Abstract
Commonly used techniques for analyzing gene expression, such as polymerase chain reaction (PCR), microarrays, and in situ hybridization, have proven invaluable in understanding RNA processing and regulation. However, these techniques rely on the use of lysed and/or fixed cells and are therefore limited in their ability to provide important spatial-temporal information. This has led to the development of numerous techniques for imaging RNA in living cells, some of which have already provided important insight into the dynamic role RNA plays in dictating cell behavior. Here we review the fluorescent probes that have allowed for RNA imaging in living cells and discuss their utility and limitations. Common challenges faced by fluorescent probes, such as probe design, delivery, and target accessibility, are also discussed. It is expected that continued advancements in live cell imaging of RNA will open new and exciting opportunities in a wide range of biological and medical applications.
Keywords: hairpin probe, oligonucleotide probe, molecular beacon, fluorescence resonance energy transfer, delivery
INTRODUCTION
Over the past decade or so, there has been increasing evidence that RNA molecules are responsible for a wide range of functions in living cells, from the physical conveyance and interpretation of genetic information, structural support for molecular machines, and regulation/silencing of gene expression, to essential catalytic roles. These functions are realized through control of the expression level and stability, both temporally and spatially, of specific RNAs in a cell. The ability to acquire complete spatial-temporal profiles of RNA synthesis, processing, and transport is therefore of critical importance to our understanding of cell function and behavior in conditions of health and disease and in response to external stimuli. This level of insight could offer unprecedented opportunities for advancement in molecular biology, disease pathophysiology, drug discovery, and medical diagnostics.
The important role RNA plays in dictating cell behavior has led to the development of numerous methods for measuring gene expression and/or measuring differences in gene expression levels between cell populations. Several of the more widely adopted methods include polymerase chain reaction (PCR) (64), Northern hybridization (or Northern blotting) (3), expressed sequence tag (EST) (1), serial analysis of gene expression (SAGE) (89), representational difference analysis (RDA) (44), differential display (43), suppression subtractive hybridization (SSH) (18), and DNA microarrays (69). These technologies, combined with the rapidly increasing availability of genomic data for numerous biological entities, present exciting possibilities for understanding human health and disease. In fact, gene expression profiling has already led to the identification of numerous pathogenic and carcinogenic sequences that are being evaluated as clinical markers for diseased states. However, the ability to detect and identify foreign or mutated nucleic acids in a clinical setting remains challenging due to the low abundance of diseased cells in blood, sputum, and stool samples, combined with the need to lyse a population of cells in order to obtain an adequate amount of genetic material for analysis. This can result in many genetic alterations being overlooked or lost.
Although each of the aforementioned RNA detection methods can provide the relative change in gene expression for a population of cells, they generally cannot provide a measure of RNA expression at the single-cell level. Under many circumstances, it is the aberrant behavior of only a few cells or the stochasticity of RNA expression within a population that may be of interest (9, 31, 35, 46, 74). Arduous techniques such as single-cell reverse transcriptase-polymerase chain reaction (RT-PCR) can provide a closer look at RNA transcripts within single cells, but the RNA must still be extracted from the actual cell and processed prior to analysis. The shortcomings associated with RNA handling have been highlighted in several recent studies, which have shown that up to 90% of transcripts can be lost during RNA purification, cDNA synthesis, and other steps required for PCR (34, 49).
Due to the complexity of single-cell RT-PCR, single-cell analysis of RNA expression (and localization) is typically carried out by in situ hybridization (ISH), whereby labeled linear oligonucleotide (ODN) probes are used to label intracellular RNA in cells that are fixed and permeabilized (5). Unbound probes are removed by washing to reduce background and to achieve specificity (14). To enhance the signal level, multiple probes targeting the same messenger RNA (mRNA) can be used (5). If a sufficient number of probes are used to target each mRNA, it has been found that individual mRNAs within single cells can be visualized, and the absolute number of RNA per cell can be quantified (24, 41, 61). Clearly, this strategy can allow for unique insight into mRNA abundance; however, RNA-ISH can provide only very limited temporal resolution of RNA expression. Further, these techniques are laborious and very time consuming, image analysis is not trivial, and heterogeneity between samples remains a possibility. Specifically, fixation agents and other supporting chemicals can have considerable effect on signal level (7) and possibly on the integrity of certain organelles, such as mitochondria. Thus, fixation of cells, by either cross-linking or denaturing agents, and the use of proteases in ISH assays may lead to an inaccurate description of intracellular mRNA expression.
To obtain detailed spatial and temporal information on RNA dynamics, including the expression, localization, degradation, and storage of RNA molecules, much effort has recently been devoted to developing nanostructured molecular probes for imaging RNA in living cells. Live cell imaging not only eliminates the need to handle RNA, but also provides an opportunity to analyze gene expression at the single-cell level without arduous fixation, permeabilization, and washing steps. Clearly, live cell imaging strategies that are capable of providing a more complete spatial-temporal profile of gene expression would be of significant value in our understanding of the role genetic processing plays in cellular function and disease. However, in order to detect RNA molecules in living cells, the RNA imaging probes must exhibit a high specificity, sensitivity, and signal-to-background ratio, especially for low abundance genes and clinical samples containing a small number of diseased cells. The imaging probes need to recognize RNA targets with high specificity, convert target recognition directly into a measurable signal, and differentiate between true- and false-positive signals. For real-time measurements of RNA expression, it is also important for the probes to associate and dissociate from target RNA with fast kinetics. For detecting genetic alterations such as mutations, insertions, and deletions, the ability to recognize single nucleotide polymorphisms (SNPs) is essential. To achieve this optimal performance, it is necessary to have a good understanding of the structure-function relationship of the probes, probe stability, and RNA target accessibility in living cells. It is also necessary to achieve efficient cellular delivery of probes with minimal probe degradation.
In the remaining sections, we review commonly used fluorescent probes for RNA detection in living cells and subsequently discuss some of the critical issues that are faced by all of these approaches, including target accessibility, fluorophore selection, and the cellular delivery of probes.
FLUORESCENT PROBES FOR LIVE-CELL RNA DETECTION
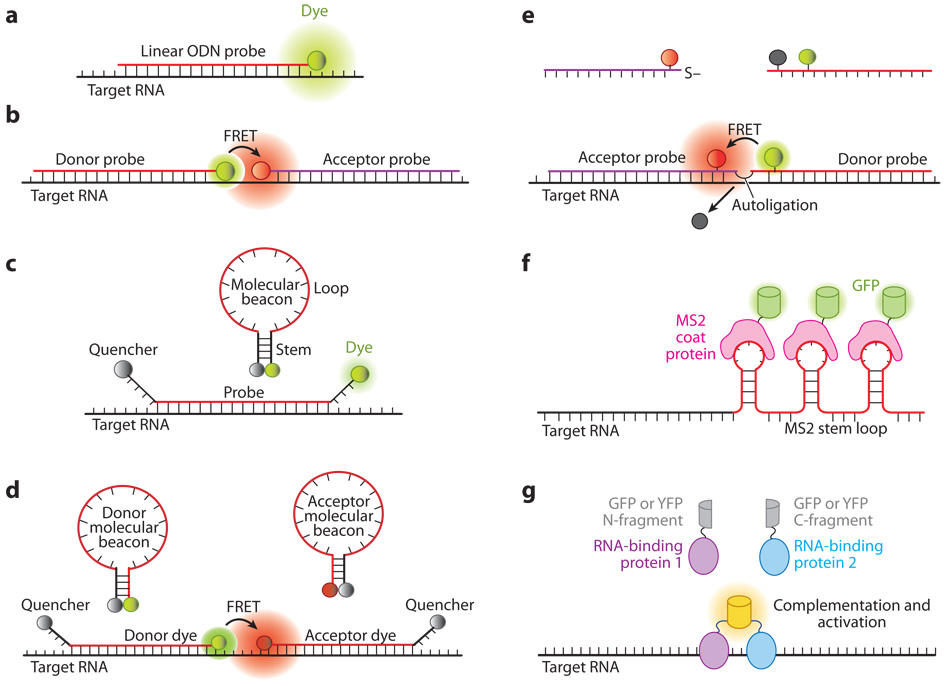
As illustrated in Figure 1, several classes of molecular probes have been developed for RNA detection in living cells, including (a) ODN probes (Figure 1a); (b) linear fluorescence resonance energy transfer (FRET) probes (Figure 1b); (c) dual-labeled oligonucleotide hairpin probes (e.g., molecular beacons, see Figure 1c); (d) dual FRET molecular beacons (Figure 1d); (e) autoligation probes (Figure 1e); and (f) probes using fluorescent proteins as reporters (Figures 1f and 1g). Although probes composed of full-length RNAs (mRNA or nuclear RNA) tagged with a fluorescent or radioactive reporter have been used to study the intracellular localization of RNA (28, 30, 32), these probes are not discussed here because they cannot be used to measure the expression level of specific endogenous RNAs in living cells.
Figure 1.
Illustrations of fluorescent probes for live-cell RNA detection. (a) Tagged linear oligonucleotide (ODN) probes. (b) Linear fluorescence resonance energy transfer (FRET) probes in which two linear ODN probes have, respectively, a donor and an acceptor fluorophore that form a FRET pair. (c) Molecular beacons are dual-labeled stem-loop oligonucleotide hairpin probes with a reporter fluorophore at one end and a quencher molecule at the other end. (d) Dual FRET donor and acceptor molecular beacons hybridize to adjacent regions on the same mRNA target, resulting in FRET signal. (e) Autoligation FRET probes. The fluorescence of the donor is initially quenched. Upon binding of the two probes to adjacent sites on the same RNA, the quencher is displaced and the ligation brings the donor and acceptor fluophores together, resulting in FRET signal. ( f ) Probes using the coat protein of the bacterial phage MS2 fused with green fluorescent protein (GFP) (MS2-GFP). The MS2-GFP complexes bind to multiple hairpin sequences in the 3′ untranslated region (UTR) of an mRNA, giving rise to a high signal compared with the background. (g) Probes based on fragment complementation of fluorescent protein. When two RNA-binding proteins, each carrying a fluorescent protein fragment, bind to adjacent sites on the same RNA, a fluorescence signal is generated owing to fragment complementation of the fluorescent protein.
Tagged Linear ODN Probes
One of the simplest methods used to visualize endogenous RNA in single living cells involves introducing fluorescently labeled antisense linear oligonucleotide probes (Figure 1a) into cells (19, 20, 50, 51, 56). In general, this approach requires the use of multiple probes targeting the same RNA transcript so that a high local concentration of hybridized probes can be seen above the high background of unbound probes (51). Alternatively, RNAs that exhibit a high local concentration within the nucleus can also be visualized, such as 28S ribosomal RNA in the nucleoli, poly(A) RNA in speckles, and U3 small nuclear RNA (snRNA) in Cajal bodies (51, 56). In addition to the limitations presented by the inability to distinguish between probes hybridized to target RNA and unbound probes, linear oligonucleotide probes cannot be used to detect RNA in the cytoplasm either, partly due to their rapid accumulation in the nucleus after delivery using microinjection. Further, linear probes may lack detection specificity because a partial match between the probe and target sequences could induce probe hybridization to RNA molecules of multiple genes. Specificity can be improved by introducing backbone and/or base modifications that improve the oligonucleotide probe’s affinity for mRNA and simultaneously shortening the probe to increase the energy penalty for a single-base mismatch; however, this will result in reduced probe selectivity. In other words, the probability of there being nontarget RNAs in the cell with the same consensus sequence increases. These shortcomings have prevented the use of fluorescently labeled oligonucleotide probes in most live-cell RNA imaging applications.
Linear FRET Probes
As shown schematically in Figure 1b, linear FRET probes utilize two linear oligonucleotides that are fluorescently labeled at their 5′ and 3′ end with donor and acceptor fluorophores, respectively, forming a FRET pair (80, 81). These probes are designed to hybridize to adjacent regions on a nucleic acid target such that the two fluorophores are brought into close proximity only when both probes are hybridized to the same RNA target. Excitation of the donor fluorophore leads to sensitized emission of acceptor fluorescence, producing a FRET signal indicative of target detection. Because a FRET signal is only generated when both probes hybridize to adjacent sequences on target RNA, this method provides a novel way to differentiate between target recognition and background fluorescence from unbound probes. This results in a significant improvement in signal-to-background ratio (i.e., fluorescent signal in the presence of target-to-fluorescent signal in the absence of target) compared with fluorescently labeled linear probes (80). The dual linear-probe approach may still exhibit a high background signal owing to direct excitation of the acceptor and emission detection of the donor fluorescence; however, careful selection of donor and acceptor fluorophores can result in a signal-to-background ratio of approximately 10:1. It has been reported that this can be increased to 20:1 by using two donor fluorophores in some instances (54). An alternative approach used to improve the sensitivity of RNA detection with linear FRET probes involved measuring the decay of acceptor fluorescence using a time-resolved method (81). Specifically, when both donor and acceptor oligonucleotides were bound to target RNA, the acceptor exhibited a significantly longer fluorescence lifetime than unhybridized probes. The fluorescence decay of the acceptor was also much slower than autofluorescence, allowing it to be easily distinguished from the background signal. It was found that time-resolved microscopy could be used to detect as few as ∼900 probe-RNA hybrids, compared with 10,000 hybrids when a conventional fluorescence microscope was used (80, 81).
A unique benefit of adopting an mRNA imaging strategy that requires the binding of two oligonucleotide FRET probes is the inherent improvement in selectivity compared with methods that utilize only a single oligonucleotide probe. Selectivity is improved because two oligonucleotides, encompassing a longer total target sequence, must bind adjacent complementary sequences on target mRNA in order to generate a FRET signal. Therefore, even if one probe binds nonspecifically to intracellular RNA, it is highly unlikely that the second probe would also bind nonspecifically to an adjacent sequence.
Conversely, a potential disadvantage of adopting an mRNA imaging strategy that requires the binding of two oligonucleotide FRET probes is the difficulty in finding large stretches of RNA with little or no secondary structure that can accommodate the hybridization of two probes. Further, it has been reported that only 13 bases are needed to identify a unique sequence in the human genome, so an improved selectivity may not be necessary in this case (88).
Finally, although linear FRET probes exhibit an improved signal-to-background ratio and improved selectivity compared with single fluorescently labeled oligonucleotide probes, they exhibit similar specificity; despite the presence of two linear probes, it remains difficult for each linear probe to distinguish targets that differ by a few bases because the difference in free energy of the two hybrids (with and without mismatch) is typically rather small. This has limited the application of linear ODN probes in biological and disease studies.
Molecular Beacons
To date, perhaps ODN hairpin probes have been the most widely adopted class of probes for live-cell RNA imaging. As illustrated in Figure 1c, typically, these probes are labeled at one end with a reporter fluorophore and at the other end with a quencher, forming molecular beacons. Although ODN hairpin probes labeled with two fluorophores that form a FRET pair have been developed (33, 95), discussions on molecular beacon designs will be the focus of this section (84). Molecular beacons are designed to form a stem-loop hairpin structure in the absence of a complementary target so that fluorescence of the fluorophore is quenched. Hybridization with the target nucleic acid opens the hairpin and physically separates the fluorophore from quencher, allowing a fluorescent signal to be emitted upon excitation (Figure 1c). This enables a molecular beacon to function as a sensitive probe with a high signal-to-background ratio. Under optimal conditions, the fluorescence intensity of molecular beacons can increase by >200-fold upon binding to their targets (84). The ability to transduce target recognition directly into a fluorescence signal with a high signal-to-background ratio has allowed molecular beacons to enjoy a wide range of biological and biomedical applications, including real-time monitoring of PCR, genotyping and mutation detection, multiple analyte detection, assaying for nucleic acid cleavage in real time, cancer-cell detection, the study of viral infection, and the monitoring of RNA expression and localization in living cells (11, 19, 38, 42, 47, 51, 53, 58, 59, 67, 73, 79, 82, 83, 90).
As an example, shown in Figure 2 are fluorescence images of the distribution of β-actin mRNA in live fibroblast cells visualized using molecular beacons with 2′-O-methyl ribonucleotide backbones (82). Each beacon was complexed with streptavidin to prevent nuclear localization of the beacon after microinjection. As shown in Figures 2a and 2b respectively, tetramethylrhodamine (TMR)-labeled β-actin mRNA-specific molecular beacons gave a strong signal (TMR channel), whereas Texas Red–labeled control molecular beacons show only a weak background signal (Texas Red channel). Figure 2c shows the ratio image obtained by dividing the fluorescence intensity of TMR by that of Texas Red at every pixel in the image, indicating clearly the localization of β-actin mRNA in fibroblast cells.
Figure 2.
Imaging the distribution of β-actin mRNA in living fibroblasts using molecular beacons. Tetramethylrhodamine (TMR)-labeled β-actin mRNA-specific molecular beacons and Texas Red–labeled control molecular beacons complexed with streptavidin were microinjected into cells. The fluorescence signals in the TMR (a) and Texas Red (b) channels were detected, and a ratio image (c) was obtained by dividing the fluorescence intensity of TMR by that of Texas Red at every pixel in the image, indicating the localization of β-actin mRNA. The color of each pixel in (c) reflects the value of the ratio, with warmer colors representing higher ratios and cooler colors representing lower ratios.
A conventional molecular beacon has four essential components: a loop, a stem, a fluorophore, and a quencher. The loop usually consists of 15–25 nucleotides and is selected to have a unique antisense targeting sequence and proper melting temperature. The stem, formed by two complementary short-arm sequences, is typically 4–6 bases long and is chosen to be independent of the target sequence (Figure 1c).
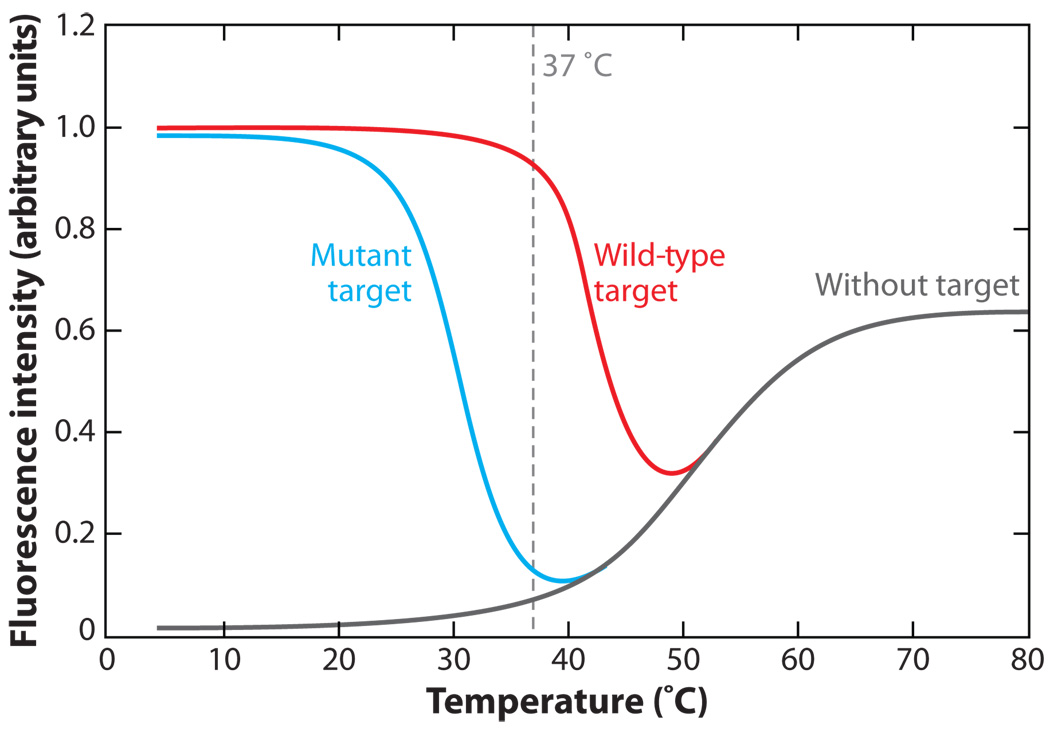
One major advantage of utilizing oligonucleotide probes with a hairpin structure is that they exhibit a higher specificity for perfectly complementary nucleic acid targets than linear ODN probes. Properly designed molecular beacons can readily discriminate between targets that differ by as little as a single nucleotide (10, 78). This level of specificity, however, requires that the loop and stem lengths and sequences be designed appropriately for the designated set of experimental conditions (77, 78). For example, in live-cell studies, it is desirable for the melting temperature of perfectly complementary hybrids to be above 37°C and the melting temperature for hybrids with single-base mismatches to be less than 37°C (Figure 3). Both of these melting temperatures will increase with increasing loop length and decreasing stem length. Eventually, if the melting temperatures are raised too high, it would not be possible to discriminate between perfectly complementary and mismatched targets under physiological conditions. Alternatively, if the melting temperatures are reduced by too much, only a small fraction of perfectly complementary target would be bound by molecular beacons under physiological conditions, which could result in false negatives. The melting temperature of a molecular beacon can be tailored by changing its stem-loop structure, as demonstrated in Figure 4a.
Figure 3.
Typical molecular beacon thermal denaturation profiles. With wild-type (complementary) targets, molecular beacons emit a maximal signal at low temperatures, indicating that the molecular beacons are bound to target; as temperature increases, the molecular beacons melt away from target. With mutant targets, the melting temperature of the molecular beacons is reduced. The difference between the wild-type target and mutant target curves over a range of temperatures represents the window of discrimination between wild-type and mutant targets. In live-cell studies, it is desirable for the melting temperature of perfectly complementary hybrids to be above 37°C and the melting temperature for hybrids with single-base mismatches to be less than 37°C.
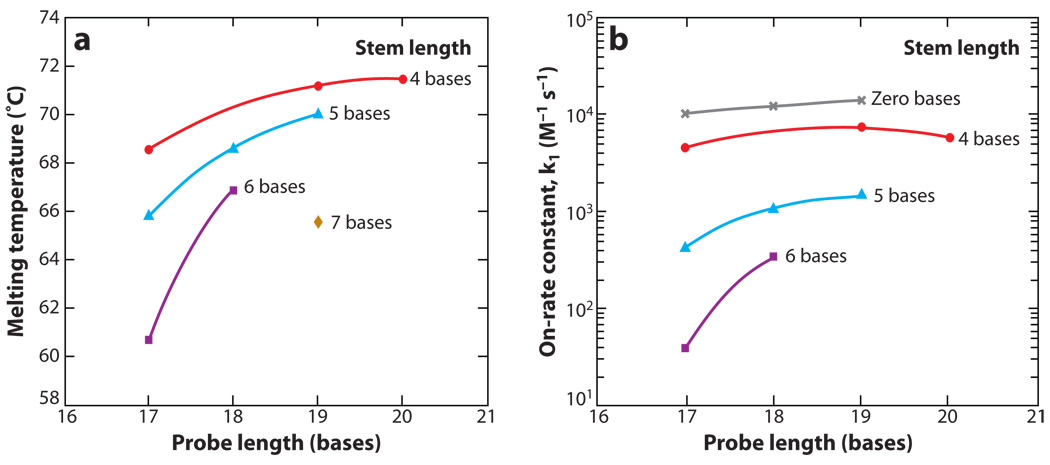
Figure 4.
Structure-function relations of molecular beacons. (a) Melting temperatures for molecular beacons with different structures in the presence of complementary targets. (b) The rate constant of hybridization k1(on-rate constant) for molecular beacons with various probe and stem lengths hybridized to their complementary targets.
In general, the specificity of a particular oligonucleotide probe can be loosely defined by the difference in melting temperature between perfectly complementary hybrids and hybrids with single-base mismatches. This window between melting temperatures is generally wider for molecular beacons compared with linear probes. This is because the energy penalty associated with unwinding the molecular beacon stem reduces binding to mismatched targets to a greater extent than to perfectly complementary targets. In experiments where the detection of SNPs is required, the specificity of molecular beacons can be improved by increasing the stem length. In other words, a longer stem provides a wider set of conditions over which molecular beacons can discriminate between two targets. This can be attributed to the enhanced stability of the molecular beacon stem-loop structure and the resulting smaller free-energy difference between closed (unbound) molecular beacons and beacon-target duplexes, which generates a condition where a single-base mismatch reduces the energetic preference of probe-target binding. A longer stem is also associated with a higher signal-to-background ratio because the more stable hairpin conformation reduces the probability of stem opening due to Brownian fluctuations and results in more efficient quenching of the fluorescent dye.
Although both the stability of the hairpin probe and its ability to discriminate targets over a wider range of temperatures increase with increasing stem length, these attributes are accompanied by a decrease in hybridization on-rate constant, as shown in Figure 4b. For example, molecular beacons with a four-base stem had an on-rate constant up to 100 times greater than molecular beacons with a six-base stem. Changing the probe length of a molecular beacon may also influence the rate of hybridization, as demonstrated by Figure 4b, but generally to a lesser extent than changing the stem length.
Recently, a variant of molecular beacons called tentacle probes has been developed that may exhibit a higher specificity and faster hybridization kinetics than conventional molecular beacons (68). Tentacle probes consist of highly specific molecular beacons (i.e., long stem and short loop) tethered to a linear oligonucleotide. The linear oligonucleotides and molecular beacons are designed to bind adjacent sites on target RNA. Therefore, the linear oligonucleotide allows for fast hybridization kinetics with target RNA, bringing the molecular beacon into close proximity with its hybridization site. This effectively increases the local concentration of target and allows for much more rapid molecular beacon hybridization. As a result, molecular beacons can be designed with a much higher specificity than would otherwise be possible. This probe design has not yet been used for live cell imaging but shows great potential.
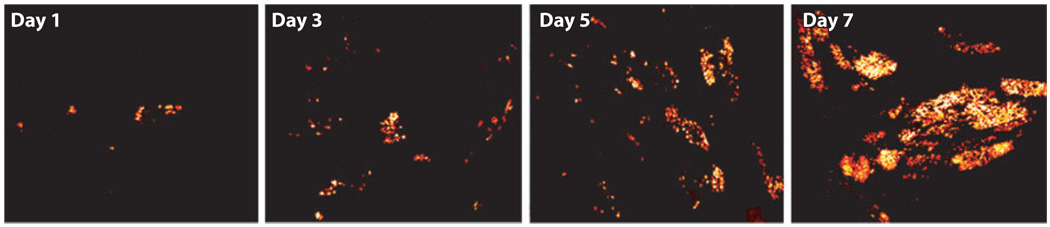
A good way to increase the signal-to-background ratio is to use multiple beacons to target the same RNA molecule. As an example, molecular beacons were designed to target a sequence in the genome of bovine respiratory syncytial virus (bRSV) that has three exact repeats (65). Shown in Figure 5 is the molecular beacon signal indicating the spreading of viral infection at days 1, 3, 5, and 7 postinfection (PI), which demonstrates the ability of molecular beacons to monitor and quantify, in real time, the viral infection process. Molecular beacons were further used to image the viral genomic RNA (vRNA) of human RSV (hRSV) in live Vero cells, revealing the dynamics of filamentous virion egress and providing insight into how viral filaments bud from the plasma membrane of the host cell (66).
Figure 5.
Live-cell fluorescence imaging of the genome of bovine respiratory syncytial virus (bRSV) using molecular beacons shows the spreading of infection in host cells at days 1, 3, 5, and 7 postinfection (PI). Primary bovine turbinate cells were infected by a clinical isolate of bRSV, CA-1, with a viral titer of 2 × 103.6 TCID50 ml−1. Molecular beacons were designed to target several repeated sequences of the gene-end-intergenic-gene-start signal within the bRSV genome, with a signal-to-noise ratio of 50–200.
In an alternative approach, multiple, optically distinct, molecular beacons can be used to visualize multiple RNAs simultaneously (i.e., multiplexing) (83, 90). This approach could be taken advantage of to highlight the orchestration between various gene expression patterns in living cells. In fact, several groups have already demonstrated the feasibility of simultaneously imaging multiple genes in single living cells (47, 58). Although multiplexing is possible for FRET-based detection schemes as well, typically the donor and acceptor fluorophores cover a significant portion of the visible spectrum, leaving little room for two additional fluorophores to participate in FRET.
A novel molecular beacon design that has been adopted for multiplexing is the wavelengthshifting molecular beacon (85). In this design, a molecular beacon contains two fluorophores: a first fluorophore that absorbs strongly in the wavelength range of the monochromatic light source and a second fluorophore that emits at the desired emission wavelength due to FRET from the first fluorophore to the second fluorophore. This allows multiple molecular beacons that emit at optically distinct wavelengths to be excited by a single light source. Wavelength-shifting molecular beacons are substantially brighter than conventional molecular beacons that contain a fluorophore that cannot efficiently absorb energy from the available monochromatic light source.
Although it has been demonstrated that molecular beacons can be used to visualize RNA in living cells, several studies have shown that the signal-to-background ratio can be significantly compromised due to probe degradation and/or nonspecific interactions (16, 42, 82, 86). In fact, it has recently been reported that when molecular beacons are introduced into living cells, there can be more than a fivefold increase in background fluorescence (15). Molecular beacons have been synthesized with numerous chemical modifications to the nucleobase, the sugar moiety, or the internucleoside linkage to prevent/slow probe degradation (39, 75); however, these modifications may not necessarily eliminate the emission of false-positive signals. For example, molecular beacons composed of 2′-O-methyl modifications have been shown to exhibit a nonspecific fluorescent signal in the nucleus of living cells (16, 51). It was found that false-positive signals could be avoided by synthesizing molecular beacons with stems composed of either l-DNA or locked nucleic acid (LNA) linkages (95, 96). In another study, it was found that the signal-to-background ratio could be improved simply by preventing nuclear localization (15). This was achieved by attaching molecular beacons to quantum dots, which were too large to pass through the nuclear pores. Surprisingly, even nuclease-vulnerable molecular beacons did not elicit a false-positive signal when they were constrained to the cytoplasm.
One area of continued debate regarding molecular beacons involves their intracellular distribution. Numerous studies have shown that molecular beacons are rapidly sequestered into the nucleus once introduced into cells; however, there have been an equal number of studies that did not observe this pattern of intracellular distribution. It is not clear whether these differences are cell-line dependent, dependent on the method of delivering molecular beacons, or due to some other variable. Nonetheless, several methods have been introduced to prevent nuclear sequestration. Specifically, molecular beacons have been conjugated to large proteins (or nanoparticles) that prevent their passage through nuclear pores, and they have been linked to tRNAs that drive nuclear export (15, 16, 48, 82).
Dual FRET Molecular Beacons
The concept of dual FRET molecular beacons is similar to that of dual FRET linear probes, except molecular beacons are used in place of the fluorescently labeled linear oligonucleotides. In other words, this approach utilizes a pair of molecular beacons labeled with a donor and an acceptor fluorophore, respectively, as illustrated in Figure 1d (67, 79). The probe sequences are chosen such that the molecular beacons hybridize to adjacent regions on a single nucleic acid target, similar to the dual FRET linear probes. The resulting FRET signal (i.e., sensitized emission of the acceptor fluorophore) upon probe hybridization serves as a positive signal, which can be readily discerned from non-FRET false-positive signals owing to probe degradation and nonspecific probe opening. Dual FRET molecular beacons, therefore, combine the low-background signal and high specificity of molecular beacons with the ability of two probe FRET assays to differentiate between true target recognition and false-positive signals. Interestingly, it has been demonstrated that upon hybridization to nucleic acid targets, dual FRET molecular beacons can provide a better signal-to-background ratio than the single molecular beacon approach when appropriate FRET pairs are selected (67, 79).
In the conventional molecular beacon design, the stem sequence is typically independent of the target sequence (Figure 1c); however, dual FRET molecular beacons can be designed such that all the bases of one arm of the stem (to which a fluorophore is conjugated) are complementary to the target sequence, thus participating in both stem formation and target hybridization (shared-stem molecular beacons) (77) (Figure 1d). The advantage of this shared-stem design is to help fix the position of the fluorophore that is attached to the stem arm, limiting its degree of freedom and increasing the FRET in the dual FRET molecular-beacon design.
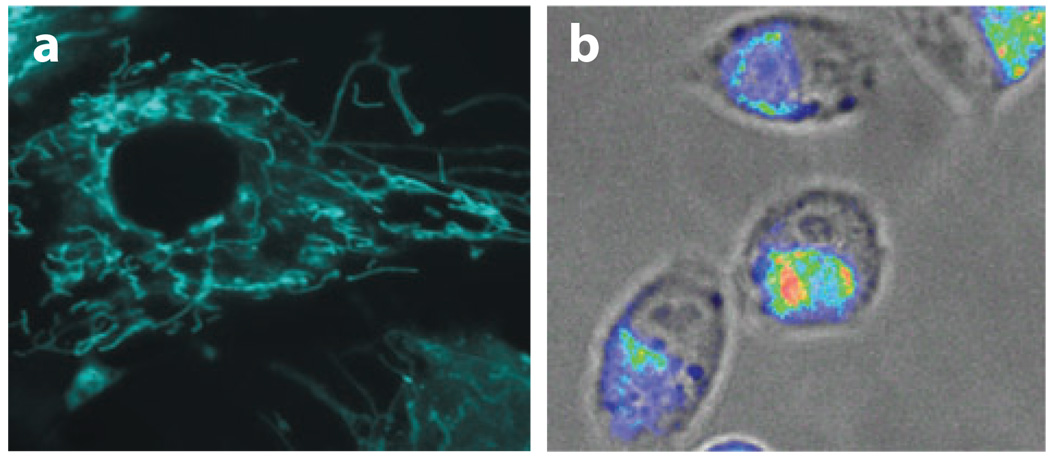
Dual FRET molecular beacons have been used to detect K-ras and survivin mRNAs in human dermal fibroblasts (HDFs) and MIAPaCa-2 cells, respectively (67). K-ras mRNAs seemed to localize with a filamentous pattern in HDF cells, whereas survivin mRNAs seemed to localize in a nonsymmetrical pattern within MIAPaCa-2 cells, often to one side of the nucleus, as shown in Figure 6. Although mRNA localization in living cells is believed to be closely related to posttranscriptional regulation of gene expression, much remains to be seen if such localization indeed targets a protein to its site of function by producing the protein on the spot. Using a dual beacon approach, the transport and localization of oskar mRNA in Drosophila melanogaster oocytes was also visualized (11). In this work, molecular beacons with 2′-O-methyl backbone were delivered into cells using microinjection, and the migration of oskar mRNAs was tracked in real time, from the nurse cells where the mRNA is produced to the posterior cortex of the oocyte where it is localized. Clearly, the direct visualization of specific mRNAs in living cells with molecular beacons will provide important insight into the intracellular trafficking and localization of RNA molecules.
Figure 6.
Imaging mRNA localization in HDF and MIAPaCa-2 cells using dual FRET molecular beacons targeting K-ras and survivin mRNA, respectively. (a) Fluorescence images of K-ras mRNA in stimulated HDF cells. Note the filamentous K-ras mRNA localization pattern. (b) A fluorescence image of survivin mRNA localization in MIAPaCa-2 cells. Note that survivin mRNAs often localized to one side of the nucleus of the MIAPaCa-2 cells.
As with other dual probes for live-cell RNA detection, although dual FRET molecular beacons exhibit an improved signal-to-background ratio and improved selectivity compared with single molecular beacons, this approach does require finding large stretches of RNA, with little or no secondary structure, that can accommodate the hybridization of two probes. This can be challenging, particularly when trying to identify a single nucleotide polymorphism, which significantly constrains the selection of target sites.
Quenched Autoligation Probes
Similar to the dual FRET molecular beacons approach, the quenched autoligation FRET (QUAL-FRET) approach combines fluorescence quenching and FRET by using two linear probes, one labeled with a FRET acceptor (e.g., Cy5) and a nucleophile on the 3′ end, and the second labeled with a FRET donor (e.g., FAM) and an electrophilic dabsyl quencher on the 5′ end (Figure 1e) (71). In the absence of a target, the fluorescence of the donor is quenched by the dabsyl quencher and the acceptor is not directly excited. Upon binding of the two probes to adjacent sites on the same RNA, the nucleophilic group displaces the dabsyl group via nucleophilic substitution reaction. This results in the formation of the ligated product, with the FRET donor and acceptor held in close proximity. Excitation of the donor fluorophore therefore generates a detectable FRET signal. The irreversibility of the ligated product is expected to provide both favorable and unfavorable attributes, depending on the application. For example, the continual expression of RNA could lead to the accumulation of ligated product and a corresponding increase in signal. This could allow for the detection of low-copy-number RNA that would not be detected otherwise. The disadvantage of using an irreversible probe is that it is not possible to monitor a downregulation in gene expression.
Because ligation is required to generate the FRET signal, a nonspecific signal is unlikely to occur even if both half probes were bound to a given nucleic acid–binding protein. This is because the reactive ends are required to be held in very close proximity for a set period of time for the nucleophilic substitution reaction to occur. Unfortunately, it is expected that this time requirement also limits the detection of some RNAs. For example, if RNAs are rapidly degraded, there may not be sufficient time for autoligation to occur.
The selectivity and specificity of quenched autoligation probes are expected to be similar to linear FRET probes; however, the signal-to-background is improved due to the quenching of donor fluorescence. In other words, fluorescence from unhybridized donor probes does not contribute to the background fluorescence in the acceptor channel even if there is a spectral overlap in donor and acceptor emission. Similar to the other two-probe methods, quenched autoligation probes also require the identification of large stretches of RNA with little or no secondary structure.
Fluorescent Protein–Based Probes
In addition to oligonucleotide probes, RNA-binding proteins (RBPs) tagged with optical reporters such as green fluorescent protein (GFP) have also been used to detect mRNA in living cells (12, 25). The feasibility of this approach was first demonstrated by coexpressing a gene that encodes the coat protein of the bacterial phage MS2 fused with GFP (GFP-MS2) and an RNA sequence containing tandem repeats of hairpin binding sites for the GFP-MS2 in the 3′ untranslated region (3′ UTR), as shown in Figure 1f (8). In this approach, the binding of multiple GFP-MS2 fusion proteins to single RNA transcripts results in a high local concentration of GFP (i.e., bright fluorescent speckles, which can be seen above the high background of unbound GFP fusions) similar to the mechanism by which tagged linear ODN probes detect intracellular RNA. The GFP-MS2 approach has been used to track the localization and dynamics of RNA in living cells with single-molecule sensitivity and has thus provided a great deal of insight into RNA processing, localization, and transport (29, 70).
Recently, a method to improve the signal-to-background ratio of fluorescent protein–based imaging of RNA was introduced, based on the concept of protein fragment complementation (PFC) (87). PFC refers to the rational dissection of a protein (or enzyme) into two inactive fragments, an N-terminal fragment and a C-terminal fragment, that fold into the complete protein and regain functionality when held in close proximity. In the case of fluorescent proteins, the complementation of the two fluorescent protein fragments results in the regeneration of an optical signal. As illustrated in Figure 1g, for RNA imaging, two RBPs are expressed in living cells, and each contains a fragment of a split GFP (or YFP). Binding of the two RBPs to adjacent sequences of an RNA drives the association of the two GFP fragments, inducing GFP complementation and activation of a fluorescence signal. An optical signal was not observed unless the target mRNA was expressed and the GFP fragments were brought into close proximity upon binding the RNA-binding site (87). Although this approach clearly improves upon the signal-to-background ratio compared with the GFP-MS2-based techniques that use full-length GFP, neither of these approaches can be used to detect endogenous RNA.
Aslightly modified PFC approach that does actually allow for the detection of endogenous RNA in living cells involves the coexpression of GFP fragments fused to genetically modified Pumilio homology domains (PUM-HD) (55). PUM-HD is composed of an array of eight elements that recognizes the RNA sequence UGUANAUA, with each element specifically recognizing a single base. The specificity of PUM-HD was altered by changing the RNA-recognizing amino acid residues within the PUM-HD elements. As a result, two different PUM-HDs were created that recognize a 16-base sequence of mitochondrial RNA encoding NADH dehydrogenase subunit 6 (ND6). Upon targeting the split GFP-PUM HD fusions into the mitochondrial matrix, it was possible to image the localization of ND6 mitochondrial RNA in real time. A significant advantage of this approach is that the probes do not have to be introduced into the cell but can be constitutively expressed. Further, the background signal is quite low because there is very little fluorescent signal unless the RNA-binding proteins are bound to the target mRNA. The split-GFP method, however, may have difficulties in tracking the dynamics of RNA expression in real time, because the reconstitution of the fluorescent protein from the split fragments typically takes 0.5–4 hours, during which the RNA expression level may change. Further, once GFP is reconstituted, it does not readily dissociate, making it difficult to monitor the downregulation of gene expression. GFP is also not particularly bright compared with most commercial fluorophores. Therefore, it can be difficult to detect faint signals above autofluorescence. Fluorescent protein– based probes are also often criticized because the binding of large bulky protein to RNA could interfere with normal RNA transport. This is particularly true when multiple GFP molecules are bound to a single RNA transcript.
COMMON DESIGN ISSUES FOR RNA-TARGETED PROBES
Target Accessibility
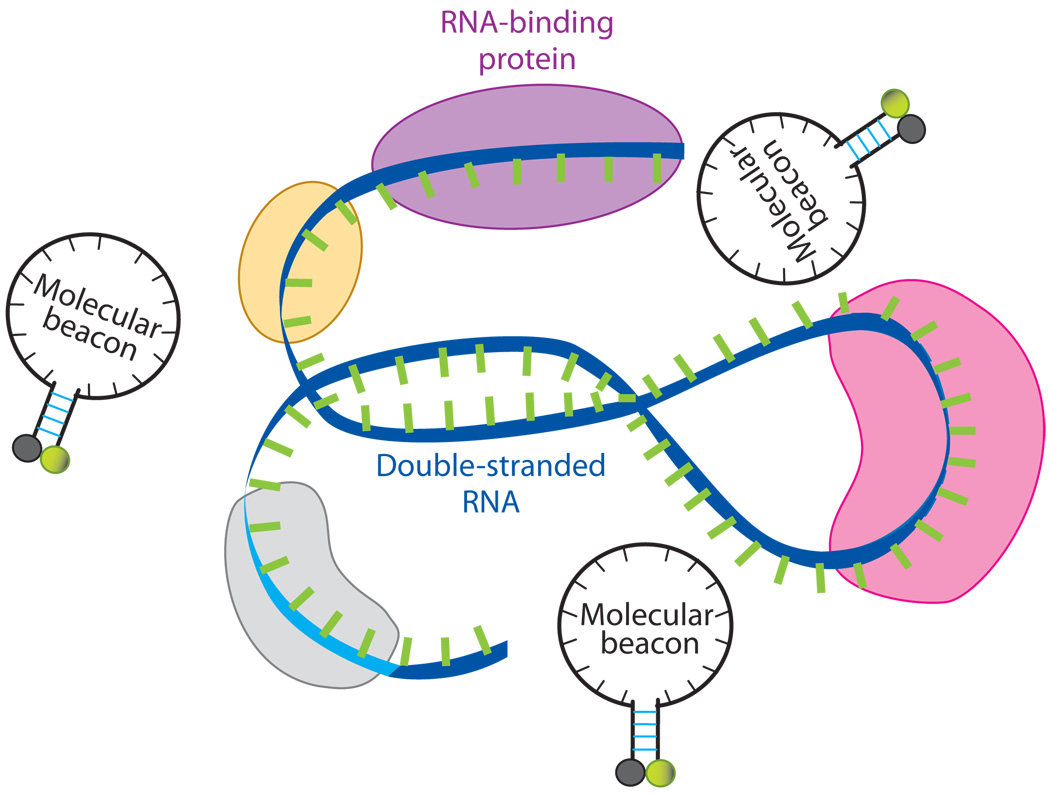
A critical issue in designing any oligonucleotide probe for live-cell RNA imaging is target accessibility. It is well known that RNA molecules within living cells are commonly associated with RNA-binding proteins, e.g., ribonucleoproteins (RNP). Further, RNA molecules form complex secondary (folded) structures (Figure 7). Therefore, when designing an oligonucleotide probe, it is preferable to avoid targeting RNA sequences that are double stranded, or occupied by RNA-binding proteins; otherwise, the probe has to compete off the RNA strand or the RNA-binding protein in order to hybridize to the target (63). Indeed, molecular beacons designed for targeting a specific RNA often show no signal when delivered to living cells (62). One difficulty in designing oligonucleotide probes is that, although predictions of mRNA secondary structure can be made using software such as Beacon Designer (www.premierbiosoft.com) and mfold (http://www.bioinfo.rpi.edu/applications/mfold/old/dna/), they are often inaccurate due to limitations of the biophysical models used and the limited understanding of protein-RNA interaction (57). Therefore, for each gene to target, it may be necessary to select multiple unique sequences along the target RNA and to have corresponding oligonucleotide probes designed, synthesized, and tested in living cells to select the best target sequence.
Figure 7.
A schematic illustration of a segment of the target mRNA with a double-stranded portion and RNA-binding proteins. A molecular beacon has to compete off an mRNA strand or RNA-binding protein(s) in order to hybridize to the target.
To identify possible oligonucleotide probe design rules, one study looked at the accessibility of BMP-4 mRNA using different beacon designs (62). Specifically, molecular beacons were designed to target the start codon and termination codon regions, the siRNA and antisense oligonucleotide probe sites identified previously, and sites that were randomly chosen. All the target sequences were determined to be unique to BMP-4 mRNA. Of the eight molecular beacons designed to target BMP-4 mRNA, it was found that only two beacons gave strong signals, one that targeted the start codon region and one that targeted the termination codon region. It was also found that, even for a molecular beacon that works well, shifting its targeting sequence by just a few bases toward the 3′ or the 5′ ends could significantly reduce the fluorescence signal, further confirming that target accessibility is extremely sensitive to the location of the targeting sequence. Although target accessibility will likely need to be tested on a case-by-case basis, these findings suggest that the start and termination codon regions may be more accessible than other locations on target RNA.
Fluorophores and Quenchers
Although any fluorophore and/or quencher can be incorporated into the oligonucleotide-based probes described above, proper selection could yield important benefits in regard to signal-to-background ratio, multiplexing, and fluorescence quantification. For example, a systematic study on a wide range of fluorophore-quencher combinations showed that the quenching efficiency (contact quenching) could vary between 57% and 98% (45). Quenching efficiencies could be further improved either by using inorganic quenchers such as gold or by incorporating multiple quenchers into a single molecular beacon (23, 94). Alternatively, several approaches that could be used to improve signal intensity include the incorporation of quantum dots or photoluminescent polymers into the molecular beacon design (36, 37, 40). Quantum dots have been reported to be brighter and more photostable than inorganic fluorophores. Further, their broad absorption and narrow emission spectra are highly amenable to multiplexing. However, initial studies have suggested that quantum dot–based molecular beacons only exhibit a signal-to-background ratio of 6:1 due to inefficient quenching. Inefficient quenching is likely to be less of an issue for photoluminescent polymers, which exhibit a superquenching effect. Specifically, when the fluorescence of any single repeat unit is quenched, then the entire polymer chain responds in the same fashion. Therefore, long polymer chains can be used to provide an amplified fluorescent signal that can be modulated by a single quencher. These polymers clearly offer great potential for RNA detection; however, it will be important to overcome the sensitivity of these polymers to nonspecific interactions if they are to be extended to live cell imaging applications (40). In addition to selecting alternative brighter fluorescent labels, a complementary approach that can be used to improve signal-to-background ratio in live cells involves simply selecting red-shifted fluorophores. This can have a significant effect on sensitivity by eliminating the interfering effects that result from autofluorescence. It is also possible to use lanthanide chelate as the donor in a dual FRET-probe assay and to perform time-resolved measurements to dramatically increase the signal-to-background ratio, thus achieving high sensitivity in detecting low-abundance genes (79).
As fluorescent imaging strategies have advanced, there has been a general trend toward more quantitative measurements of fluorescent signals, with the ultimate goal being absolute quantification. It is envisioned that the absolute quantification of fluorescence could allow the exact number of fluorophores within a compartment/cell to be quantified and could correspondingly allow the number of target genes, proteins, or enzymes to be quantified. However, factors such as nonspecific protein interactions and pH could have a dramatic effect on the fluorescence intensity of some fluorophores. Therefore, if accurate fluorescent measurements are desirable, it is necessary to select fluorescent labels that are insensitive to their environment. Recently, it has been shown that although the fluorescent intensity of a few fluorophores, e.g., fluorescein, were highly susceptible to the intracellular environment, other fluorophores, e.g., Dylight 649, Alexa647, and Alexa750, were insensitive to the intracellular environment (17).
DELIVERY OF IMAGING PROBES
One of the critical steps required for the accurate detection of RNA molecules in living cells is the efficient delivery of synthetic probes into the cytoplasm. Oligonucleotide-based probes are generally prevented from gaining access to the cytoplasm due to the barrier imposed by the cell membrane. Further, even if the probes enter the cells successfully, the efficiency of delivery in an imaging assay should be defined not only by how many probes enter the cell or how many cells have probes internalized, but also by what fraction of probes are free to hybridize intracellular RNA. This is in contrast to antisense and gene delivery applications where the reduction/increase in the level of protein expression is the final metric used to define efficiency or success. Common techniques that have been used to deliver oligonucleotide-based RNA-imaging probes into live cells include microinjection, polycationic molecules such as liposomes and dendrimers, electroporation, cell-penetrating peptides (CPPs), and streptolysin O (SLO).
Microinjection
Microinjection is, perhaps, one of the most direct methods for ensuring the delivery of oligonucleotide probes into the cytoplasm of live cells; however, an obvious limitation of this approach is the low number of cells that can be analyzed at any given time. Further, microinjection can often be damaging to the cell and can interfere with normal cell function. This has led to the use of a number of alternative methods that are thought to be more efficient and less invasive than microinjection. One such method involves the use of cationic transfection agents. Although these agents have been effective in some studies (58), others have found that many commercial transfection agents result in punctate fluorescent patterns that appear to be indicative of endosomal entrapment (53). ODN probes that enter into the endosomal/lysosomal pathway are rapidly degraded by nucleases (60). Consequently, even when transfection methods allow for endosomal/lysosomal escape, only 0.01% to 10% of the probes remain functioning (21). Further, probe delivery via the endocytic pathway typically takes 2–4 hours.
Electroporation
To avoid the deleterious effects associated with endosomal entrapment, methods such as electroporation have been used to deliver oligonucleotides directly into the cytoplasm of living cells. Although electroporation has traditionally been associated with low cell viability, recent advances in electroporation technology, such as the ability to perform electroporation in microliter-volume spaces (e.g., pipette tips), have led to a reduction in the many harmful events associated with this process, including heat generation, metal ion dissolution, pH variation, and oxide formation. This microliter-volume electroporation process is known as microporation. Recently, it was shown that microporation could lead to the uniform cytosolic distribution of oligonucleotide probes in live cells with a transfection efficiency of 93% and an average viability of 86% (16). A unique advantage of microporation is that delivery of oligonucleotide probes takes only several seconds, so cells can be analyzed immediately for RNA content. Conversely, a potential disadvantage is that most electroporation techniques involve trypsinization. Therefore, it is several hours before the cells readhere to cell-culture plate surfaces and RNA localization can be assessed.
Cell Membrane Permeabilization
Another nonendocytic delivery method is toxin-based cell membrane permeabilization. One popular reagent is streptolysin O (SLO), which is a pore-forming bacterial toxin that has been used as a simple and rapid means of introducing oligonucleotides into eukaryotic cells (4, 26, 27). SLO binds as a monomer to cholesterol and oligomerizes into a ring-shaped structure to form pores of approximately 25–30 nm in diameter, allowing the influx of both ions and macromolecules. An essential feature of this technique is that the toxin-based permeabilization is reversible. This can be achieved by introducing oligonucleotides with SLO under serum-free conditions and then removing the mixture and adding normal media with serum (4, 93). Since cholesterol composition varies with cell types, the permeabilization protocol needs to be optimized for each cell type by varying temperature, incubation time, cell number, and SLO concentration. Typically, RNA localization can be assessed 30 minutes to 2 hours following the introduction of oligonucleotide probes into cells using SLO-based delivery.
Cell-Penetrating Peptides
Cell-penetrating peptides (CPPs) have been used to introduce proteins, nucleic acids, and other biomolecules into living cells (6, 72, 91). Among the family of peptides with membrane translocating activity are antennapedia, HSV-1 VP22, and the HIV-1 Tat peptide. To date, the most widely used peptides are HIV-1 Tat peptide and its derivatives, due to their small size and high delivery efficiency. The Tat peptide is rich in cationic amino acids, which are very common in many of the cell-penetrating peptides. However, the exact mechanism for CPP-induced membrane translocation remains elusive.
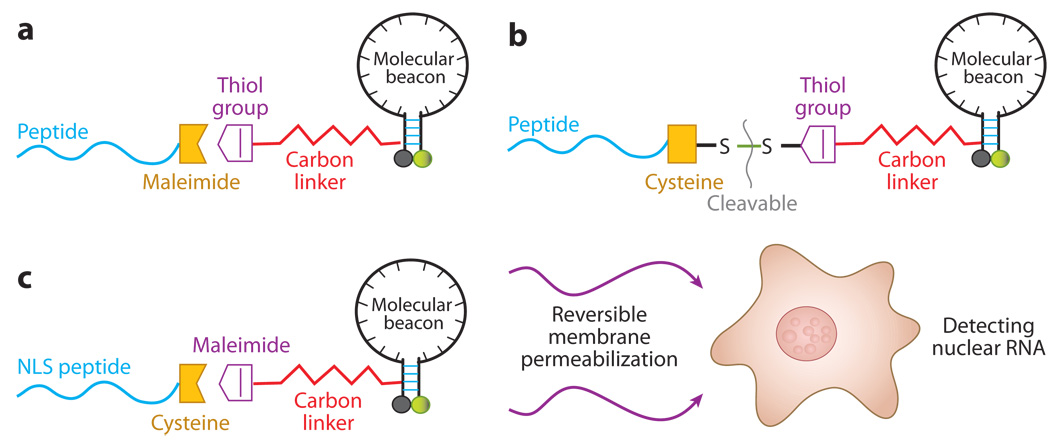
A wide variety of cargos have been delivered to living cells both in cell culture and in tissue using cell-penetrating peptides (13, 92). For example, Allinquant et al. (2) linked Antennapedia peptide to the 5′ end of DNA oligonucleotides (with biotin on the 3′ end) and incubated both peptide-linked ODNs and ODNs alone with cells. By detecting biotin using streptavidin-alkaline phosphatase amplification, it was found that the peptide-linked ODNs were internalized very efficiently into all cell compartments compared with control ODNs. No indication of endocytosis was found. Similar results were obtained by Troy et al. (76) with a 100-fold increase in antisense delivery efficiency when ODNs were linked to antennapedia peptides. Recently, Tat peptides were conjugated to molecular beacons using different linkages (Figures 8a and 8b); the resulting peptide-linked molecular beacons were delivered into living cells to target GAPDH and survivin mRNAs (53). It was demonstrated that, at relatively low concentrations, peptide-linked molecular beacons were internalized into living cells within 30 minutes with nearly 100% efficiency. Further, peptide-based delivery did not interfere with either specific targeting or hybridization-induced florescence of the probes, and the peptide-linked molecular beacons could have self-delivery, targeting, and reporting functions.
Figure 8.
A schematic of peptide-linked molecular beacons. (a) A peptide-linked molecular beacon using the thiol-maleimide linkage in which the quencher arm of the molecular beacon stem is modified by adding a thiol group, which can react with a maleimide group placed to the C terminus of the peptide to form a direct, stable linkage. (b) A peptide-linked molecular beacon with a cleavable disulfide bridge in which the peptide is modified by adding a cysteine residue at the C terminus, which forms a disulfide bridge with the thiol-modified molecular beacon. This disulfide bridge design allows the peptide to be cleaved from the molecular beacon by the reducing environment of the cytoplasm. (c) A schematic illustration of the design of a peptide-linked molecular beacon and its delivery into cell nucleus. The NLS peptide is covalently linked to the molecular beacon using a modified nucleotide in its quencher arm. The NLS-linked molecular beacons are delivered into the cytoplasm first using streptolysin O (SLO), and the NLS peptide actively transports the probes into the nucleus of a living cell.
Peptide-linked molecular beacons can also be used to target RNA molecules in the cell nucleus by attaching a nuclear localization signal (NLS) peptide to a molecular beacon. Combining the NLS-linked beacon with the SLO-based reversible membrane permeabilization (Figure 8c), molecular beacons designed to target snRNAs U1 and U2 as well as small nucleolar RNA (snoRNA) U3 were delivered into the nuclei of live HeLa cells, and the localization and colocalization (U1 and U2) of these nuclear RNAs was imaged (52). This delivery method can be used to image transcriptional and posttranscriptional processing of RNAs in the nucleus of living cells.
ENGINEERING CHALLENGES AND FUTURE DIRECTIONS
There are a number of challenges in detecting and quantifying RNA expression in living cells. In addition to issues with target accessibility, detection specificity, and probe delivery as discussed above, accurate measurements of RNA expression also require an understanding of RNA biology and probe-target interactions. It is likely that different applications have different requirements on the properties of probes. For example, rapid determination of RNA-expression level and localization requires fast probe-target hybridization kinetics, whereas long-time monitoring of gene-expression dynamics requires probes with high intracellular stability.
An important issue in living-cell gene detection using hairpin ODN probes is the possible effect of probes on normal cell function, including protein expression. As revealed in the field of antisense therapy research, complementary pairing of even short oligonucleotides to RNA can have a profound impact on protein-expression levels and even cell fate. For example, tight binding of the probe to the translation start site may block mRNA translation by preventing the binding or progression of the transcriptional machinery. Alternatively, binding of an oligonucleotide-based probe to RNA can also trigger RNase H-mediated RNA degradation. Of course, it can be argued that the probability of eliciting antisense effects with hairpin probes is very low because low concentrations of probes (<200 nM) are used for RNA detection in contrast to the high concentrations [typically 20 µM; (27)] employed in antisense experiments. Further, it generally takes four hours before any noticeable antisense effect occurs, whereas visualization of RNA with hairpin probes requires less than two hours after delivery. Nonetheless, it remains important to carry out systematic studies to examine the possible antisense effects, particularly if temporal measurements are to be made. It should be noted that the likelihood of avoiding antisense effects could be improved by incorporating chemical modifications into the probe design that do not induce RNase H activity, e.g., 2′-O-methyl modifications. It is expected that the noninvasive binding of RNA probes could also benefit from the synthesis of new chemically modified oligonucleotides that exhibit high affinity and specificity for RNA, without inducing RNase H, as well as from new computational programs that could be used to select RNA target sites devoid of secondary structure and RNA-binding proteins.
Another issue that can interfere with the accurate measurement of gene expression is the large cell-to-cell variation in probe delivery that is often observed when methods such as transfection are used. This variability could have a significant impact on the total cellular fluorescence and can lead to ambiguous findings and incorrect conclusions. For example, cells that have no or low amounts of internalized probe could easily be mistaken for cells with low gene expression, thus resulting in a false negative. Alternatively, high levels of molecular beacons could exhibit a measurable background that may be mistaken for probe hybridization, i.e., false positive. Another complication that may arise when comparing cellular fluorescence is the change in fluorescence intensity due to instrumental fluctuations. Instrumental fluctuations can result in as much as a 28% difference in fluorescent intensity from one day to the next (15). Several groups have tried to improve the quantification of fluorescence by simultaneously introducing molecular beacons and optically distinct, fluorescently labeled oligonucleotides into living cells and performing ratiometric imaging (11, 22, 47); however, even this method is sensitive to probe delivery. For example, if twice the amount of probes (i.e., molecular beacon and reference probe) is injected into a cell, then the fluorescent ratio would be reduced to half because the number of reference probes would be doubled but the number of hybridized molecular beacons (i.e., the number of RNA targets) would remain the same. Therefore, there is still the need to develop improved methods for the accurate quantification of RNA in living cells.
Perhaps the most challenging hurdle in developing fluorescent probes for RNA imaging in living cells is detection sensitivity. As noted above, the careful selection of fluorophores and quenchers can significantly improve the signal-to-background ratio; however, RNA imaging probes are thus far limited to imaging relatively abundant transcripts. Nonetheless, with the continued development of new fluorescent reporters with high extinction coefficients and quantum yields (such as quantum dots), combined with advances in charge-coupled device (CCD) camera technologies, the lower detection limit is expected to continue to improve, and single molecular detection in real time is soon expected. Single-molecule sensitivity will, of course, significantly enhance our ability to image, track, and quantify gene expression in vivo. When combined with peptide-based delivery and an endoscopic method, it may be possible to use hairpin ODN probes with nearinfrared (NIR) fluorophores to detect specific RNAs in tissue samples, animals, or even humans, thus providing a powerful tool for basic and clinical studies of human health and disease.
SUMMARY POINTS.
Fluorescently labeled, linear oligonucleotide probes can be used to image highly expressed RNAs with unique localization patterns in the nucleus, e.g., 28S ribosomal RNA in the nucleoli, poly(A) RNA in speckles, and U3 snRNA in Cajal bodies.
Two fluorescently labeled linear oligonucleotide probes that form a FRET pair and that are designed to bind adjacent regions on target RNA can be used to detect highly expressed cytoplasmic RNA. The sensitivity of this approach can be significantly improved by implementing time-resolved imaging.
Molecular beacons have been the most widely adopted method for imaging RNA expression in living cells largely because of their high signal-to-background ratio and specificity, both of which stem from their unique hairpin conformation. Molecular beacons also only require the identification of a single binding site, in contrast to FRET-based methods.
Dual FRET molecular beacons (i.e., two molecular beacons that form a FRET pair and bind adjacent regions of target RNA) retain the specificity of conventional molecular beacons but can also differentiate between true target recognition and false positives that can result from nuclease degradation and nonspecific interactions.
Quenched autoligation probes undergo an irreversible ligation upon binding RNA, which precludes them from monitoring the temporal regulation of RNA. However, this approach could provide an amplification mechanism for detecting low-copy-number transcripts as ligated products accumulate.
FP has been used to detect RNA by rationally dissecting GFP into two fragments that reassociate and regain functionality upon their binding to adjacent sites on target RNA. This method avoids concerns of probe delivery but cannot be used to monitor the downregulation of RNA because reconstituted GFP cannot readily dissociate back into a nonfluorescent form.
Methods for RNA imaging in living cells can benefit from the synthesis of new chemically modified oligonucleotides that exhibit high affinity and specificity for RNA, without inducing RNase H, as well as from new computational programs that could be used to select RNA target sites devoid of secondary structure and RNA-binding proteins.
The greatest challenge faced by live-cell RNA imaging methods is achieving a high enough sensitivity to detect low-copy-number RNAs.
ACKNOWLEDGMENTS
This work was supported by the NIH as an NHLBI Program of Excellence in Nanotechnology award (HL80711 to GB), an NCI Center of Cancer Nanotechnology Excellence award (CA119338 to GB), and two NCI grants (CA116102 and CA125088 to AT). It was also supported by the NSF (BES-0616031 to AT) and the American Cancer Society (RSG-07-005-01 to AT).
Abbreviations
- Expressed sequence tag (EST)
short portion of a gene that can often be used to identify whole genes
- Serial analysis of gene expression (SAGE)
technique to analyze the messenger RNA population in a sample of interest. Output typically provides quantitative information on the number of times each particular RNA is observed
- Representational difference analysis (RDA)
technique used to find differences in RNA expression between two different cell samples
- Suppression subtractive hybridization (SSH)
technique used to find differentially expressed genes in two different cell samples. Identifies unique genes with low expression whereas highly expressed genes are suppressed
- Single nucleotide polymorphisms (SNPs)
nucleic acid sequences that differ by a single nucleotide
- Fluorescence resonance energy transfer (FRET)
nonradiative energy transfer from one fluorescent dye the donor, to another fluorescent dye, the acceptor, when in close proximity to each other (typically <10 nm)
- Small nuclear RNA (snRNA)
class of small RNA molecules found in the nucleus of eukaryotic cells; involved in activities such as RNA splicing regulation of transcription factors, and maintaining telomeres
- Green fluorescent protein (GFP)
protein isolated from the jellyfish Aequore victorea that fluoresces green when exposed to blue light
- Cell-penetrating peptides (CPPs)
short polycationic peptides that can facilitate the cellular uptake of attached cargo (e.g. oligonucleotides, peptides, drugs, proteins, and nanoparticles)
- Streptolysin O (SLO)
pore-forming endotoxin that binds to cell membranes
Footnotes
DISCLOSURE STATEMENT
Dr. Tsourkas is a scientific advisor and partial shareholder of Vivonetics, a company that develops dual FRET-MBs for imaging gene expression. The authors have received funding from NSF (BES-0616031), NIH/NCI (1R21-CA-116102, R21-CA-125088), and the American Cancer Society (RSG-07-005-01-CCE) to develop molecular beacon conjugates and flow cytometric techniques for the analysis of gene expression.
LITERATURE CITED
- 1.Adams MD, Dubnick M, Kerlavage AR, Moreno R, Kelley JM, et al. Sequence identification of 2375 human brain genes. Nature. 1992;355:632–634. doi: 10.1038/355632a0. [DOI] [PubMed] [Google Scholar]
- 2.Allinquant B, Hantraye P, Mailleux P, Moya K, Bouillot C, Prochiantz A. Downregulation of amyloid precursor protein inhibits neurite outgrowth in vitro. J. Cell Biol. 1995;128:919–927. doi: 10.1083/jcb.128.5.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alwine JC, Kemp DJ, Parker BA, Reiser J, Renart J, et al. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- 4.Barry MA, Eastman A. Identification of deoxyribonuclease II as an endonuclease involved in apoptosis. Arch. Biochem. Biophys. 1993;300:440–450. doi: 10.1006/abbi.1993.1060. [DOI] [PubMed] [Google Scholar]
- 5.Bassell GJ, Powers CM, Taneja KL, Singer RH. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J. Cell Biol. 1994;126:863–876. doi: 10.1083/jcb.126.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker-Hapak M, McAllister SS, Dowdy SF. TAT-mediated protein transduction into mammalian cells. Methods. 2001;24:247–256. doi: 10.1006/meth.2001.1186. [DOI] [PubMed] [Google Scholar]
- 7.Behrens S, Fuchs BM, Mueller F, Amann R. Is the in situ accessibility of the 16S rRNA of Escherichia coli for Cy3-labeled oligonucleotide probes predicted by a three-dimensional structure model of the 30S ribosomal subunit? Appl. Environ. Microbiol. 2003;69:4935–4941. doi: 10.1128/AEM.69.8.4935-4941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 9.Blake WJ, Kærn M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 10.Bonnet G, Tyagi S, Libchaber A, Kramer FR. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl. Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brodsky AS, Silver PA. Identifying proteins that affect mRNA localization in living cells. Methods. 2002;26:151–155. doi: 10.1016/S1046-2023(02)00017-8. [DOI] [PubMed] [Google Scholar]
- 13.Brooks H, Lebleu B, Vives E. Tat peptide-mediated cellular delivery: back to basics. Adv. Drug Deliv. Rev. 2005;57:559–577. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Buongiorno-Nardelli M, Amaldi F. Autoradiographic detection of molecular hybrids between RNA and DNA in tissue sections. Nature. 1970;225:946–948. doi: 10.1038/225946a0. [DOI] [PubMed] [Google Scholar]
- 15.Chen AK, Behlke MA, Tsourkas A. Avoiding false-positive signals with nuclease-vulnerable molecular beacons in single living cells. Nucleic Acids Res. 2007;35:e105. doi: 10.1093/nar/gkm593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen AK, Behlke MA, Tsourkas A. Efficient cytosolic delivery of molecular beacon conjugates and flow cytometric analysis of target RNA. Nucleic Acids Res. 2008;36:e69. doi: 10.1093/nar/gkn331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen AK, Cheng Z, Behlke MA, Tsourkas A. Assessing the sensitivity of commercially available fluorophores to the intracellular environment. Anal. Chem. 2008;80(19):7437–7444. doi: 10.1021/ac8011347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dirks RW, Molenaar C, Tanke HJ. Methods for visualizing RNA processing and transport pathways in living cells. Histochem. Cell Biol. 2001;115:3–11. doi: 10.1007/s004180000214. [DOI] [PubMed] [Google Scholar]
- 20.Dirks RW, Tanke HJ. Advances in fluorescent tracking of nucleic acids in living cells. Biotechniques. 2006;40:489–496. doi: 10.2144/000112121. [DOI] [PubMed] [Google Scholar]
- 21.Dokka S, Rojanasakul Y. Novel nonendocytic delivery of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2000;44:35–49. doi: 10.1016/s0169-409x(00)00082-x. [DOI] [PubMed] [Google Scholar]
- 22.Drake TJ, Medley CD, Sen A, Rogers RJ, Tan W. Stochasticity of manganese superoxide dismutase mRNA expression in breast carcinoma cells by molecular beacon imaging. Chembiochem. 2005;6:2041–2047. doi: 10.1002/cbic.200500046. [DOI] [PubMed] [Google Scholar]
- 23.Dubertret B, Calame M, Libchaber AJ. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat. Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 24.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 25.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giles RV, Ruddell CJ, Spiller DG, Green JA, Tidd DM. Single base discrimination for ribonuclease H-dependent antisense effects within intact human leukaemia cells. Nucleic Acids Res. 1995;23:954–961. doi: 10.1093/nar/23.6.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giles RV, Spiller DG, Grzybowski J, Clark RE, Nicklin P, Tidd DM. Selecting optimal oligonucleotide composition for maximal antisense effect following streptolysin O-mediated delivery into human leukaemia cells. Nucleic Acids Res. 1998;26:1567–1575. doi: 10.1093/nar/26.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glotzer JB, Saffrich R, Glotzer M, Ephrussi A. Cytoplasmic flows localize injected oskar RNA in Drosophila oocytes. Curr. Biol. 1997;7:326–337. doi: 10.1016/s0960-9822(06)00156-4. [DOI] [PubMed] [Google Scholar]
- 29.Haim L, Zipor G, Aronov S, Gerst JE. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat. Methods. 2007;4:409–412. doi: 10.1038/nmeth1040. [DOI] [PubMed] [Google Scholar]
- 30.Huang Q, Pederson T. A human U2 RNA mutant stalled in 3′ end processing is impaired in nuclear import. Nucleic Acids Res. 1999;27:1025–1031. doi: 10.1093/nar/27.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hume DA. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood. 2000;96:2323–2328. [PubMed] [Google Scholar]
- 32.Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc. Natl. Acad. Sci. USA. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jockusch S, Marti AA, Turro NJ, Li Z, Li X, et al. Spectroscopic investigation of a FRET molecular beacon containing two fluorophores for probing DNA/RNA sequences. Photochem. Photobiol. Sci. 2006;5:493–498. doi: 10.1039/b600213g. [DOI] [PubMed] [Google Scholar]
- 34.Johnson DR, Lee PK, Holmes VF, Alvarez-Cohen L. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl. Environ. Microbiol. 2005;71:3866–3871. doi: 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kepler TB, Elston TC. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys. J. 2001;81:3116–3136. doi: 10.1016/S0006-3495(01)75949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JH, Morikis D, Ozkan M. Adaptation of inorganic quantum dots for stable molecular beacons. Sensors Actuators B-Chem. 2004;102:315–319. [Google Scholar]
- 37.Kim Y, Sohn D, Tan W. Molecular beacons in biomedical detection and clinical diagnosis. Int. J. Clin. Exp. Pathol. 2008;1:105–116. [PMC free article] [PubMed] [Google Scholar]
- 38.Kostrikis LG, Tyagi S, Mhlanga MM, Ho DD, Kramer FR. Spectral genotyping of human alleles. Science. 1998;279:1228–1229. doi: 10.1126/science.279.5354.1228. [DOI] [PubMed] [Google Scholar]
- 39.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 40.Kushon SA, Ley KD, Bradford K, Jones RM, McBranch D, Whitten D. Detection of DNA hybridization via fluorescent polymer superquenching. Langmuir. 2002;18:7245–7249. [Google Scholar]
- 41.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 42.Li JJ, Geyer R, Tan W. Using molecular beacons as a sensitive fluorescence assay for enzymatic cleavage of single-stranded DNA. Nucleic Acids Res. 2000;28:E52. doi: 10.1093/nar/28.11.e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 44.Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 45.Marras SA, Kramer FR, Tyagi S. Efficiencies of fluorescence resonance energy transfer and contactmediated quenching in oligonucleotide probes. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. USA. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medley CD, Drake TJ, Tomasini JM, Rogers RJ, Tan W. Simultaneous monitoring of the expression of multiple genes inside of single breast carcinoma cells. Anal. Chem. 2005;77:4713–4718. doi: 10.1021/ac050881y. [DOI] [PubMed] [Google Scholar]
- 48.Mhlanga MM, Vargas DY, Fung CW, Kramer FR, Tyagi S. tRNA-linked molecular beacons for imaging mRNAs in the cytoplasm of living cells. Nucleic Acids Res. 2005;33:1902–1912. doi: 10.1093/nar/gki302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitsuhashi M, Tomozawa S, Endo K, Shinagawa A. Quantification of mRNA in whole blood by assessing recovery of RNA and efficiency of cDNA synthesis. Clin. Chem. 2006;52:634–642. doi: 10.1373/clinchem.2005.048983. [DOI] [PubMed] [Google Scholar]
- 50.Molenaar C, Abdulle A, Gena A, Tanke HJ, Dirks RW. Poly(A) + RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J. Cell Biol. 2004;165:191–202. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molenaar C, Marras SA, Slats JC, Truffert JC, Lemaitre M, et al. Linear 2′ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29:e89. doi: 10.1093/nar/29.17.e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nitin N, Bao G. NLS peptide conjugated molecular beacons for visualizing nuclear RNA in living cells. Bioconj. Chem. 2008;19(11):2205–2211. doi: 10.1021/bc800322a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nitin N, Santangelo PJ, Kim G, Nie S, Bao G. Peptide-linked molecular beacons for efficient delivery and rapid mRNA detection in living cells. Nucleic Acids Res. 2004;32:e58. doi: 10.1093/nar/gnh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okamura Y, Kondo S, Sase I, Suga T, Mise K, Furusawa I. Double-labeled donor probe can enhance the signal of fluorescence resonance energy transfer (FRET) in detection of nucleic acid hybridization. Nucleic Acids Res. 2000;28:E107. doi: 10.1093/nar/28.24.e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozawa T, Natori Y, Sato M, Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat. Methods. 2007;4:413–419. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- 56.Paillasson S, Van De Corput M, Dirks RW, Tanke HJ, Robert-Nicoud M, Ronot X. In situ hybridization in living cells: detection of RNA molecules. Exp. Cell Res. 1997;231:226–233. doi: 10.1006/excr.1996.3464. [DOI] [PubMed] [Google Scholar]
- 57.Pattanayak V, Gifford LK, Lu P, Gewirtz AM. Observed vs predicted structure of fluorescent selfquenching reporter molecules (SQRM): caveats with respect to the use of “stem-loop” oligonucleotides as probes for mRNA folding. RNA. 2008;14:657–665. doi: 10.1261/rna.890408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peng XH, Cao ZH, Xia JT, Carlson GW, Lewis MM, et al. Real-time detection of gene expression in cancer cells using molecular beacon imaging: new strategies for cancer research. Cancer Res. 2005;65:1909–1917. doi: 10.1158/0008-5472.CAN-04-3196. [DOI] [PubMed] [Google Scholar]
- 59.Piatek AS, Tyagi S, Pol AC, Telenti A, Miller LP, et al. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotech. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 60.Price NC, Stevens L. Fundamentals of Enzymology: The Cell and Molecular Biology of Catalytic Proteins. New York: Oxford Univ. Press; 1999. p. 478. [Google Scholar]
- 61.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rhee WJ, Santangelo PJ, Jo H, Bao G. Target accessibility and signal specificity in live-cell detection of BMP-4 mRNA using molecular beacons. Nucleic Acids Res. 2008;36:e30. doi: 10.1093/nar/gkn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rudnick SI, Swaminathan J, Sumaroka M, Liebhaber S, Gewirtz AM. Effects of local mRNA structure on posttranscriptional gene silencing. Proc. Natl. Acad. Sci. USA. 2008;105:13787–13792. doi: 10.1073/pnas.0805781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 65.Santangelo P, Nitin N, LaConte L, Woolums A, Bao G. Live-cell characterization and analysis of a clinical isolate of bovine respiratory syncytial virus, using molecular beacons. J. Virol. 2006;80:682–688. doi: 10.1128/JVI.80.2.682-688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santangelo PJ, Bao G. Dynamics of filamentous viral RNPs prior to egress. Nucleic Acids Res. 2007;35:3602–3611. doi: 10.1093/nar/gkm246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Santangelo PJ, Nix B, Tsourkas A, Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Satterfield BC, West JA, Caplan MR. Tentacle probes: eliminating false positives without sacrificing sensitivity. Nucleic Acids Res. 2007;35:e76. doi: 10.1093/nar/gkm113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 70.Shav-Tal Y, Darzacq X, Shenoy SM, Fusco D, Janicki SM, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Silverman AP, Kool ET. Quenched autoligation probes allow discrimination of live bacterial species by single nucleotide differences in rRNA. Nucleic Acids Res. 2005;33:4978–4986. doi: 10.1093/nar/gki814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Snyder EL, Dowdy SF. Protein/peptide transduction domains: potential to deliver large DNA molecules into cells. Curr. Opin. Mol. Ther. 2001;3:147–152. [PubMed] [Google Scholar]
- 73.Sokol DL, Zhang X, Lu P, Gewirtz AM. Real time detection of DNA-RNA hybridization in living cells. Proc. Natl. Acad. Sci. USA. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl. Acad. Sci. USA. 2002;99:12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toulmé JJ, Di Primo C, Moreau S. Modulation of RNA function by oligonucleotides recognizing RNA structure. Prog. Nucleic Acid Res. Mol. Biol. 2001;69:1–46. doi: 10.1016/s0079-6603(01)69043-3. [DOI] [PubMed] [Google Scholar]
- 76.Troy CM, Derossi D, Prochiantz A, Greene LA, Shelanski ML. Downregulation of Cu/Zn superoxide dismutase leads to cell death via the nitric oxide-peroxynitrite pathway. J. Neurosci. 1996;16:253–261. doi: 10.1523/JNEUROSCI.16-01-00253.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsourkas A, Behlke MA, Bao G. Structure-function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res. 2002;30:4208–4215. doi: 10.1093/nar/gkf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsourkas A, Behlke MA, Rose SD, Bao G. Hybridization kinetics and thermodynamics of molecular beacons. Nucleic Acids Res. 2003;31:1319–1330. doi: 10.1093/nar/gkg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsourkas A, Behlke MA, Xu Y, Bao G. Spectroscopic features of dual fluorescence/luminescence resonance energy-transfer molecular beacons. Anal. Chem. 2003;75:3697–3703. doi: 10.1021/ac034295l. [DOI] [PubMed] [Google Scholar]
- 80.Tsuji A, Koshimoto H, Sato Y, Hirano M, Sei-Iida Y, et al. Direct observation of specific messenger RNA in a single living cell under a fluorescence microscope. Biophys. J. 2000;78:3260–3274. doi: 10.1016/S0006-3495(00)76862-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsuji A, Sato Y, Hirano M, Suga T, Koshimoto H, et al. Development of a time-resolved fluorometric method for observing hybridization in living cells using fluorescence resonance energy transfer. Biophys. J. 2001;81:501–515. doi: 10.1016/S0006-3495(01)75717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tyagi S, Alsmadi O. Imaging native beta-actin mRNA in motile fibroblasts. Biophys. J. 2004;87:4153–4162. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 84.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 85.Tyagi S, Marras SA, Kramer FR. Wavelength-shifting molecular beacons. Nat. Biotechnol. 2000;18:1191–1196. doi: 10.1038/81192. [DOI] [PubMed] [Google Scholar]
- 86.Uchiyama CM, Zhu J, Carroll RS, Leon SP, Black PM. Differential display of messenger ribonucleic acid: a useful technique for analyzing differential gene expression in human brain tumors. Neurosurgery. 1995;37:464–469. doi: 10.1227/00006123-199509000-00014. [DOI] [PubMed] [Google Scholar]
- 87.Valencia-Burton M, McCullough RM, Cantor CR, Broude NE. RNA visualization in live bacterial cells using fluorescent protein complementation. Nat. Methods. 2007;4:421–427. doi: 10.1038/nmeth1023. [DOI] [PubMed] [Google Scholar]
- 88.van Dam RM, Quake SR. Gene expression analysis with universal n-mer arrays. Genome Res. 2002;12:145–152. doi: 10.1101/gr.198901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 90.Vet JA, Majithia AR, Marras SA, Tyagi S, Dube S, et al. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc. Natl. Acad. Sci. USA. 1999;96:6394–6399. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wadia JS, Dowdy SF. Protein transduction technology. Curr. Opin. Biotechnol. 2002;13:52–56. doi: 10.1016/s0958-1669(02)00284-7. [DOI] [PubMed] [Google Scholar]
- 92.Wadia JS, Dowdy SF. Transmembrane delivery of protein and peptide drugs by TAT-mediated transduction in the treatment of cancer. Adv. Drug Deliv. Rev. 2005;57:579–596. doi: 10.1016/j.addr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 93.Walev I, Bhakdi SC, Hofmann F, Djonder N, Valeva A, et al. Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-O. Proc. Natl. Acad. Sci. USA. 2001;98:3185–3190. doi: 10.1073/pnas.051429498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang CJ, Lin H, Tan W. Molecular assembly of superquenchers in signaling molecular interactions. J. Am. Chem. Soc. 2005;127:12772–12773. doi: 10.1021/ja053482t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang P, Beck T, Tan W. Design of a molecular beacon DNA probe with two fluorophores. Angew Chem.Int. Ed. Engl. 2001;40:402–405. doi: 10.1002/1521-3773(20010119)40:2<402::AID-ANIE402>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]