Abstract
Objective
The present study aimed to investigate possible association of the apolipoprotein E (APOE) gene polymorphisms with age at natural menopause (ANM) in Caucasian females.
Design
Four SNPs (including two replacements, SNP3 Cys112Arg and SNP4 Arg158Cys) were genotyped in 253 randomly selected unrelated Caucasian women having experienced natural menopause. The comprehensive statistical analyses focusing on the association of the APOE gene and some environmental factors with ANM were conducted.
Results
Alcohol consumption was a significantly predictor of earlier natural menopause (P < 0.05). One SNP (rs769450) was significantly associated with ANM according to both population based and the transmission disequilibrium test (TDT) analyses (P = 0.007 and 0.046, respectively). However, no association was observed between APOE ε2,ε3, ε4 and ANM.
Conclusions
Genetic variation in the APOE gene may influence the variation in ANM in Caucasian women.
Keywords: age at natural menopause (ANM), association, APOE, polymorphism
Introduction
Natural menopause status is considered as an important cornerstone which is related to women’s psychological and physical well-being. The timing of natural menopause may serve as a marker for the process of reproductive ageing and as an indicator of postmenopausal women health risks. Earlier menopause was reported to increase the risk of several female age-related diseases, such as ovarian cancer [1,2], osteoporosis [3,4], and cardiovascular disease [5], whereas later menopause contributes to the higher risk of breast [6] and endometrial cancer [7]. Thus, knowing the determinants of age at natural menopause (ANM) may help to foreseen possible postmenopausal health complications.
Similar to other complex traits (e.g., osteoporosis, obesity, etc.), ANM is determined by both environmental and genetic factors [8–14]. Large volume of data has been accumulated about various environmental factors contributing to ANM, including smoking [8], oral contraceptive use [9], breastfeeding and alcohol consumption [10]. Several studies have reported that genetic factors could explain a large proportion of ANM variation [11–14]. The heritability of ANM was estimated to vary between 49–52% [13] and 63% [14]. To date, a number of studies have been conducted to identify genetic determinants of ANM using linkage [15,16] and candidate genes association approaches [17–22]. However, the causal genetic variants and associated physiological mechanisms underlying the variation of ANM need further studying.
One of the consequences of menopause is an average increase in total cholesterol [23]. Allelic variation of apolipoprotein E (APOE) has been shown to influence the level of serum lipids, especially in postmenopausal women [24,25]. It has been found that the effect of menopause on LDL-cholesterol level significantly varied according to the APOE genotype [26]. In addition, some previous studies reported a possible role of APOE in human reproduction, specifically association of allele ε3 with the highest fertility [27–28]. All these data suggest that APOE may contribute to the onset of menopause.
APOE has been recognized as an important candidate gene for many postmenopausal human diseases. Numerous studies have demonstrated the association between the APOE polymorphisms and Alzheimer’s disease [29], osteoporosis [30], breast cancer [31], atherosclerosis [32], and coronary disease [33]. However, only few studies about association between APOE polymorphisms and ANM have been reported thus far [20, 34].
The present study examines possible association of the APOE polymorphisms with ANM in postmenopausal Caucasian women.
Material and methods
Subjects
This study was approved by the Creighton University Institutional Review Board. All the study subjects signed informed-consent documents before entering the project. The subjects came from an expanding database being created for studies to search for genes underlying the risk of osteoporosis and obesity, which is underway in the Osteoporosis Research Center (ORC) of Creighton University. A random sample of 260 Caucasian females with natural menopause was included in the present study. All the study subjects signed informed-consent documents before entering the project. All of participants are US Caucasians of European origin and came from our previous studies [10, 35]. Information about ANM, age at menarche (AAM), current smoking habits, breastfeeding, number of pregnancies (parity), and alcohol consumption before menopause was recorded for each subject by nurse-administered questionnaire.
AAM was calculated as the age of first menstrual period, following the onset of menses minus the date of birth (in years rounded to the tenth decimal); ANM was defined as an age at the last menstrual period (years) without having menstrual periods for at least 12 consecutive months. Women with surgical menopause, a history of perimenopausal hormone therapy use before menopause and women with probable premature ovarian failure (i.e. age at menopause < 38 years, 6 subjects) were excluded from this study. The varied range for ANM in our study followed the distribution of menopausal ages in Cacausian females [14]. Thus, finally 253 postmenopausal women with the data about ANM were involved into the study.
Genotyping
SNPs were identified by searching through public databases, such as dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), JSNP (http://snp.ims.u-tokyo.ac.jp/). SNPs in the APOE gene were selected on the basis of the following criteria: (1) validation status, especially in Caucasians, (2) position in or around the gene, (3) degree of heterozygosity, i.e. minor allele frequencies (MAF) > 0.05, (4) functional relevance and importance, and (5) reported to dbSNP by various sources. Finally 4 SNPs (rs440446, rs769450, rs429358, and rs7412) were selected for the analysis.
Genomic DNA was extracted from peripheral blood using a commercial isolation kit (Gentra Systems, Inc. Minneapolis, MN, USA). DNA concentration was assessed by a DU530 UV/VIS Spectrophotometer (Beckman Coulter, Inc. Fullerton, CA, USA). The genotyping procedure and primers for the four SNP markers were detailed elsewhere [35].
Statistical analyses
Hardy-Weinberg equilibrium (HWE) analyses
The program PowerMarker [36] was used to calculate allele frequencies. The χ2 test was adopted to check correspondence of the genotype data to the Hardy-Weinberg equilibrium.
Association analyses of the four single markers
Stepwise multiple regression was used to adjust the effects of the lifestyle factors (here including smoking, breastfeeding, alcohol consumption, pregnancies and AAM) on ANM. We also analyzed the influence of amount of alcohol consumption on ANM. To investigate association between any single SNP and ANM, we employed two complementary statistical designs. First, we used the population-based association test, which has maximal power to detect association, but is susceptible to false positive results because of population stratification [37]. Population based association between each single SNP and ANM was estimated using the nonparameric test. P < 0.05 was considered significant. All the statistical analyses were performed using SPSS (v. 13.0, Inc. Chicago, IL). Then we extracted 30 pedigrees (69 subjects) from the sample and used them for the family based association analysis (transmission disequilibrium test, TDT) [38]. The Haploview software [39] was utilized for the TDT.
Association analyses for APOE ε2, ε3, ε4
Human APOE is a polymorphic protein with three common isoforms (APOE ε2, ε3, ε4), encoded by three alleles (ε2, ε3, ε4) of a single gene on chromosome 19q13.2. From the genotypes of SNP3 (rs429358, Cys112Arg Exon4) and SNP4 (rs7412, Arg158Cys Exon4), we obtained five distributions of the APOE genotype (ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4, ε4/ε4) through various combinations of any two of the three major alleles. This is a commonly used approach in population association studies of APOE with various traits [40–42]. In this part of study, our available sample contained 243 subjects excluding 10 with missing genotype data. Then we assigned subjects to three genotype groups: genotype group ε2 (ε2/ε2 + ε2/ε3+ ε2/ε4), group ε3 (ε3/ε3) and group ε4 (ε4/ε4 + ε3/ε4). Using the nonparametric test we established the contribution of the APOE genotype to the phenotypic variation of ANM. We considered two-sided P value < 0.05 as significant. SPSS 13.0 (SPSS Inc., Chicago, IL) was used for all analyses.
Results
Sample characteristics
The basic characteristics of 253 study subjects are presented in Table 1. The mean ANM of these subjects was 49.22 ± 0.24 years, ranging from 38 to 58 years. The stepwise multiple regression analysis showed significant effect of alcohol consumption on ANM (P = 0.028). The univariate regression analysis indicated similar patterns for not only alcohol consumption (P = 0.017) but also for breastfeeding (P = 0.042). Women who have drunk alcohol had, on average, 1.0 years earlier menopause than those have never had. And women which had 3–6 times/week of alcohol consumption had 1.3 years earlier menopause than that of the average ANM of others who had alcohol consumption (P = 0.034). Likewise, breastfeeding seemed to promote 1.1 years earlier menopause. Table 3 provides a summary of the confounding factors for the ANM association analyses.
Table 1.
Characteristics of women involved in the study
| Number (% of total sample) | Mean ± S.E. | |
|---|---|---|
| Age(year) | 251 ( 99.20 ) | 62.25 ± 0.62 |
| Age at natural menopause | 253 | 49.22 ± 0.24 |
| Height | 182 (71.94) | 1.62 ± 0.00 |
| Weight | 185 (73.12) | 72.96 ± 1.77 |
| Age at menarche | 247 (97.63) | 13.06 ± 0.09 |
| Smoking | 39 (15.42) | |
| Breastfeeding | 136 (53.75) | |
| Alcohol consumption | 167 (66.01) | |
| <1–2/week | 85 | |
| 1–2/week | 25 | |
| 3–6/week | 41 | |
| 7–10/week | 8 | |
| >10/week | 8 | |
| No. of pregnancies | 243 (96.05) | |
| None | 10 | |
| 1–3 | 120 | |
| 4–6 | 82 | |
| >7 | 41 |
Table 3.
Results of the univariate and multiple analyses for association between lifestyle factors and ANM
| Factor | P value for univariate analyses | P value for stepwise regression |
|---|---|---|
| Smoking | 0.139 | 0.993 |
| Alcohol consumption | 0.017 | 0.028 |
| No. of pregnancies | 0.817 | 1.000 |
| breastfeeding | 0.042 | 0.997 |
| Age at menarche | 0.768 | 0.998 |
P value <0.05 are shown in bold.
Association analyses for the single markers
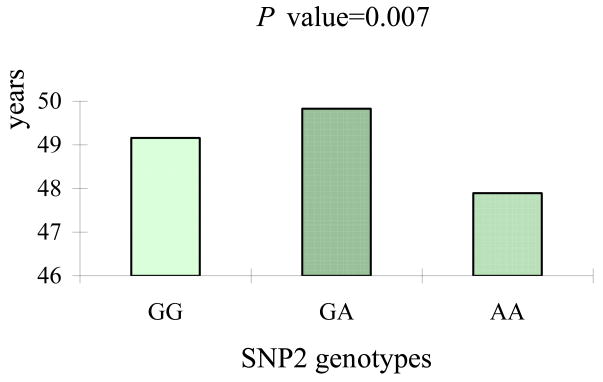
The information of the four SNPs analyzed is shown in Table 2. All of them were in Hardy-Weinberg equilibrium (P > 0.05). The minor allele frequencies (MAFs) ranged from 0.075 to 0.437. The results of four single SNP association analyses are summarized in Table 3. SNP2 (rs769450) showed significant association with ANM in the population-based analyses (P = 0.007). Consistent with these results, significant association was also observed in the family-based TDT analysis, though P value was slightly lower (0.046). As shown in Figure 1, the subjects carrying genotype AA for SNP2 had, on average, ~1.93 year earlier onset of menopause than those with the GA genotype (47.90 versus 49.83 years).
Table 2.
Information of SNPs for APOE
| SNP | SNP ID | Genomic positiona (bp) | Allele variantsb | Position in the gene | MAF |
|---|---|---|---|---|---|
| SNP1 | rs440446 | 50101007 | G/C | Intron1 | 0.331 |
| SNP2 | rs769450 | 50102284 | G/A | Intron2 | 0.437 |
| SNP3 | rs429358 | 50103781 | T/C | Cys112Arg Exon4 | 0.144 |
| SNP4 | rs7412 | 50103919 | C/T | Arg158Cys Exon4 | 0.075 |
Positions are based on the GenBank data, www.ncbi.nlm.nih.gov/SNP.
The minor allele is bold.
GenBank accession no. NC_000019.
Figure 1.
Differences in the ANM across genotypes of the SNP2.
The subjects carrying genotype AA for SNP2 had, on average, ~1.93 year earlier onset of menopause (P = 0.007) than those with the GA genotype (47.90 versus 49.83 years).
Association analyses for APOE ε2, ε3, ε4
The distribution of the APOE allele frequencies in our 243 women were as follows: 9.47% for ε2/ε3, 5.35% for ε2/ε4, 63.37% for ε3/ε3, 20.58% for ε3/ε4 and 1.23% for ε4/ε4 (Table 5). Genotypic distribution was consistent with HWE (P > 0.05). The ε2/ε2 genotype was absent in our sample. No significant difference in ANM was observed for any single genotype of the five. We did not find any significant association for ε2, ε3 and ε4 alleles either. Even when the subjects were partitioned into three genotype groups according to carrying/not carrying a particular allele, no significant difference for ANM between the groups was detected (Table 5).
Table 5.
Distribution of APOE genotypes and allele frequencies and results for relative association analyses
| Number of subjects (%) | P value | |
|---|---|---|
| ε2/ε3 | 23 (9.47) | 0.831 |
| ε2/ε4 | 13 (5.35) | 0.134 |
| ε3/ε3 | 154 (63.37) | 0.185 |
| ε3/ε4 | 50 (20.58) | 0.546 |
| ε4/ε4 | 3 (1.23) | 0.967 |
| ε2 allele | 36 (7.40) | 0.261 |
| ε3 allele | 381 (78.40) | 0.180 |
| ε4 allele | 69 (14.20) | 0.195 |
| APOE group ε2 | 36 (10.00) | 0.746 |
| Non-APOE group ε2 | 207 (90.00) | |
| APOE group ε3 | 154 (66.96) | 0.373 |
| Non-APOE group ε3 | 89 (33.04) | |
| APOE group ε4 | 53 (23.04) | 0.444 |
| Non-APOE group ε3 | 190 (76.96) |
Discussion
In the present study we performed analyses to examine association between four APOE polymorphisms and ANM. We also analyzed the probable correlation between the APOE genotypes (ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4, ε4/ε4) and ANM. In the single SNP analyses, we found a significant relationship between SNP2 (rs769450) and ANM.
ANM is affected by environmental and lifestyle factors. In the present study we found that alcohol consumption and breastfeeding were likely promoting the earlier onset of natural menopause. These two significant factors and other factors such as smoking and the number of pregnancies were discussed in our previous study [10]. We did not find any significant association between AAM and ANM that was consistent with the results of some previous studies [9,13,18,43,44]. However, other studies have reported positive [45] or inverse [46,47] association between them. This inconsistency can be partly attributed to different genetic backgrounds, environmental and lifestyle factors.
Both population- and family-based analyses suggested that SNP2 (rs769450) is significantly associated with ANM. A large body of data provide support for such association. APOE is a ligand for various cell-surface receptors including the low-density lipoprotein (LDL) receptor, the LDL-receptor related protein and the very low-density lipoprotein (VLDL) receptor [48]. It serves as a ligand for the receptor-mediated uptake of cholesterol-rich particles by hepatocytes and peripheral tissues [49]. On the one hand, APOE plays a major physiological role in the regulation of overall lipid and lipoprotein homeostasis [48,50,51]. Several studies reported association between APOE and obesity [35,52,53]. On the other hand, menopause is one of the critical periods of a woman’s life during which obesity prevalence is the highest [54]. Serum lipid level increases during hormonal transition before and after menopause [55]. As a result, postmenopausal women have significantly more fat and lower lean tissue mass than premenopausal women [56]. It is thought that at least part of the higher prevalence of cardiovascular disease in postmenopausal women may be due to increased central body fatness [57]. Postmenopausal obesity is commonly associated with many physiological disorders that results in higher mortality [58]. APOE genotype also contributes to the variation in cholesterol increase with menopause [26]. In addition, it has been found that the APOE gene could determine serum testosterone and dehydroepiandrosterone levels in postmenopausal women [59]. All these results are in favour that APOE likely contributes to the development of menopause.
Previously, Tempfer et al.[34] found that SNP4 (rs7412) was significantly (P = 0.03) associated with lower ANM in an ethnically homogenous cohort of Middle European white women. Koochmeshgi et al.[20] reported that the APOE genotype group ε4 predicted earlier age at menopause with the two other genotype groups (P = 0.049) in the Iranian population. However, we did not find any significant association between the above two genotypes and ANM in our study sample. In addition to the differences in sample size, statistical methods and confounding environmental factors, the inconsistency may be mostly attributed to differences in the ethnic background of the population. The abovementioned studies used virtually monoethnic cohorts. Specifically, in the study by Tempfer et al. [34], only women of the Austrian and German ancestry were included to avoid confounding by ethnicity, while the study sample of Koochmeshgi et al. [20] consisted only of Iranian women. Although all subjects in our study identified themselves as Caucasians, they might in fact represent different ethnic subgroups, e.g., Northern or Southern Europeans, Jews, etc. Ethnicity may be an important factor contributing to the complex traits with between-ethnic differences in prevalence [60–63] and interacting with the genetic factors underlying these traits [64–66]. However, despite the inconsistencies as regards the association of the particular polymorphisms, all above results are in favor that the APOE gene is likely associated with ANM.
Like many other studies involving in reproductive history characteristics, the data of AAM and ANM were obtained by questionnaire, which might potentially incur a recalling bias. Specifically, several studies have demonstrated that accuracy of long-term recall of AAM and self-reported ANM varied between 70% and 84% [67–70].
In the present study we did not find significant association between APOE ε2, ε3, ε4 and ANM, though the ε4 allele is recognized as the most commonly identifiable genetic risk factor in some postmenopausal disorders, such as Alzheimer’s disease [50], coronary artery disease [32], and low bone mass [30]. On the other hand, one SNP (rs769450) within the intron 2 of the APOE gene was significantly associated with ANM.
In summary, this study provides further evidence for that APOE gene may influence timing of menopause and, thus, potentially contribute to development of diseases associated with age of menopause. Further association studies utilizing denser markers within the APOE gene and larger sample size are required to better clarify the importance of the APOE gene polymorphisms for the onset of menopause.
Table 4.
The results of single SNPs association analyses
| SNP | P value for Population-based analyses | P value for TDT analyses |
|---|---|---|
| SNP1 | 0.714 | 0.317 |
| SNP2 | 0.007 | 0.046 |
| SNP3 | 0.592 | 0.132 |
| SNP4 | 0.255 | 0.257 |
P value <0.05 are shown in bold.
Acknowledgments
Investigators of this work were partially supported by grants from NIH (R01 AR050496, K01 AR02170-01, R01 AR45349-01, and R01 GM60402-01A1) and an LB595 grant from the State of Nebraska. The study also benefited from grants from National Science Foundation of China, Huo Ying Dong Education Foundation, HuNan Province, Xi’an Jiaotong University, and the Ministry of Education of China.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Cramer DW. Epidemiologic aspects of early menopause and ovarian cancer. Ann N Y Acad Sci. 1990;592:363–75. doi: 10.1111/j.1749-6632.1990.tb30347.x. [DOI] [PubMed] [Google Scholar]
- 2.Schildkraut JM, Cooper GS, Halabi S, et al. Age at natural menopause and the risk of epithelial ovarian cancer. Obstet Gynecol. 2001;98:85–90. doi: 10.1016/s0029-7844(01)01388-6. [DOI] [PubMed] [Google Scholar]
- 3.Harlow BL, Signorello LB. Factors associated with early menopause. Maturitas. 2000;35:3–9. doi: 10.1016/s0378-5122(00)00092-x. [DOI] [PubMed] [Google Scholar]
- 4.Kritz-Silverstein D, Barrett-Connor E. Early menopause, number of reproductive years, and bone mineral density in postmenopausal women. Am J Public Health. 1993;83:983–88. doi: 10.2105/ajph.83.7.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobsen BK, Knutsen SF, Fraser GE. Age at natural menopause and total mortality and mortality from ischemic heart disease: the Adventist Health Study. J Clin Epidemiol. 1999;52:303–7. doi: 10.1016/s0895-4356(98)00170-x. [DOI] [PubMed] [Google Scholar]
- 6.Velie EM, Nechuta S, Osuch JR. Lifetime reproductive and anthropometric risk factors for breast cancer in postmenopausal women. Breast Dis. 2005;24:17–35. doi: 10.3233/bd-2006-24103. [DOI] [PubMed] [Google Scholar]
- 7.de Graaff J, Stolte LA. Age at menarche and menopause of uterine cancer patients. Eur J Obstet Gynecol Reprod Biol. 1978;8:187–93. doi: 10.1016/0028-2243(78)90014-x. [DOI] [PubMed] [Google Scholar]
- 8.Kinney A, Kline J, Levin B. Alcohol, caffeine and smoking in relation to age at menopause. Maturitas. 2006;54:27–38. doi: 10.1016/j.maturitas.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Palmer JR, Rosenberg L, Wise LA, et al. Onset of natural menopause in African American women. Am J Public Health. 2003;93:299–306. doi: 10.2105/ajph.93.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dvornyk V, Long JR, Liu PY, et al. Predictive factors for age at menopause in Caucasian females. Maturitas. 2006;54:19–26. doi: 10.1016/j.maturitas.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 11.de Bruin JP, Bovenhuis H, van Noord PA, et al. The role of genetic factors in age at natural menopause. Hum Reprod. 2001;16:2014–18. doi: 10.1093/humrep/16.9.2014. [DOI] [PubMed] [Google Scholar]
- 12.Treloar SA, Do KA, Martin NG. Genetic influences on the age at menopause. Lancet. 1998;352:1084–85. doi: 10.1016/S0140-6736(05)79753-1. [DOI] [PubMed] [Google Scholar]
- 13.Snieder H, MacGregor AJ, Spector TD. Genes control the cessation of a woman’s reproductive life: a twin study of hysterectomy and age at menopause. J Clin Endocrinol Metab. 1998;83:1875–80. doi: 10.1210/jcem.83.6.4890. [DOI] [PubMed] [Google Scholar]
- 14.Murabito JM, Yang Q, Fox C, et al. Heritability of age at natural menopause in the Framingham Heart Study. J Clin Endocrinol Metab. 2005;90:3427–30. doi: 10.1210/jc.2005-0181. [DOI] [PubMed] [Google Scholar]
- 15.van Asselt KM, Kok HS, Putter H, et al. Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative trait loci influencing variation in human menopausal age. Am J Hum Genet. 2004;74:444–53. doi: 10.1086/382136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kok HS, van Asselt KM, van der Schouw YT, et al. Genetic studies to identify genes underlying menopausal age. Hum Reprod Update. 2005;11:483–93. doi: 10.1093/humupd/dmi024. [DOI] [PubMed] [Google Scholar]
- 17.Hefler LA, Grimm C, Heinze G, et al. Estrogen-metabolizing gene polymorphisms and age at natural menopause in Caucasian women. Hum Reprod. 2005;20:1422–27. doi: 10.1093/humrep/deh848. [DOI] [PubMed] [Google Scholar]
- 18.Zhang F, Xiong DH, Wang W, et al. HDC gene polymorphisms are associated with age at natural menopause in Caucasian women. Biochem Biophys Res Commun. 2006;348:1378–82. doi: 10.1016/j.bbrc.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Asselt KM, Kok HS, Peeters PH, et al. Factor V Leiden mutation accelerates the onset of natural menopause. Menopause. 2003;10:477–81. doi: 10.1097/01.GME.0000056040.51813.1A. [DOI] [PubMed] [Google Scholar]
- 20.Koochmeshgi J, Hosseini-Mazinani SM, Morteza SS, et al. Apolipoprotein E genotype and age at menopause. Ann N Y Acad Sci. 2004;1019:564–67. doi: 10.1196/annals.1297.105. [DOI] [PubMed] [Google Scholar]
- 21.Gorai I, Tanaka K, Inada M, et al. Estrogen-metabolizing gene polymorphisms, but not estrogen receptor-alpha gene polymorphisms, are associated with the onset of menarche in healthy postmenopausal Japanese women. J Clin Endocrinol Metab. 2003;88:799–803. doi: 10.1210/jc.2002-020353. [DOI] [PubMed] [Google Scholar]
- 22.Weel AE, Uitterlinden AG, Westendorp IC, et al. Estrogen receptor polymorphism predicts the onset of natural and surgical menopause. J Clin Endocrinol Metab. 1999;84:3146–50. doi: 10.1210/jcem.84.9.5981. [DOI] [PubMed] [Google Scholar]
- 23.van Beresteijn EC, Korevaar JC, Huijbregts PC, et al. Perimenopausal increase in serum cholesterol: a 10-year longitudinal study. Am J Epidemiol. 1993;137:383–92. doi: 10.1093/oxfordjournals.aje.a116686. [DOI] [PubMed] [Google Scholar]
- 24.Eichner JE, Kuller LH, Ferrell RE, et al. Phenotypic effects of apolipoprotein structural variation on lipid profiles. III. Contribution of apolipoprotein E phenotype to prediction of total cholesterol, apolipoprotein B, and low density lipoprotein cholesterol in the healthy women study. Arteriosclerosis. 1990;10:379–85. doi: 10.1161/01.atv.10.3.379. [DOI] [PubMed] [Google Scholar]
- 25.Schaefer EJ, Lamon-Fava S, Johnson S, et al. Effects of gender and menopausal status on the association of apolipoprotein E phenotype with plasma lipoprotein levels. Results from the Framingham Offspring Study. Arterioscler Thromb. 1994;14:1105–13. doi: 10.1161/01.atv.14.7.1105. [DOI] [PubMed] [Google Scholar]
- 26.Hak AE, Witteman JC, Hugens W, et al. The increase in cholesterol with menopause is associated with the apolipoprotein E genotype. A population-based longitudinal study. Atherosclerosis. 2004;175:169–76. doi: 10.1016/j.atherosclerosis.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Gerdes LU, Gerdes C, Hansen PS, et al. Are men carrying the apolipoprotein ε4- or ε2 allele less fertile than ε3ε3 genotypes? Hum Genet. 1996;98:239–42. doi: 10.1007/s004390050200. [DOI] [PubMed] [Google Scholar]
- 28.Corbo RM, Scacchi R, Cresta M. Differential reproductive efficiency associated with common apolipoprotein E alleles in postreproductive-aged subjects. Fertil Steril. 2004;81:104–7. doi: 10.1016/j.fertnstert.2003.05.029. [DOI] [PubMed] [Google Scholar]
- 29.Laws SM, Hone E, Gandy S, et al. Expanding the association between the APOE gene and the risk of Alzheimer’s disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J Neurochem. 2003;84:1215–36. doi: 10.1046/j.1471-4159.2003.01615.x. [DOI] [PubMed] [Google Scholar]
- 30.Shiraki M, Shiraki Y, Aoki C, et al. Association of bone mineral density with apolipoprotein E phenotype. J Bone Miner Res. 1997;12:1438–45. doi: 10.1359/jbmr.1997.12.9.1438. [DOI] [PubMed] [Google Scholar]
- 31.Chang SJ, Hou MF, Tsai SM, et al. Association between the apolipoprotein E genotypes and breast cancer patients in Taiwanese. Breast Cancer Res Treat. 2006;98:109–13. doi: 10.1007/s10549-005-9137-0. [DOI] [PubMed] [Google Scholar]
- 32.Davignon J, Gregg RE, Sing CF. Apolipoprotein E polymorphism and atherosclerosis. Arteriosclerosis. 1988;8:1–21. doi: 10.1161/01.atv.8.1.1. [DOI] [PubMed] [Google Scholar]
- 33.Reilly M, Rader DJ. Apolipoprotein E and coronary disease: a puzzling paradox. PLoS Med. 2006;3:e258. doi: 10.1371/journal.pmed.0030258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tempfer CB, Riener EK, Keck C, et al. Polymorphisms associated with thrombophilia and vascular homeostasis and the timing of menarche and menopause in 728 white women. Menopause. 2005;12:325–30. doi: 10.1097/01.gme.0000141760.98678.ed. [DOI] [PubMed] [Google Scholar]
- 35.Long JR, Liu PY, Liu YJ, et al. APOE and TGF-beta1 genes are associated with obesity phenotypes. J Med Genet. 2003;40:918–24. doi: 10.1136/jmg.40.12.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–29. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 37.Schork NJ, Fallin D, Thiel B, et al. The future of genetic case-control studies. Adv Genet. 2001;42:191–212. doi: 10.1016/s0065-2660(01)42023-2. [DOI] [PubMed] [Google Scholar]
- 38.Purcell S, Sham P, Daly MJ. Parental phenotypes in family-based association analysis. Am J Hum Genet. 2005;76:249–59. doi: 10.1086/427886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–65. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 40.Pertovaara M, Lehtimäki T, Rontu R, et al. Presence of apolipoprotein E epsilon4 allele predisposes to early onset of primary Sjogren’s syndrome. Rheumatology (Oxford) 2004;43:1484–7. doi: 10.1093/rheumatology/keh383. [DOI] [PubMed] [Google Scholar]
- 41.Zetterberg M, Tasa G, Palmér MS, et al. Apolipoprotein E polymorphisms in patients with primary open-angle glaucoma. Am J Ophthalmol. 2007;143:1059–60. doi: 10.1016/j.ajo.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 42.Sando SB, Melquist S, Cannon A, et al. APOE epsilon 4 lowers age at onset and is a high risk factor for Alzheimer’s disease; a case control study from central Norway. BMC Neurol. 2008;8:9. doi: 10.1186/1471-2377-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hassa H, Tanir HM, Tekin B, et al. Possible factors affecting the age at menopause among women in the central anatolian region of Turkey. Clin Exp Obstet Gynecol. 2006;33:59–60. [PubMed] [Google Scholar]
- 44.van Noord PA, Dubas JS, Dorland M, et al. Age at natural menopause in a population-based screening cohort: the role of menarche, fecundity, and lifestyle factors. Fertil Steril. 1997;68:95–102. doi: 10.1016/s0015-0282(97)81482-3. [DOI] [PubMed] [Google Scholar]
- 45.Ozdemir O, Col M. The age at menopause and associated factors at the health center area in Ankara, Turkey. Maturitas. 2004;49:211–19. doi: 10.1016/j.maturitas.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 46.Do KA, Treloar SA, Pandeya N, et al. Predictive factors of age at menopause in a large Australian twin study. Hum Biol. 1998;70:1073–91. [PubMed] [Google Scholar]
- 47.Johnston SL. Associations with age at natural menopause in Blackfeet women. Am J Hum Biol. 2001;13:512–20. doi: 10.1002/ajhb.1083. [DOI] [PubMed] [Google Scholar]
- 48.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–30. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 49.Brown MS, Goldstein JL. The receptor model for transport of cholesterol in plasma. Ann N Y Acad Sci. 1985;454:178–82. doi: 10.1111/j.1749-6632.1985.tb11856.x. [DOI] [PubMed] [Google Scholar]
- 50.Rubinsztein DC. Apolipoprotein E: a review of its roles in lipoprotein metabolism, neuronal growth and repair and as a risk factor for Alzheimer’s disease. Psychol Med. 1995;25:223–29. doi: 10.1017/s0033291700036138. [DOI] [PubMed] [Google Scholar]
- 51.Gao J, Katagiri H, Ishigaki Y, et al. Involvement of apolipoprotein E in excess fat accumulation and insulin resistance. Diabetes. 2007;56:24–33. doi: 10.2337/db06-0144. [DOI] [PubMed] [Google Scholar]
- 52.Elosua R, Demissie S, Cupples LA, et al. Obesity modulates the association among APOE genotype, insulin, and glucose in men. Obes Res. 2003;11:1502–8. doi: 10.1038/oby.2003.201. [DOI] [PubMed] [Google Scholar]
- 53.Marques-Vidal P, Bongard V, Ruidavets JB, et al. Obesity and alcohol modulate the effect of apolipoprotein E polymorphism on lipids and insulin. Obes Res. 2003;11:1200–1206. doi: 10.1038/oby.2003.165. [DOI] [PubMed] [Google Scholar]
- 54.Pavon dPI, Alameda HC, Olivar RJ. [Obesity and menopause] Nutr Hosp. 2006;21:633–37. [PubMed] [Google Scholar]
- 55.Torng PL, Su TC, Sung FC, et al. Effects of menopause on intraindividual changes in serum lipids, blood pressure, and body weight--the Chin-Shan Community Cardiovascular Cohort study. Atherosclerosis. 2002;161:409–15. doi: 10.1016/s0021-9150(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 56.Svendsen OL, Hassager C, Christiansen C. Age- and menopause-associated variations in body composition and fat distribution in healthy women as measured by dual-energy X-ray absorptiometry. Metabolism. 1995;44:369–73. doi: 10.1016/0026-0495(95)90168-x. [DOI] [PubMed] [Google Scholar]
- 57.Tchernof A, Calles-Escandon J, Sites CK, et al. Menopause, central body fatness, and insulin resistance: effects of hormone-replacement therapy. Coron Artery Dis. 1998;9:503–11. doi: 10.1097/00019501-199809080-00006. [DOI] [PubMed] [Google Scholar]
- 58.Milewicz A, Tworowska U, Demissie M. Menopausal obesity--myth or fact? Climacteric. 2001;4:273–83. [PubMed] [Google Scholar]
- 59.Zofkova I, Zajickova K, Hill M, et al. Apolipoprotein E gene determines serum testosterone and dehydroepiandrosterone levels in postmenopausal women. Eur J Endocrinol. 2002;147:503–6. doi: 10.1530/eje.0.1470503. [DOI] [PubMed] [Google Scholar]
- 60.Freedman BI, Hsu FC, Langefeld CD, et al. The impact of ethnicity and sex on subclinical cardiovascular disease: the Diabetes Heart Study. Diabetologia. 2005;48:2511–18. doi: 10.1007/s00125-005-0017-2. [DOI] [PubMed] [Google Scholar]
- 61.Loh FH, Khin LW, Saw SM, et al. The age of menopause and the menopause transition in a multiracial population: a nation-wide Singapore study. Maturitas. 2005;52:169–80. doi: 10.1016/j.maturitas.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 62.Araujo AB, Travison TG, Harris SS, et al. Race/ethnic differences in bone mineral density in men. Osteoporos Int. 2007;18:943–53. doi: 10.1007/s00198-006-0321-9. [DOI] [PubMed] [Google Scholar]
- 63.Gold EB, Bromberger J, Crawford S, et al. Factors associated with age at natural menopause in a multiethnic sample of midlife women. Am J Epidemiol. 2001;153:865–74. doi: 10.1093/aje/153.9.865. [DOI] [PubMed] [Google Scholar]
- 64.Jackson FL. Human genetic variation and health: new assessment approaches based on ethnogenetic layering. Br Med Bull. 2004;69:215–35. doi: 10.1093/bmb/ldh012. [DOI] [PubMed] [Google Scholar]
- 65.Dvornyk V, Liu PY, Long JR, et al. Contribution of genotype and ethnicity to bone mineral density variation in Caucasians and Chinese: a test for five candidate genes for bone mass. Chin Med J (Engl) 2005;118:1235–44. [PubMed] [Google Scholar]
- 66.Dvornyk V, Liu XH, Shen H, et al. Differentiation of Caucasians and Chinese at bone mass candidate genes: implication for ethnic difference of bone mass. Ann Hum Genet. 2003;67:216–27. doi: 10.1046/j.1469-1809.2003.00037.x. [DOI] [PubMed] [Google Scholar]
- 67.Casey VA, Dwyer JT, Coleman KA, et al. Accuracy of recall by middle-aged participants in a longitudinal study of their body size and indices of maturation earlier in life. Ann Hum Biol. 1991;18:155–66. doi: 10.1080/03014469100001492. [DOI] [PubMed] [Google Scholar]
- 68.Must A, Phillips SM, Naumova EN, et al. Recall of early menstrual history and menarcheal body size: after 30 years, how well do women remember? Am J Epidemiol. 2002;155:672–79. doi: 10.1093/aje/155.7.672. [DOI] [PubMed] [Google Scholar]
- 69.Clavel-Chapelon F, Dormoy-Mortier N. A validation study on status and age of natural menopause reported in the E3N cohort. Maturitas. 1998;29:99–103. doi: 10.1016/s0378-5122(98)00020-6. [DOI] [PubMed] [Google Scholar]
- 70.den Tonkelaar I. Validity and reproducibility of self-reported age at menopause in women participating in the DOM-project. Maturitas. 1997;27:117–23. doi: 10.1016/s0378-5122(97)01122-5. [DOI] [PubMed] [Google Scholar]