or type I fibrillinopathy carrying a pathogenic FBN1 mutation.
Patients, materials and methods
Out of a series of 1013 probands carrying a pathogenic FBN1 mutation recruited for a genotype–phenotype correlation study,9, 10 we extracted a subgroup of 198 probands with a mutation in exons 24–32 in order to better describe their clinical and molecular characteristics. Patients were recruited to this study during the period 1995–2005 through the framework of the Universal Mutation Database–FBN111, 12 (UMD-FBN1; http://www.umd.be), or were referred by specialized MFS clinics in their respective countries. Patients originated from 38 countries in five continents. The required clinical information included a range of qualitative and quantitative clinical parameters, including age of diagnosis, presence or absence of clinical features including cardiac, ophthalmological, skeletal, cutaneous, pulmonary and dural manifestations of the Ghent nosology.13 The ages at diagnosis and at surgery for aortic dilatation, mitral valve prolapse and regurgitation, ectopia lentis and scoliosis were also collected. Patients were classified as ‘neonatal MFS' when presenting with characteristic features of MFS including severe valvular anomalies by 4 weeks of age; ‘severe MFS' when presenting with positive Ghent criteria including the presence of ascending aortic dilatation before 10 years of age; ‘classical MFS' when Ghent criteria were positive in the remaining patients; ‘incomplete MFS' when Ghent criteria were negative in adulthood and ‘probable MFS' when Ghent criteria were negative and follow-up was limited to childhood.
The pathogenic nature of a putative mutation was assessed using recognized criteria. In brief, all nonsense mutations, all deletions or insertions (in or out of frame) were considered pathogenic; for all splice mutations the wild-type and mutant strength values of the splice sites were compared using genetic algorithms12, 14, 15 and only mutations displaying significant deviation from the normal were included. Missense mutations were considered pathogenic when at least one of the following features was found: (i) de novo missense mutation, (ii) missense mutation substituting or creating a cysteine, (iii) missense mutation involving a consensus calcium-binding residue,16 (iv) substitution of glycines implicated in correct domain–domain packing,17 (v) intrafamilial segregation of a missense mutation involving a conserved amino acid. For other missense mutations not displaying one of the above features, additional data provided by SIFT,18, 19 BLOSUM-6220 and biochemical value (http://cmgm.stanford.edu/biochem218/Projects% 202001/Yu.pdf) were gathered and analyzed using a new UMD tool21 (G Collod-Beroud, personal communication).
The phenotypes and the genotypes of the overall cohort of patients are described elsewhere.9, 10 Here, we focus on the clinical and molecular characteristics of patients with a mutation in exons 24–32. We took advantage of this large series to study the MFS presentation types associated with these mutations, the distribution of mutations in this region and the genotype–phenotype correlations.
As the prevalence of many features of MFS increases with age, and as our study included patients with different lengths of follow-up, we performed a time-to-event analysis technique to estimate a reliable cumulative probability of observation of the different manifestations of MFS. This technique could be applied for the following events: diagnosis of MFS or type I fibrillinopathy, scoliosis, ectopia lentis, aortic dilatation or dissection, mitral abnormalities, as well as surgery for these different manifestations for which the age at diagnosis was systematically collected. In all time-to-event analyses, the baseline date (time zero) was the date of birth. The time-to-event diagnosis was defined as the interval between the baseline date and the date of event observation. Patients who did not manifest the studied event during the follow-up course were censored at their last follow-up. Patients for whom the age at diagnosis of a specific manifestation was not available were excluded from these analyses. The Kaplan–Meier method22 was used to estimate the cumulative probabilities of clinical manifestations of the disease at 10, 25 and 40 years of age to describe the evolution of clinical features over time. Differences among the different types of mutation groups (different locations or type of mutations) were tested using the non-parametric log-rank test. For the other features (skeletal features other than scoliosis, skin, lung and dural involvement), for which the ages at diagnosis were not collected, age at last follow-up was the only information available concerning the time of clinical features observation. To indirectly take into account the length of patient follow-up even in this situation, we adjusted all comparisons of MFS manifestation proportions for the ages at last follow-up, categorized into 10-year age groups. These adjusted comparisons were performed using the Mantel–Haenszel test.23 We compared the phenotypic data for each exon of the region with the others. To study the effect of mutation types, we compared patients with a premature termination codon (PTC) to patients with an in-frame mutation and patients with missense mutations involving a cysteine versus other missense mutations. We also searched if the position of the substituted cystein influenced the phenotype by comparing clinical data of patient with a mutation affecting the first disulfide bond with patients with a mutation in the second or third disulfide bond and conversely. To study the effect of the position of the affected EGF-like domain relative to the TGFBP-like domain, we compared the phenotype of patients with a missense mutation in exons 25 and 26 encoding EGF-like domains 11 and 12 (located near the TGFBP-like domain) to the phenotype of patients with a missense mutation in exons 27–32 encoding EGF-like domains 13–18.
SAS™ software version 9.2 (SAS Institute Inc., Cary, NC, USA) and Stata software version 9 (Stata Corp, College Station, TX, USA) were used for all statistical analyses. To take into account multiple testing, only P-values of less than 0.001 were considered significant.
Results
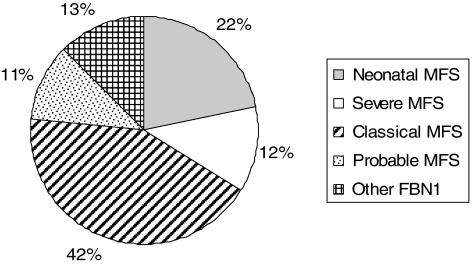
The genotype–phenotype correlation study in exons 24–32 versus other exons has been reported elsewhere.9 The MFS presentation type of patients with a mutation in exons 24–32 is summarized in Figure 1. An over-representation of neonatal and severe MFS and an under-representation of classical MFS were noted when compared to the overall series.9 Accordingly, a high percentage of sporadic cases were found (69%).
Figure 1.
Type of presentation of Marfan syndrome (MFS) in patients with an FBN1 mutation in exons 24–32 (N=198).
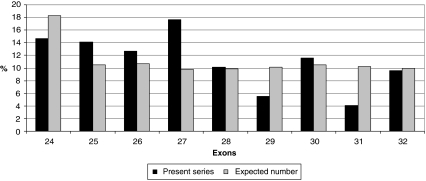
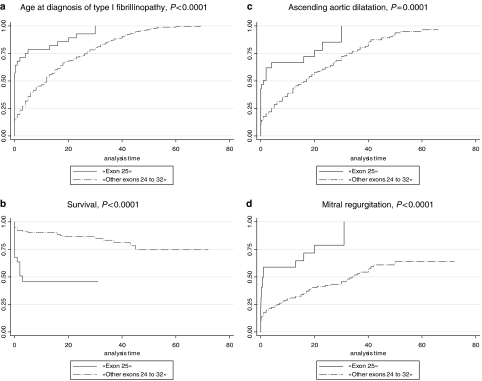
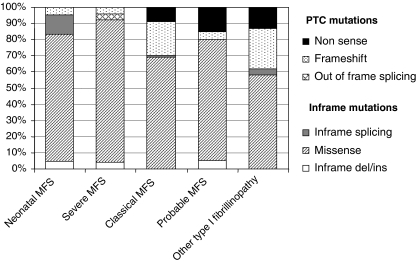
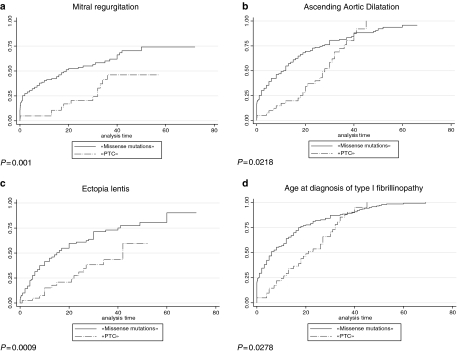
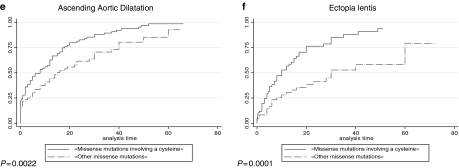
Twenty percent of the FBN1 mutations in the overall series were found in the exons 24–32 region (n=198), indicating a clustering of mutations in this region as only 14.5% was expected based on the length of genomic sequence of the gene. Figure 2 shows the number of mutations by exon, from exons 24 to 32, and, although results were nonsignificant, the clustering of mutations can be mainly explained by an excess of mutations in exons 25 and 27. An unequal distribution regarding the type of MFS presentation was found between exons of the studied region, with severe phenotypes most likely to be associated with mutations in exons 25, 26, 29, 31 and 32. Conversely, neonatal MFS was under-represented in patients with a mutation in exons 24, 27, 28 and 30 (Table 1). When comparing the probability of the different clinical features for one individual exon compared to the other exons of the region, significant results were found only for patients carrying a mutation in exon 25. Indeed, a younger age at diagnosis of MFS or type I fibrillinopathy, a higher probability of ascending aortic dilatation, mitral regurgitation, valvular surgery and scoliosis, as well as a lower chance of survival were all found when compared to patients with a mutation within other exons of the exons 24–32 region (Figure 3). These results can be explained at least in part by a higher frequency of patients with neonatal MFS in exon 25 (57%, Table 1, P<0.001). The majority of mutations was in-frame and predicted to result in an altered protein (79%), while 21% were predicted to result in a PTC. Within the 139 missense mutations, 75 involved a cysteine (54%). Twenty-five mutations affected the first disulfide bond, 11 mutations the second disulfide bond and 23 mutations the third disulfide bond. Fifty-two patients had a mutation in the EGF-like domains 11 or 12, and 96 in the EGF-like domains 13–17. Figure 4 presents the distribution of types of mutations, depending on the severity of the clinical presentation. In particular, PTCs were under-represented in patients with severe phenotypes and an absence of nonsense mutations, while missense mutations were over-represented. We questioned whether the type of mutation within the exons 24–32 region could lead to a differing clinical phenotype. Some significant results were found when patients with an exons 24–32 PTC mutation were compared with patients with an exons 24–32 missense mutation (Figure 5). Indeed, the cumulative probability of mitral insufficiency diagnosed before or at 25 years was 54% (99.9% CI=39–69%) in patients with a missense mutation in exons 24–32, compared to 20% (99.9% CI=5–53%) in patients with a PTC mutation in the same region (log-rank test P=0.001). Similarly, the cumulative probability of ectopia lentis diagnosed before or at 25 years was 61% (99.9% CI=45–77%) in patients with a missense mutation in exons 24–32, compared to 31% (99.9% CI=11–63%) in patients with a PTC mutation in the same region (log-rank test P=0.0009). Conversely, a higher frequency of pectus deformity was found in patients with a PTC mutation in exons 24–32 when compared to patients with a missense mutation in the same region (83 versus 54%, MH test P=0.001). No significant results were found for the other clinical features of the MFS spectrum. A tendency towards a higher probability of ascending aortic dilatation and a younger age at diagnosis was noted with missense mutations, although these associations were only marginally significant (P=0.0218 and 0.0278, respectively) (Figure 5). When comparing patients with missense mutations involving a cysteine to other missense mutations, significant results were found for ectopia lentis. Indeed, the cumulative probability of ectopia lentis diagnosed before or at 25 years was 76% (99.9% CI=60–89%) in patients with a missense mutation involving a cysteine in exons 24–32, compared to 41% (99.9% CI=25–63%) in patients with another missense mutation in the same region (log-rank test P=0.0001). A tendency towards a higher probability of ascending aortic dilatation was noted in patients with missense mutations involving a cysteine, although this association was only marginally significant (P=0.0022) (Figure 5). No significant differences were found when comparing clinical data of patients with a mutation affecting the first, second or third disulfide bond but the numbers were small. Significant differences were found when comparing the clinical phenotype of patients carrying a missense mutation in exons 25 and 26 encoding EGF-like domains 11 and 12 located near the TGFBP-like domain with patients carrying a missense mutation in exons 27–32 encoding EGF-like domains 13–18. Indeed, patients with a missense mutation affecting EGF-like domains 11 or 12 (n=52) have a shorter survival, a younger age at diagnosis, a higher risk of presenting a neonatal presentation, a higher risk of developing ascending aortic dilatation and a higher risk of developing mitral insufficiency than patients with a missense mutation affecting EGF-like domains 13–17 (P<0.001).
Figure 2.
Number of mutations in the ‘exons 24–32 region' for each exon (black), as compared with the number of mutations expected from the genomic sequence of the gene (grey), N=198.
Table 1. Type of MFS presentations by exon, in the exons 24–32 region.
| Exons | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Type of presentation | 24 (n=29) | 25 (n=28) | 26 (n=25) | 27 (n=35) | 28 (n=20) | 29 (n=11) | 30 (n=23) | 31 (n=8) | 32 (n=19) |
| Neonatal MFS | 10% | 57% | 28% | 11% | 5% | 27% | 13% | 25% | 21% |
| Severe MFS | 10% | 11% | 4% | 11% | 30% | 18% | 17% | 0% | 4% |
| Classical MFS | 53% | 11% | 40% | 40% | 45% | 45% | 57% | 75% | 53% |
| Probable MFS | 17% | 11% | 12% | 9% | 5% | 10% | 9% | 0% | 16% |
| Type I fibrillinopathy | 10% | 10% | 16% | 29% | 15% | 0% | 4% | 0% | 6% |
| P Fisher | 0.365 | <0.001 | 0.628 | 0.046 | 0.052 | 0.738 | 0.428 | 0.475 | 0.655 |
Figure 3.
Kaplan–Meier analyses for various clinical features in patients with a mutation in exon 25 as compared to patients with a mutation in other exons of the ‘24–32 region'. (a) Age at diagnosis of type I fibrillinopathy in exon 25 mutations versus mutations in other exons of the ‘24–32 region'. Seventy-nine percent of patients with a mutation in exon 25 (solid line) were diagnosed by 10 years (99.9% CI=51–98%) of age versus 46% (99.9% CI=35–60%) of patients with a mutation in other exons of the ‘24–32 region' (broken line) (log-rank test P<0.0001). (b) Survival of patients with exon 25 mutations versus mutations in other exons of the 24–32 region. Forty-six percent of patients with mutations within exons 25 (solid line) were alive at 10 years (99.9% CI=13–75%) compared to 90% (99.9% CI=80–96%) of patients with a mutation in other exons of the 24–32 region (broken line) (log-rank test P<0.0001). (c) Probability of ascending aortic dilatation in exon 25 mutations versus mutations in other exons of the ‘exons 24–32 region'. The cumulative probability of ascending aortic dilatation before or at 10 years was 67% (99.9% CI=44–88%) in patients with mutations within exon 25 (solid line) compared to 39% (99.9% CI=30–49%) in patients with a mutation in other exons of the ‘24–32 region' (broken line) (P=0.0001). (d) Probability of mitral regurgitation in exon 25 mutations versus mutations in other exons of the ‘exons 24–32 region‘. The cumulative probability of mitral regurgitation before or at 10 years was 59% (99.9% CI=35–84%) in patients with mutations within exon 25 (solid line) compared to 30% (99.9% CI=22–40%) in patients with a mutation in other exons of the ‘24–32 region' (broken line) (P<0.0001).
Figure 4.
Distribution of types of mutations within the exons 24–32 region depending on the clinical presentation (N=191) (seven splicing mutations could not be classified as in-frame or out of frame).
Figure 5.
Kaplan–Meier analyses for various clinical features in patients with a missense mutation in exons 24–32 compared to patients with a premature termination codon (PTC) mutation in the same region. (a) Probability of mitral insufficiency in missense mutations in exons 24–32 versus PTC mutations in the same region. The cumulative probability of mitral insufficiency diagnosed before or at 25 years was 54% (99.9% CI=39–69%) in patients with a missense mutation in exons 24–32 (solid line) compared to 20% (99.9% CI=5–53%) in patients with a PTC mutation in the same region (broken line) (P=0.001). (b) Probability of ascending aortic dilatation in missense mutations in exons 24–32 versus PTC mutations in the same region. The cumulative probability of ascending aortic dilatation before or at 25 years was 74% (99.9% CI=60–86%) in patients with a missense mutation in exons 24–32 (solid line) compared to 40% (99.9% CI=18–70%) in patients with a PTC mutation in the same region (broken line), but these results were not significant because the curves join together with follow-up (P=0.0218). (c) Probability of ectopia lentis in missense mutations in exons 24–32 versus PTC mutations in the same region. The cumulative probability of ectopia lentis diagnosed before or at 25 years was 61% (99.9% CI=45–77%) in patients with a missense mutation in exons 24–32 (solid line) compared to 31% (99.9% CI=11–63%) in patients with a PTC mutation in the same region (broken line) (P=0.0009). (d) Age at diagnosis of type I fibrillinopathy in missense mutations in exons 24–32 versus PTC mutations in the same region. Fifty-five percent of patients with a missense mutation in exons 24–32 (solid line) were diagnosed at 6 years (IQR (0.7;18)) of age versus 21 years (IQR (11;32)) of age in patients with a PTC mutation in the same region (broken line), but results of the log-rank test were not significant because the curves join together with follow-up (P=0.0278). (e) Probability of ascending aortic dilatation in missense mutations involving a cysteine in exons 24–32 versus other missense mutations in the same region. The cumulative probability of ascending aortic dilatation before or at 25 years was 83% (99.9% CI=70–92%) in patients with a missense mutation involving a cysteine in exons 24–32 (solid line) compared to 62% (99.9% CI=45–78%) in patients with another missense mutation in the same region (broken line), but these results were only marginally significant (P=0.0022). (f) Probability of ectopia lentis in missense mutations involving a cysteine in exons 24–32 versus other missense mutations in the same region. The cumulative probability of ectopia lentis diagnosed before or at 25 years was 76% (99.9% CI=60–89%) in patients with a missense mutation involving a cysteine in exons 24–32 (solid line) compared to 41% (99.9% CI=25–63%) in patients with another missense mutation in the same region (broken line) (P=0.0001).
Twenty-four mutations were recurrent. Table 2 shows the MFS presentation types in these recurrent mutations. Interestingly, some recurrent mutations lead to a similar phenotype, while others lead to different presentations.
Table 2. Recurrent mutations in the exons 24–32 region and their type of presentation.
| Mutation | Effect | Number of event | Type of presentation | References |
|---|---|---|---|---|
| c.2860C>T | p.Arg954Cys | 2 | Classical MFS (2) | Comeglio et al24; L Adès, personal communication |
| c.2953G>A | p.Gly985Arg | 2 | Classical MFS (1), probable MFS (1) | Loeys et al25, Rommel et al26 |
| c.2980G>T | p.Glu994X | 2 | Classical MFS (2) | Comeglio et al24, Karttunen et al27 |
| c.3037G>A | p.Gly1013Arg | 7 | Neonatal MFS (1), severe MFS (3), classical MFS (2), probable MFS (1) | Tiecke et al8, Comeglio et al24, Nijbroek et al28, Liu et al29, Lo et al30, Biggin et al31 |
| c.3058A>G | p.Thr1020Ala | 2 | Probable MFS (1), other type I fibrillinopathy (1) | Tiecke et al8; C Boileau, personal communication |
| c.3095G>A | p.Cys1032Tyr | 2 | Neonatal MFS (2) | Comeglio et al24, Ng et al32 |
| c.3143T>C | p.Ile1048Thr | 5 | Neonatal MFS (3), severe MFS (1), probable MFS (1) | Collod-Beroud et al11, Comeglio et al24, Lonnqvist et al33, Jacobs et al34 |
| c.3157T>C | p.Cys1053Arg | 2 | Neonatal MFS (2) | Putnam et al7, Collod-Beroud et al11 |
| c.3202T>C | p.Cys1068Arg | 2 | Neonatal MFS (1), probable MFS (1) | L Adès, personal communication; P Robinson, personal communication |
| c.3217G>A | p.Glu1073Lys | 3 | Neonatal MFS (2), probable MFS (1) | Putnam et al7, Nijbroek et al28, Loeys et al35 |
| c.3299G>T | p.Gly1100Val | 2 | Classical MFS (2) | Loeys et al35; C Boileau, personal communication |
| c.3302A>G | p.Tyr1101Cys | 6 | Classical MFS (5), probable MFS (1) | Loeys et al25, Biggin et al31, Rommel et al36, Arbustini et al37; P Robinson, personal communication; C Boileau, personal communication |
| c.3344A>G | p.Asp1115Gly | 2 | Classical MFS (1), other type I fibrillinopathy (1) | Tiecke et al8; P Robinson, personal communication |
| c.3350G>A | p.Cys1117Tyr | 2 | Severe MFS (1), classical MFS (1) | Biggin et al31, Tynan et al38 |
| c.3373C>T | p.Arg1125X | 3 | Probable MFS (1), other type I fibrillinopathy (2) | Rommel et al26; A De Paepe, personal communication; JC Hyland, personal communication |
| c.3388delC | p.His1130IlefsX32 | 4 | Classical MFS (4) | Collod-Beroud et al11, Loeys et al35 |
| c.3410G>C | p.Arg1137Pro | 2 | Classical MFS (2) | Dietz et al39 |
| c.3412T>C | p.Cys1138Arg | 2 | Severe MFS (1), classical MFS (1) | Collod-Beroud et al11; De Paepe, personal communication |
| c.3463G>A | p.Asp1155Asn | 2 | Classical MFS (1), other type I fibrillinopathy (1) | Biggin et al31, Milewicz et al40 |
| c.3511T>C | p.Cys1171Arg | 2 | Classical MFS (1), probable MFS (1) | Schrijver et al41; L Adès, personal communication |
| c.3656A>G | p.Tyr1219Cys | 2 | Classical MFS (2) | Arbustini et al37 |
| c.3668G>A | p.Cys1223Tyr | 2 | Severe MFS (1), classical MFS (1) | Hewett et al42, Sood et al43 |
| c.3725G>A | p.Cys1242Tyr | 3 | Severe MFS (1), classical MFS (1), probable MFS (1) | Kainulainen et al6, Schrijver et al41; P Robinson, personal communication |
| c.3976T>C | p.Cys1326Arg | 2 | Neonatal MFS (1), severe MFS (1) | Comeglio et al24, Schrijver et al41 |
Discussion
Here, we further delineate the clinical and molecular characteristics of the so-called ‘neonatal exons 24–32 region' from the data of a large series in which the phenotype of 1013 probands with MFS and other type I fibrillinopathies were collected. We confirm that the region encompassing exons 24 to 32 is associated with more severe phenotypes than the other exons of the gene. Indeed, one-third of the patients with a mutation within this region had neonatal or severe MFS, as compared to 6% in the other regions.9
We showed previously that the presence of a mutation in exons 24–32 was predictive of a severe cardiovascular phenotype even in non-neonatal phenotypes,9 but it is unknown if the location of the mutation is the only cause of the phenotypic severity. Genotype–phenotype correlation analyses can be complicated by the fact that both the location and the type of a mutation are critical in producing a severe phenotype and these data are often studied independently. For this reason, we looked for clinical differences between patients with different mutation types within this region. A higher probability of mitral regurgitation and ectopia lentis, as well as a lower frequency of pectus deformity were found in patients with a missense mutation within this region when compared to patients with a PTC mutation. Also, a higher probability of ectopia lentis was found in patients with a missense mutation involving a cysteine within this region when compared to patients with other missense mutations. These results were highly superposable to those obtained for all the regions of the FBN1 gene,9 showing that, beside the predominant role of the location of the mutations within the exons 24–32 region, the type of mutation is also important.
We showed previously that the distribution of the mutation types in exons 24–32 is different from the distribution found in other exons of the gene. Indeed, mutations leading to PTC are under-represented, contrasting with an over-representation of in-frame mutations.9 Here, we show that PTC mutations are under-represented in the severe MFS phenotype. In particular, nonsense mutations have never been described in association with neonatal and severe MFS presentations. In contrast, in the overall cohort, we showed that patients with an FBN1 PTC mutation had a more severe skeletal and skin phenotype than patients with an in-frame mutation.9 Therefore, it is not known whether the absence of nonsense mutations in the neonatal and severe phenotypes, as well as the under-representation of PTC mutations in these phenotypes, could be explained by early lethality or by a milder effect on phenotype of PTC mutations within this region. In searching for differences in various clinical system involvements between PTC and in-frame mutations within this region, there were no emerging clues for this region regarding the dominant negative versus haploinsufficiency pathogenesis models and no evidence to support a differential mechanism for the phenotypic and genotypic differences within the exons 24–32 region and other regions of the gene. Recent data have highlighted the complexity of the pathogenicity of FBN1 mutations, with some mutations acting as dominant negative, and others as haploinsufficiency secondary to different effects on trafficking.44, 45, 46, 47 However, mutation data accumulated by diagnostic laboratories worldwide are generally not associated with mRNA and protein studies. Therefore, no data are available to assess the true effect of PTC mutations and whether they are submitted to nonsense-mediated RNA decay or they give rise to truncated peptides of various sizes. Until more information is available, the true impact of PTC mutations on microfibril formation can only be speculated.
The clustering of mutations with an excess of mutations in exons 24–32 has been postulated before.8, 48 This hypothesis is confirmed in this study and might explain the high proportion of sporadic cases. The same clustering of mutations in exons 24–34 of the FBN2 gene in patients with congenital contractural arachnodactyly (OMIM 121050)7, 49, 50 is in favor of a critical role of this region in both fibrillin-1 and -2. The domains encoded by exons 25–36 in fibrillin-1 are found midway through the protein and constitute the longest stretch of EGF-like domains in the protein. Exon 24 encodes an eight-cysteine domain found immediately amino-terminal to this stretch of EGF-like domains. Schrijver et al41 reported that the position of an affected EGF-like domain relative to an eight-cysteine domain could be related to the severity of the phenotype. In keeping with this report, we queried for possible differences in clinical presentation in patients carrying a missense mutation in exons 25 and 26, versus exons 27–32. We found a significantly more severe presentation in the patients with mutations in exons 25 and 26 that encode EGF-like domains 11 and 12. Furthermore, exon 25 was associated with a higher frequency of neonatal presentations and a higher probability of ascending aortic dilatation compared with mutations in other exons within this region. This exon encodes EGF-like domain 11, which is immediately downstream from the eight-cysteine domain. Interestingly, this relative location is conserved between fibrillin-1 and transforming growth factor β1-binding protein (LTBP).51 LTBP plays a role in the assembly and secretion of TGFβ1 and is thought to target TGFβ1 to particular extracellular matrix sites, thus controlling the production and structure of the extracellular matrix, along with affecting cell growth, morphology and differentiation.52, 53 The homology of fibrillin-1 and LTBP raises the possibility that disruption of the extracellular targeting of the action of TGFβ1 during development underpins the more severe phenotype. Alternatively, mutations in this region of fibrillin-1 may be more disruptive to microfibril formation. Although mutations throughout the FBN1 gene have been shown to disrupt fibrillin-1 incorporation into microfibrils, exons 24–32 may encode a region of fibrillin-1 with a unique function in the multimerization of the protein into stable microfibrils. In contrast to microfibrils formed by classic MFS fibroblasts, the fibrils formed by neonatal MFS show not only an apparent decrease in fibrillin accumulation, but are also short, fragmented and frayed.24 Therefore, alterations in this region of the protein may have a significant and specific effect on microfibril formation, implying a unique role of this region in microfibril formation.
Finally, the study of recurrent mutations was of interest. The majority of these recurrent mutations were only found in two instances. Three mutations were represented in five instances or more. Although the c.3302A>G mutation was generally associated with the classical MFS, the c.3037G>A mutation led to different phenotypes, ranging from neonatal to classical MFS. Five mutations responsible for a neonatal MFS phenotype in some patients were also found in other patients with another MFS type of presentation (c.3037G>A, c.3143T>C, c.3202T>C, c.3217G>A and c.3976T>C). These data give further emphasis to the clinical variability in FBN1 mutations and strongly argue for the role of modifier genes or the existence of a digenic mechanism to explain neonatal MFS.
In conclusion, even if the exons 24–32 location of mutations appears as a major cause of the severity of the phenotype in patients harboring a mutation in this region, other factors such as the type of mutation or modifier genes might also be involved. These data could be helpful in understanding the role of the central region of the FBN1 gene in disease pathogenicity.
Acknowledgments
We thank H Plauchu (Lyon, France), D Halliday (Oxford, UK), HC Dietz (Baltimore, USA), I Kaitila (Helsinki, Finland), S Davies (Cardiff, Wales) and T Uyeda (Irosaki, Japan) for their participation in the study. This study was supported by a grant from the French ministry of health (PHRC 2004), GIS maladies rares 2004, Bourse de la Société Francaise de Cardiologie, Fédération Française de Cardiologie 2005 and ANR-05-PCOD-014. BC and BL are respectively a research fellow and a senior clinical investigator of the Fund for Scientific Research – Flanders. AC and PC thank the Marfan Trust and the Bluff Field Charitable Fund for support.
References
- Pyeritz RE. Marfan syndrome: current and future clinical and genetic management of cardiovascular manifestations. Semin Thorac Cardiovasc Surg. 1993;5:11–16. [PubMed] [Google Scholar]
- Judge DP, Dietz HC. Marfan's syndrome. Lancet. 2005;366:1965–1976. doi: 10.1016/S0140-6736(05)67789-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karttunen L, Raghunath M, Lonnqvist L, Peltonen L. Compound-heterozygous Marfan patient: two defective fibrillin alleles result in a lethal phenotype. Am J Hum Genet. 1994;55:1083–1091. [PMC free article] [PubMed] [Google Scholar]
- De Vries BBA, Pals G, Odink R, Hamel BCJ. Homozygosity for a FBN1 missense mutation: clinical and molecular evidence for recessive Marfan syndrome. Eur J Hum Genet. 2007;15:930–935. doi: 10.1038/sj.ejhg.5201865. [DOI] [PubMed] [Google Scholar]
- Milewicz DM, Duvic M. Severe neonatal Marfan syndrome resulting from a de novo 3-bp insertion into the fibrillin gene on chromosome 15. Am J Hum Genet. 1994;54:447–453. [PMC free article] [PubMed] [Google Scholar]
- Kainulainen K, Karttunen L, Puhakka L, Sakai L, Peltonen L. Mutations in the fibrillin gene responsible for dominant ectopia lentis and neonatal Marfan syndrome. Nat Genet. 1994;6:64–69. doi: 10.1038/ng0194-64. [DOI] [PubMed] [Google Scholar]
- Putnam EA, Cho M, Zinn AB, Towbin JA, Byers PH, Milewicz DM. Delineation of the Marfan phenotype associated with mutations in exons 23–32 of the FBN1 gene. Am J Med Genet. 1996;62:233–242. doi: 10.1002/(SICI)1096-8628(19960329)62:3<233::AID-AJMG7>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Tiecke F, Katzke S, Booms P, et al. Classic, atypically severe and neonatal Marfan syndrome: twelve mutations and genotype–phenotype correlations in FBN1 exons 24–40. Eur J Hum Genet. 2001;9:13–21. doi: 10.1038/sj.ejhg.5200582. [DOI] [PubMed] [Google Scholar]
- Faivre L, Collod-Beroud G, Loeys BL, et al. Effect of mutation type and location on clinical outcome in 1013 probands with Marfan syndrome or related phenotypes with FBN1 mutations: an international study. Am J Hum Genet. 2007;81:454–466. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faivre L, Collod-Beroud G, Child A, et al. Contribution of molecular analyses in diagnosing Marfan syndrome and type I fibrillinopathies: an international study of 1009 probands. J Med Genet. 2008;45:384–390. doi: 10.1136/jmg.2007.056382. [DOI] [PubMed] [Google Scholar]
- Collod-Beroud G, Le Bourdelles S, Adès L, et al. Update of the UMD-FBN1 mutation database and creation of an FBN1 polymorphism database. Hum Mutat. 2003;22:199–208. doi: 10.1002/humu.10249. [DOI] [PubMed] [Google Scholar]
- Beroud C, Hamroun D, Collod-Beroud G, Boileau C, Soussi T, Claustres M. UMD (Universal Mutation Database): 2005 update. Hum Mutat. 2005;26:184–191. doi: 10.1002/humu.20210. [DOI] [PubMed] [Google Scholar]
- De Paepe A, Devereux RB, Dietz HC, Hennekam RC, Pyeritz RE. Revised diagnostic criteria for the Marfan syndrome. Am J Med Genet. 1996;62:417–426. doi: 10.1002/(SICI)1096-8628(19960424)62:4<417::AID-AJMG15>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Dietz HC, Pyeritz RE. Mutations in the human gene for fibrillin-1 (FBN1) in the Marfan syndrome and related disorders. Hum Mol Genet. 1995;4:1799–1809. doi: 10.1093/hmg/4.suppl_1.1799. [DOI] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapathy P, Shapiro MB, Harris NL. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Downing A, Knott V, Werner J, Cardy C, Campbell ID, Handford PA. Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders. Cell. 1996;85:597–605. doi: 10.1016/s0092-8674(00)81259-3. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frédéric MY, Lalande M, Boileau C, et al. UMD-predictor, a new prediction tool for nucleotide substitution pathogenecity; application for four genes: FBN1, FBN2, TGFBR1 and TGFBR2. Hum Mut (submitted) [DOI] [PubMed]
- Kaplan E, Meier P. Non parametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- Comeglio P, Johnson P, Arno G, et al. The importance of mutation detection in Marfan syndrome and Marfan-related disorders: report of 193 FBN1 mutations. Hum Mutat. 2007;28:928. doi: 10.1002/humu.9505. [DOI] [PubMed] [Google Scholar]
- Loeys B, Nuytinck L, Delvaux I, De Bie S, De Paepe A. Genotype and phenotype analysis of 171 patients referred for molecular study of the fibrillin-1 gene FBN1 because of suspected Marfan syndrome. Arch Intern Med. 2001;161:2447–2454. doi: 10.1001/archinte.161.20.2447. [DOI] [PubMed] [Google Scholar]
- Rommel K, Karck M, Haverich A, et al. Identification of 29 novel and nine recurrent fibrillin-1 (FBN1) mutations and genotype-phenotype correlations in 76 patients with Marfan syndrome. Hum Mutat. 2005;26:529–539. doi: 10.1002/humu.20239. [DOI] [PubMed] [Google Scholar]
- Karttunen L, Ukkonen T, Kainulainen K, Syvänen A-C, Peltonen L. Two novel fibrillin-1 mutations resulting in premature termination codons but in different mutant transcript levels and clinical phenotype. Hum Mutat. 1998. pp. S34–S37. [DOI] [PubMed]
- Nijbroek G, Sood S, McIntosh I, et al. Fifteen novel FBN1 mutations causing Marfan syndrome detected by heteroduplex analysis of genomic amplicons. Am J Hum Genet. 1995;57:8–21. [PMC free article] [PubMed] [Google Scholar]
- Liu WO, Oefner PJ, Qian C, Odom RS, Francke U. Denaturing HPLC-identified novel FBN1 mutations, polymorphisms, and sequence variants in Marfan syndrome and related connective tissue disorders. Genet Test. 1997–98;1:237–242. doi: 10.1089/gte.1997.1.237. [DOI] [PubMed] [Google Scholar]
- Lo IF, Wong RM, Lam FW, Tong TM, Lam ST. Missense mutations of the fibrillin-1 gene in two Chinese patients with severe Marfan syndrome. Chin Med J. 2001;114:473–476. [PubMed] [Google Scholar]
- Biggin A, Holman K, Brett M, Bennetts B, Ades L. Detection of thirty novel FBN1 mutations in patients with Marfan syndrome or a related fibrillinopathy. Hum Mutat. 2004;23:99. doi: 10.1002/humu.9207. [DOI] [PubMed] [Google Scholar]
- Ng DK, Chau KW, Black C, Thomas TM, Boxer M. Neonatal Marfan syndrome: a case report. J Paediatr Child Health. 1999;35:321–323. doi: 10.1046/j.1440-1754.1999.00355.x. [DOI] [PubMed] [Google Scholar]
- Lonnqvist L, Child A, Kainulainen K, Davidson R, Puhakka L, Peltonen L. A novel mutation of the fibrillin gene causing ectopia lentis. Genomics. 1994;19:573–576. doi: 10.1006/geno.1994.1110. [DOI] [PubMed] [Google Scholar]
- Jacobs AM, Toudjarska I, Racine A, Tsipouras P, Kilpatrick MW, Shanske A. A recurring FBN1 gene mutation in neonatal Marfan syndrome. Arch Pediatr Adolesc Med. 2002;156:1081–1085. doi: 10.1001/archpedi.156.11.1081. [DOI] [PubMed] [Google Scholar]
- Loeys B, De Backer J, Van Acker P, et al. Comprehensive molecular screening of the FBN1 gene favors locus homogeneity of classical Marfan syndrome. Hum Mutat. 2004;24:140–146. doi: 10.1002/humu.20070. [DOI] [PubMed] [Google Scholar]
- Rommel K, Karck M, Haverich A, Schmidtke J, Arslan-Kirchner M. Mutation screening of the fibrillin-1 (FBN1) gene in 76 unrelated patients with Marfan syndrome or Marfanoid features leads to the identification of 11 novel and three previously reported mutations. Hum Mutat. 2002;20:406–407. doi: 10.1002/humu.9075. [DOI] [PubMed] [Google Scholar]
- Arbustini E, Grasso M, Ansaldi S, et al. Identification of sixty-two novel and twelve known FBN1 mutations in eighty-one unrelated probands with Marfan syndrome and other fibrillinopathies. Hum Mutat. 2005;26:494. doi: 10.1002/humu.9377. [DOI] [PubMed] [Google Scholar]
- Tynan K, Comeau K, Pearson M, et al. Mutation screening of complete fibrillin-1 coding sequence: report of five new mutations, including two in 8-cysteine domains. Hum Mol Genet. 1993;2:1813–1821. doi: 10.1093/hmg/2.11.1813. [DOI] [PubMed] [Google Scholar]
- Dietz H, Cutting GR, Pyeritz RE, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- Milewicz DM, Michael K, Fisher N, Coselli JS, Markello T, Biddinger A. Fibrillin-1 (FBN1) mutations in patients with thoracic aortic aneurysms. Circulation. 1996;94:2708–2711. doi: 10.1161/01.cir.94.11.2708. [DOI] [PubMed] [Google Scholar]
- Schrijver I, Liu W, Brenn T, Furthmayr H, Francke U. Cysteine substitutions in epidermal growth factor-like domains of fibrillin-1: distinct effects on biochemical and clinical phenotypes. Am J Hum Genet. 1999;65:1007–1020. doi: 10.1086/302582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett DR, Lynch JR, Child A, Sykes BC. A new missense mutation of fibrillin in a patient with Marfan syndrome. J Med Genet. 1994;31:338–339. doi: 10.1136/jmg.31.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood S, Eldadah ZA, Krause WL, McIntosh I, Dietz HC. Mutation in fibrillin-1 and the Marfanoid-craniosynostosis (Shprintzen-Goldberg) syndrome. Nat Genet. 1996;12:209–211. doi: 10.1038/ng0296-209. [DOI] [PubMed] [Google Scholar]
- Whiteman P, Handford PA. Effective secretion of recombinant fragments of fibrillin-1: implications of protein misfolding for the pathogenesis of Marfan syndrome and related disorders. Hum Mol Genet. 2003;12:727–737. doi: 10.1093/hmg/ddg081. [DOI] [PubMed] [Google Scholar]
- Eldadah ZA, Brenn T, Furthmayr H, Dietz HC. Expression of a mutant human fibrillin allele upon a normal human or murine genetic background recapitulates a Marfan cellular phenotype. J Clin Invest. 1995;95:874–880. doi: 10.1172/JCI117737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge DP, Biery NJ, Keene DR, et al. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J Clin Invest. 2004;114:172–181. doi: 10.1172/JCI20641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mátyás G, Alonso S, Patrignani A, et al. Large genomic fibrillin-1 (FBN1) gene deletions provide evidence for true haploinsufficiency in Marfan syndrome. Hum Genet. 2007;122:23–32. doi: 10.1007/s00439-007-0371-x. [DOI] [PubMed] [Google Scholar]
- Robinson PN, Booms P, Katzke S, et al. Mutations of FBN1 and genotype-phenotype correlations in Marfan syndrome and related fibrillinopathies. Hum Mutat. 2002;20:153–161. doi: 10.1002/humu.10113. [DOI] [PubMed] [Google Scholar]
- Park ES, Putnam EA, Chitayat D, Child A, Milewicz DM. Clustering of FBN2 mutations in patients with congenital contractural arachnodactyly indicates an important role of the domains encoded by exons 24 through 34 during human development. Am J Med Genet. 1998;78:350–355. [PubMed] [Google Scholar]
- Frederic M, Monino C, Marschall C, et al. The FBN2 gene: New mutations, Locus Specific DataBase (UMD-FBN2) and genotype-phenotype correlations Hum Mut 2008(in press) [E-pub ahead of print]. [DOI] [PubMed]
- Kanzaki T, Olofsson A, Moren A, et al. GF-beta 1 binding protein: a component of the large latent complex of TGF-beta 1 with multiple repeat sequences. Cell. 1990;61:1051–1061. doi: 10.1016/0092-8674(90)90069-q. [DOI] [PubMed] [Google Scholar]
- Taipale J, Miyazono K, Heldin CH, Keski-Oja J. Latent transforming growth factor-beta 1 associates to fibroblast extracellular matrix via latent TGF-beta binding protein. J Cell Biol. 1994;124:171–181. doi: 10.1083/jcb.124.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley DM. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]