Abstract
The Notch signalling pathway plays multiple and important roles in mammals. However, several aspects of its action, in particular the precise mapping of its sites of activity, remain unclear. To address this issue, we have generated a transgenic line carrying a construct consisting of a nls-lacZ reporter gene under the control of a minimal promoter and multiple RBP-Jκ binding sites. Here we show that this transgenic line, we named NAS for Notch Activity Sensor, displays an expression profile that is consistent with current knowledge on Notch activity sites in mice, even though it may not report on all these sites. Moreover, we observe that NAS transgene expression is abolished in a RBP-Jκ deficient background indicating that it indeed requires Notch/RBP-Jκ signalling pathway activity. Thus, the NAS transgenic line constitutes a valuable and versatile tool to gain further insights into the complex and various functions of the Notch signalling pathway.
Keywords: Animals; Arteries; embryology; metabolism; Base Sequence; Chromosomal Proteins, Non-Histone; genetics; Female; Gene Expression; Genes, Reporter; Heart; embryology; Immunoglobulin J Recombination Signal Sequence-Binding Protein; genetics; metabolism; Lac Operon; Male; Mice; Mice, Transgenic; embryology; physiology; Models, Biological; Molecular Sequence Data; Myocardium; metabolism; Promoter Regions, Genetic; Receptors, Notch; metabolism; physiology; Signal Transduction; Somites; metabolism; Transgenes
Introduction
The Notch signalling pathway plays a fundamental role in numerous developmental processes in metazoans (Artavanis-Tsakonas et al., 1995; Artavanis-Tsakonas et al., 1999; Lai, 2004). Depending on the context, signalling through the Notch pathway impinges on a wide variety of cellular responses such as binary cell fate decisions, stem cell maintenance, induction of differentiation, proliferation or apoptosis. Accordingly, genetic alterations of Notch pathway components have been implicated in various pathologies in humans, including cancer (Garg et al., 2005; Gridley, 2003; Joutel and Tournier-Lasserve, 1998; Radtke and Raj, 2003).
Notch is a large single spanning transmembrane protein composed of an extracellular domain containing epithelial-growth-factor-like repeats and an intracellular domain (NICD) containing ankyrin motifs, a transactivation domain and nuclear localisation signals. Interaction of the extracellular domain of Notch receptors with membrane-bound ligands encoded by Delta and Serrate/Jagged family genes triggers the proteolytic cleavage of the Notch receptor and the release of NICD from the membrane. NICD then translocates into the nucleus where it interacts with the DNA-binding protein RBP-Jκ/CBF1/Su(H). RBP-Jκ and co-factors assemble into a complex that activates the expression of target genes, while in the absence of activated Notch, RBP-Jκ recruits repressor complexes to the cis-regulatory region of the Notch target genes.
Numerous studies have been performed in order to decipher the in vivo functions of the Notch pathway in vertebrates. In particular, genetic studies in mice have pointed to many roles for the Notch pathway: in somite formation and patterning (reviewed in (Aulehla and Herrmann, 2004; Weinmaster and Kintner, 2003)), vasculature development and arterial identity acquisition (reviewed in (Shawber and Kitajewski, 2004)), neurogenesis (reviewed in (Yoon and Gaiano, 2005)) or lymphoid progenitors differentiation (reviewed in (Radtke et al., 2004)). To dissect out the cellular responses directly controlled by the Notch pathway, it is important to identify cells in which the pathway is activated at a given time during development and in adult tissues. However, acquisition of such knowledge is hampered by several limitations. First, the Notch pathway involves a complex set of proteins. In rodents for example, there are four Notch receptors, five ligands of the Delta/Serrate/Jagged family and two recently identified new ligands, DNER and F3 contactin (Eiraku et al., 2005; Hu et al., 2003). The corresponding genes are expressed at many sites and exhibit complex and often overlapping expression patterns. Second, Notch signalling activity is modulated by numerous accessory proteins acting either at the extracellular side (such as members of the Fringe family, (Johnston et al., 1997)) or at the intracellular side (such as Mind Bomb or Neuralized, (Le Borgne et al., 2005)) and is therefore highly context-dependent (Artavanis-Tsakonas et al., 1995; Schweisguth, 2004). Third, the crosstalk between the Notch pathway and other signal-transduction pathways (see for example (Dahlqvist et al., 2003; Gustafsson et al., 2005)) make the outcome of Notch activation difficult to predict. As a consequence, Notch pathway activity cannot be simply deduced from the expression profiles of its components. Thus, activation of direct target genes could represent a more reliable readout of Notch activity. The best characterized Notch target genes are members of the Hairy/enhancer of split (Hes) family that encode basic helix-loop-helix transcription factors (Iso et al., 2003b). However, several additional direct target genes have been recently characterised (see for example (Anthony et al., 2005; Krebs et al., 2003; Raya et al., 2003; Reizis and Leder, 2002)), suggesting that visualization of Notch activity using this approach may necessitate the prior identification of the target genes activated in the system under study.
As an alternative approach, we generated a RBP-Jκ/Notch transgenic reporter mouse that we named NAS for Notch pathway Activity Sensor transgenic mouse. In this mouse line, identification of lacZ expressing cells or tissue is expected to reflect the distribution of transcriptionally active RBP-Jκ/NICD nuclear complexes and therefore potentially those cells or tissues where Notch signalling is at work. Here, we describe the lacZ expression pattern in NAS mice and show that it is consistent with various previously identified sites of Notch pathway activity.
Results and discussion
Generation of a RBP-Jκ/Notch reporter mouse
To assess the activity of the RBP-Jκ/Notch signalling pathway in the whole organism, we generated an RBP-Jκ-dependent transgenic reporter mouse. The Escherichia coli lacZ reporter gene was chosen because it is sensitive and allows detection at the single cell level. A modified version of the lacZ gene, coding for a nuclear β-gal, was used in order to discriminate transgene activity from the cytoplasmic non-specific activity observed in certain cells. To drive the expression of the “nlacZ” gene, we used the well-characterized TP1 promoter, which consists of 12 multimerized RBP-Jκ binding motifs upstream from a minimal promoter (Kato et al., 1997; Minoguchi et al., 1997) (fig. 1). The TP1 promoter is transactivated in a RBP-Jκ dependent manner by the activated forms of the four mammalian Notch receptors (Kato et al., 1996; Shimizu et al., 2002) and thus, has been often used to read out Notch pathway activity in various ex vivo studies (see for example (Gupta-Rossi et al., 2001; Joutel et al., 2004)) as well as in a recent in vivo study (Kohyama et al., 2005). The resulting construct, TP1-nlacZ, was microinjected in mouse zygotes and two transgenic lines were established. Transgene expression was monitored by X-gal staining of E9.5 to E17.5 embryos and 1 to 4 weeks old mice. No transgene expression was observed in one of the two lines, therefore it was discarded. In the other line, restricted and dynamic expression of nlacZ gene was observed (fig. 2a–d). At a given stage, transgenic embryos exhibited identical profiles of expression. However, some variation in the intensity of X-gal staining was observed between embryos of the same litter (fig. 2c), which is likely to reflect genetic background heterogeneity amongst littermates. This is further substantiated by our preliminary observation that backcrossing twice this transgenic line onto C57BI/6 background reduces X-gal staining variability between litter mates (data not shown). In the rest of the manuscript, we will describe the main features of the expression profile of this line. Because this profile is consistent with sites where activity of the Notch signalling pathway was predicted from previous studies (see below), we named this transgenic line NAS for Notch pathway Activity Sensor transgenic line.
Figure 1. Rationale of a Notch pathway activity sensor transgene.
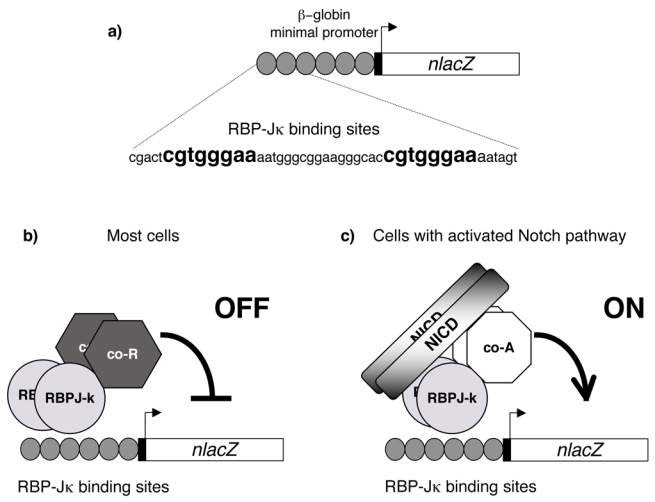
(a) Schematic representation of the TP1-nlacZ transgene. The nlacZ gene is under the control of the β-globin minimal promoter (black rectangle) preceded by six copies of a 50 bp long fragment (grey circle) from the promoter region of the Epstein-Barr virus TP1 gene. This fragment contains two RBP-Jκ binding sites (indicated in bold) and thus the TP1-nlacZ construct harbours 12 binding sites, (b, c) In absence of activated Notch (b), RBP-Jκ might recruit co-repressor complexes (co-R) to the TP1-nlacZ transgene which therefore should not be transcribed. When a cell receives signals through Notch receptor, RBP-Jκ/NICD, and co-factors (co-A) assemble into an activation complex leading to the transcription of the nlacZ gene.
Figure 2. TP1-nlacZ expression pattern during somitogenesis and in heart and arteries.
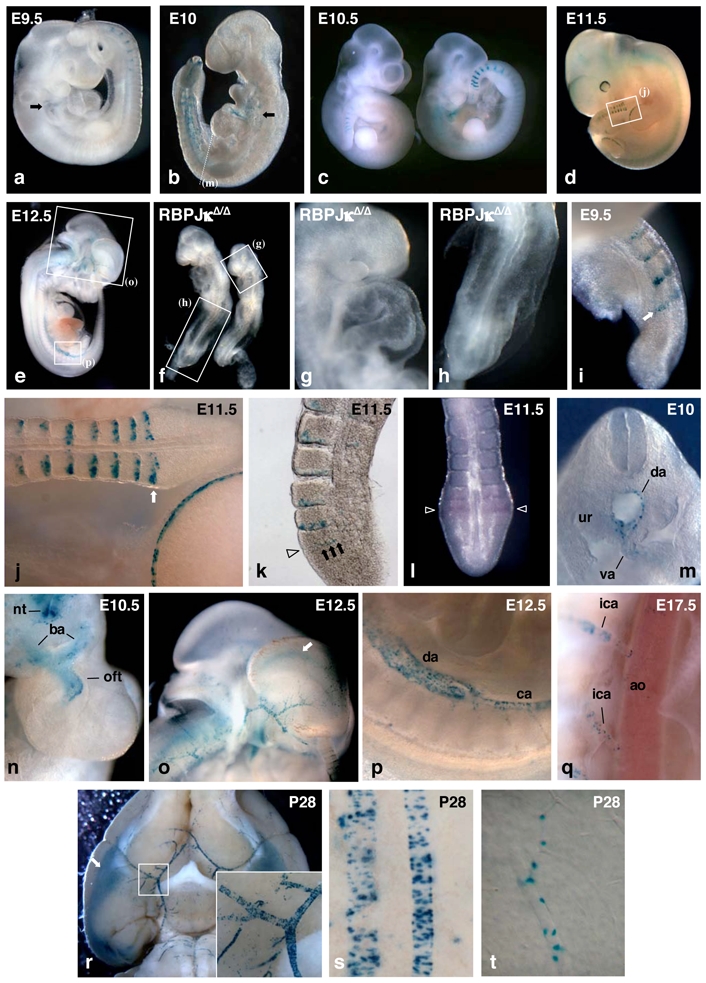
Whole mount X-gal staining of embryos of the following ages: E9.5 (a), E10 (b), E10.5 (c), and E11.5 (d). Arrows in (a,b) point to lacZ expressing cells in the cardiac region, (e) Internal view of a E12.5 NAS embryo cut in half, (f) RBP-JκΔ/Δ deficient embryos recovered 10 days after mating do not express the TP1-nlacZ transgene. Magnified views of regions indicated by boxes in (f) show the absence of IacZ expressing cells in cardiac (g) and tail (h) regions. Detail of the staining of the tail of E9.5 (i) and E11.5 (j, boxed region in (d)) NAS embryos. Arrows point to the band of β-gal positive cells in the PSM (k) Vibratome thick section of X-gal stained E11.5 tail showing a row of NAS expressing cells in the PSM (arrows) at the time when cleft starts to form (arrowhead). (l) Whole mount in situ hybridization of E10.5 tail showing that IacZ mRNA is detected essentially in the boundary forming region of the PSM (arrowhead). (m) Section in the trunk region (arrow in (b)) of E10 embryo showing TP1-nlacZ expression in the walls of dorsal aorta (da) and vitelline artery (va). ur: urogenital ridge. (n) Upper view of the heart region of E10.5 NAS embryo from which the head has been cut off. TP1-nlacZ expression is found in the distal portion of the outflow tract (oft) and in the branchial arch arteries (ba). nt: neural tube, (o–p) Enlarged views corresponding to boxed regions in (e) showing TP1-nlacZ expression in dorsal aorta (da) and caudal artery (ca) (p) and in head arteries (o) of E12.5 embryo. Arrow in (o) points to X-gal staining in the region of cortical hem (q) Detail of the X-gal staining of E17.5 NAS embryo. TP1-nlacZ expression is barely detectable in the dorsal aorta (ao) while it is clearly visible in the intercostal arteries (ica) branching directly from it. (r) Ventral view of dissected brain from a 4 weeks old NAS mouse showing the staining in cerebral arteries and in entorhinal cortex (arrow). Inset shows a magnified view of the branching of the right sylvian artery to the Willis polygon, (s–t) Details of pial arteries (s) and cerebral cortex microvessel (t) X-gal staining from 4 weeks old NAS mouse.
TP1-nlacZ transgene expression is dependent on the presence of RBP-Jκ
It was essential to demonstrate that lacZ expression in the NAS transgenic line indeed results from activity of the Notch/RBP-Jκ pathway. To that end, we transferred TP1-nlacZ transgene into a RBP-Jκ null background. RBP-Jκ Δ deleted allele was produced by Cre mediated recombination of a conditional RBP-Jκf floxed allele (Han et al., 2002). Mice carrying the TP1-nlacZ transgene in a RBP-Jκ Δ/+ background were intercrossed and embryos were recovered at 10 days post-coitum shortly before lethality due to the absence of RBP-Jκ function (Oka et al., 1995). As previously described for RBP-Jκ knock-out embryos, RBP-JκΔ/Δ deficient embryos exhibited severe growth retardation and many abnormalities including defective somitogenesis, incomplete turning of the body axis, microcephaly and pericardiac oedema (fig. 2f–h). Importantly, TP1-nlacZ transgene expression was completely abolished in RBP-JκΔ/Δ deficient embryos (fig. 2f – h, n=4). In RBP-JκΔ/+ and RBP-Jκ+/+ embryos of the same litter, TP1-nlacZ transgene expression was observed in the heart and tail regions (fig. 2b and see below), while no lacZ expressing cells were detected in the corresponding regions of RBP-JκΔ/Δ embryos (fig. 2g–h). This shows that in vivo TP1-nlacZ transgene activation is mediated by RBP-Jκ, at least at this stage. Moreover, RBP-Jκ is expected to function as a transcriptional represser of TP1-nlacZ transgene in absence of Notch activity (fig. 1b). We find that depletion of RBP-Jκ does not lead to transgene derepression that would have resulted in widespread NAS activation.
lacZ expression during sonitogenesis
Notch signalling plays important roles in the process of somitogenesis. Hence, deficiency in core components (Notch1, Delta-like (DII) 1, DII3, Presenilin-1, Kuzbanian and RBP-Jκ) as well as in targets and modulators (Hes7, Mesp2 and Lunatic Fringe) of the Notch pathway all results in perturbations of somite fomation and patterning (for review see (Aulehla and Herrmann, 2004; Weinmaster and Kintner, 2003)). Interestingly, the NAS reporter transgene is expressed during somitogenesis. At E9.5, a stripe of X-gal positive cells was detected in the posteriormost region of newly formed somites (fig. 2a, i). In the presomitic mesoderm (PSM), a band of lacZ expressing cells was also observed at the boundary-forming region of the prospective somite (fig. 2i arrow). A similar expression profile was observed during somitogenesis of older embryos (fig. 2c, d, j). On E11.5 tail transverse sections, β-gal positive cells were mainly found at the posterior border of the new somites (fig. 2k). In the PSM TP1-nlacZ transgene expression was detected before a cleft was formed (arrowheads on fig. 2k), consistent with the reported critical role of Notch signalling pathway in inter-somites boundary formation (Barrantes et al., 1999; Conlon et al., 1995; Hrabe de Angel is et al., 1997; Huppert et al., 2005; Kusumi et al., 1998; Oka et al., 1995; Swiatek et al., 1994). X- gal staining was observed in several somites (usually 8 to 12). Owing to the dynamic nature of somitogenesis (a new somite is formed every two hours in the mouse) and to perdurance of β-gal protein, such X-gal staining might overlook the dynamic nature of NAS transactivation (as previously described for LfnglacZ allele, (Zhang and Gridley, 1998)). To address this possibility, we looked by in situ hybridization at the distribution of lacZ mRNA in the tail of E10. 5–11.5 embryos. A single band of lacZ transcripts was detected in PSM in the cleft-forming area (arrowheads fig. 2l) indicating that it is the main site of TP1-nlacZ transactivation and that β-gal stability is most likely responsible for the staining observed in the more anterior somites. Such activation is likely to ensue from signalling through Notch1 receptor since accumulation of activated Notch1 at the cleft forming region between somites S-1 and S0 has been recently visualized using an antibody raised against a novel NICD epitope generated by a-secretase cleavage of Notch1 (Huppert et al., 2005; Morimoto et al., 2005). It should be however noted that cyclical production of activated Notch1 detected in these studies in a more posterior region of PSM using the same antibody is not reflected by a detectable NAS reporter transgene transactivation.
lacZ expression in the heart and the arterial vascular system
The Notch pathway is involved in multiple aspects of cardio-vascular development including vascular remodelling, arterial/venous specification, maturation of vascular smooth muscle cells and epithelial - mesenchymal transition during heart development (for review see (Shawber and Kitajewski, 2004)).
Human Notch genes are linked to Alagille’s Syndrome, a developmental disorder with vascular defects, to CADASIL, a cerebral arteriopathy as well as to congenital aortic valve disease (Garg et al., 2005; Joutel and Tournier- Lasserve, 1998). β-gal expression was observed in the embryonic and postnatal vasculature. Interestingly, TP1-nlacZ transgene showed arterial specific expression, as do many components of the Notch pathway (reviewed in (Iso et al., 2003a; Shawber and Kitajewski, 2004)). In the case of placental and yolk-sac arteries however, we noted that, contrary to components of the Notch pathway, no lacZ expression was observed, and this at all embryonic stages analysed. In the developing vasculature, transgene expression was first detected in the caudal region of the dorsal aorta and in vitelline and umbilical arteries at E9.5–E10 (fig. 2b, m). Transverse sections in the trunk region showed a patchy X-gal staining distributed around the aorta close to the lumen (fig. 2m). Expression was also observed in the pericardiac region. At E9.5–E10, scattered staining was found in the third branchial arch (arrows in fig. 2a, b). Then at E10.5, β-gal positive cells were detected in the region lining the lumen of the upper part of the cardiac outflow tract and of the branchial arch arteries directly branching from it (fig. 2n). Sections of E10.5 showed that transgene expression in the heart was restricted to the endocardium of the outflow tract (data not shown). In the following days, lacZ expression progressed to iliac arteries and major vessels of the head (fig. 2o). At E12.5, X-gal staining was observed throughout dorsal aorta, being more intense in the caudal region (fig. 2e, p). At E17.5, transgene expression was almost absent from the abdominal aorta but was detected in intercostal arteries branching directly from it (fig. 2q) as well as in many other small arteries of the embryo. At this stage, as well as in postnatal mice, the distribution and ovoid shape of X-gal positive nuclei associated with vessels was evocative of smooth muscle cells (SMCs). Spirales of positive nuclei encircling various vessels, such as tail or pial arteries, were detected (fig. 2r, s). Many cerebral microvessels were similarly decorated with X-gal positives nuclei, which likely correspond to pericytes (fig. 2t).
lacZ expression in other sites
In addition to the two mains sites described above, restricted and dynamic expression of TP1-nlacZ transgene was observed in several other tissues. For example, during limb development, scattered β-gal expressing cells were first detected at E10, in the surface ectoderm of the ventral side of forelimb buds (fig. 3a). β-gal expressing cells then accumulated at the tip of the limb bud and transgene expression became progressively restricted to the apical ectodermal ridge (AER) (fig. 3b–c; see also fig. 2j). This expression pattern is in agreement with previous evidence that Jagged2 is initially expressed throughout the limb ectoderm and then restricted to the AER soon after its formation and that Notch1 is weakly expressed in the AER at E10.5 (Jiang et al., 1998; Sidow et al., 1997; Valsecchi et al., 1997). In addition, both spontaneous and targeted Jagged2 mutants display abnormal thickening of the AER (Jiang et al., 1998; Sidow et al., 1997) suggesting that Notch signalling in the limb ectoderm might be regulating the number of AER progenitor cells. Demonstration that Notch signalling acts in the ectoderm at an early stage of limb development has been recently provided through conditional mutagenesis (Pan et al., 2005).
Figure 3. TP1- nlacZ expression pattern in developing limbs, central nervous system and teeth.
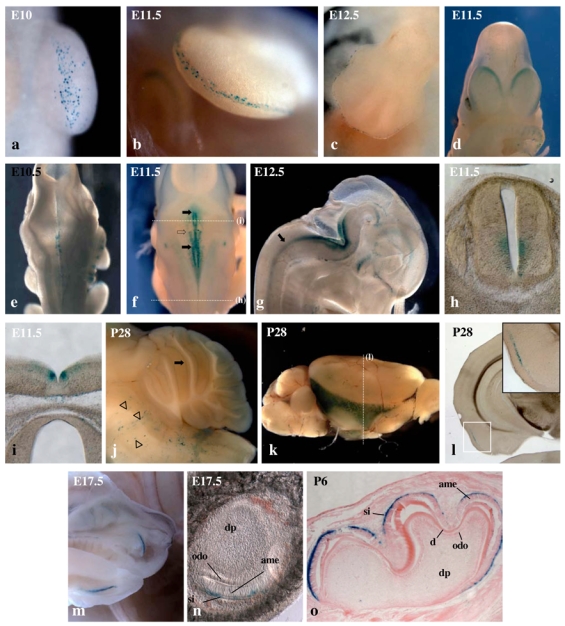
(a–c) Detail of the staining of the forelimb bud of E10 (a), E11.5 (b) and E12.5 (c) NAS embryos. (d) Frontal view of E11.5 NAS embryo showing labelling of the medio-caudal portion of cortical hemispheres. (e–f) Dorsal views showing X-gal staining in the hindbrain of E10.5 (e) and E11.5 (f) NAS embryos. Black arrows in (f) indicate the anterior and posterior limits of the medial bilateral stripe and the open arrow indicates the anterior limit of the lateral stripe. (g) Internal view of the head of E12.5 NAS embryo cut in half. Arrow points to a stripe of lacZ expressing cells in the neural tube, (h–i) Transverse sections of E11.5 NAS embryo in the trunk (h) and hindbrain (i) regions. Positions of sections along the antero-posterior embryonic axis are indicated by dotted lines in (f). (j) Md-sagittal view of cerebellum from 4 weeks old mouse showing TP1-nlacZ expression in Purkinje cell layer (arrow). Stripes of X-Gal labeled cells in the bulb (arrowheads) correspond to cerebral microvessels. (k) Whole mount X-gal staining of dissected brain from 4 weeks old NAS mouse. (I) Coronal section at the level indicated by a dotted line in (k) showing layer restricted expression of TP1-nlacZ in entorhinal cortex. Inset shows a magnified view of the TP1-nlacZ expressing region. (m) Internal view of the head of E17.5 embryo cut in half showing TP1-nlacZ expression in the labial surface of developing incisors. (n) Vibratome thick frontal section of X-gal stained E17.5 incisor bub showing NAS expression in stratum intermedium (si) layer, ame: amel oblast layer, odo: odontoblast layer, dp: dental papilla. (o) Microtome section of P6 developing molar. d: dentine.
Restricted transgene expression was also observed in the developing central nervous system (CNS). In the forebrain, expression was detected in the caudal third of the medial wall of E11.5 and E12.5 cortical hemispheres (fig. 3d and 2o). This region includes the cortical hem which is important for telencephal on patterning. In the hindbrain, X-gal labelled cells accumulated on both sides of ventral midline structures of E10.5 embryos (fig. 3e). At E11.5, β-gal positive cells were organized into well defined stripes (fig. 3f, i). A first bilateral stripe close to the floor plate was observed in the hindbrain (black arrows in fig. 3f and fig. 3i). A second stripe was observed in the hindbrain in a more lateral position (fig. 3f, empty arrow indicates its anterior limit).
This stripe extended posteriorly into most of the neural tube of E11.5 (fig. 3f, h) and E12.5 (fig. 3g, arrow) embryos. Sections indicated that TP1-nlacZ transgene is expressed in the ventricular zone (fig. 3h, i) as are Notch receptors and ligands in the midgestation embryonic mammalian CNS (Lindsell et al., 1996; Myat et al., 1996). Interestingly, bilateral stripes of expression in the ventricular zone of the hindbrain and spinal cord have been reported for chick Delta-1 and Jagged-1 (Myat et al., 1996) and rodent jagged-1, delta-1, 3 and 4 (Benedito and Duarte, 2005; Dunwoodie et al., 2002; Lindsell et al., 1996) suggesting that signalling via different Notch receptors and ligands might be involved in the production of distinct neural phenotypes. A detailed analysis will be necessary to establish the precise relations between the TP1-nlacZ transgene and the expression profiles of components of the Notch pathway. In postnatal brains, weak transgene expression, besides the vessels associated cells, was observed in the Purkinje cell layer of cerebellum (fig. 3j, arrow). A strong layer-specific X-gal staining was also detected in a ventrolateral region of the cortex, which is likely to correspond to the entorhinal cortex (fig. 3k and arrow in fig. 2r). Sections indicated that transgene expression was restricted to an intermediate cortical layer (fig. 3l).
Another site of expression was the developing tooth. At E17.5, a strong staining was observed on the labial surface of the incisor buds (fig. 3m). Sections indicated that transgene expression was restricted to stratum intermedium cells underlying tall columnar ameloblasts secreting the enamel matrix (fig. 3n). Similar observations were made on postnatal developing molar (fig. 3o). Hence, in developing teeth, the expression profile of the TP1-nlacZ transgene is again consistent with previously reported Notch components expression patterns (Harada et al., 1999; Mtsiadis et al., 1998). Signalling between stratum intermedium cells expressing Notch and ameloblasts expressing Jagged and Delta has been proposed to be essential for the maintenance of the differentiated state of ameloblasts (Harada et al., 1999). No obvious transgene expression was observed in some sites where RBP-Jκ/Notch signalling has been previously shown to be active. For example, lymphoid organs were not labelled by X-gal staining, while a role for Notch during lymphoid precursor differentiation has been clearly established (reviewed in (Radtke et al., 2004). The reasons that may account for this discrepancy are yet unknown but a possibility would be that the TP1-nlacZ construct cannot be transactivated in certain tissues because of position effect silencing. The structure of the transgene itself may also confer some quantitative or qualitative restrictions to its activation by the RBP-Jκ/Notch pathway. In the promoter region of Notch target genes, spacing and orientation of RBP-Jκ binding sites as well as their association with other DNA binding sites modulate the transcriptional response mediated by activated Notch receptors (Cave et al., 2005; Ong et al., 2005). Hence, mouse Hes-1 promoter contains paired high affinity RBP-Jκ binding sites in a head-to-head orientation and changing their orientation abrogates its Notch mediated activation in cultured cells (Ong et al., 2005). TP1-nlacZ transgene is composed of twelve head-to-tail high affinity RBP-Jκ binding sites joined to a minimal promoter and, therefore, is very different from promoters of direct Notch target genes. It is thus possible that, in certain cellular contexts, low level of activated Notch receptor would not be sufficient to ensure efficient transgene activation, while in synergy with other transcription factors, it would activate endogenous Notch target genes.
Concluding remarks
In conclusion, the NAS mice permit the readout of the activity of RBP-Jκ/Notch signalling pathway in vivo in various cell types. Having demonstrated that transgene expression is dependent on the presence of functional RBP-Jκ we investigated lacZ expression and found a striking coincidence between NAS reporter transgene activation and a number of known sites of activity of components of the Notch pathway. Altogether, these data strongly suggest that the NAS transgenic mouse line is a bona fide sensor of Notch activity in several tissues. This conclusion is further substantiated by the recent demonstration that TP1-nlacZ expression in arteries is abolished in Notch3 deficient mice, while it is maintained in other sites (Monet et al., manuscript in preparation). The observation that only part of the TP1-nlacZ expression profile is mediated by Notch3 receptor, highlights the ability of NAS reporter transgene to respond to signalling through the different Notch receptors. Moreover, the analysis of TP1-nlacZ expression in Notch3−/− mice has permitted us to clearly establish that the function of Notch3 in arterial SMCs is mediated through a RBP-Jκ-dependent pathway, although its action is independent of Hes/Hrt genes (Monet et al. manuscript in preparation).
The use of reporter constructs comprising multiple transcription factor binding sites joined to a minimal promoter has proven to be very useful to study other pathways such as for example the Wnt/β-cateni n/LEF pathway (DasGupta and Fuchs, 1999; Maretto et al., 2003) or signalling through NF-kB (Schmidt-Ullrich et al., 1996). Due to the artificial structure of the TP1-nlacZ promoter region, it was anticipated that transgene expression profile might not report on all Notch activity. This is the case, however our first survey of the lacZ expression profile in NAS mice indicates that a number of known sites of Notch activity express the transgene.
It should be noted that a more extensive analysis of TP1-nlacZ expression might reveal other sites of Notch activity. Generation and characterization of other Notch reporter transgenic lines carrying various RBP-Jκ binding sites and different reporter genes configuration shall also prove to be useful for extensive visualization of Notch activity in vivo. In this respect, it should be mentioned that sorting of Notch responsive haematopoietic stem cells was recently achieved thanks to the use of a transgenic line harbouring four copies of RBP-Jκ binding sites linked to EGFP (Duncan et al., 2005). However the expression profile of this line apart from the adult bone marrow has not been reported so far. Finally, comparison of TP1-nlacZ expression profiles with that of transgenic lines carrying a reporter gene under the control of regulatory regions of direct Notch target genes might also help to define the relative contribution of RBP-Jκ/NICD complexes to the transcriptional regulation of individual target genes.
In addition to its use in the description of Notch pathway activity during normal development and in adult tissues, NAS transgenic mice may be useful in addressing other important biological questions. For example, it should help for genetic or drug screens for Notch signalling modulators in mice. It should also help to evaluate in vivo the crosstalks between Notch and other signalling pathways or to fine tune the involvement of the former in various pathogenic processes (tumorigenesis, developmental anomalies, etc…). Future work using the NAS transgenic line will certainly help to gain new insights into the complex functions of the Notch signalling pathway in the mouse.
Methods
Construction of the RBP-nlsLacZ transgene and production of transgenic mice
TP1-nlacZ construct was derived from plasmid pGa981-6 (Minoguchi et al., 1997) and plasmid pSKTNLSLACZ (a gift from S. Tajbakhsh). The hexamerized 50 bp EBNA2 response element of the TP-1 promoter in front of the minimal β-globin promoter was recovered from pGa981-6 plasmid and put in front of the nlsLacZ gene from pSKTNLSLACZ coding for a nuclear β-galactosidase (βgal) (fig. 1a). StuI-NotI linearized construct was microinjected into (C57BL/6xSJL/J)2 fertilized eggs, which were then transferred to pseudo-pregnant females. Transgenic embryos and animals were identified by PCR and Southern blot analysis of tail or placental DNA using lacZ oligonucleotides (lacZ1 : 5′-GTC GTT TTA CAA CGT CGT GAC T-3′; lacZ2 : 5′-GAT GGG CGC ATC GTA ACC GTG C-3′) and probe as described in (Cohen-Tannoudji et al., 1992). Stable lines, harbouring single integration of the transgene, were established by crossing transgenic founders with (C57BL/6xSJL/J)F1 mice. Embryos were recovered from mating between N1 or N2 transgenic males with wild-type females. The age of the embryos was determined according to the appearance of the vaginal plug (day 0.5) and confirmed by morphological criteria.
Whole mount β-galactosidase staining
Embryos as well as organs dissected from postnatal mice were fixed for 30–60 minutes at room temperature in either 4% paraformaldehyde or 2% paraformaldehyde and 0.2% glutaraldehyde in PBS. From E12.5 onwards, embryos and organs were cut in halves after 30 min in the fixative solution in order to improve the penetration of fixative and staining solutions and then returned to the fixative solution for another 30 min. After fixation, samples were rinsed several times with PBS and incubated overnight at 32 °C in X-Gal staining solution (PBS containing 0.01% Tween20, 2mM MgCl2, 4mM K4Fe(CN)6, 4mM K3Fe(CN)6, and 4mg/ml of X-gal (Invitrogen)). Then, samples were rinsed several times with PBS, postfixed in 4% paraformaldehyde and stored at 4°C in a 1/1 mixture of 4% paraformaldehyde and glycerol. Pictures of embryos and organs were taken using a SMZ1500 stereomicroscope (Nikon) equipped with an AxioCam color (Zeiss).
Production of RBP-Jκ null embryos
RBP-Jκ f/f males, carrying an exons 6 and 7-floxed RBP-Jκ allele (Han et al., 2002), were crossed to ZP3-Cre females (de Vries et al., 2000) in order to produce mice carrying a deleted RBP-JκΔ allele, which were then crossed with TP1-nlacZ transgenic mice. Embryos were collected from intercrosses between RBP-JκR/+, TP1-nlacZ transgenic mice and monitored individually for β-galactosidase activity. Genotype of each embryo was determined by PCR analysis of placental DNA using lacZ and RBP-Jκ primers (WT1: 5′-GTT CTT AAC CTG TTG GTC GGA ACC-3′ ; WT2: 5′-GTC TGA GGC TTG ATG TTC TGT ATT GC-3′ ; G1: 5′-GTG GCA AAG CCC TTA AAA AT-3′ ;-2: 5′-GAG ATA GAC CTT GGT TTG TT-3′).
Embryo in situ hybridization and histology
Whole mount RNA in situ hybridization was carried out according to (Wilkinson and Nieto, 1993). Embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C, dehydrated in methanol and stored at −20°C. RNA probes were labeled with digoxigenin and visualized with BM Purple according to the manufacturer’s instruction (Roche Biosciences). The lacZ hybridization probe used included the full ORF. For histology, X-gal reacted samples were either embedded in 4% agarose and cut at 50 tm thickness using a Leica VT1000 S Vibratome or embedded in paraffin and cut at 12 am thickness using a Leica RM 2155 microtome. Some sections were counterstained with eosin.
Acknowledgments
We thank T. Honjo and K. Tanigaki (Kyoto, Japan) for the RBP-Jκflox mice and the pGa981-6 plasmid, B. Knowles (Bar Harbor, USA) for the ZP3/Cre transgenic line and S. Tajbakhsh for the pSKTNLSLACZ plasmid. We also thank C. Brou for helpful discussions, J. Artus, J.L. de la Pompa, S. Tajbakhsh and M. Wassef for critical reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique, the Association pour la Recherche contre le Cancer (ARC), the Institut Pasteur GPH07 on stem cells and the Agence National de la Recherche (contract n° JC05_41835). C. S. and M. M. received grants from CNRS (“Bourse de Doctorat pour les Ingénieurs”) and the “Ministére de l’Education Nationale, de la Recherche et de la Technologie” respectively, and S.C. received grants from the Pasteur-Negri-Weizmann Council and the ARC.
References
- Anthony TE, Mason HA, Gridley T, Fishell G, Heintz N. Brail lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev. 2005;19:1028–1033. doi: 10.1101/gad.1302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Herrmann BG. Segmentation in vertebrates: clock and gradient finally joined. Genes Dev. 2004;18:2060–2067. doi: 10.1101/gad.1217404. [DOI] [PubMed] [Google Scholar]
- Barrantes IB, Elia AJ, Wunsch K, Hrabe de Angelis MH, Mak TW, Rossant J, Conlon RA, Gossler A, de la Pampa JL. Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr Biol. 1999;9:470–480. doi: 10.1016/s0960-9822(99)80212-7. [DOI] [PubMed] [Google Scholar]
- Benedito R, Duarte A. Expression of D ll4 during mouse embryogenesis suggests multiple developmental roles. Gene Expr Patterns. 2005;5:750–755. doi: 10.1016/j.modgep.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Cave JW, Loh F, Surpris JW, Xia L, Caudy MA. A DNA transcription code for cell-specific gene activation by notch signaling. Curr Biol. 2005;15:94–104. doi: 10.1016/j.cub.2004.12.070. [DOI] [PubMed] [Google Scholar]
- Cohen-Tannoudji M, Morello D, Babinet C. Unexpected position-dependent expression of H-2 and beta 2-microglobulin/lacZ transgenes. Mol Reprod Dev. 1992;33 :149–159. doi: 10.1002/mrd.1080330206. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaum e AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Dahlqvist C, Blokzijl A, Chapman G, Falk A, Dannaeus K, Ibanez CF, Lendahl U. Functional Notch signaling is required for BMP4-induced inhibition of myogenic differentiation. Development. 2003;130:6089–6099. doi: 10.1242/dev.00834. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, Reya T. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS. Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene DII3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development. 2002;129:1795–1806. doi: 10.1242/dev.129.7.1795. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, Kengaku M. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;8:873–880. doi: 10.1038/nn1492. [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12(Spec No 1):R9–13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev Cell. 2005;9:617–628. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabe de Angelis M, Mclntyre J, 2nd, Gossler A. Maintenance of somite borders in mice requires the Delta homologue DII1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, Takeda Y, Chia W, Sankar N, Ng YK, Ling EA, Maciag T, Small D, Trifonova R, Kopan R, Okano H, Nakafuku M, Chiba S, Hirai H, Aster JC, Schachner M, Pallen CJ, Watanabe K, Xiao ZC. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–175. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Huppert SS, Ilagan MX, De Strooper B, Kopan R. Analysis of notch function in presomitic mesoderm suggests a gamma-secretase- independent role for presenilins in somite differentiation. Dev Cell. 2005;8:677–688. doi: 10.1016/j.devcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vase Biol. 2003a;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003b;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SH, Rauskolb C, Wilson R, Prabhakaran B, Irvine KD, Vogt TF. A family of mammalian Fringe genes implicated in boundary determination and the Notch pathway. Development. 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- Joutel A, Monet M, Domenga V, Riant F, Tournier- Lasserve E. Pathogenic mutations associated with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy differently affect Jagged1 binding and Notch3 activity via the RBP/JK signaling Pathway. Am J Hum Genet. 2004;74:338–347. doi: 10.1086/381506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A, Tournier-Lasserve E. Notch signalling pathway and human diseases. Semin Cell Dev Biol. 1998;9:619–625. doi: 10.1006/scdb.1998.0261. [DOI] [PubMed] [Google Scholar]
- Kato H, Sakai T, Tamura K, Minoguchi S, Shirayoshi Y, Hamada Y, Tsujimoto Y, Honjo T. Functional conservation of mouse Notch receptor family members. FEBS Lett. 1996;395:221–224. doi: 10.1016/0014-5793(96)01046-0. [DOI] [PubMed] [Google Scholar]
- Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- Kohyama J, Tokunaga A, Fujita Y, Miyoshi H, Nagai T, Miyawaki A, Nakao K, Matsuzaki Y, Okano H. Visualization of spatiotemporal activation of Notch signaling: live monitoring and significance in neural development. Dev Biol. 2005;286:311–325. doi: 10.1016/j.ydbio.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Krebs LT, Iwai N, Nonaka S, Welsh IC, Lan Y, Jiang R, Saijoh Y, O’Brien TP, Hamada H, Gridley T. Notch signaling regulates left- right asymmetry determination by inducing Nodal expression. Genes Dev. 2003;17:1207–1212. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi DC, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES. The mouse pudgy mutation disrupts Delta homologue DII3 and initiation of early somite boundaries. Nat Genet. 1998;19:274–278. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci USA. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoguchi S, Taniguchi Y, Kato H, Okazaki T, Strobl LJ, Zimber-Strobl U, Bornkamm GW, Honjo T. RBP-L, a transcription factor related to RBP-Jκ. Mol Cell Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis TA, Hirsinger E, Lendahl U, Goridis C. Delta-notch signaling in odontogenesis: correlation with cytodifferentiation and evidence for feedback regulation. Dev Biol. 1998;204:420–431. doi: 10.1006/dbio.1998.9092. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Takahashi Y, Endo M, Saga Y. The Mesp2 transcription factor establishes segmental borders by suppressing Notch activity. Nature. 2005;435:354–359. doi: 10.1038/nature03591. [DOI] [PubMed] [Google Scholar]
- Myat A, Henrique D, Ish-Horowicz D, Lewis J. A chick homologue of Serrate and its relationship with Notch and Delta homologues during central neurogenesis. Dev Biol. 1996;174:233–247. doi: 10.1006/dbio.1996.0069. [DOI] [PubMed] [Google Scholar]
- Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, Honjo T. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Collaboration between discrete protein domains and binding site architecture on the promoter determine target selectivity of vertebrate notch proteins. J Biol Chem. 2005 doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- Pan Y, Liu Z, Shen J, Kopan R. Notch1 and 2 cooperate in limb ectoderm to receive an early Jagged2 signal regulating interdigital apoptosis. Dev Biol. 2005;286:472–482. doi: 10.1016/j.ydbio.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- Raya A, Kawakami Y, Rodriguez-Esteban C, Buscher D, Koth CM, Itoh T, Morita M, Raya RM, Dubova I, Bessa JG, de la Pompa JL, Belmonte JC. Notch activity induces Nodal expression and mediates the establishment of left- right asymmetry in vertebrate embryos. Genes Dev. 2003;17:1213–1218. doi: 10.1101/gad.1084403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 2002;16:295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Memet S, Lilienbaum A, Feuillard J, Raphael M, Israel A. NF-kappaB activity in transgenic mice: developmental regulation and tissue specificity. Development. 1996;122:2117–2128. doi: 10.1242/dev.122.7.2117. [DOI] [PubMed] [Google Scholar]
- Schweisguth F. Notch signaling activity. Curr Biol. 2004;14:R129–138. [PubMed] [Google Scholar]
- Shawber CJ, Kitajewski J. Notch function in the vasculature: insights from zebrafish, mouse and man. Bioessays. 2004;26:225–234. doi: 10.1002/bies.20004. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Saito T, Kumano K, Hamada Y, Hirai H. Functional diversity among Notch1, Notch2, and Notch3 receptors. Biochem Biophys Res Commun. 2002;291:775–779. doi: 10.1006/bbrc.2002.6528. [DOI] [PubMed] [Google Scholar]
- Sidow A, Bulotsky MS, Kerrebrock AW, Bronson RT, Daly MJ, Reeve MP, Hawkins TL, Birren BW, Jaenisch R, Lander ES. Serrate2 is disrupted in the mouse limb-development mutant syndactylism. Nature. 1997;389:722–725. doi: 10.1038/39587. [DOI] [PubMed] [Google Scholar]
- Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- Valsecchi C, Ghezzi C, Ballabio A, Rugarli EI. JAGGED2: a putative Notch ligand expressed in the apical ectodermal ridge and in sites of epithelial- mesenchymal interactions. Mech Dev. 1997;69:203–207. doi: 10.1016/s0925-4773(97)00146-9. [DOI] [PubMed] [Google Scholar]
- Weinmaster G, Kintner C. Modulation of notch signaling during somitogenesis. Annu Rev Cell Dev Biol. 2003;19:367–395. doi: 10.1146/annurev.cellbio.19.111301.115434. [DOI] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 1993;225:361–373. doi: 10.1016/0076-6879(93)25025-w. [DOI] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Zhang N, Gridley T. Defects in somite formation in lunatic fringe-deficient mice. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]


