Abstract
In cultured spinal neurons, NMDA receptors are absent from excitatory synapses under basal conditions, but can be made to appear at excitatory synapses following blockade of excitatory synaptic activity. The activity dependent synaptic localization of NMDA receptors is critically dependent on both the gradual, global accumulation of the NR2A and NR2B subunits and on a rapid, surface redistribution phase that is primed by the accumulation of NR2A and NR2B and inhibited by synaptic activity. Global changes in NR2A and NR2B accumulation and heterogeneous increases in synaptic NMDA receptor localization can also result from inhibitors of proteasomal processing, from manipulations of proteasomal subunit composition and from media conditioned by neurons undergoing synaptic scaling. While proteasomal processing is a mechanism shared with AMPA receptor scaling in cultured spinal neurons, diffusible factors, heterogeneity, and a rapid surface redistribution phase appear to be unique to activity dependent synaptic NMDA receptor localization.
Keywords: NMDA, Scaling, Plasticity, Activity, Synapse, Glutamate
Introduction
The mechanisms governing glutamate receptor aggregation at central synapses is the subject of intense investigation (Li and Sheng 2003). In addition to the mechanisms which regulate their localization during the formation of synapses, glutamate receptors are also dynamically regulated during experience dependent plasticity (Malinow 2003) and as part of the homeostatic response of neurons to global changes in synaptic input, a process termed synaptic scaling (Lissin et al., 1998; Turrigiano et al., 1998; OBrien et al., 1998; Ehlers, 2003). A growing body of work has implicated synaptic scaling as a necessary component for working memory (Renart et al., 2003) and activity dependent refinement of cortical connectivity during development (Desai et al., 2002). In cultured spinal neurons, global alterations of AMPA receptor half-life were first described as a potential mechanism for synaptic scaling (OBrien et al., 1998). Subsequent work by Ehlers (2003) and Burbea et al., (2002) showed that changes in receptor half-life are mediated by proteasomes.
While the effect of a global scaling of synaptic AMPA receptors on the homeostatic maintenance of postsynaptic spiking rates (with preservation of relatively weighted synaptic inputs) is straight-forward (Turrigiano et al., 1998), the activity dependent regulation of other synaptic proteins such as NMDA receptors have a more indirect linkage to postsynaptic spike output. Theoretical work (Wang, 2001) has suggested that NMDA receptors, due to their slower kinetics, may be crucial for the maintenance of networks operating in the alpha and beta frequency range, and the well documented role of NMDA receptors in initiating second messenger systems (Vanhoutte and Bading, 2003; Ehlers 2003), may also lead to changes in postsynaptic spike output.
Studies of the effect of chronic changes in synaptic activity on synaptic NMDA receptor accumulation have yielded inconsistent results (Watt et al., 2000; Ehlers, 2003; Lissin et al., 1998). In cultured spinal neurons, nearly 100% of excitatory synapses have clustered surface AMPA receptors (O’Brien et al., 1997) with little synaptic NMDA receptor clustering (O’Brien et al., 1997). The preferential clustering of AMPA receptors at spinal synapses in vitro occurs in spite of high level expression of the ubiquitous NMDA receptor subunit NR1 (Mi et al., 2002) and robust chemosensitivity to exogenous NMDA (O’Brien et al., 1997).
In this paper we show that the lack of basal synaptic NMDA receptor expression in cultured spinal neurons is a function of basal glutamate receptor activation. NMDA receptors become localized to excitatory synapses with blockade of AMPA and NMDA receptors, a phenomenon which can be mimicked in part by the addition of inhibitors of proteasomal processing and by media conditioned by neurons undergoing synaptic scaling. Evidence is presented that the global accumulation of NR2A and NR2B is regulated robustly by synaptic activity and is necessary for synaptic localization of NMDA receptors. Moreover, an additional, rapid phase of synaptic receptor localization involving surface translocation of NR2A and NR2B containing NMDA receptors is also present and modulated by synaptic activity. The time course, the role of soluble factors, the heterogeneity of the effect, and the rapid, activity dependent phase distinguish activity dependent synaptic NMDA receptor localization from AMPA receptor scaling.
Results
The Effect of Glutamate Receptor Activation on Synaptic NMDA Receptor Localization
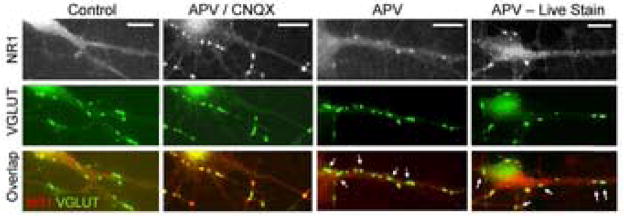
As shown in Figure 1 and Table 1A, control ventral spinal neurons showed little evidence of clustered NMDA receptors at excitatory synapses, identified by antibodies to the vesicular glutamate transporter (VGLUT), in spite of the presence of clustered post-synaptic MAGUK (membrane-associated guanylate kinase) proteins, identified with a pan-PDZ monoclonal, at nearly all excitatory synapses (Table 1A). Almost all excitatory synapses also have AMPA receptor clusters under these same conditions (OBrien et al., 1997). Treatment of cultured spinal neurons with APV plus CNQX (Figure 1 and Table 1A) or TTX (not shown) for 36 to 48 hrs resulted in a significant increase in the number and intensity (Table 1B) of synaptic NMDA receptor clusters but caused no change in the number of VGLUT or PDZ positive synapses or in the intensity of post synaptic MAGUK protein associated immunofluorescence (designated as PDZ fluorescence) at those synapses.
Figure 1. Synaptic Activity Regulates the Localization of NMDA Receptors.
Ventral spinal neurons were treated with the indicated pharmacologic agents for 36 to 48 hrs. Excitatory synapses were identified by staining with antibodies to the presynaptic vesicular glutamate transporters VGLUT1 and VGLUT2. Synaptic clustering of NR1 could be brought out by APV plus CNQX or to a lesser extent by APV. Live staining indicates a neuron transfected with an N-terminal myc tagged NR1 construct and stained live with anti myc prior to fixation. Arrows indicate the presence of NR1 immunonegative synapses on neurons with nearby NR1 immunopositive synapses. Scale bars = 10 μm.
Table 1. Synaptic Activity Regulates Synaptic NMDA Receptor Localization.
– Ventral Spinal Cultures were grown in the presence of the indicated agents for 36 to 48 hrs. In A, each value represents data from approximately 200 to 250 neurons gathered over 4 experiments and is expressed as the mean of 4 experiments +/− SD. PDZ refers to the number of VGLUT positive synapses associated with clusters of postsynaptic PDZ domain containing MAGUK proteins. In B, only synapses deemed immunopositive for the indicated protein were analyzed. All immunopositve synapses in a cell-body centered 100x field were analyzed. Each data point in B represents the mean of the means of a minimum of 4 experiments, each sampling 10–20 neurons.
| A | |||
|---|---|---|---|
| Synaptic Clusters/Neuron |
|||
| NR1 | VGLUT | PDZ | |
| Control | 2.3 +/− 1.9 | 13.6 +/− 4.0 | 13.7 +/− 6.1 |
| APV (0.3) | 6.6 +/− 3.7* | 13.1 +/− 3.9 | 13.0 +/− 2.2 |
| APV (0.1) | 6.1 +/− 2.4* | 14.3 +/− 2.6 | 14.3 +/− 3.7 |
| APV + CNQX | 14.3 +/− 3.2** | 15.0 +/− 3.8 | 14.8 +/− 5.2 |
| B | |||
| Mean Synaptic NR1 | Mean Synaptic GluR2 | Mean Synaptic PDZ | |
| Fluorescence Intensity | Fluorescence Intensity | Fluorescence Intensity | |
| Control | 537 +/− 211 | 5686 +/− 1002 | 10421 +/− 2655 |
| APV (0.3 mM) | 3508 +/− 918* | 6030 +/− 1118 | 11717 +/− 2170 |
| APV (0.1 mM) | 3143 +/− 1144* | 5325 +/− 1394 | 10009 +/− 3941 |
| APV + CNQX | 6402 +/− 1027** | 9051 +/− 1581** | 12247 +/− 3011 |
p<0.02 compared to control and APV / CNQX treated cultures.
p<0.02 compared to control and APV treated cultures.
Blocking NMDA receptors with APV alone also resulted in an increase in the mean number of synaptic NMDA receptor clusters per cell compared to controls (Figure 1Table 1), but, in contrast to APV and CNQX, left about half of all excitatory presynaptic terminals with no discernable postsynaptic NR1 clusters (Arrows in Figure 1). We saw no evidence that APV was acting by modulating AMPA receptor function, as no increase in synaptic GluR2 immunofluorescence was seen in the presence of APV (Table 1B), implying that both AMPA and NMDA receptors are involved in the full expression of activity dependent synaptic NMDA receptor localization.
Studies of neurons transfected with N-terminal myc-epitope tagged versions of NR1 indicated that cell surface NMDA receptors responded to synaptic inhibitors in a manner similar to the total pool of NMDA receptors. As shown in Figure 1, neurons transfected with an N-terminal myc epitope tagged NR1 construct, grown for 36 hrs in APV and stained live with anti-myc antibodies, showed the same heterogeneous pattern of activity dependent synaptic NMDA receptor localization as that determined by C-terminal antibodies. Quantitatively, 178 of 396 VGLUT positive synapses from 33 transfected neurons grown for 36 hours in APV had clustered surface synaptic NR1 staining while the remainder did not (Figure 1; arrows in overlap). In contrast, under control conditions only 42 of 379 VGLUT positive synapses had detectable surface synaptic NR1 clusters while neurons from cultures treated with APV and CNQX had synaptic NR1 clusters in 293 of 354 VGLUT positive synapses on 23 neurons. Each of these differed from APV treated cultures at the 0.01 level.
NMDA Receptor Subunit Accumulation Correlates with Synaptic Localization
When APV and CNQX were added to cultured spinal neurons, a robust increase in the accumulation of the NMDA receptor subunits NR2A and NR2B was seen (Figure 2A) along with a more modest increase in NR1. NMDA receptor subunit accumulation peaked in the first 24 hrs following blockade of excitatory synaptic transmission and then decayed over the next 72 hrs in the presence of continued receptor blockade (Figure 2A and 2B). In contrast, GluR2 expression peaked at 72 hrs. Untreated cultures showed no change in NR2A, NR2B or GluR2 accumulation over the same period (not shown). The time course for the synaptic localization of NMDA receptors in response to activity blockade, calculated by the mean synaptic fluorescence intensity for NR1 (Figure 2C), corresponded closely with the incremental accumulation of NR2A and NR2B. Activity blockade caused little change in the accumulation of NR2C, NR2D, PSD-95 or Chapsyn-110 (Table 2). APV also increased the accumulation of NR2A and NR2B but to a lesser extant than APV plus CNQX (Table 2).
Figure 2. The Effect of Synaptic Activity on NMDA Receptor Subunit Accumulation.
In A, immunoblots of cultures solubilized at different times after exposure to APV and CNQX were probed with the indicated antibodies. In B, the quantitative results of 4 such experiments are expressed as the mean +/− S.D of 4 experiments. In C, the mean synaptic fluorescence intensity for NR1 and GluR2 obtained over 6 experiments are shown at the indicated time points after APV and CNQX addition. In D, equivalent amounts of mRNA from cultures 36 hrs after addition of the indicated agents were run along with an equivalent amount of mRNA from untreated cultured hippocampus neurons. The arrows in D indicate the position of the 28S and 18S ribosomal RNA bands. In E, biotinylated surface NR1 was streptavidin immunoprecipitated and probed with a pan-NR1 antibody as a function of time after biotinylation. The APV and CNQX treated samples were diluted 1/1.5 so that the signal at t=0 would be equivalent to the control sample.
* p<0.05 compared to prior time point for the same subunit in a paired comparison.
Table 2. Quantitative Assessment of Synaptic Activity and Proteasomal Inhibition on NMDA Receptor Subunit Accumulation.
High density ventral spinal neuron cultures were grown in 60 mm dishes for 7–8 days and were then treated with the indicated agents for 36 hrs. Cultures were solubilized in M-Per and processed for immunoblotting as described in Methods, or they had their RNA extracted for northern blotting. For immunoblotting, equivalent percentages of a dish were run on the gel, while for northern blots, 2 mg of polyA RNA (n=1) or 5 mg of total RNA (n=1) were run. Protein accumulation data is drawn from four separate experiments while the RNA data is from 2 experiments. Data are expressed as an increase/decrease relative to control for each experiment.
| Relative Expression Levels after 36 hrs | ||||
|---|---|---|---|---|
| APV/CNQX | APV | Lactacystin | MG132 | |
| 0.3mM/20μM | 0.3 mM | 1 μM | 10 μM | |
| NR1-C2 | 2.4+/−0.3*** | 1.7+/−0.3*** | 1.9+/−0.5*** | |
| NR1-C2′ | 2.3+/−0.3*** | 1.7+/−0.3*** | 1.6+/−0.3*** | |
| NR2A | 7.2+/−2.0* | 3.6+/−1.9** | 6.5+/−1.3* | 5.8+/−1.8* |
| NR2B | 8.7+/−1.6* | 4.4+/−1.3** | 7.9+/−1.8* | 6.0+/−1.5* |
| GluR2 | 1.7+/−0.4* | 1.8+/−0.2* | 1.7+/−0.3* | |
| NR2C | 0.9+/−0.1 | 1.1+/−0.1 | 0.9+/−0.2 | |
| NR2D | 0.9+/−0.1 | 1.2+/−0.1 | 1.0+/−0.2 | |
| Stargazin | 1.1+/−0.2 | 1.2+/−0.3 | 0.9+/−0.3 | |
| Chapsyn110 | 0.9+/−0.1 | 1.1+/−0.1 | 1.0+/−0.1 | |
| Tubulin | 1.0+/−0.1 | 1.0+/−0.2 | 0.9+/−0.1 | |
| PSD-95 | 1.2+/−0.3 | 1.3+/−0.4 | 1.1+/−0.2 | |
| Synaptophysin | 1.4+/−0.2*** | 1.3+/−0.3 | 1.2+/−0.2 | |
| VGLUT1 | 1.3+/−0.3 | 1.1+/−0.1 | 1.1+/−0.2 | |
| VGLUT2 | 1.5+/−0.2*** | 1.2+/−0.2 | 1.3+/−0.3 | |
| NR1 mRNA | 1.1+/−0.04 | |||
| NR2A mRNA | 0.9+/−0.03 | |||
| NR2B mRNA | 1.0+/−0.11 | |||
p<0.05 compared to control in a paired comparison
p<0.05 compared to control but not compared to APV/CNQX treated cultures in a paired comparison
p<0.05 compared to control in a paired comparison but loses significance compared to control for this single experiment when the p value is multiplied by the number of comparisons made (14) and was not replicated in separate independent experiments.
The Role of NR2A and NR2B in Activity Dependent Synaptic Localization
Previous work (Mi et al., 2004; Rao and Craig 1997; Barria and Malinow 2002) has shown that NR2A and/or NR2B accumulation plays a crucial role in the synaptic localization of NMDA receptors while NR2C and NR2D accumulation does not. In order to directly implicate the increased expression of NR2A and NR2B in the activity dependent synaptic localization of NMDA receptors, we used a series of previously characterized dominant-negative NMDA receptor mutants, whose disruptive effect is mediated by saturating sites essential for synaptic NMDA receptor localization (OBrien et al., 2002). These constructs express well in neurons and are distributed throughout the dendritic tree of transfected neurons (not shown). As shown in Figure 3A and 3B, the effect of glutamate receptor blockade on synaptic NMDA receptor localization could be abrogated by overexpression of the soluble C-terminus of NR2A and NR2B but not by overexpression of NR2ACTΔ10, a C-terminal NR2A construct lacking the terminal IESDV motif responsible for binding to the PDZ domains of synaptic MAGUK proteins. Overexpression of C-terminal constructs from NR1 (C2 and C2′), NR2C and NR2D had no effect on activity regulated synaptic NMDA receptor localization.
Figure 3. Dominant Negative NR2 Mutants Disrupt Activity Dependent Synaptic NMDA Receptor Localization.

Ventral spinal neurons were transfected with constructs expressing the indicated NMDA receptor subunit C-terminal tails, along with GFP (insert), and were then exposed to APV and CNQX for 36 hrs. Control is a pCMV-LacZ vector. In A and B, the NR2A C-terminus is shown to disrupt the APV and CNQX induced synaptic localization of NR1, while the NR2A C-terminus minus its terminal 10 amino acids (NR2ACTΔ10) did not. Small residual clusters of NR1 can sometimes be seen in NR2ACT transfected neurons (arrows). A total of 10 – 20 GFP positive neurons per transfection were analyzed for each construct and each value is derived from a minimum of 3 experiments. Scale bar = 10 μm.
* p<0.01 compared to control transfection in a paired comparison. This remains significant (p<0.03) when corrected for the total number of constructs compared (7).
No Evidence for Increased Transcription of NR2A and NR2B mRNA During Activity Dependent Synaptic NMDA Receptor Localization
The increase in NR1, NR2A and NR2B protein accumulation observed in the presence of APV and CNQX was not accompanied by an increased accumulation of the mRNA for these subunits (Figure 2D; Table 2). Moreover, a 14 hr incubation with the transcription inhibitor actinomycin D (longer incubations were non-specifically toxic) did not alter the accumulation of the NR2A subunit in response to APV and CNQX (Table 3B). Also consistent with a post-translational regulation of activity dependent synaptic NMDA receptor localization, the surface half-life of NMDA receptors, measured by quantitating the decay of biotinylated surface NR1 (Figure 2E), increased from 13.2 +/− 3.4 hrs (control) to 27.8 +/− 5.9 hrs following exposure to APV and CNQX (n=6 experiments; p<0.002).
Table 3. The Effect of Conditioned Media on Synaptic Glutamate Receptor Localization.
A, media conditioned by cultures exposed to APV & CNQX, CM (50% final concentration; vol/vol) or containing the indicated pharmacologic agents, was added to naïve cultures for 48 hrs with 1/2 media changes every 12 hrs (standard conditions) or added for 36 hrs with complete media changes every 8 hrs (designated as “8hr”). The number of synaptic clusters of the indicated proteins, as well as their mean synaptic fluorescence intensity, was determined in a series of 4 or 5 experiments. Control CM refers to media taken from spinal neurons grown under standard conditions and treated as described in Methods. Control refers to neurons fed with standard media on the same schedule as the conditioned media experiments. In B, similarly treated high density cultures were solubilized in M-Per and processed for immunoblotting or had their RNA extracted for northern blotting after 48 hrs except in the case of GluR1 where an additional 96 hr time point was obtained. Each data point in B represents results from four to 10 separate experiments while the RNA data is from 2 experiments. Data are expressed as an increase/decrease relative to control for each experiment. The data for actinomycin D (10 μM) represents only a 14 hour incubation in conditioned media in order to maximize the health of the cultures.
| A | |||||||
|---|---|---|---|---|---|---|---|
| Synaptic Clusters Cell |
Mean Synaptic Intensity (x103) |
||||||
| NR1 | GluR1 | PDZ | NR1 | GluR1 | PDZ | ||
| Control | 3.1 +/− 2.0 | 15.3 +/−3.9 | 13.7 +/− 2.2 | 0.3 +/− 0.2 | 4.5 +/− 1.3 | 10.6 +/− 2.3 | |
| APV/CNQX | 14.8 +/− 4.0* | 15.8 +/− 3.9 | 14.9 +/− 2.8 | 5.6 +/− 1.2* | 8.4 +/− 1.9* | 9.5 +/− 1.5 | |
| Control CM | 2.3 +/− 1.0 | 14.0 +/− 4.3 | 14.3 +/− 3.6 | 0.3 +/− 0.2 | 5.2 +/− 2.0 | 8.1 +/− 1.6 | |
| CM | 7.3 +/− 2.4** | 13.9 +/− 4.2 | 15.3 +/− 3.7 | 3.3 +/− 0.9** | 5.5 +/− 1.8 | 8.8 +/− 1.0 | |
| Concentrated CM (5x) | 9.5 +/− 4.3** | 15.9 +/− 5.2 | - | 3.7 +/− 0.9** | 4.5 +/− 0.9 | - | |
| CM + APV/CNQX | 16.8 +/− 3.4* | 17.3 +/− 4.0 | - | 6.0 +/− 1.3* | 7.8 +/− 1.6* | - | |
| APV/CNQX (8 hr) | 10.4 +/− 2.7** | 15.7 +/− 3.4 | - | 2.9 +/− 0.6** | 7.5 +/− 1.3* | - | |
| CM + APV/CNQX (8 hr) | 16.2 +/− 4.5* | 15.9 +/− 4.2 | - | 5.9 +/− 1.4* | 8.1 +/− 2.0* | - | |
| B | |||||||
|---|---|---|---|---|---|---|---|
| Protein Levels (Relative to Control) |
mRNA Levels (Relative to Con) |
||||||
| NR1 | NR2B | NR2A | GluR1 | GluR1 (96 hrs) | NR2B | NR2A | |
| APV/CNQX | 2.4 +/− 0.3* | 6.9+/− 1.5* | 7.6 +/− 1.7* | 2.5 +/− 0.8* | 3.2 +/− 0.6* | ||
| Control CM | 1.2 +/− 0.2 | 1.1 +/− 0.1 | 0.8 +/− 0.2 | 0.9 +/− 0.1 | 0.9 +/− 0.1 | ||
| CM | 1.8 +/− 0.4* | 5.3 +/− 1.4** | 4.9 +/− 2.2** | 1.2 +/− 0.2 | 1.1+/− 0.2 | 1.0 +/− 0.1 | 0.9 +/− 0.1 |
| CM (5X) | - | 7.2 +/− 2.6* | 6.8 +/− 1.1* | 1.2 +/− 0.5 | |||
| CM + APV/CNQX | 7.9 +/− 2.0* | 8.6 +/− 2.4* | 2.4 +/− 0.7* | ||||
| APV/CNQX (8 hr) | 4.4 +/− 1.7** | 5.0 +/− 2.3** | 2.2 +/− 0.6* | ||||
| CM + APV/CNQX (8hr) | 7.5 +/− 2.3* | 8.2 +/− 2.7* | 2.2 +/− 0.4* | ||||
| CM (14 hr) | 1.8+/− 0.3 | ||||||
| CM + Actinomycin D (14 hr) | 2.1+/− 0.4 *** | ||||||
| APV/CNQX (14 hr) | 2.6+/− 0.7 | ||||||
| APV/CNQX + Actinomycin D (14 hr) | 3.1+/− 1.1 *** | ||||||
| Actinomycin D (14hr) | 0.9+/− 0.2 | ||||||
p<0.05 compared to control in a paired comparison.
p<0.05 compared to control and APV/CNQX treated cultures in a paired comparison.
p<0.01 compared to actinomycin D alone
Media Conditioned by Neurons Experiencing Synaptic Activity Blockade Modulates Synaptic NMDA Receptor localization
While activity mediated synaptic glutamate receptor localization could be due solely to postsynaptic depolarization (Leslie et al., 2001), it is equally possible that activity modulates the pre or postsynaptic secretion of additional signaling molecules. To investigate this, we collected the media from neurons grown in the presence of APV and CNQX for 48 hrs, dialyzed it to remove residual APV and CNQX, and added it to naïve cultures every 12 hrs for a total of 36–48 hrs. As shown in Figure 4A and Table 3A, conditioned media (CM), but not media taken from neurons growing under control conditions (Control CM), caused a significant increase in synaptic NMDA receptor localization compared to untreated cultures (Control) but had no effect on synaptic AMPA receptor or MAGUK protein localization, either when expressed as the number of synaptic clusters per neuron, or as the amount of protein specific immunofluorescence per synapse.
Figure 4. Conditioned Media Induces Synaptic Localization of NMDA Receptors.

In A, neurons grown in the presence of media conditioned by neurons undergoing activity dependent synaptic NMDA receptor localization (1x; 50% (vol / vol) dilution) also show synaptic NMDA receptor localization. Control media consisted of media conditioned by neurons in the absence of APV and CNQX. Residual excitatory synapses without synaptic NMDA clusters are indicated with arrows. In B, the effect of media conditioned by spinal neurons exposed to APV and CNQX on the accumulation of NR2A, NR2B and GluR1 is shown using immunoblots of the indicated synaptic proteins after a 48 hr incubation in 1x CM at a 50% (vol / vol) dilution. For comparison, neurons treated directly with APV and CNQX or with conditioned media plus picrotoxin and strychnine are also shown. In C, the effect of different concentrations of conditioned media as well the effect of concentrating the conditioned media on NR2A accumulation is shown. The data is expressed +/− S.D (n=3). The percentage of conditioned media indicated represents the percentage of the total media in which the neurons were grown which is conditioned, either 1X or 5X. The 40% conditioned media is significantly different from the 10% conditioned media at the 0.01 level for both concentrated and unconcentrated forms.
* p < 0.05 compared to 1x conditioned media at the same dilution
** p <0.05 compared to 1x conditioned media at the same dilution and p <0.05 compared to APV and CNQX treatment
Similar to activity blockade, conditioned media taken from neurons grown in the presence of APV and CNQX for 48 hrs (CM) but not from control cultures (Control CM) caused a significant increase in NR2A and NR2B subunit accumulation, with no effect on GluR1 accumulation (Figure 4B,C and Table 3B). No change in NR2A or NR2B mRNA was seen and the effect was not abrogated by actinomycin D (Table 3B).
Combining APV and CNQX with conditioned media for a 36 hr incubation caused no increase in the maximal synaptic NR1 localization or NR2A and NR2B accumulation over APV and CNQX alone (Table 3A). However, performing a complete media change with fresh unconditioned media containing APV and CNQX every 8 hrs for a total of 36 hrs (designated APV/CNQX 8 hr) significantly abrogated the effect of APV and CNQX on synaptic NR1 localization (Table 3A) as well as on NR2A and NR2B accumulation (Table 3B) but had no effect on GluR1 localization or accumulation. When conditioned media was included with APV and CNQX in these frequent change experiments (designated CM + APV/CNQX 8 hr), the full effect of activity blockade was reproduced. These observations suggest that conditioned media and activity blockade share a mechanism that can be saturated and that activity blockade requires the presence of diffusible extracellular factors. In addition to its mechanistic implications, these results also suggest that NMDA and AMPA receptor scaling are regulated by different mechanisms. Members of the family of neurotrophins do not mediate the effect of conditioned media (See Supplemental Data).
When the conditioned media was concentrated 5 fold (by volume), a further increase in NR2A accumulation was seen, which, at higher doses, mimicked the effect of APV and CNQX on NR2A and NR2B accumulation (Figure 4C, Table 3B). Importantly, even though the levels of NR2A and NR2B could be increased by conditioned media to levels observed in cultures treated with APV and CNQX, synaptic NR1 localization still fell short of that seen with APV and CNQX (Table 3A). As detailed below, we feel this is likely due to the continued presence of synaptic activity in conditioned media treated cultures.
The Role of Proteasomal Processing in NMDA Receptor Localization
As shown in Figure 5A and Table 4A, we found that the increase in NR1, NR2A and NR2B accumulation caused by APV and CNQX or conditioned media could be quantitatively mimicked by the addition of inhibitors of proteasomal processing (Lee and Goldberg 1998; Myung et al., 2001) such as lactacystin (1 μM) and MG132 (10 μM). These same proteasomal inhibitors also caused an increase in the synaptic localization of NMDA receptors (Figure 5B and Table 4B). Other synaptic and non synaptic proteins, aside from glutamate receptors, were only modestly affected by the 36 hr incubation with lactacystin and MG132, a pattern similar to that evoked by APV/CNQX incubation (Table 2). The lysosomal inhibitor leupeptin used at 50 μM had no effect on NR2A or NR2B accumulation or on synaptic NR1 localization (Table 4). Combining proteasomal inhibition with synaptic blockade had no additive effect or NR2A and NR2B accumulation (Table 4A) but significantly increased synaptic NMDA receptor localization compared with proteasomal inhibitors alone (Table 4B).
Figure 5. Proteasomal Inhibitors Induce the Synaptic Localization of NMDA Receptors.

Ventral spinal neurons were treated with the indicated agents for 36 hrs. In A, total and surface (streptavidin immunoprecipitated) immunoblots from the treated cultures were probed with the indicated antibodies. In B, synaptic clusters of NR1 induced by lactacystin (1 μM), lactacystin plus TTX (10 μM), and lactacystin plus picrotoxin and strychnine (P&S; 100 μM and 2 μM respectively) are shown. Note the VGLUT positive synapses without NR1 clusters in lactacystin treated cultures (arrows in overlap). In C, surface (streptavidin immunoprecipitated) NR1 and GluR1 are shown in cultures grown under control conditions and following 48 hrs of exposure to picrotoxin and strychnine. In D, histograms of the number of VGLUT positive synapses per neuron which are NR1 or GluR2 positive are displayed. Neurons were exposed to lactacystin. The NR1 data is from 452 neurons, the GluR2 data from 247 neurons. Scale bars = 10 μm.
Table 4. Regulation of NMDA Receptor Subunit Accumulation and Localization.
In A, quantitative analyses of total and surface NMDA receptor subunit levels from spinal cultures treated with the indicated pharmacological agents for 36 hrs are shown. Data are expressed relative to control for each experiment and represent a minimum of 4 experiments. Each result is significantly different from control at the 0.02 level. In B, the mean synaptic fluorescence intensity for NR1 at synapses judged immunopositive for NR1 is displayed from a minimum of 3 experiments.
| A | ||||||||
|---|---|---|---|---|---|---|---|---|
| MG132 | Lactacystin | Lactacystin + P&S |
Lactacystin +TTX |
APV +CNQX |
Lactacystin +APV/CNQX |
Conditioned Media (5x) |
Leupeptin | |
| Total NR1 | 2.9+/−0.6 | 2.6+/−0.4 | 2.5+/−0.5 | 3.0+/−0.4 | 2.7+/−0.7 | - | - | - |
| Total NR2A | 6.0+/−1.9 | 6.6+/−2.4 | 7.2+/−2.3 | 6.4+/−1.8 | 7.7+/−2.1 | 8.0+/−3.3 | 6.9+/−2.1 | 1.2+/−0.2** |
| Total NR2B | 5.4+/−1.3 | 8.3+/−3.1 | 7.8+/−2.6 | 9.0+/−3.3 | 6.9+/−1.9 | 7.6+/−2.5 | 6.4+/−2.4 | 0.9+/−0.2** |
| Total GluR1 | 1.8+/−0.4 | 1.7+/−0.3 | - | - | 2.0+/−0.3 | - | - | |
| Surface NR1 | - | 1.4+/−0.2* | 0.6+/−0.2** | 2.5+/−0.7** | 3.4+/−0.5 | - | - | |
| Surface NR2A | - | 4.8+/−1.8* | 2.0+/−1.1** | 11.4+/−4.7** | 13.1+/−5.0 | 14.0+/−4.2 | 4.8+/−1.5* | - |
| Surface NR2B | - | 5.7+/−2.0* | 2.5+/−1.4** | 9.9+/−2.8** | 12.5+/−3.6 | 11.3+/−3.8 | 3.7+/−1.2* | - |
| B | ||||||||
| MG132 | Lactacystin | Lactacystin +P&S | Lactacystin +TTX | APV +CNQX | Lactacystin +APV/CNQX | Conditioned Media (5x) | Leupeptin | |
| NR1(+) Synapses per Neuron | 7.8+/−2.4* | 6.9+/−3.8* | 3.0+/−2.1** | 13.1+/−6.7** | 16.0+/−3.8 | 15.2+/−4.1 | 8.0+/−3.3* | 2.3+/−1.9** |
| NR1 Fluorescence per Synapse (x103) | 4.4+/−1.3* | 5.1+/−1.0* | 2.0+/−0.9** | 7.4+/−1.4** | 8.5+/−1.3 | 8.9+/−1.6 | 4.2+/−1.4* | - |
p<0.01 compared to APV/CNQX treated neurons in a paired comparison.
p<0.02 compared to lactacystin treated neurons in a paired comparison.
Like conditioned media, proteasomal inhibitors could increase NR2A, NR2B and NR1 levels to that seen with blockade of excitatory synaptic activity (Table 4A), but their effect on synaptic localization of NMDA receptors was incomplete both qualitatively (Figure 5B arrows) and quantitatively (Table 4B).
Proteasome Subunit Composition Effects Synaptic NMDA Receptor Localization
The importance of proteasomes in synaptic NMDA receptor localization was revealed not only by the use of proteasome inhibitors but also by examining the effect of altering proteasome subunit composition on synaptic NMDA receptor localization. As shown in Figure 6A and 6B, the proteasomal subunit LMP2, which alters proteasomal processing in lymphocytes (Van den Eynde and Morel 2001), is distributed in a reticulated pattern throughout the dendrites of transfected neurons and significantly alters synaptic NMDA receptor localization in neurons grown in the presence of APV and CNQX. No change was noted in the synaptic distribution of PDZ domain containing proteins under similar conditions. In contrast overexpression of the C2 subunit, which is expressed constitutively in spinal neurons, had no effect on synaptic NMDA receptor accumulation in the presence of activity blockade (Figure 6B). Expression of the LMP2 subunit is very low in spinal cultures but always detectable (Figure 6C). In the presence of APV and CNQX LMP2 becomes undetectable (n=3 repetitions). No change was noted in expression of C2. The very low expression of LMP2 in spinal neurons makes it doubtful that it has a physiological role in the process described in this paper. These results do however further highlight the potential role of proteasomes in this process. Interestingly, the expression of LMP2 goes up in aged human brains and in some disease states (Mishto et al., 2006) raising the possibility of an alteration of the process of activity dependent synaptic receptor localization with aging.
Figure 6. Proteasome subunit composition alters synaptic NMDA receptor localization.

In A, cultured spinal neurons were transfected with an equal amount of LMP2 and LMP2-GFP and cultured for an additional 36hrs in the presence of APV and CNQX. Images of the LMP2-GFP and NR1 signals are shown in isolation and superimposed on the VGLUT signal. A high power image of the dendritic LMP2-GFP signal is shown in the insert. Arrows indicate excitatory synapses without NR1 accumulation. In B, results of a series of 3 transfection experiments for each of the indicated constructs is shown. For each transfection experiment, 25 transfected neurons were analyzed. Data is expressed as the mean +/− SD of the mean for the 3 experiments per construct. All results reflect the number of clusters seen on neurons grown in the presence of APV and CNQX for 36 hrs. In C, immunoblots of HEK293 cells transfected with the indicated constructs (non GFP tagged) as well as non transfected spinal neurons grown in the presence or absence of APV and CNQX for 36 hrs are shown after probing with antibodies to C2 (left) or LMP2 (right). The results shown were replicated in 2 other similar experiments. Arrows in C indicate the position of the 25 kD molecular weight marker.
* p<0.01 compared to eGFP or C2 transfected neurons.
A Spatially Heterogeneous Pattern of Synaptic NMDA Receptor Localization is Seen Following Exposure to Proteasomal Inhibitors or Conditioned Media
In a series of 452 individual neurons from lactacystin treated cultures, the percentage of VGLUT positive synapses per individual neuron which were NR1 positive was 51.1 +/− 19%, using a blinded assay and a forced choice (yes/no) paradigm. A histogram of these results is shown in Figure 5D. For conditioned media treated neurons, the number was 61.1 +/− 24%. Since this result is derived by generating a histogram of the percentage of excitatory synapses which were NR1 positive on each of the 452 neurons, the small size of the standard deviation and the shape of the histogram in Figure 5D indicates that the process reflects heterogeneity within synapses on individual neurons and is not due to 2 populations of neurons, one responsive and one unresponsive to lactacystin or conditioned media.
In contrast to the results obtained with NR1, the mean percentage of VGLUT positive synapses which are GluR2 positive, expressed on a per neuron basis, is 65.1 +/− 45% in control cultures, 67.9 +/− 41% in APV/CNQX treated cultures, and 71 +/− 40% in lactacystin treated cultures (Figure 5D), based on a minimum of 200 neurons for each condition (p<0.002 compared to NR1 heterogeneity). The large standard deviation in these latter 3 experiments and the shape of the histograms in Figure 5D is due to the fact that GluR2 positive synapses in spinal neurons are distributed among two populations of neurons (OBrien et al., 1997). One group of neurons, reflecting 64% of the total population, has GluR2 localization at 90–100% of its excitatory synapses. A second group of neurons (36% of the population) has almost no GluR2 immunopositive synapses.
Quantitative Immunofluorescence Also Reveals Heterogeneity in Synaptic NMDA Receptor Localization in Cultures Exposed to Lactacystin and Conditioned Media
A separate method for expressing the synaptic NR1 heterogeneity seen in the presence of lactacystin or conditioned media is by plotting a histogram of individual synaptic NR1 fluorescence intensities at all the excitatory synapses on a series of neurons. In Figure 7 (A–D) we display the histograms of synaptic NR1 fluorescence intensities from all the excitatory synapses on a series of 30–40 consecutive neurons from a single coverslip for each experimental condition. In Figure 7E, the mean synaptic fluorescence intensity for four such experiments plus the standard deviations of those means is shown. The data is divided into all the VGLUT positive synapses and into those judged to be NR1 immunopositive (Figure 7F). As shown, the coefficient of variation (SD/Mean) of individual synaptic NR1 fluorescence intensities in the lactacystin (1 μM) and conditioned media (5x) treated cultures is twice as high as that seen in APV and CNQX treated cultures and significantly greater that that seen for synaptic AMPA receptor clusters under similar conditions, reflecting the significant heterogeneity in the response of individual synapses to the lactacystin induced increase in NR2A and NR2B accumulation. Adding TTX (10 μM) alters the pattern of synaptic NR1 accumulation in lactacystin treated cultures (Figure 7E), approaching that seen in APV and CNQX treated cultures. This observation argues that ongoing synaptic or spiking activity is the source of the heterogeneity seen in lactacystin treated cultures.
Figure 7. Heterogeneity in Individual Synaptic NR1 Fluorescence Intensities Induced by Pharmacological Manipulation of Ventral Spinal Neurons.

In A–D, histograms of individual synaptic NR1 fluorescence intensities, representing all the VGLUT positive synapses visible in a single 100x field, are displayed from a series of 30 – 40 consecutive neurons per coverslip per condition. In each case the coverslip was treated with the indicated pharmacological agents or conditioned media for 36 hrs. Note that the 0 – 0.5 bin is divided into 2 groups. Above the histograms are the mean (+/− S.D.) of the individual synaptic fluorescence intensities for that experiment and the coefficient of variation of those individual synaptic fluorescence intensities. In E and F, the mean +/− S.D of the mean for a minimum of 3 experiments similar to those displayed in the histogram and the coefficient of variation of the individual synaptic fluorescence intensities for those same experiments are displayed. In E, all VGLUT (+) synapses were included in the analysis. In F, only those judged to be NR1 immunopositive (using blinded forced choice) were analyzed. For GluR1 data in E, cultures were treated with the indicated agents for 48 hrs.
* p<0.02 compared APV plus CNQX treated cultures.
** p<0.02 compared to lactacystin treated cultures
When only the NR1 immunopositive synapses are considered (using a forced choice (yes/no) paradigm on blinded coverslips), those synapses judged to be NR1 immunopositive are still significantly smaller in lactacystin and conditioned media treated cultures compared to APV/CNQX treated cultures (Figure 7F). The coefficient of variation of the synaptic NR1 fluorescence intensities in lactacystin and conditioned media treated cultures is not significantly different from those in APV/CNQX treated cultures, implying that most of the heterogeneity in excitatory synapses as a whole results from a large population of NR1 immunonegative synapses.
Synaptic Heterogeneity is not a Detection Threshold Phenomenon
The increased coefficient of variation seen in lactacystin and conditioned media treated cultures is not simply a threshold effect due to the lower mean synaptic NR1 fluorescence intensities in these cultures. Prior work with synaptic AMPA receptor accumulation in the presence of picrotoxin and strychnine showed mean synaptic fluorescence intensities in the range of 3500 units but a mean coefficient of variation of only 0.68 (OBrien et al., 1998). Moreover, in experiments done as part of the time course for synaptic NMDA receptor accumulation following activity blockade (Figure 2) the 12 hour time point resulted in a mean synaptic NR1 immunofluorescence of 2145 units but a coefficient of variation of only 0.68 +/− 0.17 (n=6 experiments).
NMDA Receptor Surface Accumulation Correlates with Synaptic Localization
While the total accumulation of NR2A and NR2B subunit protein induced by proteasomal inhibitors and maximal doses of conditioned media was not different from that induced by APV and CNQX (Table 4A), the synaptic localization of NR1, expressed either as the number of NR1 immunopositive excitatory synapses per neuron or as the amount of synaptic NR1 immunofluorescence (Table 4B) differed significantly between the APV/CNQX treated cultures and those treated with proteasomal inhibitors or conditioned media.
Unlike total receptor levels however, the surface accumulation of NR1, NR2A and NR2B correlated closely with synaptic NMDA receptor localization (Figure 5A, Table 4A and 4B). While lactacystin clearly increased surface NMDA receptors, the amount of surface accumulation was also further influenced by synaptic activity. This was most easily seen when proteasomal inhibitors were combined with picrotoxin (100 μM) and strychnine (2 μM) to increase overall synaptic excitation and when TTX was used to block synaptic activity. The synaptic localization of NMDA receptors always tracked the surface rather than the total accumulation of NR1, NR2A and NR2B.
In contrast to the activity regulated surface accumulation of NMDA receptors observed in cultures with high levels of expression of NR2A and NR2B (Figure 5A), basal NR1 surface expression was not diminished by chronic incubation with picrotoxin and strychnine (Figure 5C; 0.93 +/− 0.17, n=4, p=0.7), consistent with prior results showing a constitutive, non-synaptic, NR2C and NR2D mediated pathway for surface NR1 accumulation under these conditions (Mi et al., 2004). Surface GluR1 accumulation, in contrast, was significantly affected by picrotoxin and strychnine under the same conditions (Figure 5C; 0.61 +/− 0.15, p=0.03).
Preincubation with Proteasomal Inhibitors or Conditioned Media Reveals a Second, Rapid, Activity - Dependent Redistribution Process
When lactacystin or conditioned media primed (36 hrs) cultures were briefly treated with APV and CNQX (30 minutes), the number of excitatory synapses with postsynaptic NR1 clusters and the amount of NMDA receptors at individual excitatory synapses rapidly increased compared to cultures treated only with lactacystin or conditioned media (Table 5A). No effect on AMPA receptor localization was seen. The rapid synaptic localization of NMDA receptors was accompanied by increased surface NMDA receptor subunit accumulation but no change in total levels (Table 5B). No change was noted in surface AMPA receptor subunit accumulation. A brief exposure to APV and CNQX in the absence of the prolonged priming incubation had no effect (not shown). These results suggest a two step process for chronic activity regulated synaptic NMDA receptor localization, a slow component, dependent on the accumulation of NR2A and NR2B and a second, rapid component, involving surface translocation and synaptic localization.
Table 5. Acute Regulation of NMDA Receptor Subunit Accumulation and Localization.
A, the number of synapses immunopositive for the indicated protein as well as the mean synaptic fluorescence intensity for that protein is displayed after a 36 hr priming incubation with lactacystin (1 μM) or conditioned media (5x; 50% vol/vol) and following a subsequent secondary (30 min) exposure to APV and CNQX. Data represent a minimum of 3 experiments. B, quantitative analyses of total and surface NMDA receptor subunit levels from neurons treated with a 36 hr priming incubation with lactacystin (1 μM) and following a subsequent secondary (30 min) exposure to APV and CNQX. Data represent a minimum of 4 experiments.
| A | |||
|---|---|---|---|
| Primary Incubation (36 hr) | Secondary Incubation (30 min) | ||
| Lactacystin | Lactacystin + APV/CNQX | Lactacystin Only | |
| NR1 Positive Synapses | 8.4+/−3.3 | 14.6+/−5.0* | 8.8+/−2.7 |
| VGLUT Positive Synapses | 15.8+/−2.8 | 16.7+/−3.5 | 16.3+/−2.8 |
| NR1 Synaptic Fluorescence (x103) | 4.0+/−0.8 | 7.2+/−1.5* | 3.7+/−1.1 |
| GluR1 Synaptic Fluorescence (x 103) | 10.1+/−3.6 | 8.8+/−2.5 | 9.7+/−2.2 |
| Primary Incubation (36 hr) | Secondary Incubation (30 min) | ||
| Conditioned Media (5x) | CM (5x) + APV/CNQX | CM (5x) Only | |
| NR1 Positive Synapses | 10.9+/−2.8 | 14.6+/−3.0* | 8.8+/−2.7 |
| VGLUT Positive Synapses | 17.3+/−3.6 | 17.0+/−2.9 | 16.3+/−3.4 |
| NR1 Synaptic Fluorescence (x103) | 5.6+/−1.4 | 8.5+/−2.3* | 4.9+/−1.0 |
| GluR1 Synaptic Fluorescence (x103) | 9.3+/−2.5 | 9.6+/−1.7 | 8.5+/−2.8 |
| B | |||
| Primary Incubation (36 hr) (Relative to Baseline) | Secondary Incubation (30 min) (Relative to 1° 36 hr Incubation) | ||
| Lactacystin | Lactacystin + APV/CNQX | Lactacystin Only | |
| Total NR1 | 2.3+/−0.8 | 0.9+/−0.2 | 1.0+/−0.1 |
| Total NR2A | 6.6+/−2.7 | 1.1+/−0.1 | 1.0+/−0.2 |
| Total NR2B | 5.8+/−2.4 | 1.1+/−0.2 | 0.9+/−0.1 |
| Total GluR1 | 1.7+/−0.7 | 1.0+/−0.1 | 1.1+/−0.2 |
| Surface NR1 | 1.5+/−0.3 | 3.5+/−0.9* | 0.8+/−0.2 |
| Surface NR2A | 5.3+/−1.9 | 3.2+/−0.5* | 1.2+/−0.3 |
| Surface NR2B | 6.0+/−1.8 | 2.7+/−0.4* | 1.1+/−0.2 |
| Surface GluR1 | 2.3+/−0.6 | 1.1+/−0.2 | 1.2+/−0.2 |
p<0.02 compared to primary incubation.
Discussion
Synaptic NMDA Receptor Localization is Regulated by Chronic Changes in Synaptic Activity
Several stages appear crucial to activity dependent synaptic NMDA receptor localization in spinal neurons. The first involves a slow global accumulation of NR2A and NR2B, influenced by synaptic activity and conditioned media, and mediated by proteasomes. In stage 2, NMDA receptors, as part of NR1/NR2 oligomers (Mi et al 2004), are directed to the surface. This stage is also regulated by synaptic activity. Moreover, it appears that the particular NR2 subunit to which NR1 is complexed determines the ability of synaptic activity to regulate this stage. Under basal conditions in which NR2A and NR2B accumulation is low, chronic or acute changes in synaptic activity have little effect on surface accumulation of NR1. Prior work had shown that under these conditions NR1 is complexed to NR2C and NR2D and is constitutively directed to the surface but not the synapse (Mi et al., 2004).
The final stage of activity dependent synaptic NMDA receptor localization involves an interaction between the terminal PDZ binding domain on the C-terminus of NR2A and NR2B and synaptic MAGUK proteins (Kim and Sheng 2004). Importantly, the synaptic localization of MAGUK proteins does not appear to be regulated by synaptic activity.
Synaptic Localization of NMDA Receptors is Dependent on NR2A and NR2B Accumulation and Proteasomal Activity
Previously, we (Mi et al., 2004) and others (Rao and Craig 1997; Barria and Malinow 2002) have shown that the synaptic localization of NMDA receptors is dependent on the NR2A and NR2B subunit (although see Mu et al., (2003) for a different mechanism). In the current paper we show that synaptic activity regulates the accumulation of NR2A and NR2B, which in turn mediates synaptic NMDA receptor localization. Dominant negative mutants showed that the C-terminal tail of either NR2A or NR2B could block the activity dependent localization of NMDA receptors, while similar mutants constructed from other NMDA receptor subunits were without effect. Like others (Mi et al., 2004; Mori et al., 1998; Steigerwald et al., 2000), we have found that the C-terminal IESDV motif on NR2A and NR2B is crucial for synaptic localization of NMDA receptors.
The increase in NR2A and NR2B subunit accumulation in response to activity blockade or proteasome inhibition is accompanied by an increase in surface NR1 accumulation and an increase in NMDA receptor half-life. The time course for NR2A and NR2B accumulation is similar to the slow phase of activity dependent synaptic NMDA receptor localization and likely underlies this phase. The preexisting synaptic MAGUK protein clusters, which are resistant to modification by changes in synaptic activity, provide the appropriate scaffolding for the NR2A and NR2B subunits to mediate their effect on NMDA receptor localization through an interaction between their PDZ binding domains and the terminal IESDV of NR2A and NR2B.
The demonstration that inhibitors of proteasomal but not lysosomal processing can both reproduce and saturate the effects of synaptic blockade confirms Ehlers’ (2003) contention that proteasomal function is pivotal in connecting changes in synaptic activity to changes in the localization of synaptic proteins. Further, the demonstration that overexpressing specific proteasomal subunits can alter synaptic NMDA receptor accumulation reinforces this notion. The LMP2 subunit has been shown to alter the protein specificity of proteasomes in lymphocytes (Groettrup et al., 2001) and elucidating the role of proteasome subunit composition in neurons will now be a priority. Moreover, detecting the precise ubiquitination sites on the NMDA receptor complex that control the NR2 dependent process will also be important (Kato et al., 2005). While our current work demonstrates the importance of proteasomes, our prior work (Mi et al., 2004) has shown that any mechanism which increases NR2A or NR2B will increase synaptic NMDA receptor accumulation. While proteasomal effects on NR2 subunit accumulation are key to NMDA receptor subunit accumulation and subsequent synaptic localization, it is unlikely that proteasomes can account for all aspects of synaptic NMDA receptor localization. In particular, the marked dissociation between total NR2A and NR2B levels, surface NR1 accumulation, and synaptic NMDA receptor localization in the presence of differing amounts of ambient synaptic activity argue that NR2A and NR2B accumulation is necessary but not sufficient for synaptic NMDA receptor localization. The fact that proteasomal inhibition alone can increase surface and synaptic NMDA receptor localization in the presence of differing ambient synaptic conditions suggest that the accumulation of the NR2A and NR2B subunits alone can increase synaptic NR1 localization independent of differences in local synaptic activity, although the latter can influence the amount of synaptic localization. This is consistent with our prior observations that simply overexpressing NR2A or NR2B can increase synaptic NMDA receptor accumulation (Mi et al., 2004).
NMDA Receptor Localization is Mediated in Part by Diffusible Substances in Conditioned Media
The demonstration that media from neurons grown in the presence of synaptic blockade can increase the synaptic localization of NMDA receptors implies that certain features of activity regulated synaptic NMDA receptor localization are not dependent on postsynaptic depolarization. Similarly, Rutherford et al (1998) have shown that BDNF blocks synaptic AMPA receptor scaling in cortical cultures, while Stellwagen and Malenka (2006) have recently shown that TNFα, secreted by glia, can upregulate AMPA receptor scaling. Neither BDNF nor TNFα had an effect on NMDA receptors but others have shown that other neurotrophins can modulate the expression of NMDA receptor subunits (Small et al., 1998; Choi et al., 2004). The specificity of the conditioned media for certain NMDA receptor subunits is reminiscent of the role of neuregulins at the neuromuscular junction (Loeb, 2003). The demonstration that soluble factors can interact with local synaptic activity to induce heterogeneities in receptor localization at the neuromuscular junction may be an important model for our system as well (Zhang and Poo, 2002). Our inability to demonstrate an effect of conditioned media on AMPA receptor scaling may reflect an effect of the growth factor rich media in which our neurons are grown, or may represent intrinsic differences between spinal and cortical neurons.
A Rapid Surface Accumulation of NMDA receptors Accompanies Activity -Dependent Synaptic NMDA Receptor Localization
In spite of the role played by NR2A and NR2B accumulation in synaptic NMDA receptor localization, there is not a direct relationship between the total accumulation of these subunits and global synaptic localization, The disconnect appears to be related to a second effect of synaptic activity on inhibiting surface (or synaptic) NMDA localization. This effect is rapidly reversed once residual synaptic activity is suppressed. This is similar to the pattern of AMPA receptor insertion/subtraction seen in LTP and LTD (Malenka and Bear 2004; Rumpel et al., 2005), but in contrast to those mechanisms, rapid NMDA receptor insertion in spinal neurons is induced by activity blockade. We do not yet have data on whether either surface translocation effect is as rapid as that seen in LTP/LTD and whether it is a localized or perisynaptic phenomenon.
Differences between Activity - Dependent Synaptic NMDA Receptor Localization and Synaptic AMPA receptor Scaling and Potential Implications
That NMDA receptors undergo activity dependent synaptic localization is clear from this and other work (Lissin et al., 1998; Ehlers 2003; Watt et al., 2000). What is not clear is the relationship between the activity-induced regulation of NMDA receptors and that of AMPA receptors. Our data suggests that in spinal neurons the 2 classes of glutamate receptors have different time courses for activity-dependent synaptic localization (Figure 2) as well as different mediators (Figure 4). In addition, the effect of synaptic activity on the acute surface translocation of NMDA receptor subunits, independent of tetanic stimulation, appears unique to NMDA receptors (Table 5).
Another important aspect of activity-dependent synaptic NMDA receptor localization is the heterogeneity in synaptic NMDA receptor localization seen in the presence of proteasomal inhibitors and conditioned media. This heterogeneity was reflected morphologically in the appearance of excitatory synapses with NMDA receptor clusters next to synapses without them, as well as in a large coefficient of variation of individual synaptic NR1 fluorescence intensities. The cause of the heterogeneity is not clear but could be related to differences in the molecular properties of the synapses or to differences in local synaptic activity. The fact that the excess heterogeneity is eliminated by TTX implies that the source of the heterogeneity lies in the frequency of, or response to, presynaptic glutamate release. It should be pointed out that in the current study heterogeneities in synaptic NMDA receptor localization were brought out by relatively artificial circumstances. However, variations in synaptic activity as may exist in vivo may make these heterogeneities manifest under more physiologic conditions. Given that AMPA receptor scaling is assumed to be a homogeneous process, preserving the relative weights of individual synapses (Turrigiano and Nelson 2004), the possibility of localized heterogeneities in synaptic NMDA receptor regulation and differing mechanisms for activity dependent synaptic NMDA and AMPA receptor accumulation suggests that the ratio of AMPA to NMDA receptors at individual synapses may be neither fixed nor static with important implications for local variation in the threshold for synaptic plasticity (Bear, 2003).
Experimental Methods
Cell Cultures
Low and high-density ventral spinal neurons from E15 rat embryos were grown as described (OBrien et al., 1997). When pharmacologic agents were added, the growth media was completely changed to fresh glial conditioned media with the indicated agents. Half the media was then changed every 24 hrs with media containing the pharmacologic agents at a 1x concentration. APV and CNQX were used at 0.1 to 0.3 mM and 10 to 20 μM respectively. Picrotoxin and strychnine were used at 200 μM and 2 μM respectively, leupeptin at 50μM, TTX at 10 μM, lactacystin at 1 μM, MG132 at 10 μM and actinomycin D at 10μM.
Conditioned Media
10 cm dishes containing high-density spinal or glial cultures were switched to 6 ml of fresh growth media with or without APV (.3 mM) and CNQX (20 μM). The media was left in place for 48 hrs with a 50% change at 24 hrs. Control conditioned media was derived from cultures grown for 48 hrs in fresh media without APV and CNQX. After collection, the control neuronal media had APV and CNQX added to a final concentration of 1mM and 60 μM respectively. The media were dialyzed overnight against several changes of complete growth media with 10 mM HEPES buffer at 4 degrees using a 1 kDa cutoff dialysis membrane (Spectra/Por). The dialyzed media was then frozen, thawed, filtered through a 0.2 μM filter and used in a 1:1 dilution with preexisting growth media. The conditioned or control media was refreshed every 12 hrs with a 50% change for 36 to 48 hrs.
Neuronal Transfections
Spinal neurons were transfected after 7 or 8 days in vitro as described (Mi et al., 2004). A GFP expression construct was included with the test construct at a 1:4 ratio. In experiments looking at the surface localization of NR1, N-terminal tagged myc-NR1-1A (C2 cassette) was transfected into neurons alone and surface staining assayed as described below. In experiments with the proteasomal subunits LMP2 or C2, neurons were transfected with an equal mixture of GFP tagged and untagged LMP2 or C2 and assayed for synaptic NR1 clusters 36 hrs after transfection. Neurons were grown in the presence of APV and CNQX during that time.
Immunohistochemistry
Neuronal cultures were fixed and processed for immunocytochemistry as described (Mi et al., 2004). Live immunostaining prior to fixation was carried out for 30 minutes at 37 degrees in MEM plus APV (100 μM) and CNQX (10 μM). Synaptic localization of NR1 and GluR2 was performed by colocalizing the combined polyclonal VGLUT1 and VGLUT2 antibodies (each at 1:1000) with the monoclonal pan NR1 antibody (S3C11) at 0.5 μg/ml or the GluR2 specific monoclonal (Chemicon; MAB 397) at 1 μg/ml. For conditioned media experiments we tabulated synaptic GluR1 rather than GluR2 clusters using a C-terminal polyclonal GluR1 antibody (OBrien et al., 1997).
Clustering Assay
The number of synaptic glutamate receptor clusters on individual neurons was determined by staining for VGLUT1/2 (polyclonal) and NR1 or GluR2 monoclonals and then observing their correlation in a blinded assay. The technical details of the assay have been published (OBrien et al., 2002). In all cases, the numbers expressed are the mean +/− S.D. of the mean of 3 or 4 separate experiments.
In experiments looking at the effect of dominant-negative mutations on endogenous synaptic NR1 clustering, serial GFP positive neurons were identified in a blinded assay and the number of VGLUT (Blue) associated NR1 clusters (Red) determined. In experiments looking at the surface localization of NR1, N-terminal myc-tagged NR1-1A (C2 cassette) was transfected into neurons. Cultures were stained live with mouse monoclonal anti myc (0.5 μg/ml, 30 minutes at 37°), fixed in sequential paraformaldehyde and methanol and then stained with polyclonal anti VGLUT followed by rhodamine anti mouse and FITC anti rabbit. Sequential, surface myc-positive neurons were identified and scored for synaptic NR1 as described above.
Quantitation of Proteins at Individual Synapses
Quantitative estimates of synaptic NR1, GluR1, GluR2 or PDZ immunofluorescence were performed as described (5). In most cases, except when specifically designated, we analyzed synaptophysin or VGLUT associated immunopositive clusters of the indicated protein (i.e. synapses that appeared immunonegative were not analyzed). Each data point represents all the synapses visible in a single (cell centered) 100 x visual field from a minimum of 30 randomly and sequentially selected neurons taken from a minimum of 3 separate experiments. The data is expressed (unless otherwise noted) as the mean of the means of the separate experiments +/− the SD of those means.
Antibodies
VGLUT1 and VGLUT2 antibodies were purchased from Synaptic Systems and were used by diluting each 1:1000 in the same tube for immunochemistry and 1:1000 (individually) for western blotting. The pan-PDZ monoclonal antibody K28/86.2 was purchased from Upstate Biotech and used at 2 μg/ml. This antibody recognizes the PDZ domains on PSD-95, Chapsyn-110 and SAP 97. Other antibodies used in this study have been described before (OBrien et al., 1997; Mi et al., 2002). For NR1 half-life experiments, biotinylated and streptavidin immunoprecipitated material was probed with the pan-NR1 monoclonal 05-432 (Upstate). This same pan NR1 monoclonal was used for most quantitative assessments of NR1 levels (immunoblots) unless otherwise specified. Antibodies to the proteasomal subunits C2 (#3325) and LMP2 (#3328) were purchased from abcam and used for immunoblotting at 1:500 and 1:100 respectively.
Immunoblotting and Quantitation of Relative Protein Levels
Immunoblots were performed as described (Mammen et al., 1997). For receptor half-life experiments, cell surface proteins were biotinylated and streptavidin immunoprecipitated as a function of time (Mammen et al., 1997). The volume of sample loaded for each half-life experiment was adjusted such that all time points would be within the linear range of the film. Serial dilutions of the t = 0 sample were run on each gel to provide a standard curve for each experiment. Material from APV and CNQX treated cultures was diluted so that the total NR1 signal at t=0 would be equivalent to the control samples. All proteins were visualized by ECL and analyzed using a Molecular Dynamics Personal Densitometer SI. To calculate half-lives, we plotted the natural logarithm of the percentage of protein remaining as a function of time. Quantitation of protein levels in total cell material was performed by generating a standard curve for each film using serial dilutions of either the control or APV/CNQX sample such that individual band intensities could be expressed as a percentage of the control or APV and CNQX treated sample.
Northern Blots
Total RNA or mRNA was extracted using Totally RNA (Ambion) or Oligotex Direct (Qiagen) respectively and processed for Northern blotting and quantitation as described (Mi et al., 2004), running 5 μg of total RNA or 2 μg mRNA per lane. Probes consisted of a 1.2 kb Pst1 fragment of rat NR2A, an 800 bp PCR fragment (3677–4470) of rat NR2B, and a 1.4 kB Xho1 – Hind3 fragment of NR1 labeled with 32P by random priming.
Constructs
All constructs used in this study have been described (Mi et al., 2004) except for the following: The cytoplasmic tail of NR2B, consisting of the terminal 256 amino acids of the rat NR2B C-terminus, was inserted in-frame with an N-terminal myc tag into the EcoR1 and Not1 sites of pCMV-Myc. NR2BCTΔ10 was similar to NR2BCT, minus the final 10 amino acids. Rat NR2CCT was generated from the terminal 248 amino acids of the rat NR2C C-terminus, inserted in frame with an N-terminal myc tag using the EcoR1 and Not1 sites of pCMV-Myc. NR2DCT and NR2DCTΔ10 were generated from the terminal 322 amino acids of the rat NR2D C-terminus minus the terminal 10 amino acids for NR2DCTΔ10. Mouse LMP and LMP2-GFP (after Reits et al., 1997) was obtained by RT PCR from mouse brain polyA RNA using the primers (SENSE) GCTAGCCTTGCAGGGATGCTGCGGGCAGGAGCACC and (ANTISENSE) ACCGGTGGCTCATCGTAGAATTTTGGCAGCTC (LMP2-GFP) or (ANTISENSE) ACCGGTGGTCATCACTCATCGTAGAATTTTGGCAGCTC (LMP2), TA cloned into PCR4 (Invitrogen), digested with Nhe1 and Age1, subcloned into pEGFPN1 (Clontech) and sequenced completely. Mouse Proteasome subunit C2 was obtained by RT PCR from mouse brain polyA RNA using the primers (SENSE) GCTAGCCTTGCAGGGATGTTTCGAAACCAGTATGACAA and (ANTISENSE) ACCGGTGGATGTTCCATTGGTTCATCGG (C2-GFP) or (ANTISENSE) ACCGGTGGTTATTAATGTTCCATTGGTTCATCGG (C2) and subcloned similar to LMP2.
Statistical Comparisons
Comparisons between the number of clusters per neuron under different experimental conditions or between the intensity of those synaptic clusters was performed using a paired t-test of the means of a series of at least 3 separate experiments, each including all the appropriate and indicated conditions. Comparisons between total or surface subunit protein accumulation were performed by comparing the relative increase or decrease of the indicated subunits compared to control over a minimum of three experiments each including all the appropriate and indicated conditions using a paired t-test. For the most part no corrections were made for multiple comparisons as the results were neither post-hoc nor independent (i.e. an increase in NR2A would be expected to be associated with a change in NR1, or an increase with activity blockade would be expected to be accompanied by a decrease with activity augmentation). Moreover the results are physiologically plausible. Lastly, almost all the significantly different results mentioned in the text (with the notable exceptions mentioned in the legend to Table 2 and Figure 3) were either replicated in non-overlapping experiments, or resulted in confirmatory data in downstream experiments.
Supplementary Material
Acknowledgments
Supported by NIH Grant R01NS037694 and the Robert Packard Center for ALS Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barria A, Malinow R. Subunit-specific NMDA receptor trafficking to synapses. Neuron. 2002;35:345–53. doi: 10.1016/s0896-6273(02)00776-6. [DOI] [PubMed] [Google Scholar]
- Bear MF. Bidirectional synaptic plasticity: from theory to reality. Philos Trans R Soc Lond B Biol Sci. 2003;358:649–55. doi: 10.1098/rstb.2002.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbea M, Dreier L, Dittman JS, Grunwald ME, Kaplan JM. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C. elegans. Neuron. 2002;35:107–20. doi: 10.1016/s0896-6273(02)00749-3. [DOI] [PubMed] [Google Scholar]
- Choi SY, Hwang JJ, Koh JY. NR2A induction and NMDA receptor-dependent neuronal death by neurotrophin-4/5 in cortical cell culture. J Neurochem. 2004;88:708–16. doi: 10.1046/j.1471-4159.2003.02187.x. [DOI] [PubMed] [Google Scholar]
- Desai NS, Cudmore RH, Nelson SB, Turrigiano GG. Critical periods for experience-dependent synaptic scaling in visual cortex. Nat Neurosci. 2002;5:783–9. doi: 10.1038/nn878. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin- system. Nat Neurosci. 2003;6:231–42. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Groettrup M, van den Broek M, Schwarz K, Macagno A, Khan S, de Giuli R, Schmidtke G. Structural plasticity of the proteasome and its function in antigen processing. Crit Rev Immunol. 2001;21(4):339–58. [PubMed] [Google Scholar]
- Kato A, Rouach N, Nicoll RA, Bredt DS. Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. PNAS. 2005;102:5600–5. doi: 10.1073/pnas.0501769102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–81. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Lee DH, Goldberg AI. Proteasome inhibitors: valuable new tools for cell biologists. Trends in Cell Biology. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- Leslie KR, Nelson SB, Turrigiano GG. Postsynaptic depolarization scales quantal amplitude in cortical pyramidal neurons. J Neurosci. 2001;21:RC170:1–6. doi: 10.1523/JNEUROSCI.21-19-j0005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Sheng M. Some assembly required: the development of neuronal synapses. Nat Rev Mol Cell Biol. 2003;4:833–41. doi: 10.1038/nrm1242. [DOI] [PubMed] [Google Scholar]
- Lissin DV, Gomperts SN, Carroll RC, Christine CW, Kalman D, Kitamura M, Hardy S, Nicoll RA, Malenka RC, von Zastrow M. Activity differentially regulates the surface expression of synaptic AMPA and NMDA glutamate receptors. Hippocampus Proc Natl Acad Sci U S A. 1998;95:7097–102. doi: 10.1073/pnas.95.12.7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JA. Neuregulin: an activity-dependent synaptic modulator at the neuromuscular junction. J Neurocytol. 2003;32:649–64. doi: 10.1023/B:NEUR.0000020640.84708.35. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Malinow R. AMPA receptor trafficking and long-term potentiation. Philos Trans R Soc Lond B Biol Sci. 2003;358:707–14. doi: 10.1098/rstb.2002.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammen AL, Huganir RL, O’Brien RJ. Redistribution and stabilization of cell surface glutamate receptors during synapse formation. J Neurosci. 1997;17:7351–8. doi: 10.1523/JNEUROSCI.17-19-07351.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi R, Tang X, Sutter R, Xu D, Worley P, O’Brien RJ. Differing mechanisms for glutamate receptor aggregation on dendritic spines and shafts in cultured hippocampal neurons. J Neurosci. 2002;22:7606–16. doi: 10.1523/JNEUROSCI.22-17-07606.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi R, Sia GM, Rosen K, Tang X, Moghekar A, Black JL, McEnery M, Huganir RL, O’Brien RJ. AMPA receptor-dependent clustering of synaptic NMDA receptors is mediated by Stargazin and NR2A/B in spinal neurons and hippocampal interneurons. Neuron. 2004;44:335–49. doi: 10.1016/j.neuron.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Mishto M, Bellavista E, Santoro A, Stolzing A, Ligorio C, Nacmias B, Spazzafumo L, Chiappelli M, Licastro F, Sorbi S, Pession A, Ohm T, Grune T, Franceschi C. Immunoproteasome and LMP2 polymorphism in aged and Alzheimer’s disease brains. Neurobiol Aging. 2006;27(1):54–66. doi: 10.1016/j.neurobiolaging.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Mori H, Manabe T, Watanabe M, Satoh Y, Suzuki N, Toki S, Nakamura K, Yagi T, Kushiya E, Takahashi T, Inoue Y, Sakimura K, Mishina M. Role of the carboxy-terminal region of the GluR epsilon2 subunit in synaptic localization of the NMDA receptor channel. Neuron. 1998;21:571–80. doi: 10.1016/s0896-6273(00)80567-x. [DOI] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD. Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron. 2003;40:581–94. doi: 10.1016/s0896-6273(03)00676-7. [DOI] [PubMed] [Google Scholar]
- Myung J, Kim KB, Crews CM. The ubiquitin-proteasome pathway and proteasome inhibitors. Med Res Rev. 2001;21(4):245–73. doi: 10.1002/med.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien RJ, Mammen AL, Blackshaw S, Ehlers MD, Rothstein JD, Huganir RL. The development of excitatory synapses in cultured spinal neurons. J Neurosci. 1997;17:7339–50. doi: 10.1523/JNEUROSCI.17-19-07339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien RJ, Kamboj S, Ehlers MD, Rosen KR, Fischbach GD, Huganir RL. Activity-dependent modulation of synaptic AMPA receptor accumulation. Neuron. 1998;21:1067–78. doi: 10.1016/s0896-6273(00)80624-8. [DOI] [PubMed] [Google Scholar]
- O’Brien R, Xu D, Mi R, Tang X, Hopf C, Worley P. Synaptically targeted Narp plays an essential role in the aggregation of AMPA receptors at excitatory synapses in cultured spinal neurons. J Neurosci. 2002;22:4487–98. doi: 10.1523/JNEUROSCI.22-11-04487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Craig AM. Activity regulates the synaptic localization of the NMDA receptor in hippocampal neurons. Neuron. 1997;19:801–12. doi: 10.1016/s0896-6273(00)80962-9. [DOI] [PubMed] [Google Scholar]
- Reits EA, Benham AM, Plougastel B, Neefjes J, Trowsdale J. Dynamics of proteasome distribution in living cells. EMBO J. 1997;16:6087–94. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renart A, Song P, Wang XJ. Robust spatial working memory through homeostatic synaptic scaling in heterogeneous cortical networks. Neuron. 2003;38:473–85. doi: 10.1016/s0896-6273(03)00255-1. [DOI] [PubMed] [Google Scholar]
- Rumpel S, LeDoux J, Zador A, Malinow R. Postsynaptic receptor trafficking underlying a form of associative learning. 2005;308:83–88. doi: 10.1126/science.1103944. [DOI] [PubMed] [Google Scholar]
- Rutherford LC, Nelson SB, Turrigiano GG. BDNF has opposite effects on the quantal amplitude of pyramidal neuron and interneuron excitatory synapses. Neuron. 1998;21:521–30. doi: 10.1016/s0896-6273(00)80563-2. [DOI] [PubMed] [Google Scholar]
- Small DL, Murray CL, Mealing GA, Poulter MO, Buchan AM, Morley P. Brain derived neurotrophic factor induction of N-methyl-D-aspartate receptor subunit NR2A expression in cultured rat cortical neurons. Neurosci Lett. 1998;252:211–4. doi: 10.1016/s0304-3940(98)00587-4. [DOI] [PubMed] [Google Scholar]
- Steigerwald F, Schulz TW, Schenker LT, Kennedy MB, Seeburg PH, Kohr G. C-Terminal truncation of NR2A subunits impairs synaptic but not extrasynaptic localization of NMDA receptors. J Neurosci. 2000;20:4573–81. doi: 10.1523/JNEUROSCI.20-12-04573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen D, Malenka RC. Synaptic scaling mediated by glial TNF-α. Nature. 2006;440:1054–1059. doi: 10.1038/nature04671. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- Van den Eynde BJ, Morel S. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr Opin Immunol. 2001;13(2):147–53. doi: 10.1016/s0952-7915(00)00197-7. [DOI] [PubMed] [Google Scholar]
- Vanhoutte P, Bading H. Opposing roles of synaptic and extrasynaptic NMDA receptors in neuronal calcium signalling and BDNF gene regulation. Curr Opin Neurobiol. 2003;13:366–71. doi: 10.1016/s0959-4388(03)00073-4. [DOI] [PubMed] [Google Scholar]
- Wang XJ. Synaptic reverberation underlying mnemonic persistent activity. Trends Neurosci. 2001;24:455–63. doi: 10.1016/s0166-2236(00)01868-3. [DOI] [PubMed] [Google Scholar]
- Watt AJ, van Rossum MC, MacLeod KM, Nelson SB, Turrigiano GG. Activity coregulates quantal AMPA and NMDA currents at neocortical synapses. Neuron. 2000;26:659–70. doi: 10.1016/s0896-6273(00)81202-7. [DOI] [PubMed] [Google Scholar]
- Zhang X, Poo MM. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:549–50. doi: 10.1016/s0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.