Abstract
In this work, it is shown that the incorporation of an 8-deuteroguanine (G*) moiety in DNA-oligomers allows for direct determination at 77 K of (i) the location of holes (i.e., the radical site) within dsDNA at specific base sites, even within stacks of G, as well as (ii) the protonation state of the hole at that site. These findings are based on our work and demonstrate that selective deuteration at C-8 on guanine moiety in dGuo results in an ESR signal from the guanine cation radical (G*•+) which is easily distinguishable from that of the undeuterated guanine cation radical (G•+). G*•+ is also found to be easily distinguishable from its conjugate base, the N1-deprotonated radical, G*(−H)•. Our ESR results clearly establish that at 77 K (i) one-electron oxidized guanine in double stranded DNA-oligomers exists as the deprotonated neutral radical G(−H)• as a result of facile proton transfer to the hydrogen bonded cytosine, and (ii) the hole is preferentially located at the 5′-end in several ds DNA-oligomers with a GGG sequence.
Introduction
Hole transfer through DNA has fostered intensive cross disciplinary interest as a result of both its fundamental significance and growing practical uses.1 – 4 Experimental studies employing a variety of techniques including, UV-visible excitation followed by gel electrophoresis,2 – 4 γ or X-ray irradiation followed by electron spin resonance in DNA systems including single crystals of DNA-oligonucleotides (ESR),1, 3, 5, 6 cyclic voltametry,2, 3, 7, 8 and time-resolved techniques such as, pulse radiolysis and flash photolysis 1(d), 2, 9–13,14 have established unequivocally that during charge transfer processes in double stranded DNA, guanine is the primary site of “hole” localization i.e., site of electron loss. These results are in agreement with the theoretically predicted ionization potential value of guanine in the G:C base pair system. 15 – 17
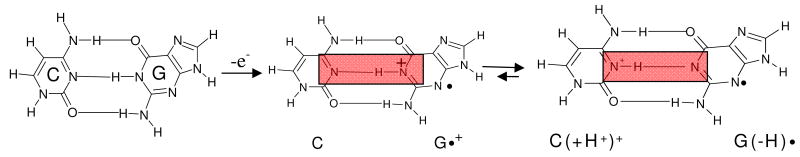
The efficiency (rate and extent) of the hole transfer processes between guanine moieties in DNA, is controlled by two important proton transfer processes: (i) the intra-base proton transfer process within the G•+:C base pair, and (ii) the proton transfer processes between the G•+:C base pair and the surrounding water. 1 – 6, 9 – 11, 16, 17 Intra-base pair proton transfer leads to separation of charge from spin (see scheme 1) and thus, these intra-base pair proton transfer processes have been proposed to slow or stop the hole transfer.1, 5, 6, 9 – 13 Such proton transfer processes will strongly depend on the pKa of the ion-radical involved.9 – 13 The pKa of a cation radical is invariably found to be lower than the parent compound. For example, the pKa of guanine (N1-H) is reported as 9.6 in aqueous solution at ambient temperature, while the corresponding pKa of the guanine cation radical (G•+) is ca. 3.9.9, 10 Based upon the pKa values of G•+ (ca. 3.9) and C(H+) (ca. 4.5), Steenken had estimated the equilibrium constant for this intra-base proton transfer process, Keq, between (C:G•+) and (C(+H+):G(−H)•) to be ca. 2.5. 9 – 11 This value of Keq suggests that at ambient temperature, the base pair (G•+:C) in DNA would have only partial proton transfer and both cation radical (30%) and deprotonated neutral radical (70%) forms would exist. There exists extensive discussion in the literature, 1 – 6, 9 – 17 regarding the protonation state of the base pair (C:G•+). Theoretical studies have treated this issue of intra-base proton transfer along the N-H bond (scheme 1) in the gas phase at the standard state (298 K, 1 atm) and have shown that ΔS (proton transfer) = ca. 0 cal/mol/K, and ΔG (proton transfer) = 1.4 kcal/mol. 15 – 17 However, until now, no definitive experiments had been performed to show the evidence of intra-base proton transfer in the C:G•+ base pair as shown in scheme 1.
Scheme 1.
Schematic representation of prototropic equilibria in the G•+:C pair
It is well-established that GG and GGG sites are more easily oxidized than isolated guanine sites.2 – 4 Thus, during oxidative hole transfer processes in DNA, GG and GGG sites are hot spots for localization of the hole.2 – 4 A number of experimental, and theoretical studies suggest that there is a marked preference for hole localization at the 5′-G site in GG and GGG sequences.2 – 4, 18 – 24 In GGG sequences, there was evidence for a small extent of hole localization at the central G. 2 – 4, 18 – 24 However, these experimental studies were carried out in aqueous solution at ambient temperature. Therefore, the final site for the hole localization was determined after the temperature-dependent hopping of the hole and subsequent reactions at sites where the hole is stabilized, e.g., via the nucleophilic addition of H2O at C8 in the guanine moiety followed by an one-electron oxidation leading to the formation of 8-oxo-guanine (8-oxo-G) moieties. 1(e), 5(b)–(e), 6, 25b On the basis of nuclear reorganization energy (NRE) values, it can be theoretically expected, in part, that the hole could be preferentially located to a single guanine in a stacked guanine environment. The formation G•+ in the G:C base pair has been shown to have a higher NRE value (0.34 eV) than that for the formation of A•+ in A:T (0.10 eV). 15 – 17, 26 Consequently, the G•+:C base pair has a significantly different structure than the neutral G:C; while, the A•+:T base pair has a near identical structure to neutral A:T. Therefore, guanine stacks are predicted to readily undergo reorganization and localize the charge (i.e., hole) to a single G; whereas, adenine stacks would tend to delocalize the hole. Of course, solvent polarization effects and the effects of surrounding ions limit the extent of delocalization within the stack. 23, 26 – 28 However, there are no direct experimental results identifying the actual location of the hole in stretches of guanine in a ds DNA oligomer prior to the temperature-dependent hopping and subsequent reactions e.g., 8-oxo-G formation, while recent work in our laboratory has given evidence for hole delocalization in A stacks even at low temperature range (77 – 154 K) 26 where the temperature-dependent hopping of the hole does not occur.25
Thus, in stacked environments, there are two very important questions regarding oxidative hole formation in DNA prior to any subsequent reactions: (i) What is the protonation state of the hole? (ii) Where is the site of radical localization in (G)n sequences?
We have recently found that selective incorporation of deuterium (D) at C-8 on guanine in dGuo [8-D-dGuo = G*] reduces the C-8-H proton hyperfine coupling of ca. 8 G to the C-8-D coupling of ca. 1.2 G thereby narrowing the spectral width and improving spectral resolution.29, 30 Owing to the collapse of the anisotropic C-8-H hyperfine coupling, the ESR signal from the 8-D-dGuo cation radical (G*•+), became easily distinguishable from that of the cation radical (G•+) in the undeuterated dGuo.29, 30 Furthermore, the ESR spectrum of G*•+ is also distinguishable from the N1-deprotonated neutral radical, G*(−H)•, owing to their different lineshape, linewidth, and g⊥ values.29 Hence, in this work, we incorporate 8-D-dGuo (G*) into sequences of ss and ds DNA (e.g., TG*T, and d[G*CG*CG*CG*C]2), and based on their ESR spectra, directly provide evidence of the protonation state of the hole at specifically labeled guanine sites in these sequences. Moreover, owing to clear and observable difference in the ESR spectra of G*•+ and G•+ as well as that of G*(−H)• and G(−H)•, we find that a hole on the G* is easily distinguishable from a hole on G. Thus, we demonstrate that this novel method provides for the direct determination of the location of holes within dsDNA at specific guanine sites, even within stacks of G at 77 K (i.e., before the temperature-dependent hopping of the hole) as well as their state of protonation.
Materials and methods
Compounds
2′-Deoxyguanosine (dGuo) and lithium chloride (99% anhydrous, SigmaUltra) were obtained from Sigma Chemical Company (St Louis, MO). Potassium persulfate (crystal) was purchased Mallinckrodt, Inc. (Paris, KY). 8-D-dGuo (G*) 29, 30 was synthesized according to the procedure of Huang et al. 31(a) by dissolving dGuo in D2O in the presence of triethylamine (TEA) (Fischer Scientific, NJ) and by warming the solution at 55 – 60° C for 16 h. Employing a Bruker 200 MHz NMR, in G*, the degree of deuteration was found to be as >96% by 1D signal integration.
The phosphoramidite of G* was synthesized by Glen Research, VA. Starting with this phosphoramidite of G*, the ds DNA sequences d[G*CG*CG*CG*C]2, d[G*GGCCC]2, d[GG*GCCC]2, d[GGG*CCC]2, were synthesized (Midland Certified, TX) and desalted and dried for this work. Moreover, again employing the phosphoramidite of G*, the ds DNA oligomers d[TG*GGCCCA]2, d[TGG*GCCCA]2, d[TGGG*CCCA]2 were synthesized, desalted, HPLC-purified, and dried (Midland Certified, TX). The unlabeled ds DNA oligomers, viz. d[GCGCGCGC]2 and d[GGGCCC]2 which were desalted and dried, were also obtained from Midland Certified, TX,. All the compounds were used without any further purification. Our analyses showed (see supporting information Figure S1) that the extent of G* incorporation in each of the DNA-oligomers which were not HPLC-purified, e.g., TG*T, d[G*CG*CG*CG*C]2, d[G*GGCCC]2, d[GG*GCCC]2, and d[GGG*CCC]2 – were found to be ca. 80%. On the other hand, HPLC-purified oligos were found to retain ca. 50% G* instead of ca. 80% (see supporting information Figure S1). These values were accounted for in the analyses of the ESR spectra.
Preparation of solutions of dGuo and G*
Following our earlier works with DNA-nucleosides and nucleotides, the solutions of dGuo and G* were prepared by dissolving ca. 3 mg of the compound in 1 mL of 7.5 M LiCl in D2O along with ca. 5 mg K2S2O8 which serves as an electron scavenger 25, 26 thereby eliminating the probability of formation of anion radicals in our system.
Preparation of solution of DNA-oligomers
As per our works with DNA-oligomers,1e, 26, 33, 34 about 1.5 mg of each DNA-oligomer was dissolved in 0.35 mL of 7.5 M LiCl in D2O at room temperature and the solutions were incubated for denaturation at 60 – 70°C for 10 min. Subsequently, these solutions were slowly annealed to room temperature for re-naturation and ca. 2.5 – 3 mg K2S2O8 was added to each solution as an electron scavenger.
pD adjustments
Following our previous work29, 30 with dGuo and G* and, with dAdo,26, 32 pH/pDs of the solutions of dGuo and G* have been adjusted by quick addition of adequate amount (in μL) of 1 M NaOH or 1 M HCl in D2O under ice-cooled condition. pH papers were employed for pD measurements of these solutions.
In our experimental system (i.e., in homogenous aqueous glasses at low temperature), it has been observed that radicals show similar properties as found in aqueous solutions at room temperature e.g., the pKa of G•+ (dGuo) has been found to be ca. 5 in aqueous glasses at 150 K 1e, 29, 30 and this value is just ca. 1 to 1.5 units higher than the pKa value of 3.9 of G•+ in aqueous solution at room temperature. 9 – 11 This modest difference in the pKa value of G•+ results likely from the high ionic strength and low temperature for these glassy systems.
The dsDNA-oligomers in homogenous aqueous glasses at low temperature are reported to be in the B-conformation.31(b)
Glassy sample preparation
According to our previous works, homogenous solutions (7.5 M LiCl D2O) of dGuo, G*, and of the DNA-oligomers were thoroughly bubbled with nitrogen to remove oxygen. 25, 26, 29, 30, 32 – 34 Transparent glassy samples were then prepared drawing the solution into 4 mm Suprasil quartz tubes (cat. no. 734-PQ-8, WILMAD Glass Co., Inc., Buena, NJ) followed by cooling to 77 K. We note that these samples at low temperatures are not crystalline solids but are glassy homogeneous super-cooled solutions – which, upon annealing, soften to allow for molecular migration and solution phase chemistry. 25, 26, 29, 30, 32 – 34 All of these glassy samples are stored at 77 K in Teflon containers in the dark.
γ-Irradiation and storage of irradiated samples
The Teflon containers containing the glassy samples of dGuo, G*, and of DNA-oligomers were placed in a 400 ml Styrofoam dewar under liquid nitrogen (77 K). We have performed γ-irradiation of these glassy samples with the help of a model 109-GR 9 irradiator containing a shielded 60Co source. Following our work,1e glassy samples of dGuo, G*, and of all the oligonucleotides containing number of bases up to 4 were γ-irradiated (60Co) with an absorbed dose of 2.5 kGy at 77 K. For glassy samples of oligonucleotides containing number of bases higher than 4, the absorbed dose was 5 kGy at 77 K. 34 These γ-irradiated samples were stored at 77 K in the dark.
Formation of one-electron oxidized dGuo, G*, and DNA-oligomers via thermal annealing and storage of these samples
Following our previous works, the one-electron oxidized dGuo and the one-electron oxidized DNA-oligomers were produced via annealing in the dark in a variable temperature assembly (Air Products) in the temperature range 100 – 160 K employing cooled nitrogen gas. 34 We have annealed samples for 15 – 30 min in the dark at 155±2 K which softens the aqueous (D2O) glass facilitating molecular migration of Cl2•-. Cl2•- one-electron oxidizes the solute with concomitant decrease of the ESR spectrum of Cl2•- with simultaneous increase in ESR spectrum of one-electron oxidized guanine moiety in dGuo, G* or DNA-oligomers. 1e, 25, 26, 29, 30, 32 – 34 After annealing, we have immersed these samples immediately in liquid nitrogen (77 K) and have stored in Teflon containers at 77 K in the dark before carrying out ESR studies.
Electron Spin Resonance
The ESR spectra of these γ-irradiated and subsequently annealed samples were recorded at 77 K and at 40 dB (20 μW) using a Varian Century Series ESR spectrometer operating at 9.2 GHz with an E-4531 dual cavity, 9-inch magnet and with a 200 mW klystron. For field calibration, we have used Fremy's salt (g (center of the spectrum) = 2.0056, AN = 13.09 G) as per our previous works. 1e, 25, 26, 29, 30, 32 – 34
Analyses of ESR spectra
Following our previous work, 26 each spectrum has been stored in a 1000-point array with field calibration marks from the 3 ESR lines of the Fremy's salt. We have estimated the fraction that a particular radical contributes to the overall ESR spectrum by employing doubly integrated areas of first derivative ESR spectra. The fractional contribution of the radicals in each experimental ESR were obtained by use of programs developed in our laboratory (ESRADSUB, ESRPLAY) by fitting benchmark spectra to ESR signal of interest via linear least squares analysis. 25, 26, 34 In these analyses an error function (R) is employed as a test for goodness of fit from the sum of the differences between the squares in heights of the experimentally obtained spectrum and that of the simulated spectrum obtained employing the benchmark spectra (R = [{ (ΣYexp(i))2 } − { (ΣYsim(i))2 }]). To estimate the error limits in our analyses, nonoptimized fits were generated and the value of the error (R) was plotted against the fraction of the contributing radical benchmark spectra (see supporting information Figure S2 as an example). On this basis, we have estimated the error limit of our analyses as ±10%.
Following our previous work, we have subtracted a small singlet “spike” from irradiated quartz at g = 2.0006 from the recorded spectra before analyses.
Results and Discussion
The organization of this paper is as follows: (i) We first present evidence that the ESR spectra of pairs, G•+ vs. G*•+, G(−H)• vs. G*(−H)•, and G*•+ vs. G*(−H)• are each distinguishable from each other in nucleotides. (ii) We then show these differences in ESR spectra allow the determination of the protonation state of one-electron oxidized guanine in ss and ds DNA-oligomers. (iii) Finally, recognizing that the hole in ds DNA is in the form of G(−H)•, we then use the spectra of G(−H)• and G*(−H)• obtained from the ds DNA-oligomers as benchmarks to analyze the spectra of the ds DNA-oligomers containing GGG sequences and assign the position of the hole in these oligomers.
Distinction between (G•+ and G*•+) and (G(−H)• and G*(−H)•) in nucleosides
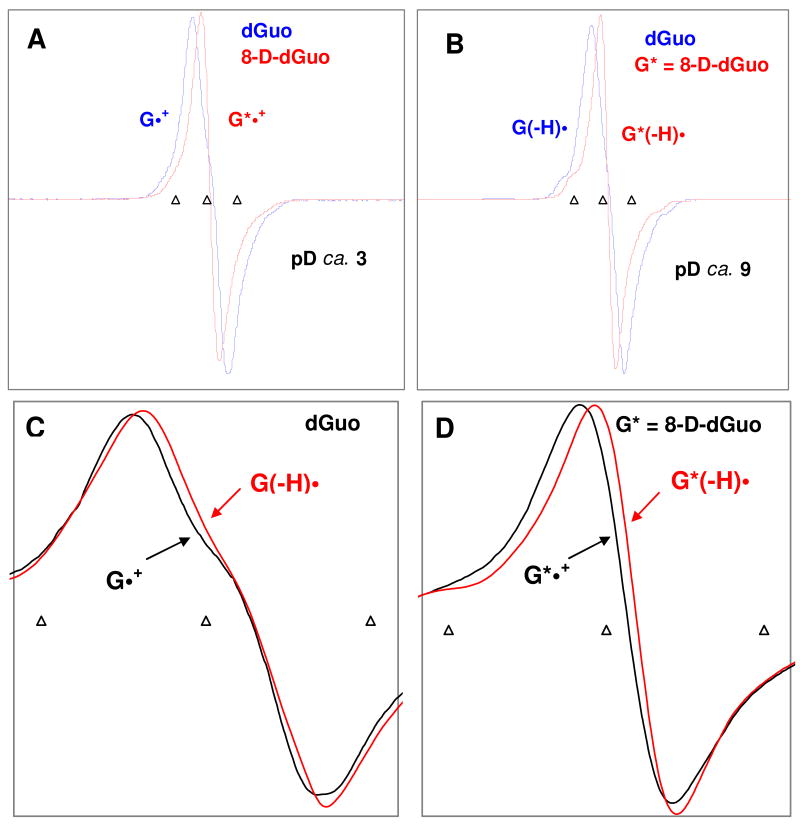
Following the procedure of Huang et al.,31(a) we have synthesized 8-D-dGuo (G*) in our laboratory.29, 30 The radical species, G*•+ and G*(−H)•, have been produced from G* in aqueous glassy samples (7.5 M LiCl/D2O) via one-electron oxidation of Cl2•- at pDs ca. 3 and 9 respectively. 29, 30 The ESR spectra in Figure 1A show a comparison of the guanine cation radical for dGuo, G•+(blue), with that of the cation radical of 8-D-dGuo, G*•+(red), while Figure 1B shows a comparison for the corresponding deprotonated species G(−H)• or G*(−H)• in D2O.
Figure 1.
ESR spectra found for (A) guanine cation radicals (G•+, G*•+) and (B) deprotonated guanine neutral radicals (G(−H)•, G*(−H)•). These radicals were produced by Cl2•– oxidation of dGuo (3 mg/mL) (blue) and of G* (3 mg/mL) [G* = 8-D-dGuo, 96%D] (red) at pDs ca. 3 and ca. 9 in 7.5 M LiCl glasses in D2O in the presence of K2S2O8 (5 mg/mL) as an electron scavenger. 29, 30 The one-electron oxidation by Cl2•- was carried out via thermal annealing at 155 K in the dark. Figures (A) and (B) clearly show that deuteration at C8, replaces the C-8-H proton hyperfine coupling from ca. 8 G in G•+ and in its N1-deprotonated neutral radical, G(−H)• with a ca. 1.2 G (C-8-D coupling) thereby narrowing the ESR spectral width and improving the spectral resolution in the deuterated species, G*•+ and G*(−H)•. Figure (C) shows that, in dGuo, G•+ (black) is not clearly distinguishable from G(−H)•; whereas Figure (D) represents that, in G*, owing to the smaller C-8-D coupling, G*•+ (black) is readily distinguishable from its N1-deprotonated neutral radical, G*(−H)• (red), because of their different lineshapes, linewidths and g⊥ values. All spectra were recorded at 77 K. The three triangular calibration markers in this figure and in the other figures represent positions of Fremy's salt resonances (the central marker is at g = 2.0056, and each of three markers is separated from one another by 13.09 G).
The comparisons clearly demonstrate that reduction of the dGuo C-8-H proton hyperfine coupling from ca. 8 G in G•+ and G(−H)• to ca. 1.2 G (C-8-D coupling) on deuteration narrows the ESR spectral width and improves spectral resolution in the deuterated species, G*•+ and G*(−H)• in accord with our previous work.26, 29, 30 It is evident from Figures 1A and B that, owing to the substantially smaller C-8-D hyperfine couplings, the ESR signal from G*•+ becomes easily distinguishable from that of G•+. Further, as a result of the C8-H proton hyperfine coupling, the ESR spectrum of G•+ in dGuo is not clearly distinguishable from that of its N1-deprotonated neutral radical, G(−H)• (see Figure 1C); whereas, the spectrum of G*•+ is clearly distinguishable from its N1-deprotonated neutral radical, G*(−H)• because of different lineshapes, linewidths and g⊥ values (2.0045 (G*•+) vs. 2.0041 (G*(−H)•))29 (see Figure 1D). These differences in the ESR spectra of one-electron oxidized guanine monomers, formed the basis of our ESR spectral studies by employing G* incorporated into DNA-oligomers to elucidate the protonation state of the hole and its localization.
Protonation state of G•+ in ss and in ds DNA oligomers
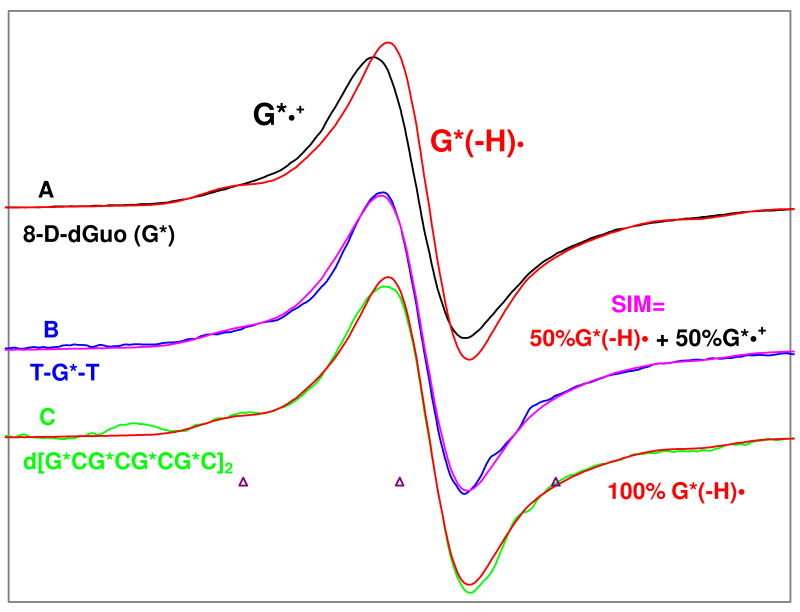
The ESR spectra of G*•+ (black) and its N1-deprotonated radical, G*(−H)• (red) both from the nucleoside, dGuo, from Figure 1D are repeated in Figure 2A and are employed as benchmarks for the analyses of the subsequent spectra of ssDNA-oligomer (Figure 2B) and dsDNA-oligomer (Figure 2C).
Figure 2.
ESR spectra of: (A) G*•+ (black) and G*(−H)• (red) obtained from glassy (7.5 M LiCl in D2O) samples of G* [G* = 8-D-dGuo, 96% D] (3 mg/mL) 29, 30. (B) one-electron oxidized ss oligomer, TG*T, (4.5 mg/mL) in blue after subtraction of 20% of the G(−H)• spectrum (to account for 80% 8-deuteration level (see supporting information Figure S1). In Figure B, the match between the experimentally observed spectrum of one-electron oxidized TG*T (blue) with that of the simulated (pink) spectrum obtained using the spectra of G*•+ and G*(−H)• shown in (A) as benchmarks indicate that for one-electron oxidized TG*T at pD 5, G*•+ and G*(−H)• are present in equal amounts. (C) Spectrum of the one-electron oxidized ds DNA oligomer d[G*CG*CG*CG*C]2 (2 mg/mL) after subtraction of 20% of the G(−H)• spectrum (to account for 80% 8-deuteration level (see supporting information Figure S1), again with G*(−H)• from A in red superimposed. The match of green and red spectra in (C) clearly shows that, one-electron oxidized guanine in ds DNA oligomer d[G*CG*CG*CG*C]2 exists as G*(−H)•. One-electron oxidation of the monomer and the DNA-oligomers were carried out via thermal annealing at 155 K. All spectra were recorded at 77 K. Lineshape and g value differences between G*•+ and G*(− H)• allow for this analysis.
In Figures 2B and 2C, we present the ESR spectra of one-electron-oxidized DNA-oligomers TG*T (blue) (a model for ss DNA) and d[G*CG*CG*CG*C]2 (green) (a model for ds DNA) respectively at pD ca. 5 in 7.5 M LiCl in D2O after reaction with Cl2•-. One-electron oxidation of the DNA-oligomers by Cl2•- was carried out via annealing the glassy aqueous samples in the dark at ca.155 K.
In Figure 2B, the ESR spectrum (blue) of one-electron oxidized TG*T at the native pD (ca. 5) of 7.5 M LiCl glass/D2O after accounting for the fact that TG*T had incorporation of G* at an 80% level (see supporting information Figure S1) is presented. Over this spectrum, we superimpose the best fit spectrum (violet) consisting of 50% G*•+ and 50% G*(−H)• weighting of double integrated spectra (Figure 2A). The fit shows that one-electron oxidized TG*T consists of both G*•+ and G*(−H)• in near equal amounts. The supporting information Figure (Figure S2) gives evidence that the estimates have a 10% uncertainty.
One-electron oxidized d[G*CG*CG*CG*C]2 results in an ESR spectrum (Figure 2C) that is identical with g⊥ values, and anisotropic N-couplings as the spectrum for G*(−H)• in 8-D-dGuo after accounting for the fact that the deuteration in the DNA-oligomer d[G*CG*CG*CG*C]2 was at an 80% level (see supporting information Figure S1). Thus the spectrum shown in Figure 2C is the spectrum obtained after subtraction of 20% of the G(−H)• spectrum. This result clearly indicates that one-electron oxidized G in this ds DNA environment exists as G(−H)•.
Thus, these results show that we are able to distinguish the protonation states of one-electron oxidized G in both ss and dsDNA based on their ESR spectra alone. In ss DNA, the one-electron oxidized guanine exists as an equal mixture of G•+ and G(−H)•. In ds DNA, the base pair (G•+:C) in DNA exist as G(−H)• or, more accurately, as C(+H+):G(−H)• (Scheme 1).
Site of hole localization at G in a ds DNA-oligomer
Employing ESR spectral studies, our aim is to directly identify the site of hole localization in a series of ds DNA-oligomers of various sequences. As discussed there are no reported direct experimental results identifying the precise location of the hole (or, the radical site) in GGG containing DNA oligos. Here, ESR work is presented in Figure 3 that show the site of hole localization can be identified in such ds DNA-oligomers.
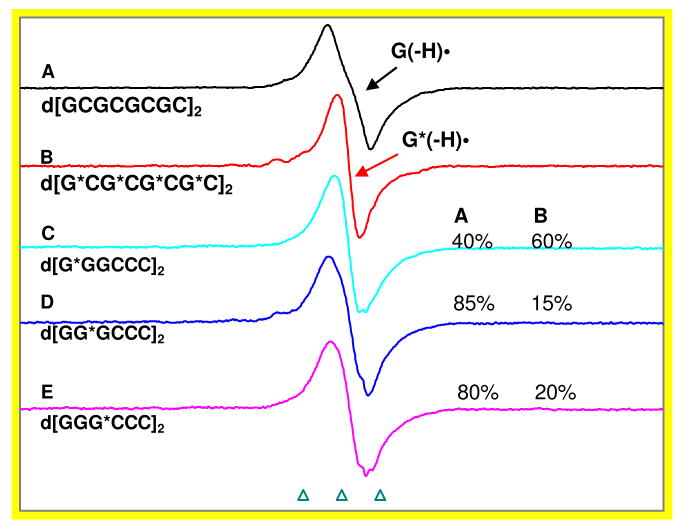
Figure 3.
Spectra A and B are the benchmark spectra of G(−H)• and G*(−H)• (G* = 8-D-dGuo). These spectra are for the glassy samples of d[GCGCGCGC]2 and d[G*CG*CG*CG*C]2 and are assigned to G(−H)• and G*(−H)•. Note that the spectrum in B is 80% 8-deuterated. This spectrum is used as the basis function as it self corrects in our analysis. Spectra C to E are the one-electron oxidized d[GGGCCC]2 with G* moieties at 5′- end, middle and at the 3′-end of the G stack respectively. Analyses using G(−H)• and G*(−H)• spectra show that the site of hole localization is approximately 60% at the 5′-end. We have expressed the radical percentages to ±10% relative error (see materials and methods, and supporting information Figure S2). All the spectra were recorded at 77 K and the spectra of one-electron oxidized oligomers shown in Figures (A) to (E) were obtained by thermal annealing in the dark at 155 K for 20 – 30 min.
We have already assigned the spectrum of one-electron oxidized d[G*CG*CG*CG*C]2 to G*(−H)• (see supporting information Figure S1A and Figure 2C which was corrected for the 80% deuteration level, Figure 3B shows the uncorrected spectrum corresponding to Figure 2C). We also assign the spectrum (Figure 3A) of the one-electron oxidized ds DNA-oligomer d[GCGCGCGC]2 to G(−H)•. This assignment is clearly established by the very close similarities in spectral parameters (overall hyperfine splitting, anisotropic nitrogen hyperfine couplings, lineshape, and the center of the spectrum) in one-electron oxidized d[GCGCGCGC]2 and G(−H)• spectrum29 from dGuo (see supporting information Figure S3 for evidence). We also find that the ESR spectra of one-electron oxidized ds DNA-oligomers d[GCGCGCGC]2 and d[GGGCCC]2 are identical (see supporting information Figure S4). Thus, it can be concluded that irrespective of the occurrence of the guanine moiety in the ds DNA sequence as continuous stretch (e.g., GGG) or as a single G unit, one-electron oxidation of guanine moiety in ds DNA would lead to G(−H)• formation. Figure 3A and B, therefore, show ESR spectra of G(−H)• and G*(−H)• spectra obtained in the glassy aqueous samples of d[GCGCGCGC]2 and d[G*CG*CG*CG*C]2 respectively. These spectra are used below as benchmarks for G(−H)• and G*(−H)• to analyze the fractional compositions of G(−H)• and G*(−H)• in the ESR spectra shown in Figures 3C to E.
In Figures 3C to E, we show the ESR spectra of one-electron oxidized d[GGGCCC]2 with G* moieties at the 5′- end, in the middle, and at the 3′-end of the GGG sequence.
Analyses of spectra in Figure 3C to 3E shown in Table 1 demonstrate that the hole localizes mainly at the 5′-G end (ca. 60%) with lesser amounts at the middle and at the 3′-end (ca. 15 – 20 %).
Table 1. Hole localization on guanine as G(-H)• in one-electron oxidized d[GGGCCC]2, and d[TGGGCCCA]2 by selective substitution of 8-D-dGuo (i.e., G*).
| Ds DNA oligomer | Radical percentages | ||
|---|---|---|---|
| Nature | Name | G*(−H)• | G(−H)• |
| 5′-T and 3′-A | d[TG*GGCCCA]2 | 55 | 45 |
| d[TGG*GCCCA]2 | 30 | 70 | |
| d[TGGG*CCCA]2 | 20 | 80 | |
| 5′-OH and 3′-OH | d[G*GGCCC]2 | 60 | 40 |
| d[GG*GCCC]2 | 15 | 85 | |
| d[GGG*CCC]2 | 20 | 80 | |
We note here that the percentages of G(−H)• and G*(−H)• (see Table 1, and Figure 3) are obtained independently and separately for each DNA-oligomer. Thus the hole localization on G* in each of the three positions in GGG (G*GG, GG*G and GGG*) give independent assessments of the distribution. The fact that the totals of G*(−H)• on the three sites vary from 95 to 105% (vs. 100% expected) gives some confidence in the method.
One-electron oxidation of the isotopically substituted ds DNA oligomers viz. d[TG*GGCCCA]2, d[TGG*GCCCA]2, and d[TGGG*CCCA]2 that were HPLC purified, were carried out as well. The ESR spectra of these one-electron oxidized ds DNA oligomers were recorded at 77 K and these spectra were found to be very similar to the corresponding ESR spectra shown in Figures in 3(C) to (E) (see supporting information Figure S5). Using the G*(-H)• spectrum from HPLC-purified d[G*CG*CG*CG*C]2 and G(-H)• spectrum from d[GCGCGCGC]2 as benchmarks, the analyses of radical percentages in these oligomers shows a clear preference for localization of the hole at the 5′-G end (see Table 1). Note that owing to the further HPLC purification the level of deuteration dropped to 50% for these oligos which was accounted for in our analyses (see supporting information Figure S1). We also note here that by employing G*(-H)• spectrum from G* (96% deuterated) and G(-H)• spectrum from dGuo as benchmarks, we have obtained percentages of G*(-H)• and G(-H)• in these ds DNA-oligomers (see supporting information) and these percentages are found to be very similar to those shown in Table 1.
Therefore, the radical percentages of G(−H)• and G*(−H)• in the GGG sequence are similar for ds DNA-oligomers with and without single nucleotide extension at the ends, 5′-T and 3′-A (see Table 1). As a result, we conclude that the presence of 5′-T and 3′-A units in the ds DNA oligomers with GGG stretches did not alter the preferential location of the hole at the 5′-G site in these sequences.
Conclusion
(i) Sites of hole localization and protonation states of one-electron oxidized guanine in DNA-oligomers
Our work directly finds that the hole is preferentially localized at the 5′-end in GGG sequences at 77 K. Our results also show that the hole is not fully localized to the 5′ G site. Previous experimental results obtained in aqueous solution at room temperature 2 – 4, 13b,c, 18–21 had previously suggested that in GGG sequences the 5′-G would likely be the preferential site of hole localization. In those experiments, it was the preferential formation of 8-oxo-G at the 5′-end of the GGG sequences in ds DNA-oligomers after photo-oxidation that suggested the preferential hole localization at the 5′-end. 18 – 21 In agreement theoretical calculations point out that in a GGG sequence, the 5′-end is the most stable site.22,24 Further, owing to significant NRE values,15 – 17 guanine stacks are not predicted to share charge and are expected to readily undergo reorganization so as to localize the charge (i.e., hole) on a single G. Our experiments show no evidence for delocalization of charge as it would be expected to markedly alter the ESR spectra. Any comparison of 77K hole localization with 8-oxo-G formation of course depends on the kinetics of hole conversion to 8-oxo-G formation being independent of the base sequence. Experiments show no such dependence of 8-oxo-G formation on the base sequence.35 As a consequence, the agreement between our results regarding sites of hole localization at 77 K with those results 18 – 21 found at room temperature for the distribution of the formation of 8-oxo-G is reasonable.
(ii) Protonation State of one-electron oxidized guanine in DNA-oligomers
We find that one-electron oxidized guanine is entirely the proton transferred radical (C(+H+):G(−H)•) at 77 K in ds DNA. At ambient temperatures, a prototropic equilibrium between G•+:C and C(+H+):G(−H)• would be established that would still favor the proton transferred species, as it is the thermodynamically stable state (see Introduction). This equilibrium has been proposed by a number of workers as a gating switch to hole transfer along the DNA strand.1 – 6 The presence of G•+:C would also allow for nucleophilic addition of H2O that leads to the formation of 8-oxo-G. It is well established that one-electron oxidized guanine must remain in its cation radical form to undergo water addition.5(b) – (e),25 Thus, Steenken's proposition9 – 11 that (C(+H+):G(−H)•) would be the dominant species at ambient temperature is supported by our work.
(iii) Prototropic equilibrium of one-electron oxidized guanine in DNA-oligomers
Previous theoretical calculations carried out for the base pair (G•+:C) in the gas phase at standard state (298 K, 1.0 atm), 15 – 17 predict that ΔS (proton transfer) = ca. 0 cal/mol/K, and ΔG (proton transfer) = 1.4 kcal/mol. However, our ESR studies at low temperature clearly demonstrate that the base pair (G•+:C) exists as (C(+H+):G(−H)•) i.e., the free energy, ΔG, for proton transfer within the base pair (G•+:C) is negative. This disagreement between our experimental results and previous theoretical work 15, 16 is small in magnitude and could easily arise from the neglect of stacking and water interactions in the gas phase calculations. 26, 28
(iv) Why isn't the hole fully localized at the 5′-end?
It is clear from the results reported here, and others that the 5′-end is the dominant site for hole localization, 18 – 24 although a significant fraction of the holes are also localized at the middle and 3′- end (Table 1). This indicates that the energetic preference for the 5′-end is small and that variations in the local environment such as counter ion placement or structural variations36 are sufficient to stabilize the hole at the other sites. We note that there is no evidence for delocalization of the hole in the GGG stacks. This is in contrast to stacks of A for which some delocalization of hole is theoretically predicted via charge-resonance interaction.26 This difference in the behavior of holes in guanine vs. adenine stacks has been theoretically attributed to the lack of significant nuclear reorganization on formation of the adenine cation radical.16, 26 This allows holes in A stacks to be shared whereas in G stacks the larger reorganization energy causes localization to a single base.
(v) Advantage of the technique
The most important feature of incorporation of G* (8-deuteroG) in oligos is the clear resolution of the ESR spectra of one-electron oxidized ds DNA oligomers with G* at specific sites in GGG. Thus we can, not only identify directly the protonation state of the G moiety, but also the site of localization of the hole in DNA sequences having stretches of Gs.
Supplementary Material
Supporting Information available: Figure S1: Analyses for deuteration levels in the G* labeled ds DNA-oligomers. Figure S2: Estimation of relative error regarding fitting of the experimentally observed spectrum using benchmarks. Figure S3: ESR spectrum of the guanine neutral radical (G(−H)•) from dGuo and d[GCGCGCGC]2. Figure S4: ESR Spectra of the one-electron oxidized dsDNA-oligomers: d[GGGCCC]2 and d[GCGCGCGC]2. Figure S5: ESR Spectra of the one-electron oxidized ds DNA-oligomers with and without single nucleotide extension at the ends, 5′-T and 3′-A. Analyses: Analyses of the ESR spectra of ds DNA-oliomers shown in Figure S5 using guanine neutral radical (G(−H)•) from dGuo and guanine neutral radical (G*(−H)•) from G* [G* = 8-D-dGuo, 96% deuterated]
Acknowledgments
This work was supported by the NIH NCI under grant no. R01CA045424. A. A. thanks Dr. Hugh Mackie (Glen Research, VA) for synthesis of the phosphoramidite of 8-D-dGuo. The authors thank Prof. A. W. Bull for critical review and helpful suggestions.
References
- 1.Bernhard WA. Adv Radiat Biol. 1981;9:199–280.Becker D, Sevilla MD. Adv Radiat Biol. 1993;17:121–180.Ward JF. Cold Spring Harb Symp Quant Biol. 2000;65:377–382. doi: 10.1101/sqb.2000.65.377. and references therein. von Sonntag C. Free-radical-induced DNA Damage and Its Repair. Springer-Verlag; Berlin, Heidelberg: 2006. pp. 335–447.Becker D, Adhikary A, Sevilla MD. In: Charge Migration in DNA: Physics, Chemistry and Biology Perspectives. Chakraborty T, editor. Springer-Verlag; Berlin, Heidelberg, New York: 2007. pp. 139–175.
- 2.O'Neill MA, Barton JK. In: Charge Transfer in DNA: From Mechanism to Application. Wagenknecht HA, editor. Wiley-VCH Verlag GmbH & Co KGaA; Weiheim: 2005. pp. 27–75. [Google Scholar]
- 3.Schuster GB, editor. Topics in Current Chemistry. Springer-Verlag; Berlin, Heidelberg: 2004. Long Range Charge Transfer in DNA. I and II. [Google Scholar]
- 4.Giese B. Ann Rev Biochem. 2002;71:51–70. doi: 10.1146/annurev.biochem.71.083101.134037. [DOI] [PubMed] [Google Scholar]
- 5.(a) Sevilla MD, Becker D, Yan M, Summerfield SR. J Phys Chem. 1991;95:3409–3415. [Google Scholar]; (b) Spalletta RA, Bernhard WA. Radiation Research. 1992;130:7–14. [PubMed] [Google Scholar]; (c) Weiland B, Hüttermann J. Int J Radiat Biol. 1998;74:341–358. doi: 10.1080/095530098141483. [DOI] [PubMed] [Google Scholar]; (d) Weiland B, Hüttermann J. Int J Radiat Biol. 1999;75:1169–1175. doi: 10.1080/095530099139647. [DOI] [PubMed] [Google Scholar]; (e) Debije MG, Bernhard WA. J Phys Chem B. 2000;104:7845–7851. [Google Scholar]
- 6.(a) Bernhard WA, Close DM. In: Charged Particle and Photon Interactions with Matter Chemical, Physicochemical and Biological Consequences with Applications. Mozumdar A, Hatano Y, editors. Marcel Dekkar, Inc.; New York, Basel: 2004. pp. 431–470. [Google Scholar]; (b) Close DM. In: Radiation Induced Molecular Phenomena in Nucleic Acid: A Comprehensive Theoretical and Experimental Analysis. Shukla MK, Leszczynski J, editors. Springer-Verlag; Berlin, Heidelberg, New York: 2008. pp. 493–529. [Google Scholar]
- 7.Kelley SO, Boon EM, Barton JK, Jackson NM, Hill MG. Nucleic Acids Res. 1999;27:4830–4837. doi: 10.1093/nar/27.24.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson NM, Hill MG. Current Opinion Chem Biol. 2001;5:209–215. doi: 10.1016/s1367-5931(00)00192-7. [DOI] [PubMed] [Google Scholar]
- 9.Steenken S. Chem Rev. 1989;89:503–520. [Google Scholar]
- 10.Steenken S. Free Radical Res Commun. 1992;16:349–379. doi: 10.3109/10715769209049187. [DOI] [PubMed] [Google Scholar]
- 11.Steenken S. Biol Chem. 1997;378:1293–1297. [PubMed] [Google Scholar]
- 12.Kobayashi K, Tagawa S. J Am Chem Soc. 2003;125:10213–10218. doi: 10.1021/ja036211w. (b) [DOI] [PubMed] [Google Scholar]
- 13.(a) Kobayashi K, Yamagami R, Tagawa S. J Phys Chem B. 2008;112:10752–10757. doi: 10.1021/jp804005t. [DOI] [PubMed] [Google Scholar]; (b) Lee YA, Durandin A, Dedon PC, Geacintov NE, Shafirovich V. J Phys Chem B. 2008;112:1834–1844. doi: 10.1021/jp076777x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Shafirovich V, Dourandin A, Geacintov NE. J Phys Chem B. 2001;105:8431–8435. [Google Scholar]
- 14.Stemp EDA, Arkin MR, Barton JK. J Am Chem Soc. 1997;119:2921–2925. [Google Scholar]
- 15.(a) Bertran J, Oliva A, Rodriguez-Santiego L, Sodupe M. J Am Chem Soc. 1998;120:8159–8167. [Google Scholar]; (b) Hutter M, Clark T. J Am Chem Soc. 1996;118:7574. [Google Scholar]
- 16.Li XF, Sevilla MD. Adv Quantum Chem. 2007;52:59–87. [Google Scholar]
- 17.Kumar A, Sevilla MD. In: Radiation Induced Molecular Phenomena in Nucleic Acid: A Comprehensive Theoretical and Experimental Analysis. Shukla MK, Leszczynski J, editors. Springer-Verlag; Berlin, Heidelberg, New York: 2008. pp. 577–617. [Google Scholar]
- 18.Hall DB, Holmlin RE, Barton JK. Nature. 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 19.Arkin MR, Stemp EDA, Pulver SC, Barton JK. Chem Biol. 1997;4:389–400. doi: 10.1016/s1074-5521(97)90129-0. [DOI] [PubMed] [Google Scholar]
- 20.(a) Gasper SM, Schuster GB. J Am Chem Soc. 1997;119:12762–12771. [Google Scholar]; (b) Liu CS, Schuster GB. J Am Chem Soc. 2003;125:6098–6102. doi: 10.1021/ja029333h. [DOI] [PubMed] [Google Scholar]
- 21.Núñez ME, Hall DB, Barton JK. Chem Biol. 1999;6:85–97. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 22.Saito I, Nakamura T, Nakatani K, Yoshioka Y, Yamaguchi K, Sugiyama H. J Am Chem Soc. 1998;120:12686–12687. [Google Scholar]
- 23.Conwell EM, Basko DM. J Am Chem Soc. 2001;123:11441–11445. doi: 10.1021/ja015947v. [DOI] [PubMed] [Google Scholar]
- 24.Senthilkumar K, Grozema FC, Guerra CF, Bickelhaupt FM, Siebbeles LDA. J Am Chem Soc. 2003;125:13658–13659. doi: 10.1021/ja037027d. [DOI] [PubMed] [Google Scholar]
- 25.(a) Shukla LI, Adhikary A, Pazdro R, Becker D, Sevilla MD. Nucleic Acids Res. 2004;32:6565–6574. doi: 10.1093/nar/gkh989. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gervasio FL, Laio A, Iannuzzi M, Parrinello M. Chem Eur J. 2004;10:4846–4852. doi: 10.1002/chem.200400171. [DOI] [PubMed] [Google Scholar]
- 26.Adhikary A, Kumar A, Khanduri D, Sevilla MD. J Am Chem Soc. 2008;130:10282–10292. doi: 10.1021/ja802122s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conwell EM, Bloch SM, McLaughlin PM, Basko DM. J Am Chem Soc. 2007;129:9175–9181. doi: 10.1021/ja0691472. [DOI] [PubMed] [Google Scholar]
- 28.Conwell EM. Proc Natl Acad Sci USA. 2005;102:8795–8799. doi: 10.1073/pnas.0501406102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adhikary A, Kumar A, Becker D, Sevilla MD. J Phys Chem B. 2006;110:24171–24180. doi: 10.1021/jp064361y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adhikary A, Malkhasian AYS, Collins S, Koppen J, Becker D, Sevilla MD. Nucleic Acids Res. 2005;33:5553–5564. doi: 10.1093/nar/gki857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(a) Huang X, Yu P, LeProust E, Gao X. Nucleic Acids Res. 1997;25:4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) O'Neill MA, Barton JK. J Am Chem Soc. 2004;126:13234–13235. doi: 10.1021/ja0455897. [DOI] [PubMed] [Google Scholar]
- 32.Adhikary A, Becker D, Collins S, Koppen J, Sevilla MD. Nucleic Acids Res. 2006;34:1501–1511. doi: 10.1093/nar/gkl026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adhikary A, Kumar A, Sevilla MD. Radiation Research. 2006;165:479–484. doi: 10.1667/rr3563.1. [DOI] [PubMed] [Google Scholar]
- 34.Adhikary A, Collins S, Khanduri D, Sevilla MD. J Phys Chem B. 2007;111:7415–7421. doi: 10.1021/jp071107c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. J Am Chem Soc. 1999;121:9423–9428. [Google Scholar]
- 36.(a) Henderson PT, Jones D, Hampikian G, Kan YZ, Schuster GB. Proc Natl Acad Sci USA. 1999;96:8353–8358. doi: 10.1073/pnas.96.15.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Joseph J, Schuster GB. J Am Chem Soc. 2006;128:6070–6074. doi: 10.1021/ja060655l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information available: Figure S1: Analyses for deuteration levels in the G* labeled ds DNA-oligomers. Figure S2: Estimation of relative error regarding fitting of the experimentally observed spectrum using benchmarks. Figure S3: ESR spectrum of the guanine neutral radical (G(−H)•) from dGuo and d[GCGCGCGC]2. Figure S4: ESR Spectra of the one-electron oxidized dsDNA-oligomers: d[GGGCCC]2 and d[GCGCGCGC]2. Figure S5: ESR Spectra of the one-electron oxidized ds DNA-oligomers with and without single nucleotide extension at the ends, 5′-T and 3′-A. Analyses: Analyses of the ESR spectra of ds DNA-oliomers shown in Figure S5 using guanine neutral radical (G(−H)•) from dGuo and guanine neutral radical (G*(−H)•) from G* [G* = 8-D-dGuo, 96% deuterated]