Abstract
NifB-co, a Fe-S cluster produced by the enzyme NifB, is an intermediate on the biosynthetic pathway for the iron molybdenum cofactor (FeMo-co) of nitrogenase. We have used Fe K-edge extended x-ray absorption fine structure (EXAFS) spectroscopy together with 57Fe nuclear resonance vibrational spectroscopy (NRVS) to probe the structure of NifB-co while bound to the NifX protein from Azotobacter vinelandii. EXAFS analysis of the NifX:NifB-co complex yields Fe-S distances of 2.26 Å and Fe-Fe distances of 2.66 Å and 3.74 Å. Search profile analyses reveals the presence of a single Fe-N (or C, O) interaction at 2.04 Å, implying that the interstitial light atom proposed to be present in FeMo-co is already inserted into NifB-co. NRVS reveals strong bands from Fe-S bending and stretching modes peaking around 270, 315, 385, and 408 cm−1. Additional intensity at ~185 – 200 cm−1 is interpreted as a set of cluster ‘breathing’ modes similar to those seen for the FeMo-cofactor. The strength and location of these modes also strongly suggest that the FeMo-co interstitial light atom seen in the crystal structure is already in place in NifB-co. Both the EXAFS and NRVS data are best simulated using a Fe6S9X trigonal prism structure analogous to the 6Fe core of FeMo-co, although a 7Fe structure made by capping one trigonal 3S terminus with Fe cannot be ruled out. This implies that the interstitial light atom is already present at an early stage in FeMo-co biosynthesis prior to the incorporation of Mo and R-homocitrate.
Keywords: Nitrogenase, NifB-co, FeMo-co, FeV-co, FeFe-co, NifB, NifX, NafY, FeMo-co biosynthesis, iron-sulfur protein, SAM-radical, x-ray absorption spectroscopy, XAS, extended x-ray absorption fine structure, EXAFS, near-edge spectrum, 57Fe, nuclear resonance vibrational spectroscopy, NRVS, Mössbauer, synchrotron radiation, normal mode
Introduction
All life depends on the input of the element nitrogen into the biosphere by biological nitrogen fixation performed only by diazotrophic microorganisms. These organisms achieve N2 fixation using at least one member of the nitrogenase family of metalloenzymes, with the Mo-dependent enzymes being the most widely distributed and best studied although V-dependent and Fe-only forms are also well known. The active site of Mo-Nitrogenase, which is contained within the so-called MoFe protein, is a remarkable [7Fe-9S-Mo-X-homocitrate] cluster, called the iron-molybdenum cofactor (FeMo-co). This cluster both binds the substrate N2 and catalyzes the chemically difficult reduction of the N2 triple bond to form NH3 1. The V-Nitrogenases and Fe-Nitrogenases contain similar cofactors where the Mo is thought to be substituted by V or Fe, forming FeV-co and FeFe-co respectively 2.
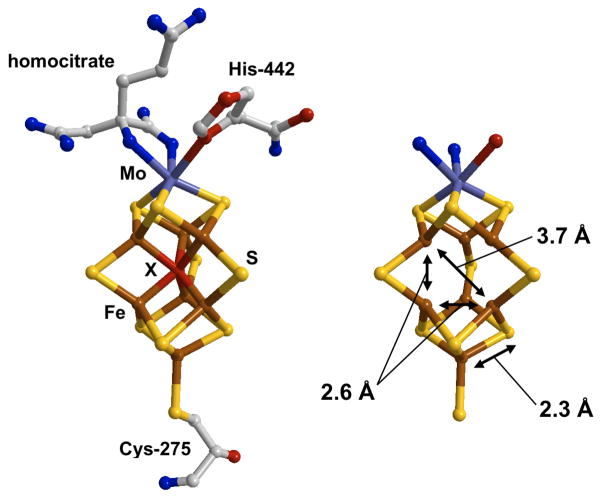
The structure of FeMo-co (figure 1) can be viewed as two Fe-S cubane clusters, each missing one bridging S, making [3FeMo-3S] and [4Fe-3S] that are joined with three extra bridging S so that 6 of the 7 Fe atoms of FeMo-co form a trigonal prism core. The cluster is anchored to the Mo-nitrogenase polypeptide by a cysteine at the Fe terminus and by a histidine at the Mo end. The Mo is also coordinated by R-homocitrate, a bidentate carboxylate ligand that is essential for N2 reduction activity. Much interest has been generated by the most recent crystal structure for Mo-nitrogenase 3. It was proposed that, after correction for Fourier artifacts, the electron density at the center of the FeMo-co could be assigned to a single light interstitial atom, namely C, N or O 3. As the element of this atom is unknown it is commonly denoted as “X”. The presence of this atom has been supported by theoretical studies 4 as well as recent analyses of nuclear resonance vibrational spectroscopy (NRVS) data 5.
Figure 1.
The structure of FeMo-co. (Left) The structure as determined by x-ray crystallography on the Mo-Nitrogenase enzyme (Protein Data Bank: 1M1N 3). (Right) The Mo-7Fe-9S core showing the 2.3 Å Fe-S, 2.6 Å short Fe-Fe and 3.7 Å long Fe-Fe distances probed by EXAFS spectroscopy. Color code: Fe (brown), sulfur (yellow), interstitial atom X (red). The central atom has been omitted in (b) for clarity.
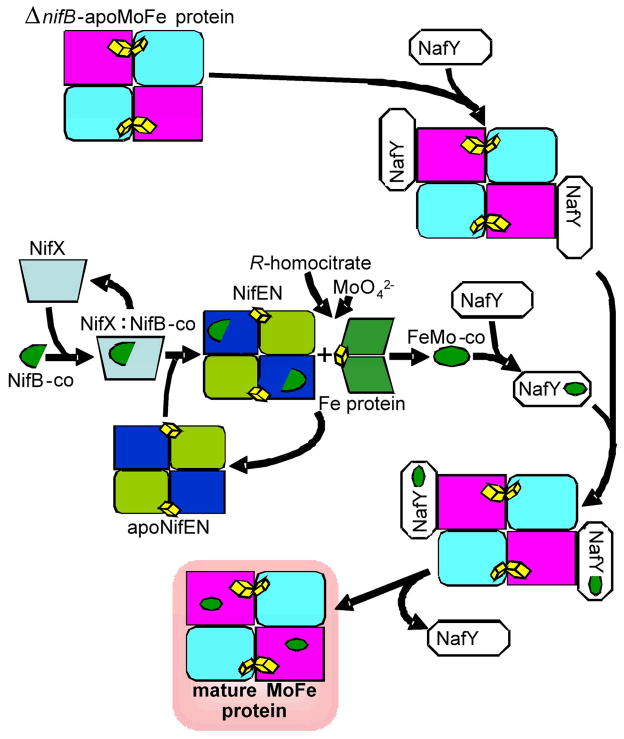
The genetic basis for the Mo-dependent nitrogenase is the nif gene cluster, which not only contains the structural genes encoding the nitrogenase component proteins, but importantly also codes for enzymes and chaperones which cooperate to biosynthesize the FeMo-co and the other unique metallo-cofactors found in the nitrogenase systems. These Nif proteins make up these biosynthetic pathways by performing an intriguing and complex series of bioinorganic reactions using Fe and S together with Mo or V. Figure 2 illustrates our understanding of the Nif catalyzed biosynthetic pathways for Mo-nitrogenase. However, the detailed chemistry of these pathways is currently undetermined 6.
Figure 2.
Proposed scheme for FeMo-co biosynthesis showing the proposed roles of NifB, NifX, NifEN, NifH, NafY and the MoFe protein of Mo-nitrogenase.
A key metabolic intermediate in the biosynthesis of FeMo-co is NifB-co, a low-molecular weight Fe-S cluster that is the product of S-adenosyl methionine mediated radical (SAM-radical) chemistry catalyzed by the 108 kDa dimeric enzyme NifB 7. In addition to being a precursor for synthesis of FeMo-co, NifB-co is also hypothesized to be the precursor for the synthesis of FeV-co and FeFe-co, suggesting that NifB-co forms the core portion that is common among all three nitrogenase active site cofactors. 6 The essential natures of NifB-co and the SAM-radical enzyme NifB are highlighted by the inability for diazotrophic growth upon deletion of the nifB gene in N2 fixing organisms such as Azotobacter vinelandii or Klebsiella pneumoniae. Genetic disruption of FeMo-co biosynthetic steps downstream of NifB also has deleterious effects on diazotrophic growth, but causes the cellular accumulation of NifB-co. The NifB-co Fe-S cluster can be isolated from a nifN mutant strain of K. pneumoniae. Isolated NifB-co can be used in vitro to complement a cellular extract from a ΔnifB strain, which is incapable of FeMo-co synthesis, and permits the synthesis of FeMo-co producing a functional nitrogenase. Isolated NifB-co does not contain Mo required for FeMo-co synthesis, but is thought to contain the majority of the Fe and S components of FeMo-co. NifB-co is further processed and converted to FeMo-co by the combined activities of NifEN and NifH. The NifB-co precursor binds to NifEN8 and is subjected to further modifications that may include the addition of more Fe, the addition of more S, and the attachment of Mo and R-homocitrate 6.
In addition to the key Nif proteins such as NifB, NifEN and NifH, additional nif and non-nif encoded proteins enhance the efficiency of FeMo-co biosynthesis by binding and stabilizing the Fe-S cluster intermediates of the pathway or the protein components. One such protein, NafY, is a 26 kDa protein that tightly binds FeMo-co (Kd ~ 60 nM) to form the NafY:FeMo-co complex 9. Another, NifX, is a 17 kDa protein that is able to bind isolated NifB-co with high affinity (Kd ~ 1 μM) 8 and the resulting NifX:NifB-co complex can donate NifB-co to the form of NifEN that is “uncharged”, that is without precursor already bound 8. Interestingly, NifX can also extract a different FeMo-co precursor from “charged” NifEN. We have recently characterized this Fe-S cluster – which we have termed the “VK-cluster” – and shown that it is spectroscopically distinct from NifB-co, and that it can act as a precursor for in vitro FeMo-co synthesis 8. This chemistry led to the hypothesis that NifX serves as an intracellular carrier for NifB-co or other FeMo-co biosynthetic intermediates.
Little is known about the molecular composition and structure of NifB-co, how it is assembled by NifB, or its use as a substrate by NifEN. Clearly, structural information on the NifB-co Fe-S cluster is required to understand of the FeMo-co biosynthetic pathway and the pertinent enzyme mechanisms. Recently, some structural information was obtained on a NifEN bound FeMo-co precursor using EXAFS difference spectroscopy of charged and uncharged NifEN 10 and these authors identified structural elements of partially assembled FeMo-co. However it is not clear whether this study addressed bound NifB-co, the VK cluster or some other FeMo-co precursor. Hence, in order to fully elucidate the bioinorganic chemistry underpinning diazotrophy, it is necessary to uncover the structural nature of NifB-co.
In this paper we use Fe K-edge extended x-ray absorption fine structure (EXAFS) spectroscopy, a well-developed technique 11, together with the more novel 57Fe nuclear resonance vibrational spectroscopy (NRVS) 12, 13 to probe the structure of NifB-co bound to NifX from A. vinelandii. We use measurements on the NafY:FeMo-co complex as a control to help calibrate and interpret the NifX:NifB-co results. The two techniques are shown to be highly complementary. EXAFS spectroscopy is essentially x-ray absorption spectroscopy (XAS) that is focused on the analysis of the extended structure above an x-ray edge. This generates the number, distance and element of the atoms immediately surrounding the absorbing atom. By contrast, 57Fe NRVS exploits vibrational structure of the inelastically scattered radiation that is concomitant with Mössbauer absorption 12. NRVS analysis results in a vibrational spectrum between approximately 10 – 1000 cm−1 that is a direct measure of the motion of the resonant 57Fe nucleus along the direction of the incident x-ray beam for the observed normal modes.
Experimental
Protein Purification and Sample Preparation
NifX was overexpressed in E. coli and purified using recently described procedures 8. NifB-co was produced in K. pneumoniae UN1217 and purified using a modified larger scale version of the procedure described by Shah and coworkers 14. All manipulations of the protein and sample preparation were performed under strict anaerobic conditions using crimp-sealed vials and gas-tight syringes or inside an anaerobic (<5 ppm O2) glove box (Coy) with a 95% N2/5% H2 environment.
Two different NifX:NifB-co samples were prepared. The first contained 56Fe (natural abundance) and was studied by EXAFS spectroscopy only. This was prepared by binding NifB-co (1.2 μmol total Fe) to phenyl sepharose equilibrated in 50 mM MOPS, pH 7.2, 2 mM Na2S2O4, washed free of detergent, and then eluted by applying 13 mg of purified NifX. The NifB-co eluted from the phenyl sepharose resin bound to NifX as a complex and was concentrated by ultrafiltration to ~1 mM in protein and loaded into Lucite cells (dimensions: 3 × 20 × 2 mm). The sample was prepared without use of a glassing agent such as 10% glycerol as it was feared that this could disrupt the cluster. The second sample contained the 57Fe necessary for NRVS studies and this was studied by both NRVS and EXAFS. Incorporation of 57Fe was achieved by replacing the regular source of Fe with 11 μM 57Fe (98%, Isotec) in the K. pneumoniae growth medium. A sarkosyl membrane extract containing 57Fe NifB-co was prepared from 190 g of 57Fe enriched cells and was applied to a column of Sephacryl 200 (S200), which NifB-co binds. The column was washed free of all proteins and then washed with detergent free buffer. Pure and metal free NifX (4 mg) was applied to the NifB-co bound S200 column and the NifB-co was eluted as the NifX:(57Fe)NifB-co complex. The NifX:(57Fe)NifB-co samples were concentrated to ~5 mM in protein by ultrafiltration and loaded into 3 × 7 × 1 mm (interior dimensions) Lucite cuvettes. The final Fe concentration, as estimated from the Fe K-emission and NRVS resonance peak count rates was approximately 30 mM, consistent with the sample preparation and a stoichiometry of 6–7 Fe per NifX molecule.
The NafY:FeMo-co complex was prepared using a modification of previously described procedures 9. FeMo-co was prepared from A. vinelandii MoFe protein15 and A. vinelandii NafY was purified 9 as previously described. To prepare the sample of NafY:FeMo-co for EXAFS, 1.4 μmol of NafY was diluted with 50 mM MOPS, pH 7.4, 2 mM Na2S2O4 in a stirred ultrafiltration cell fitted with a YM10 membrane (Millipore). FeMo-co (0.7 μmol total), dissolved in NMF solution, was added to the diluted NafY in several portions to limit the NMF concentration (<3%), with repeated concentration and dilution with every addition of FeMo-co. The NMF concentration was decreased by an additional round of dilution and concentration to a volume of 550 μL. The solution of NafY:FeMo-co was then passed through a Sephadex-10 size exclusion column (1 × 15 cm) to remove residual NMF, concentrated to 60 μL and frozen in a Lucite cell (dimensions: 3 × 20 × 2 mm). The sample for NRVS was prepared using essentially the same method using 57Fe MoFe protein and 3 × 7 × 1 mm Lucite cuvettes
X-ray Absorption Spectroscopy
Fe K-edge x-ray absorption data were measured at Stanford Synchrotron Radiation Laboratory beamline 9-3, with a Si 220 double-crystal monochromator and two rhodium-coated mirrors: one flat premonochromator mirror for harmonic rejection and vertical collimation and one toroidal postmonochromator mirror for focusing. Fluorescent x-rays were measured using a 30-element Ge fluorescence detector (Canberra Industries), fitted with Soller slits and a Mn filter to minimize the relative contribution of scattered radiation. An Oxford Instruments CF1208 liquid helium cooled sample cryostat was used to maintain the sample temperature at 9 K. The x-ray energy was calibrated using the first and major inflection point of a standard Fe foil set as 7112 eV. This was measured at the same time as the sample spectrum using two ion chambers positions downstream of the cryostat. In order to minimize the effect of x-ray photoreduction, the sample was moved after each scan so that the beam irradiated a different spot on the sample for each scan – typically a total of 3 – 4 spots were available for each sample and 5 – 11 scans were collected requiring reuse of each spot. The Fe K-edge structure and position were monitored to ensure no photochemistry.
EXAFS data analysis
EXAFS data were analyzed using the EXAFSPAK software suite 16. As the samples contained no glassing agent, it was first necessary to rigorously screen the scans from the individual detector elements to eliminate any diffraction artifacts. Curve fitting used the EXAFSPAK program OPT, with single-scattering phase and amplitude functions calculated using FEFF 7.0 11. The NafY:FeMo-co complex was used as a reference sample to provide experimentally determined estimates of Debye-Waller factors.
NRVS Measurements
NRVS is a synchrotron radiation technique where a highly monochromatic (ΔE ~ 1 meV) x-ray beam is scanned through the nuclear resonance 12, 17. In addition to the familiar ‘zero phonon’ (recoil-free) Mössbauer resonance, there are additional transitions corresponding to nuclear excitation combined with excitation (Stokes) or de-excitation (anti-Stokes) of vibrational modes. The only requirement for NRVS intensity is motion of the resonant nucleus (in this case 57Fe) along the direction of the incident X-ray beam in the given normal mode. The details of NRVS theory have been detailed elsewhere 12, 18. Essentially, the single phonon NRVS excitation probability S1(ν̄) is directly proportional to the Fe partial vibrational density of states (PVDOS) DFe (ν̄) 19. The PVDOS function DFe (ν̄) arises from convolution of a lineshape ℒ (ν̄ − ν̄α) with the Fe mode composition factor, , which represents the fraction of kinetic energy due to Fe for normal modes α at resonant energies ν̄α.18 20
57Fe NRVS spectra were recorded using published procedures 12 at beamline BL09XU at SPring-8, Japan 21. The flux was ~3 × 109 in a 1.1 meV bandwidth (1 meV ≡ 8.0655 cm−1). During NRVS measurements, the samples were maintained at ~ 20 K using a liquid He cryostat. Exact temperatures for individual spectra were calculated using the ratio of anti-Stokes to Stokes intensity according to: S(−E) = S(E)exp(−E/kT). Spectra were recorded between −20 meV and 80 meV in 0.25 meV steps. Nuclear fluorescence and Fe K fluorescence (from internal conversion) were recorded using an APD array 22. Each scan took about 40 min, and all scans were added and normalized to the intensity of the incident beam. The reported NRVS spectrum represents the average of 16 scans.
Normal Mode Calculations
Normal mode calculations for NifB-co candidate structures were conducted with a Urey-Bradley force field using a modification of program ‘Vibratz’ 23. A QR algorithm was used for finding eigenvectors 24. The structures used the distance constraints from the EXAFS analysis. The mode composition factor ejα2 for mode α and atom j was calculated by 13, 18, 25:
| (1) |
where mj and rjα are the mass of atom j and its eigenvector in mode α respectively. For comparison with the experimental spectra, Gaussian functions were calculated centered around each ωα, the frequency of mode α, using an 7 cm−1 full width at half maximum and an amplitude proportional to ejα2. The partial vibrational density of states for atom j was then obtained by summing these Gaussian functions over all modes.
Results
We have measured EXAFS and NRVS spectra of the NifX:NifB-co complex. We have also measured analogous data for NafY:FeMo-co. As this protein complex contains the completed FeMo-co center it provides an excellent control sample for the interpretation of the NifX:NifB-co data. We present first the EXAFS and near-edge spectra, together with their analysis, followed by the NRVS results.
EXAFS and Near-Edge Spectra
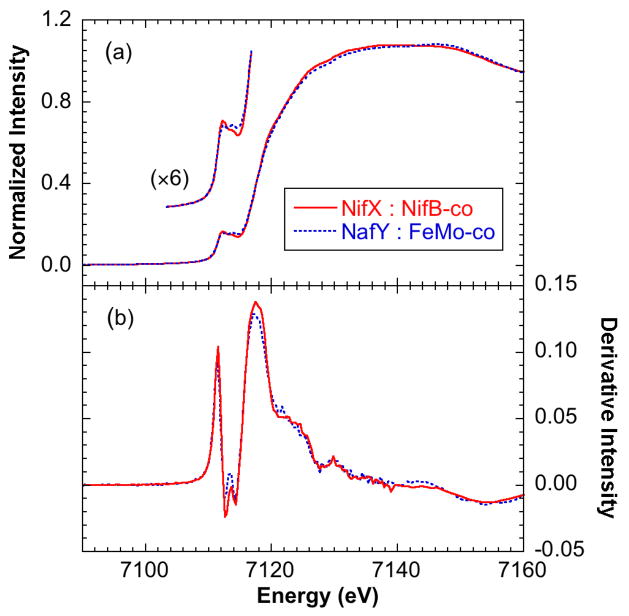
The two samples, NifX:(56Fe)NifB-co and NifX:(57Fe)NifB-co, gave very similar x-ray absorption spectra. The data presented here were obtained from the 57Fe sample that was also used for NRVS measurements. Figure 3 shows the Fe K-edge near-edge spectrum and first derivative from NifX:NifB-co, and compares these with those from NafY:FeMo-co known to be in the resting S = 3/2 Na2S2O4 reduced state. Both spectra exhibit the low-intensity broad rounded edge typical of iron-sulfur clusters together with small pre-edge features at ~ 7112.2 eV that arise from 1s → 3d transitions. The data sets from the NifX:NifB-co and NafY:FeMo-co are almost superimposable, with only subtle differences in both the pre-edge and edge regions. This indicates that the oxidation states and ligand environments of the Fe atoms in NifX:NifB-co and NafY:FeMo-co are very similar.
Figure 3.
Fe K-edge near-edge spectra of NifX:NifB-co (solid line) compared with NafY:FeMo-co (broken line). (a) Absorption spectra and (b) first derivative spectra.
We also searched the Mo K-edge region for evidence of cluster bound Mo in NifX:NifB-co. These measurements (not shown) conclusively demonstrated the absence of any measurable Mo in NifX:NifB-co, corroborating previous chemical analyses that indicated NifB-co does not contain Mo14.
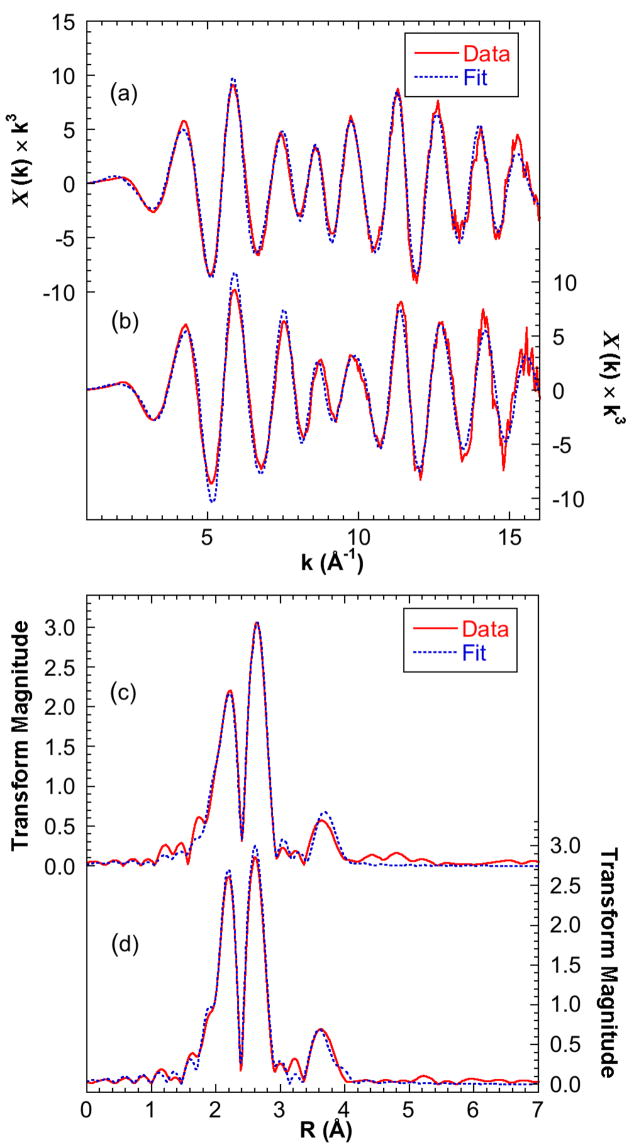
Figure 4 presents the EXAFS spectrum and Fourier transform for NifX:NifB-co together with an example fit and compares these data to those from NafY:FeMo-co. Initial inspection of the EXAFS for NifX:NifB-co suggests a strong similarity to the well-examined EXAFS of FeMo-co 26, and comparisons between these spectra are key to interpreting and understanding the differences between FeMo-co and its biosynthetic precursor, NifB-co. Curve fitting analysis reveals that the two main features in the Fourier transform in figure 4c correspond to a set of S ligands at ~2.3 Å and a set of Fe next nearest neighbors at ~2.6 Å. Importantly, a further set of Fe-Fe interactions at 3.7 Å is also clear. These interactions are very similar to those well known from FeMo-co (figure 4d) and their origin in this cofactor is illustrated in figure 1. The crystallographically determined structure of FeMo-co in MoFe protein can be viewed as a trigonal prism of 6 Fe atoms, with each of the 9 sides bridged by S, and each triangular S-bridged face capped with either Fe or Mo. The 2.3 Å Fe-S interaction is normal for Fe-S bonds. The “short” Fe-Fe interactions at ~ 2.6 Å are the sides of the trigonal prism as well as the distance of the capping Fe from its neighboring core Fe. The “long” 3.7 Å Fe-Fe distance arises from interactions across the square faces of the [6Fe-9S] trigonal prism. Such an interaction has so far only been observed in Fe-S centers for FeMo-co, and it can therefore be viewed as a strong indicator for the presence of the [6Fe-9S] core in NifB-co.
Figure 4.
Fe K-edge EXAFS of NifX:NifB-co (solid line) with fits taken from the 6Fe model (broken line) compared with data and fit of NafY:FeMo-co. (a) Unsmoothed k3-weighted EXAFS of NifX:NifB-co (b) Unsmoothed k3-weighted EXAFS of NafY:FeMo-co (c) NifX:NifB-co Fourier transforms. (d) NafY:FeMo-co Fourier transforms. Fitting parameters are given in table 1. A deconvolution of the NifX:NifB-co fit is shown in the supplementary information.
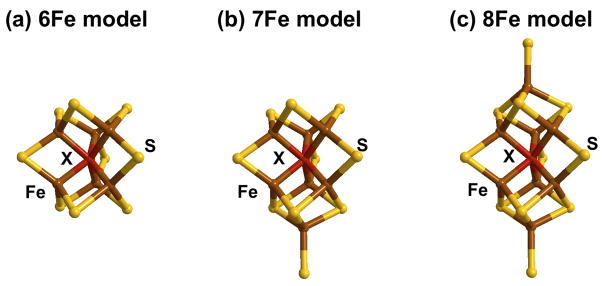
In order to rationalize these data, we constructed three working models, containing 6 Fe, 7 Fe and 8 Fe atoms, all based on the FeMo-co structure (figure 5). The 6Fe model is simply a D3h symmetry [6Fe-9S] trigonal prismatic core similar to that of FeMo-co. The 7Fe and 8Fe models were formed by capping one or both of the trigonal 3S faces of the [6Fe-9S] core. With coordination numbers constrained by these models, the short Fe-S, short Fe-Fe, and long Fe-Fe distances and Debye-Waller factors were then optimized to fit the experimental data. The resulting values for each model are summarized in table 1 and the quality of the fits is illustrated in figure 4, which includes the 6Fe model fit. All three models give reasonable fits with fitted short Fe-Fe and long Fe-Fe distances about 0.02 Å larger than those of NafY:FeMo-co, but the 6Fe D3h model gives the best fit to the experimentally observed data. As expected, the fitting process compensates for increasing the iron content of the cluster by changing the highly correlated Debye-Waller factor. All the Debye-Waller factors for each fit are within the sensible range for their bond types; that for the short Fe-Fe interaction for the 6Fe cluster at 0.0032 Å2 is exactly the minimum reasonable value30. This low value suggests a highly symmetric cluster with little variation between the 9 short Fe-Fe distances and supports the 6Fe D3h model for NifB-co. We note that the 6Fe and 7Fe models are more consistent with the values from the completed FeMo-co compared to the 8 Fe model as well as giving the better fit.
Figure 5.
Proposed models for NifB-co as described in the text. (a) 6Fe (b) 7Fe (c) 8Fe. Color code: Fe (brown), sulfur (yellow), interstitial atom X (red).
Table 1.
EXAFS curve fitting parameters for the NifX:NifB-co spectrum compared to those measured for NafY:FeMo-co. N = number of backscattering atoms. R = distance. σ2 = Debye-Waller factor. ΔE0 = threshold energy. F = fit quality defined as √[Σ(χo − χc)2 k6/Σχo2 k6] (χo = observed EXAFS; χc = calculated EXAFS). The backscatter atom numbers (N) were fixed to the values expected for the models while R, σ2 and ΔE0 were floated. Threshold energies (ΔE0) were constrained to be the same for all components. The estimated uncertainties in R, σ2 and ΔE0 were less than ± 0.01 Å, ± 0.0001 Å2 and ± 0.4 eV respectively. Phase and amplitude functions were calculated using FEFF 7.0. The scale factor used was 0.85. The Fe-X interaction was simulated assuming X = N; assuming X = C or O gave similar results but with different Debye-Waller factors 30.
| NafY:FeMo-co |
6Fe Model |
7Fe Model |
8Fe Model |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | R (Å) | σ2 (Å2) | N | R (Å) | σ2 (Å2) | N | R (Å) | σ2 (Å2) | N | R (Å) | σ2 (Å2) | |
| Fe-S | 3.10 | 2.251 | 0.00328 | 3.00 | 2.263 | 0.00424 | 3.00 | 2.264 | 0.00404 | 3.00 | 2.263 | 0.00383 |
| Fe-Fe (short) | 3.40 | 2.627 | 0.00456 | 3.00 | 2.655 | 0.00323 | 3.43 | 2.655 | 0.00385 | 3.75 | 2.654 | 0.00431 |
| Fe-Fe (long) | 1.70 | 3.726 | 0.00562 | 2.00 | 3.740 | 0.00646 | 1.71 | 3.742 | 0.00533 | 1.50 | 3.742 | 0.00436 |
| Fe-X(N) | 0.85 | 2.000 | 0.00199 | 1.00 | 2.046 | 0.00272 | 1.00 | 2.043 | 0.00240 | 1.00 | 2.037 | 0.00200 |
| Fe-Mo | 0.40 | 2.717 | 0.00350 | |||||||||
| ΔE0 (eV) | −12.66 | −11.99 | −11.81 | −12.00 | ||||||||
| F | 0.242 | 0.214 | 0.237 | 0.256 | ||||||||
We investigated whether introducing a small additional longer Fe-S or Fe-Fe interactions could significantly improve the quality of the fits. For Fe-Fe, we rationalized this search by postulating various movements of a single Fe away from the center of the trigonal prism. We found that we could improve the fit by including a small additional Fe-Fe interactions at ~2.9 Å and more significantly a split long Fe-Fe with a contribution at almost 4.0 Å which could be rationalized in terms of the distortion of a single Fe atom. However, at these distances the EXAFS from both these increased Fe-Fe interactions are almost exactly out of phase with the corresponding shorter distances, and hence there are significant risks of false minima with these distances in the curve fitting. In addition, in these fits, the Fe-Fe Debye-Waller factors are close to or below the reasonable minimum. Hence, at this time we believe the structure is best modeled by the fits in table 1.
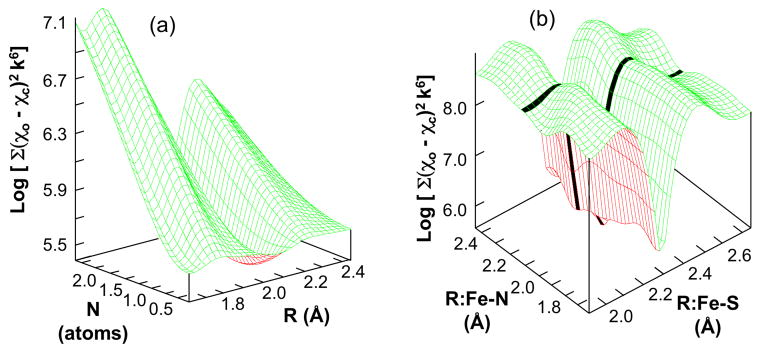
We also investigated whether the EXAFS data supports the presence of a single interstitial light atom “X” within the 6Fe trigonal prism. As noted above, the most recent crystal structure for Mo-nitrogenase 3 proposed such a C, N or O atom at the center of FeMo-co and we also note that our analysis of the NafY:FeMo-co EXAFS spectrum presented in figure 4 and table 1 readily accommodates this light atom. Clearly it is of key biosynthetic interest to know whether this atom is present in NifB-co. To address the potential presence of the interstitial atom, a multi-dimensional EXAFS “search profile” was conducted for different distances, numbers, and atomic numbers of an additional light atom. Figure 6 illustrates two results for a Fe-N interaction in the 6Fe model. The results for the number versus distance search for a Fe-X(N) interaction (figure 6a) shows a well-defined minimum at ~ 1 atom at 2.06 Å. Similarly, a search profile comparing the Fe-S distance and Fe-X(N) distance (figure 6b) clearly shows two independent troughs, with weaker Fe-X(N) trough intersecting the deep Fe-S interaction. Both these profiles clearly support the presence of a single central light atom. To be thorough, multiple curve fits both with and without a single central light atom were trialed. In every case the fit was significantly improved by including the central atom. In addition, we note that very similar profiles were obtained from the 7Fe and 8Fe models (data not shown). We emphasize that the contributions of the Fe-X(N) interaction to the fits in table 1 are significant, about 6% of the total EXAFS (see supplementary information). We therefore conclude that the EXAFS data demonstrate the presence of a one Fe-X (X = C, N or O) interaction for each of the 6 Fe of the trigonal prism of NifB-co, consistent with the presence of a single C, N, or O interstitial atom 31.
Figure 6.
Search profiles from fitting the Fe K-edge EXAFS of NifX:NifB-co 6Fe model. (a) 3D plot of the search profile of number (N) versus distance (R) for the proposed Fe-N interaction showing the minimum at ~ 2.0 Å and 1 atom (χo = observed EXAFS; χc = calculated EXAFS). (b) 3D plot of the search profile of Fe-N distance versus Fe-S distance. The dark line in (b) highlights the trough due to the Fe-N interaction.
In summary, analysis of the EXAFS and near-edge spectra shows that NifB-co in NifX:NifB-co is comprised of a 6Fe trigonal prism core very similar in geometry to that in FeMo-co. It is also clear that the interstitial atom is already present in NifB-co precursor for FeMo-co. The current EXAFS analysis does not resolve whether or not NifB-co also contains an additional capping Fe (Fe1 in PDB file 1M1N).
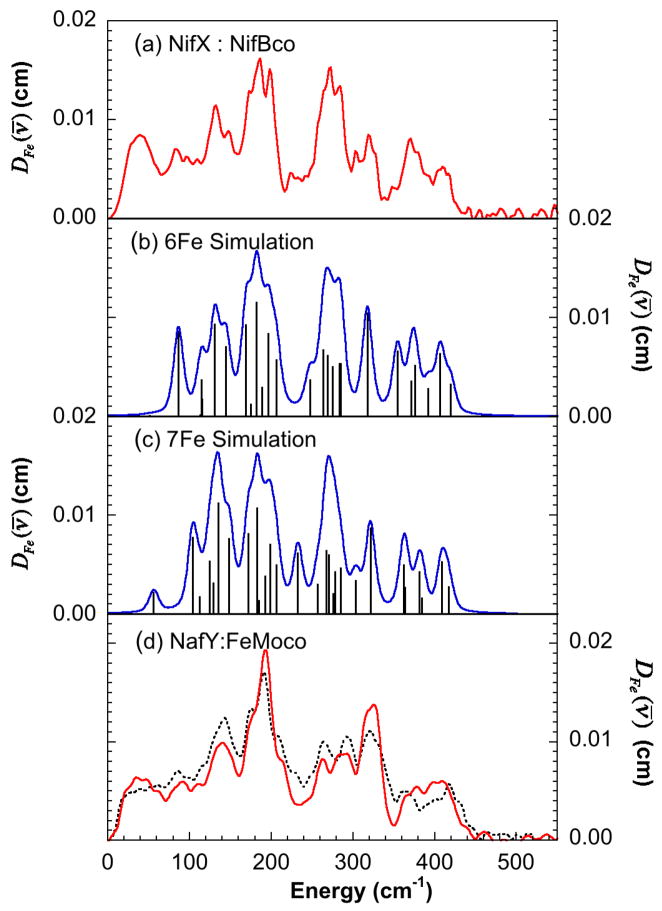
The NRVS-derived 57Fe PVDOS for NifX:NifB-co is presented in figure 7 together with calculated spectra, the spectrum of NafY:FeMo-co and the previously reported spectrum of FeMo-co isolated in N-methyl-formamide (NMF). As expected, the NafY:FeMo-co NRVS PVDOS is very similar to that previously observed for the Mo-nitrogenase and isolated FeMo-co with an almost one-to-one peak correspondence (figure 7d). The NifX bound NifB-co NRVS PVDOS spectrum in figure 7a is also similar to those of the completed cofactor, showing strong well-defined features in both the Fe-S stretching and bending regions. This is in contrast with the spectra from other Fe-S clusters where the strongest features are in the Fe-S stretching region and individual bending modes are not well resolved 27, 28. The strongest PVDOS features occur in a broad band between ~183 and 198 cm−1. This feature has been only previously observed by NRVS in Fe-S clusters of nitrogenase and isolated FeMo-co (figure 7d) and is considered to arise from the shaking modes of the c3 symmetric Fe6X core and is thus dependent on the presence of the interstitial light atom 5. A doublet at 271 cm−1 and 283 cm−1 apparently corresponds to a similar feature observed at 267 cm−1 and 290 cm−1 in the NRVS spectrum of a [4Fe-4S] cluster model compound which has been assigned to the Fe-Sb A1 stretching mode under D2d symmetry 28. There are additional peaks at 315 cm−1 and 364 cm−1, but their overall width suggests they are composed of multiple unresolved bands. There also are clear features up to 408 cm−1, which are reminiscent of the B2ub stretch modes in [2Fe-2S] clusters 27. In the lower frequency region, there are S-Fe-S bend modes around 140 cm−1 and significant NRVS intensity occurs between ~40 cm−1 and 85 cm−1 that presumably corresponds to cluster torsional modes and large-scale motion of the protein skeleton.
Figure 7.
NRVS data and Urey-Bradley force field simulations for NifX:NifB-co. (a) Data from NifX:NifB-co (b) Simulation using the 6Fe model (c) Simulation using the 7Fe model (d) Data from NafY:FeMo-co (solid line) compared with FeMo-co isolated in NMF (broken line) 5. The sticks in the simulated spectra show the calculated normal modes. The simulated spectra were obtained by broadening the stick spectrum with 7 cm−1 Gaussian lineshapes.
In order to quantitatively interpret the NRVS spectrum, we undertook an empirical normal mode analysis using a Urey-Bradley force field. Cartesian coordinates for the Fe environments in NifB-co were derived from the distances in the EXAFS D3h model. Initial force constants were taken from values used in the nitrogenase bound FeMo-co simulation 5, then adjusted to match the main peaks in the experimental spectrum. As with the EXAFS analysis, the 6Fe, 7Fe and 8Fe models in figure 5 were trialed. Figures 7b and 7c represent the calculated spectra obtained from the 6Fe and 7Fe models and compare them with the experimental spectrum in figure 6a. Table 2 presents the calculated force constants. We note that to get a good fit in the 6Fe model it was necessary to differentiate the S atoms bridging the trigonal faces of the cluster (and which become tri-coordinate in the completed FeMo-co) from those S bridging the two trigonal sides of the core. Despite extensive effort, it proved not possible to obtain a reasonable fit for the 8Fe model without using physically unreasonable force constants (data not shown).
Table 2.
NRVS force constants used to simulate the NifB-co spectrum. SU = trigonal face bridging S (3 coordinate S atoms in 7Fe or 8Fe models). FeT = terminal Fe.
| Mode | 6Fe model | 7Fe model | |
|---|---|---|---|
| Stretching (mdyn/Å) | Fe-SU | 1.136 | 1.200 |
| Fe-S | 1.065 | 0.960 | |
| Fe-N | 0.265 | 0.300 | |
| Fe-Fe | 0.225 | 0.155 | |
| FeT-S | 0.960 | ||
|
| |||
| sBending (mdyn Å/rad2) | S-Fe-S | 0.130 | 0.140 |
| S-Fe-SU | 0.455 | 0.388 | |
| S-Fe-N | 0.179 | 0.140 | |
| SU-Fe-N | 0.453 | 0.400 | |
| Fe-N-Fe | 0.056 | 0.250 | |
| Fe-S-Fe | 0.536 | 0.450 | |
| Fe-SU-Fe | 0.556 | 0.450 | |
| S-FeT-S | 0.120 | ||
| Fe-S-FeT | 0.120 | ||
|
| |||
| Nonbonding (mdyn/Å) | S–S | 0.00 | 0.00 |
| S–N | 0.270 | 0.250 | |
| S–SU | 0.047 | 0.118 | |
| SU--N | 0.280 | 0.224 | |
|
| |||
| stretch-stretch stretch-bend (mdyn/rad) | FeSU/FeSU | 0.114 | 0.074 |
| FeS/FeS | 0.124 | 0.070 | |
| FeN/FeN | 0.265 | 0.136 | |
| FeTS/FeTS | 0.113 | ||
Examination of figure 7 shows good agreement between the experimental spectrum and the simulations from both the 6Fe and 7Fe models, especially over the strongest PVDOS region between 150 and 210 cm−1. However, the 6Fe model gave a better fit, not only at this lower frequency region, but also over the Fe-S stretching region between 250 to 420 cm−1. We note that modes below 100 cm−1 mostly involve cluster torsional modes and large-scale motion of the protein skeleton, and hence it is not possible to simulate this region with a cluster based model alone 29. We also attempted fits using models without the interstitial light atom, causing the strong broad band between 183 and 198 cm−1 to be absent from the fit. As mentioned above, we note that we have previously assigned the analogous feature in FeMo-co as being characteristic of the interstitial light atom. We investigated routes to recover intensity in this region and found that one way was to use very large bending force constants for the Fe centered modes. This, however, is physically unrealistic and hence we conclude that the NRVS data also supports the presence of a light interstitial atom at the center of the 6Fe trigonal prism.
In summary, comparison of the NifX:NifB-co NRVS with the NRVS of FeMo-co and other Fe-S clusters, shows that the NifX bound NifB-co has a pattern of bands similar to those of Mo-nitrogenase and NMF isolated FeMo-co NRVS. This includes the strong signal near 190 cm−1 that is diagnostic of the interstitial light atom within the 6Fe cage. Urey-Bradley force constant simulations support this observation and demonstrate that the experimental data is most consistent with the 6Fe model shown in figure 5, although the 7Fe model cannot be completely excluded.
Discussion and Conclusions
FeMo-co is the product of a series of bioinorganic reactions performed by Nif proteins present in diazotrophic organisms. The complexities of synthesizing FeMo-co are underscored by the fact that a number of inorganic chemists have attempted to synthesize FeMo-co in vitro, but have so far not been successful. Knowledge of the structure of the NifB-co intermediate will greatly aid in understanding how nature accomplishes this chemical feat and may lead to better inorganic synthetic routes to manufacture complex Fe-S cluster moieties. To provide information for a structural model of FeMo-co, NifB-co bound to its likely intracellular carrier protein NifX has been analyzed using the combination of two highly complementary synchrotron based spectroscopic techniques: EXAFS spectroscopy and NRVS 32.
Taken together, the EXAFS and NRVS data show that NifB-co bound to NifX is likely to consist of a 6 Fe atom cage having D3h symmetry (figure 5a), although the corresponding 7 Fe model with a single capping Fe (figure 5b) cannot be absolutely excluded. The Fe K-edge EXAFS of NifX:NifB-co shows that NifB-co is more complex than a simple Fe4S4 cubane, but is also distinct from FeMo-co. The observation of the long distance Fe-Fe interactions at ~3.7 Å, supported by the curve-fitting analysis, strongly suggests that NifB-co contains a cluster resembling the 6 Fe central cage portion of FeMo-co. In addition, search profile analyses clearly indicate that the spectrum contains a Fe-X interaction at ~ 2 Å consistent with the presence of an interstitial light atom within the 6Fe cage. Measurements in the Mo K-edge region conclusively demonstrate the absence of Mo in the cluster. The Fe K-near-edge data indicates that the overall oxidation states and ligand environments of the Fe atoms in both NifX:NifB-co and NafY:FeMo-co are very similar. This is consistent with NifB-co comprising a substantially assembled FeMo-co core in the correct oxidation state. Presumably, it also indicates that capping the 6Fe core in figure 5a with a Mo or Fe atom does not significantly affect the Fe near-edge spectrum. These conclusions are corroborated by NRVS analysis of the same sample of NifX:(57Fe)NifB-co. The similarity of the NRVS spectrum of NifX:NifB-co to those of the completed FeMo-co is striking, especially as it includes the band at ~ 190 cm−1 which in FeMo-co has been assigned to modes involving the interstitial light atom within the 6Fe trigonal prism core. Vibratz simulation of the NRVS spectrum confirms this assignment. The analyses favor the 6Fe model over the 7Fe model, and clearly exclude the 8Fe model. To gain further insight into chemistry of NifB-co and how it is transformed by the FeMo-co biosynthetic pathway, it will be necessary to use EXAFS and NRVS to probe the structures of other biosynthetic precursors of the FeMo-co, such as the VK-cluster. These studies are underway and will be presented elsewhere.
The conclusion that NifB-co contains a Fe6X structure, which in turn becomes the central cage portion and interstitial atom of FeMo-co, rules out that the finished FeMo-co may be assembled without the X atom, which would be instead inserted at a late stage in the biosynthesis, such as initial enzyme turnover. This result is consistent with the suggestions that the subsequent enzymes involved in the FeMo-co biosynthetic pathway, NifEN and NifH, function to complete the cluster by adding the capping atoms of Mo, and perhaps the Fe, as well introducing the R-homocitrate ligand to the already constructed Fe6S9X core. Most interesting, however, is the implication that the function of the NifB enzyme is to perform the reaction that synthesizes this unique structure. The possible mechanistic details of this enzymatic reaction are intriguing. NifB is a SAM-radical enzyme, and it is reasonable to postulate a reaction that generates the Fe-S framework using SAM as a co-substrate. However, the origin of the interstitial atom and the chemistry of its insertion remain real unknowns and establishing this chemistry will be the next major challenge in understanding the biosynthesis of the unique and remarkable bioinorganic catalyst, FeMo-co.
Acknowledgments
This work was funded by the National Institutes of Health (NIH) under grants GM-65440 (SPC), EB-001962 (SPC), and 35332 (to PWL). The Advanced Biological and Environmental X-ray Facility (ABEX) is supported by the U.S. Department of Energy Office of Biological and Environmental Research (DOE OBER). Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the DOE Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the DOE OBER, and the NIH, National Center for Research Resources, Biomedical Technology Program. Portions of this work were also carried out at SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) under Proposal # 4032LD3-NP.
References and Footnotes
- 1.Igarashi RY, Seefeldt LC. Critical Reviews in Biochemistry and Molecular Biology. 2003;38:351–384. doi: 10.1080/10409230391036766. [DOI] [PubMed] [Google Scholar]
- 2.Eady RR. Chemical Reviews. 1996;96:3013–3030. doi: 10.1021/cr950057h. [DOI] [PubMed] [Google Scholar]
- 3.Einsle O, Tezcan FA, Andrade SLA, Schmid B, Yoshida M, Howard JB, Rees DC. Science. 2002;297:1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 4.Hinnemann B, Nørskov JK. Journal of the American Chemical Society. 2003;125:1466–1467. doi: 10.1021/ja029041g. [DOI] [PubMed] [Google Scholar]; Hinnemann B, Nørskov JK. Journal of the American Chemical Society. 2004;126:3920–3927. doi: 10.1021/ja037792s. [DOI] [PubMed] [Google Scholar]
- 5.Xiao Y, Fischer K, Smith MC, Newton W, Case DA, George SJ, Wang H, Sturhahn W, Alp EE, Zhao J, Yoda Y, Cramer SP. Journal of the American Chemical Society. 2006;128(23):7608–7612. doi: 10.1021/ja0603655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubio LM, Ludden PW. Journal of Bacteriology. 2005;187(2):405–414. doi: 10.1128/JB.187.2.405-414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curatti L, Ludden PW, Rubio LM. Proc Natl Acad Sci U S A. 2006;103(14):5297–5301. doi: 10.1073/pnas.0601115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez JA, Igarashi RY, Soboh B, Curatti L, Dean DR, Ludden PW, Rubio LM. Molecular Microbiology. 2007;63:177–192. doi: 10.1111/j.1365-2958.2006.05514.x. [DOI] [PubMed] [Google Scholar]
- 9.Rubio LM, Singer SW, Ludden PW. Journal of Biological Chemistry. 2004;279:19739–19746. doi: 10.1074/jbc.M400965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbett MC, Hu Y, Fay AW, Ribbe MW, Hedman B, Hodgson KO. Proceedings of the National Academy of Sciences. 2006;103:1238–1243. doi: 10.1073/pnas.0507853103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehr JJ, Albers RC. Reviews of Modern Physics. 2000;72:621–654. [Google Scholar]
- 12.Sturhahn W. Journal of Physics of Condensed Matter. 2004;16:S497–S530. [Google Scholar]
- 13.Scheidt WR, Durbin SM, Sage JT. Journal of Inorganic Biochemistry. 2005;99:60–71. doi: 10.1016/j.jinorgbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Shah VK, Allen JR, Spangler NJ, Ludden PW. Journal of Biological Chemistry. 1994;269(2):1154–1158. [PubMed] [Google Scholar]
- 15.Shah V, Brill W. Proceedings of the National Academy of Sciences. 1977;74:3429. doi: 10.1073/pnas.74.8.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.George GN, George SJ, Pickering IJ. EXAFSPAK: a Suite of Computer Programs for Analysis of X-ray Absorption Spectra. http://ssrl.slac.stanford.edu/exafspak.html.
- 17.Alp E, Sturhahn W, Toellner TS, Zhao J, Hu M, Brown DE. Hyperfine Interactions. 2002;144/145:3–20. [Google Scholar]
- 18.Leu BM, Zgierski MZ, Wyllie GRA, Scheidt WR, Sturhahn W, Alp EE, Durbin SM, Sage JT. Journal of the American Chemical Society. 2004;126(13):4211–4227. doi: 10.1021/ja038526h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturhahn W, Toellner TS, Alp EE, Zhang X, Ando M, Yoda Y, Kikuta S, Seto M, Kimball CW, Dabrowski B. Physical Review Letters. 1995;74:3832–3835. doi: 10.1103/PhysRevLett.74.3832. [DOI] [PubMed] [Google Scholar]
- 20.Sturhahn W. Hyperfine Interactions. 2000;125:149–172. [Google Scholar]
- 21.Yoda Y, Yabashi M, Izumi K, Zhang XW, Kishimoto S, Kitao S, Seto M, Mitsui T, Harami T, Imai Y, Kikuta S. Nuclear Instruments & Methods in Physics Research Section A-Accelerators Spectrometers Detectors & Associated Equipment. 2001;467:715–718. [Google Scholar]
- 22.Kishimoto S, Yoda Y, Seto M, Kitao S, Kobayashi Y, Haruki R, Harami T. Nuclear Instruments & Methods In Physics Research Section A. 2004;513:193–196. [Google Scholar]
- 23.Dowty E Shape Software. Physics and Chemistry of Minerals. 1987;14:67–79. http://www.shapesoftware.com/
- 24.Engeln-Mueller G, Uhlig F. Numerical Algorithms with C. Springer-Verlag; 1996. [Google Scholar]
- 25.Sage JT, Durbin SM, Sturhahn W, Wharton DC, Champion PM, Hession P, Sutter J, Alp EE. Physical Review Letters. 2001;86(21):4966–4969. doi: 10.1103/PhysRevLett.86.4966. [DOI] [PubMed] [Google Scholar]
- 26.Cramer SP, Hodgson KO, Gillum WO, Mortenson LE. Journal of the American Chemical Society. 1978;100(11):3398–3407. [Google Scholar]; Christiansen J, Tittsworth RC, Hales BJ, Cramer SP. Journal of the American Chemical Society. 1995;V117(N40):10017–10024. [Google Scholar]
- 27.Smith MC, Xiao Y, Wang H, George SJ, Coucovanis D, Koutmos M, Sturhahn W, Alp EE, Zhao J, Cramer SP. Inorganic Chemistry. 2005;44:5562–5570. doi: 10.1021/ic0482584. [DOI] [PubMed] [Google Scholar]
- 28.Xiao Y, Koutmos M, Case DA, Coucouvanis D, Wang H, Cramer SP. Dalton Transactions. 2006;2192–:2201. doi: 10.1039/b513331a. [DOI] [PubMed] [Google Scholar]
- 29.Sainz G, Jakoncic J, Sieker LC, Stojanoff V, Sanishvili N, Asso M, Bertrand P, Armengaud J, Jouanneau Y. Journal of Biological Inorganic Chemistry. 2006;11:235–246. doi: 10.1007/s00775-005-0069-2. [DOI] [PubMed] [Google Scholar]
- 30.Debye-Waller factors are frequently used (as they are here) as floating parameters to optimize the best fit to EXAFS data. They arise from the variation of distances for the given scatterer-backscatterer pair, and hence have contributions from both molecular vibrations and unresolved structural variation. Clearly the smallest possible Debye-Waller factor for a given interaction is that arising from the vibrational component alone. For Fe-Fe interactions in a Fe-S cluster, this can be estimated from a protein sample containing a single Fe-Fe interaction, such as a [2Fe-2S] containing protein. Such measurements indicate a Debye Waller of 0.0034 Å2 so for this work we assign a value 0.0032 Å2 as the “reasonable minimum”.
- 31.It is interesting to note that the fitted Debye-Waller factor was most reasonable with a N interstitial atom compared with that for a C or O atom. For example, for the 6Fe model we obtained Fe-C: 0.00061 Å2, Fe-N: 0.00272 Å2, Fe-O: 0.00620 Å2. A reasonable minimum value for a Fe-light-atom interaction is ~ 0.002 Å2.
- 32.It is worth noting that one of the potential advantages of complementary EXAFS and NRVS studies is that a proper understanding of the normal mode structure of a complex clusters like FeMo-co or NifB-co enables the potential use of NRVS derived vibrational modes to estimate and constrain further the EXAFS Debye-Waller factors.