Abstract
Alzheimer’s disease (AD) genetics may be one of the most prolifically published areas in medicine and biology. There are nearly 200 reviews on this topic since 1991, when the first report on an autosomal dominant mutation in the amyloid precursor protein gene (APP) came out. Three early-onset AD genes with causative mutations (APP, PS1, PS2) and one late-onset AD susceptibility gene (ApoE) exist with ample biological, genetic and epidemiological data and essentially universal acceptance about their roles in AD. Evidence from family and twin studies suggest a significant genetic component underlying AD which is not explained by the known genetic risk factors. The past 10 years in AD genetics research have led to ten independent whole genome linkage and association studies with implications for multiple genomic areas for harboring AD susceptibility genes. To date, there are about 900 papers reporting associations between variations in more than 350 genes spread over 23 autosomes. One hundred years after the first published article on Alzheimer’s disease, much is known about the pathophysiology of this disease, however much more remains to be discovered about its etiology. This review summarizes the evidence for the genetic component in AD, identification of the early-onset familial AD genes and ApoE, and the current state of knowledge for additional AD susceptibility loci and alleles. The future directions for genetic research in Alzheimer’s disease as a common and complex condition are also discussed.
Keywords: Alzheimer’s disease, genetics, complex disease
Introduction
Public impact of AD as a common disorder
AD is the most common adult neurodegenerative disease, with estimated 10–30% prevalence by age 85 or older and incidence of 6–8 % in the same age group [reviewed in 1]. With about 3–4 million affected individuals in the United States, 350,000 new cases per year (range: 200,000–600,000) and average annual cost per patient varying between $10,400 to $34,517 [2,3], AD is an epidemic with major health, social, and economic impact. In the absence of any interventions to delay its onset, which is the current status, the prevalence of AD is expected to reach 8.64 million (range: 4.37 to 15.4 million) by the year 2047 [3]. Brookmeyer et al. estimated that the 50-year projected prevalence of AD could decrease by 380,000 individuals, if there were an intervention to delay disease onset by a mere 6 months, corresponding to annual savings of nearly $18 billion after 50 years. Although this study was restricted to the epidemiological data from four studies in the United States only, with cost estimates from a single region in the US, the message is clear: the personal, social and economic impact of AD will continue to expand with the status quo of management approaches.
Role of environment in AD
A multitude of environmental factors, concomitant health conditions and life events have been proposed to be associated with risk for developing AD [1,3–5]. These proposed risk factors can be arbitrarily divided into the following categories: 1) non-modifiable (e.g. age, ethnicity, perinatal conditions, early-life development and growth), 2) socio-economically modifiable (e.g. socio-economic conditions, environmental enrichment, cognitive reserve, diet), 3) medically modifiable (obesity, hyperlipidemia, hypertension, diabetes). While epidemiologic studies assessing non-genetic risk factors can provide useful insights into the origins of AD, their interpretation and establishment as causative factors rather than mere associations are usually problematic, given the natural lack of an experimental setting, difficulty in assessing early-life events for a disease of the elderly population (retrospective rather than prospective studies) and the concomitant existence of multiple common conditions (such as hypertension, diabetes and AD). There are exceptions such as the Lazarov et al. study [6], which showed that transgenic mice co-expressing two familial AD genes with mutations (APPswe and PS1ΔE9) and raised in an “enriched environment” had less AD pathology reflected as reduced brain Aβ levels compared to trangenic mice raised in “standard environment”. Interestingly, of the transgenic mice in the “enriched environment”, those that were more physically active had the most significant reductions in brain Aβ levels. The authors also identified elevated activity of Neprilysin, an Aβ-degrading protease, in the environmentally enriched transgenic mice, as well as differential gene expression, suggesting gene-environment interactions and a possible role of exercise and environmental enrichment for protection against AD. Others determined that environmental enrichment improved cognition in the same transgenic mouse model of AD [7]. Though there is discrepancy between these studies regarding the effects of environmental enrichment on brain Aβ levels [6.7], which is beyond the scope of this review, both studies are important as they provide a paradigm to test the effects of environment and gene-environment interactions in an experimental setting. Despite such advances, exploring the effects of environment on a common, complex condition such as AD remains a challenging task, only surpassed in difficulty by the prospect of modifying environmental factors to decrease disease risk.
AD has a significant genetic component
Given the public impact of common, complex disorders such as AD and the challenges associated with studying and more importantly modifying environmental factors, much focus has been placed on genetic investigations. The rationale for studying genetics of any disease is multiple-fold: 1) Uncovering the underlying genetics leads to an understanding about disease pathophysiology. 2) Genetic risk factors may be modifiable, unlike many environmental risk factors. 3) Molecules encoded by genetic risk factors can be drug targets. 4) Genetic risk factors may provide clues into modifiable environmental risk factors. 5) Genetic variants and molecules identified through genetic studies can serve as biomarkers to identify at risk populations for disease prevention. 6) Knowledge about genetic risk factors in individuals may allow for personalized medicine in the future.
The first reports suggesting a genetic component for AD were published merely 2–3 decades ago, about 70–80 years after Dr. Alois Alzheimer’s publication on the very first case of this disease [8]. Early reports focused on the invariable development of AD-like disease in Down’s syndrome patients after age 40, the increased risk of disease in family members of AD patients with disease onset <65 years and the autosomal dominant inheritance pattern in the rare families with early-onset forms of this disease [9]. Systematic analyses geared towards estimation of the genetic component of AD can be grouped into the following categories: 1) Familial aggregation; 2) Transmission pattern and 3) Twin studies. These studies are discussed below:
1) Familial aggregation studies
Familial aggregation studies have revealed that having a first-degree relative with AD increased one’s risk for developing AD significantly. Breitner et al. [10] investigated 379 first-degree relatives of 79 probands in a longitudinal study of AD and found that the cumulative incidence of AD among relatives increased strikingly with age to 49% by age 87 in comparison to an incidence of < 10% for controls. Importantly, they did not observe an appreciable difference between the risks to relatives of presenile-onset (AD age of onset < 65 years) versus senile-onset AD probands (AD age of onset > 65 years), suggesting the existence of a genetic origin for both early-onset AD (EOAD) and late-onset AD (LOAD). They emphasized that their results should not be interpreted as evidence for autosomal dominant transmission in LOAD, but as a rationale for pursuing genetic studies in LOAD as well as EOAD. Farrer et al. [11] independently assessed 70 families with one or more AD subjects using survival analysis and determined evidence of different transmission patterns for EOAD (defined as mean onset age <58 years in their study) and LOAD (>58 years). Offspring of AD subjects had an estimated lifetime risk of 53% in EOAD families whereas this was 86% in LOAD families. These results supported an autosomal dominant transmission pattern for EOAD, whereas LOAD likely had a more heterogeneous transmission possibly with both genetic and environmental contributions. Thus, despite the differences in the risk-to-relative estimates, it was evident from these early studies that both EOAD and LOAD had a transmissible component.
More recent longitudinal familial aggregation studies based on much larger datasets supported the earlier findings of familial aggregation and inheritance of AD. As part of the Multi-Institutional Research in Alzheimer Genetic Epidemiology (MIRAGE) project, Lautenschlager et al. [12] estimated the risk to 12,971 first-degree relatives to 1,694 AD probands (mean age at onset 69.8) using survival analysis procedures. They found this risk to be 39.0% ± 2.1% by age 96 years, which is approximately twice the estimated cumulative incidence of AD in the general population, establishing the substantial genetic component affecting this disorder. Furthermore, the lifetime risk for LOAD (≥65 years) estimated separately was still high at 38.7±2.4%, though somewhat less than that for EOAD (39.5±4.1%). Given that the cumulative risk to first-degree relatives in autosomal dominant disorders is expected to be 50%, it was deduced from this study that the lifetime risk among relatives did not support a simple autosomal dominant inheritance pattern of disease, unlike some of the earlier familial aggregation studies, and that AD likely had a more complicated transmission pattern such as additive, multifactorial or polygenic inheritance. This study found a lower lifetime risk estimate compared to other studies [10, 11]. The difference was attributed to inflated estimates resulting from missing information especially in older age groups in other studies. MIRAGE study, which included ~400 subjects aged ≥90 determined decreased risk of AD in this oldest age group, raising the interesting hypothesis that not only are there genetic risk factors for AD, but also protective genes at play in the healthy, elderly population.
Recent analysis of the MIRAGE data [13] comparing risk to first degree relatives in the African-American (255 probands) vs. the white population (2339 probands) indicated a higher cumulative risk by age 85 for the former (43.7±3.1% vs. 26.9±0.8%, p<0.001). However, risk to the spouses was also found to be higher in the African-American population indicating that the risk attributable to familial aggregation was similar in the two ethnic groups. Given that the risk to first degree relatives was higher than that for spouses in all ApoE4 strata, this study concluded that there exist substantial heritable component to AD in both African-American and white populations, which is different from ApoE. Other familial aggregation studies controlling for ApoE genotype also found similar evidence supporting the presence of factors other than ApoE accounting for risk of AD [14–16]. In these studies, presence of an ApoE ε4 allele in the proband increased AD susceptibility in the relatives. However, there was also increased AD risk to relatives of patients without an ApoE ε4 allele; and that the estimated risk to AD relatives was higher than their predicted ApoE ε4 carrier status, implicating familial factors other than ApoE for risk of developing AD.
2) Transmission pattern studies
Although familial aggregation studies using survival analyses methods provided evidence of a transmissible factor accounting for risk of AD, these approaches did not necessarily distinguish a genetic factor from a transmissible environmental factor. To address this issue and to determine the mode of inheritance of genetic factors, segregation studies were pursued in AD families. Using segregation analysis of 232 AD families (age-at-onset 42–86 years), Farrer et al. [17] determined that the model of inheritance with the best fit was autosomal dominant genetic factor plus a multifactorial component. In this study, the models for a single major locus, multifactorial component only, no genetic susceptibility and autosomal recessive transmission were all rejected. These findings could be explained by the existence of multiple genetic and environmental risk factors for AD as well as heterogeneity with some families harboring a major risk gene and others multifactorial transmissible factors. Segregation analysis of Dutch AD families of early-onset (<65 years) yielded similar results implicating heterogeneity in EOAD [18]. Rao et al. [19] performed segregation analysis on a total of 401 AD families, 68 of which were early-onset (age at onset of AD cutoff = 65). They determined that stratification of the families according to the age of-onset always yielded a better model fit, an indication of the etiological heterogeneity between EOAD and LOAD families. AD in early-onset families was found to have an autosomal dominant transmission pattern. However all transmission models tested were rejected in the LOAD families, suggesting the possibility of heterogeneity or a complex genetic mechanism such as oligogenic inheritance. Thus, results from independent segregation analyses provided further support for genetic risk factors for both EOAD and LOAD; implicated multiple genetic and environmental factors especially for LOAD; and suggested a role for autosomal dominant transmission for at least some EOAD families.
More recently, Warwick Daw et al. [20] utilized oligogenic segregation analysis to study 75 AD families ascertained through a proband with LOAD and composed of 742 subjects, to estimate the number of trait loci affecting age-at-onset in LOAD. They accounted for effects of ApoE and sex in their study and determined that there exist four or more quantitative trait loci (QTLs) in addition to ApoE which contributed to age-at-onset in LOAD. They estimated that these loci had an effect size equal to or greater than that of ApoE, which itself accounts for 7–9% of the total variance for age-at-onset. These studies clearly provided evidence for a significant genetic component for AD, with a complex mode of inheritance for LOAD and provided a clear rationale for search of novel LOAD genes.
3) Twin studies
Twin studies particularly from Scandinavian twin registries have been instrumental in establishing the genetic component for AD [21–23]. These studies compare the concordance of disease in monozygotic (MZ) twins, who share 100% of their genetic material, and dizygotic (DZ) twins, who on average share 50% thereof. For a disease that is entirely due to genes, the lifetime concordance for MZ twins is expected to approach 100%, allowing for lesser estimates due to diagnostic inaccuracies and inherent late-onset nature of certain diseases (such as AD). The same estimate for DZ twins would be expected to approach 50%, the same estimate for sibpairs, or slightly more, secondary to possibly increased environmental sharing in DZ twins compared to other sibpairs. Given that MZ and DZ twins are assumed to share similar intrauterine and rearing environments [24], concordance rates significantly higher for MZ than DZ twins, can be attributed to the effect of shared genes. The uses and potential pitfalls of twin studies in AD have previously been reviewed [24, 25]. In a study of the Finnish Twin Cohort [21], monozygotic twins (MZ, n=51) had significantly higher cumulative incidence of AD than did dizygotic twins (DZ, n=43), whereas there was no difference between the incidence of vascular or mixed dementia between the MZ and DZ twins. There were higher pairwise disease concordance rates between the MZ than the DZ twins both for AD (18.6% vs. 4.7%) and vascular dementia (18.2% vs. 6.7%). Bergem et al. [22] found a significantly higher pairwise concordance rate for AD in MZ (78%) vs. DZ twins (39%) but no difference for vascular dementia (MZ 17% vs. DZ 25%). By using tetrachoric correlations, they estimated the heritability (proportion of total variance due to genes) for AD to be 60%. The largest AD twin study to date was based on the Swedish Twin Registry analyzing 392 twin pairs who had clinical diagnoses [23]. The MZ twins’ concordance of AD in this study was also higher than that of DZ twins, confirming the general conclusions of earlier twin studies. This study, unlike earlier twin studies, adjusted their findings for age and also addressed the issue of sex-difference on genetic factors by utilizing both same-sex and opposite-sex twins. They concluded that the age-adjusted heritability of AD was estimated to be 58–79% (varying based on model utilized) and that there was no significant gender-difference for either the prevalence or heritability of AD after controlling for age. Gatz et al. [23] also discussed the discrepancy in the concordance and heritability estimates between the different twin studies and highlighted differences between ascertainment, sample size, follow-up period, diagnostic methods, and age of analysis of subjects. Regardless of the individual parameter estimates obtained from these studies, the ultimate conclusion is unifying and support a significant genetic component for AD estimated at 60–80%.
Twin studies have also been utilized to study familial aggregation and the influence of ApoE on the genetic component of AD. In a study of 94 twin pairs who were either members of the National Academy of Sciences Registry of Aging Twin Veterans (44 pairs) or were volunteers (50 pairs), Steffens et al. [26] determined that concordance for AD among twins was significantly associated with higher rate of AD among first degree relatives (21%) compared with first degree relatives of discordant twins (9.5%). They also assessed the effect of ApoE genotype in the twins for development of AD and determined that the presence of ApoE ε4 genotype in the twin pair was associated with increased likelihood of AD in the first degree relatives. There was one family in this study concordant for AD and without ApoE ε4 with highest degree of AD in the first degree relatives (4/6 relatives), suggesting presence of additional AD genes instrumental at least in this family. Given that the ApoE status was not determined or inferred for the AD relatives in this study, and the small numbers of non-ε4 positive concordant twins, it is not possible to accurately estimate the contribution of genes other than ApoE for development of AD in first degree relatives of AD twins. Bergem and Lannfelt [27] determined that in a collection of Norwegian twin pairs, the frequency of the ApoE ε4 allele did not differ between concordant and discordant dizygotic twin pairs, though the presence of this allele was associated with an earlier age at onset. These findings from twin studies suggest that while ApoE accounts for risk and earlier age at onset of AD (the two phenotypes are interdependent for late-onset diseases), this gene does not explain all of the genetic risk for AD.
The collective evidence from familial aggregation, segregation and twin studies for AD, suggest a significant genetic component for this disease. The next section discusses the identification of the three known early-onset familial AD (EOFAD) genes.
EOAD Genetics
1) Initial Identification of the EOFAD Genes
a) Amyloid precursor protein (APP)
Genetic linkage studies followed by candidate gene analysis or positional cloning have historically been successful in EOFAD and led to the identification of the three known EOFAD genes. Amyloid precursor protein (APP) gene on chromosome 21 is the first one found to have a mutation which causes EOFAD [28]. It is a good example of how linkage analysis coupled with candidate gene approach led to the successful identification of one of the EOFAD genes.
The identification of an association between Trisomy 21 (Down’s Syndrome = DS) and AD led researchers to focus on chromosome 21 as the possible locus for an AD gene. The first report that discovered the link between DS and AD dates back to the 19th century when Fraser and Mitchell reported “precipitated senility” as a cause of death in DS patients [29]. In studies that were conducted half a century later, autopsies of DS patients who died after the age of 40 revealed AD-like histopathology, namely senile plaques and neurofibrillary tangles [30, 31, reviewed in 32]. During the early 1980’s researchers discovered that the “amyloid protein” isolated from the brains of AD patients had the same biochemical properties and amino acid sequence as that isolated from DS brains [33, 34]. Because of the clinical, histopathological, and biochemical association of DS with AD and the existence of an extra copy of chromosome 21 in DS, researchers started to focus on chromosome 21 as the possible location for an AD susceptibility gene. In 1987, St George-Hyslop et al. found linkage to a locus on chromosome 21 in extended families with the autosomal dominant form of EOAD [35]. In two related articles, Goldgaber et al. [36] and Tanzi et al. [37] reported the mapping of the amyloid beta protein found in the brains of both AD and DS patients to a region on chromosome 21 that is near the newly identified AD locus. After several reports that refuted linkage of autosomal dominant EOFAD to Aβ or to chromosome 21 in general [38–40], Goate et al., first found evidence of linkage to chromosome 21 in EOFAD families [41], and then identified a missense mutation in the APP gene segregating with AD in some of the families they analyzed [28]. The mutation occurred in exon 17 of the APP gene partially encoding for the Aβ peptide and led to a valine to isoleucine change at amino acid 717 (Val717Ile) corresponding to the transmembrane domain of the protein. It was thus predicted that this first EOFAD mutation (a.k.a. London mutation) would lead to AD through its effects on Aβ. These predictions were subsequently proven to be accurate through functional analyses discussed below. Since then, according to the Alzheimer Disease & Frontotemporal Dementia Mutation Database (http://www.molgen.ua.ac.be/ADMutations), 26 other mutations have been identified within APP from 74 EOFAD families. Many of these mutations were functionally assessed for their effects on APP processing [42]. These studies provided strong support for the “amyloid hypothesis” discussed in detail in the related section of this issue [43]. The prevalence and clinical phenotype of the EOFAD APP mutations are discussed in subsequent sections.
b) Presenilins (PSEN1, PSEN2)
Linkage analysis and positional cloning studies led to the identification of the second EOFAD gene, PSEN1 on chromosome 14. In 1992, Schellenberg et al. reported the results from a genome search they conducted in non-Volga-German kindreds with EOFAD where they found significant linkage to a locus on chromosome 14 [44]. This finding was confirmed by the positive linkage results obtained by other groups to the same region of chromosome 14 in EOFAD families [45–47]. Sherrington et al. [48] cloned AD3 in 1995, later known as PSEN1, by identifying a minimal cosegregating region on the chromosome 14 linkage area and determining the transcripts within this region. They found that one of the transcripts corresponded to a novel gene harboring mutations that segregated with EOFAD in the families they studied. Subsequent studies described the structure of the PSEN1 gene and identified new PSEN1 mutations segregating with EOFAD [49–51]. To date 157 pathogenic PSEN1 mutations have been identified in 347 EOFAD families (http://www.molgen.ua.ac.be/ADMutations), making PSEN1 mutations the most common known genetic cause of EOFAD.
The third EOFAD gene, PSEN2, was identified as a result of linkage studies accompanied by homology analysis of the transcripts in the linkage region. In 1995 Levy-Lahad et al. [52] reported significant linkage to a region on chromosome 1 in EOFAD kindreds of Volga-German origin, where linkage to chromosomes 14 and 21 have previously been excluded [40, 44]. They identified a candidate gene in this region that showed sequence homology to PSEN1 and that had a segregating mutation resulting in an asparagine to isoleucine substitution (Asn141Ile) [53]. Concurrently, Rogaev et al. [54] independently identified the same PSEN2 N141L mutation in the affected probands of Volga-German families. They also identified a second missense mutation in this gene (Met239Val) segregating in an extended EOFAD pedigree of Italian origin. Unlike the case with PSEN1, PSEN2 mutations are a rare cause of EOFAD. With 11 mutations identified from 19 pedigrees, most PSEN2 mutations are thought to be private to pedigrees [http://www.molgen.ua.ac.be/ADMutations, 51, 55, 56].
2) Impact of autosomal dominant mutations for population risk of AD
Compared with late-onset form of AD (LOAD), EOAD is exceedingly rare. Campion et al. [57] defined EOAD as age of onset <61 and assessed the population based prevalence of this disease in Rouen, France. They determined that the prevalence of EOAD was 41.2 per 100,000 people at risk (defined as ages 41–60). Their estimations were similar to those obtained from earlier population based prevalence estimates of EOAD which ranged between 18.2–45.2 per 100,000 inhabitants [58–60]. Campion et al. further defined autosomal dominant EOAD (ADEOAD) as having 3 EOAD cases in three generations and determined the prevalence of ADEOAD to be 5.3 people/100,000 at risk. They then performed mutational analysis of the three known EOAD genes in the ADEOAD families they identified in the prevalence study combined with ADEOAD families referred to their laboratory. In their screen of 34 ADEOAD families, PSEN1 mutations were identified in 56%, APP mutations in 15% and PSEN2 mutations in none of the families. Other screens of EOAD identified PSEN1 mutations at a frequency of 18–56% [50, 62], with different estimates between the studies attributed to the different selection criteria for the EOAD families analyzed. Others identified APP variations at a frequency of 13% in EOAD [63]. According to the Alzheimer Disease & Frontotemporal Dementia Mutation Database (http://www.molgen.ua.ac.be/ADMutations), of the EOFAD mutations identified, PSEN1 mutations account for the majority (81%), followed by APP (14%) and with PSEN2 mutations identified only in a handful of families (6%).
In summary, EOAD accounts for 6–7% of all AD (57, 64) and EOFAD (or ADEOAD) accounts for 13% of EOAD. Given that mutations in APP, PSEN1 or PSEN2 can explain up to 71% of the autosomal dominant transmission pattern in EOFAD, these genes account for about 0.5% of all AD. Although these are essentially negligible figures in terms of population attributable risk posed by the known EOFAD mutations, their discoveries were, nonetheless, monumental in terms of understanding the pathophysiology of AD discussed briefly below and in detail elsewhere in this issue [43]. Importantly, 50% or more of EOAD are not explained by the known EOFAD mutations, indicating this aggressive form of AD could possibly harbor as yet unknown genetic factors. Furthermore, existence of LOAD families with an apparent autosomal dominant pattern of transmission suggests the presence of other Mendelian mutations with less aggressive phenotypes [65, 66]. In a recent whole genome scan of 12 AD families with an autosomal dominant pattern of inheritance, Giedraitis et al. found evidence of a shared 40 cM haplotypic region on chromosome 8p in affected individuals from two families with age at onset 54–75 [67]. Analyses of single, extended families with autosomal dominant transmission patterns and members with either EOAD or LOAD in multiple generations have shown linkage to numerous chromosomal regions, some of which have not been identified in prior whole genome linkage scans of AD. Thus, analyses of EOAD families and large LOAD families with Mendelian transmission patterns may emerge as a way to identify other genetic causes of AD. Though the population-based impact of such discoveries are expected to be small, they can provide new insights to the pathogenesis of AD and be instrumental in the development of drugs, novel quantitative phenotypes and biomarkers for prevention and treatment of more common forms of this devastating condition.
3) Impact of autosomal dominant mutations for pathophysiology of AD
Aβ, the major proteinaceous component of senile plaques (SPs) in AD brains is processed from APP, by the cleavage of two secretases: β-secretase and γ-secretase [70–73]. Processing of APP by β-secretase and α-secretase precludes the formation of the Aβ peptide and instead leads to the production of the shorter, non-fibrillogenic and non-amyloidogenic P3 peptide [74]. The EOFAD mutations identified in the APP gene all reside close to the secretase cleavage sites [75]. Analysis of plasma Aβ from affected families and the secreted Aβ from cell lines transfected with mutant forms of APP all showed an increase in the production of Aβ42± an increase in Aβ40; increased Aβ42/Aβ40 ratios or enhanced Aβ protofibril formation for all functionally assessed APP mutations [42, 76–80]. Finally trangenic mice carrying the mutant APP were shown to have elevated Aβ, amyloid plaques and correlative memory deficits, abnormalities that are reminiscent of AD [81]. PSEN mutations, similarly have been shown to increase the production of Aβ [76]. Moreover transgenic mice overexpressing a mutant form of the PSEN1 have been shown to have increased Aβ42(43) in their brains [82], providing further support for the amyloid cascade hypothesis as the mechanism of AD. Knock-out studies have determined that endogenous wild-type PSEN1 and PSEN2 are required for the γ-secretase cleavage of APP [83]. The finding that all of the functionally tested EOFAD mutations affect Aβ metabolism that is reflected in plasma and/or fibroblasts studied from EOFAD patients, presymptomatic mutation-carriers and transgenic mice; and the evidence that PSENs are a key component of the γ-secretase complex suggest a common pathological pathway for AD, involving Aβ [42, 76–84].
Thus, though exceedingly rare, the identification of the autosomal dominant mutations in the three known EOFAD and the subsequent functional studies led to the development of the amyloid cascade hypothesis. Though the exact steps involved in the cascade of events in this pathophysiologic mechanism are as yet unknown and debatable, it has clearly set the stage for the ongoing drug discovery efforts for Alzheimer’s disease [reviewed in 85–87].
LOAD Genetics
1) Initial identification of ApoE as a LOAD susceptibility gene
Failure for multiple research groups to replicate the initial linkage reports to chromosome 21 [35] in various independent collections of AD families led to the hypothesis that AD may be a heterogeneous disorder [39–40, 88]. In 1991, Pericak-Vance et al., reported the results of a genome search they conducted in both early and LOAD families, where they found linkage to chromosome 21 in their EOAD families, as well as a novel locus on chromosome 19 [89]. The novel locus seemed to have an especially strong effect in the LOAD families (defined as age at onset >60). Given the previous association between an ApoCII allele and AD reported by Schellenberg et al. [90], initially this gene on chromosome 19 emerged as a candidate for LOAD.
Concurrently, a group studying the change of lipids in AD brains by using an antibody to ApoE, a protein having a special relevance to nervous tissue, unexpectedly found that ApoE immunoreactivity was associated with amyloid in both senile plaques and cerebral vessels and neurofibrillary tangles [91]. Given that the gene for ApoE mapped to the same locus identified in the Pericak-Vance et al. linkage study [89], researchers started investigating the genetic and biological association of ApoE with AD. In 1993, Strittmatter et al. [92] showed in an in-vitro assay that ApoE binds Aβ with high avidity and that ApoE ε4, a particular allelic form of ApoE, is found at a higher frequency in LOAD when compared to unrelated age-matched controls. Subsequently Corder et al. [93] demonstrated a dose-dependent increase in risk and a decrease in age-at-onset for LOAD, in ApoE ε4 carriers. This study which assessed 42 LOAD families found a hazard ratio of 2.84 for carriers of a single ApoE ε4 allele and 8.07 for those with the ApoE ε4/ε4 genotype.
Since then, multiple large, population or clinic-based studies established the effect of ApoE ε4 as genetic a risk factor for LOAD [94–100]. Table 1 depicts a list of studies arbitrarily defined as having a large number of participants; and either performed on population-based samples or multi-center collections. This table is not meant to depict the exhaustive list of all studies testing for association between Alzheimer’s disease risk and ApoE. Clearly, there are a large number of such studies in literature as a simple search on PubMed (www.pubmed.org) using the keywords “ApoE, association, Alzheimer, risk” yields an impressive 647 articles to date (January 18, 2007). Instead, Table 1 shows a collection of the studies with the largest sample sizes, which led to the establishment of ApoE as a susceptibility gene for AD. Several conclusions can immediately be drawn from this table: 1) ApoE ε4 allele increases risk for AD in a dose-dependent manner; 2) The risk conferred by ApoE ε4 in predominantly or entirely Caucasian populations is unequivocal with largely overlapping odds ratios (ORs) and confidence intervals; 3) Studies in the African-American and Hispanic populations yield conflicting results.
TABLE 1.
Studies of ApoE association in AD series.
| Study (ref) | Study Type | Ethnicity/Source | ApoE 3/4 OR (95%CI) |
ApoE 4/4 OR (95%CI) |
Number of Subjects |
|---|---|---|---|---|---|
| Myers et al., 1996 [94] | Population-based cohort | Predominantly Caucasian (Italian-American; Framingham, MA, USA) | 3.7 (1.9–7.5) | 30.06 (10.7–84.4) | 68 AD, 962 CON |
| Farrer et al., 1997 [95] | Meta-analysis (population-based) | Caucasians | 2.7 (2.2–3.2) | 12.5 (8.8–17.7) | 4858 (total) |
| Bickeboller et al., 1997 [96] | Population-based case-control | Caucasians (France) | 2.2 (1.5–3.5) | 11.2 (4.0–31.6) | 417 AD, 1030 CON |
| Slooter et al., 1998 [97] | Population-based cohort | Caucasians (Rotterdam, Netherlands) | 1.8 (1.0–3.1) | 6.2 (1.4–28.2) | 97 AD, 997 CON |
| Tang et al., 1998 [98] | Population-based cohort | Caucasians (New York City, NY, USA) | 2.5 (1.1–6.4)* | 23 AD, 214 CON | |
| Breitner et al.,1999*** [99] | Population-based case-control | Predominantly Caucasian (Cache County, UT, USA) | 4.77 (2.71–8.40)** | 8.52 (3.24–22.39) | 230 AD, 698 CON |
| Farrer et al., 1997 [95] | Meta-analysis (population/clinic-based) | African-Americans | 1.1 (0.7–1.8) | 5.7 (2.3–14.1) | 235 AD, 240 CON |
| Tang et al., 1998 [98] | Population-based cohort | African-Americans (New York City, NY, USA) | 1.0 (0.6–1.6)* | 53 AD, 128 CON | |
| Graff-Radford et al., 2002 [100] | Clinic-based, multi-center case-control | African-Americans | 2.6 (1.8–3.7) | 10.5 (5.1–21.8) | 338 AD, 301 CON |
| Evans et al., 2003 [101] | Population-based cohort | African-Americans (Chicago, IL, USA) | 1.02 (0.39–2.68)* | 362 (total) | |
| Murrell et al., 2006 [102] | Population-based cohort | African-Americans (Indianapolis, IN, USA) | 2.32 (1.41–3.82) | 7.19 (3.00–17.29) | 162 AD, 318 CON |
| Farrer et al.,1997[95] | Meta-analysis (population/clinic-based) | Hispanics | 2.2 (1.3–3.4) | 2.2 (0.7–6.7) | 261 AD, 267 CON |
| Tang et al., 1998 [98] | Population-based cohort | Hispanics (New York City, NY, USA) | 1.1 (0.7–1.6)* | 145 AD, 516 CON | |
| Farrer et al., 1997 [95] | Meta-analysis (population/clinic-based) | Japanese | 5.6 (3.9–6.0) | 33.1 (13.6–80.5) | 336 AD, 1977 CON |
ApoE ε3/ε4 and ε4/ε4 odds ratios (OR) are depicted in reference to the ε3/ε3 genotypes except as follows:
Subjects with one or more ApoE ε4 allele vs. ApoE ε3/ε3 genotype.
Subjects with one ApoE ε4 allele vs. ApoE ε3/ε3 genotype.
Reference group is subjects with ApoE ε2/ε2, ε2/ε3 and ε3/ε3 genotypes.
Using these studies as a starting point, the population impact of ApoE, possible ethnicity differences, role of ApoE in the diagnosis of AD and as a premorbid predictor for its development are discussed in the following sections.
2) Impact of ApoE for population risk of AD
In a population based prospective study of the predominantly Caucasian Framingham cohort, Myers et al. [94], determined that the cumulative incidence of AD increased in a dose dependent fashion where 55% of the ApoE ε4/ε4 group developed AD by age 80, compared with 27% of the ApoE ε3/ε4 and 9% of the ApoE ε3/ε3 groups by age 85 years. Despite the small number of ApoE ε4/ε4 individuals (n=16), this study clearly demonstrated that unlike the autosomal dominant EOFAD mutations, ApoE ε4 is a susceptibility and not a deterministic factor for the development of AD. Furthermore, since only 10% of the individuals with one or more copies of ApoE ε4 developed AD (positive predictive value 0.1), with specificity 0.81 and sensitivity 0.49, ApoE ε4 is unlikely to be useful for the diagnosis of AD. Similar dose dependent increases in the incidence or prevalence of AD were determined in other population-based prospective cohort [97, 98] or case-control studies [95, 96, 99].
Farrer et al., performed meta-analysis of 40 different clinic, population or autopsy-based studies and assessed the effects of sex, age and ethnicity on the ApoE association with AD [95]. They reported results from population-based and clinic-based Caucasian samples, separately (Table 1 depicts their population-based results only), which showed largely overlapping ORs for the ApoE genotypes in these two different study types. Importantly, this study showed an age-dependent effect for the risk conferred by ApoE ε4, such that this allele was associated with risk of AD across all age groups between 40–90 years, but the effect was strongest for ages 60–75, with a decline after age 70. Similar age-specific effects were also detected by others [96, 99, 100]. This age specificity with declines in the risky effect of ApoE ε4 in the older age groups could be due to a number of reasons: 1) ApoE ε4 may decrease age at onset of AD, lead to more aggressive form of disease and a shorter survival; 2) There may be genetic and non-genetic factors which render certain individuals relatively invulnerable to AD, and promote longevity; 3) There may be genetic, non-genetic factors which interact with ApoE ε4 in an age-dependent manner.
The studies in non-Caucasian populations did not yield as consistent results for the association between AD and ApoE [95, 98, 100]. The meta-analysis study by Farrer et al. determined that the ApoE ε3/ε4 association is non-significant; and that for the ApoE ε4/ε4 genotype had lower, though overlapping, OR estimates in the African-American population, compared with the Caucasian population from that study. Farrer et al. emphasized the presence of heterogeneity between these two ethnic groups in terms of ApoE. Tang et al. found that the presence of an ApoE ε4 allele did not pose significant risk to the African-Americans or Hispanics in their longitudinal cohort from New York City [98]. The African-Americans, Hispanics and Caucasians in their study had similar cumulative risk for AD in the presence of ApoE ε4; whereas in its absence the cumulative risk to the African-Americans and Hispanics were 4 and 2 times higher, respectively, even after adjusting for education and family history of AD. The authors concluded that these results provided evidence for the presence of other genetic and non-genetic risk factors for AD in these two ethnic groups. Another longitudinal study on an African-American cohort from Chicago found similar non-significance for ApoE ε4 association with AD [101], whereas a longitudinal cohort from Indianapolis [102] and a multi-center case-control study from southeastern United States [100] determined significant ApoE ε4 risk for AD, in clear contrast. A recent review discusses these opposing results in the African-American population as well as potential reasons for this discrepancy in this literature [103]. The ApoE ε4 allele frequencies are higher in controls from those studies which fail to show significant association [98, 101], compared to those which do [100, 102], suggesting different underlying population structure. Indeed, the ApoE ε4 allele control frequencies from the non-significant studies are similar to those seen in the Yoruban-African population from Nigeria [104], where AD association with ApoE is also lacking. The incidence of AD is also lower in the Yoruban-African population [104], in comparison to the African-American populations from studies both confirming and refuting ApoE-AD association [98, 101–103]. Thus, the lack of association despite high incidence rates in the cohorts from Chicago and New York suggest a) the presence of factors which prevent development of AD in the controls from these cohorts with the ApoE ε4 allele; b) the presence of risk factors other than ApoE which lead to the development of AD in non-ApoE ε4 carriers; c) the presence of gene x gene, gene x environment interactions specific to or more prominent in certain populations; d) the presence of population admixture which lead to false negative results; e) a combination of one or more of these.
The association studies in Hispanics have similarly been equivocal [95, 98]. More recent studies have demonstrated significant ApoE association and age-at-onset effect in Caribbean Hispanics with familial (but not sporadic) LOAD [105, 106], suggesting role of additional genes and other gene x ApoE effects in this population. The discrepancies between the ApoE ε4 frequencies and AD associations between Hispanic populations of different source and geography imply the effects of genetic and possibly environmental heterogeneity in this ethnic group, as well [107].
These findings highlight several important issues regarding AD and genetic studies of complex diseases in general: 1) Large studies from multiple centers are required to dissect complex genetics; 2) These studies need to control for potential population substructure arising from genetic heterogeneity; 3) There likely exist multiple genetic and non-genetic factors accounting both for risk of and protection from AD. Analysis of a variety of ethnic groups and utilizing their genetic and environmental differences may prove to be highly useful in the detection of these factors.
The population attributable risk for AD or dementia due to the ApoE ε4 allele was estimated to be between 20–70% [97, 99]. These estimates provide further evidence for the existence of additional genetic factors underlying the risk of AD.
3) Role of ApoE for the diagnosis of AD and as a premorbid marker
Clinicopathologic series where role of ApoE in diagnosing AD is estimated did not support a rationale for its genotyping in the clinical setting [108, 109]. Mayeux et al. assessed the accuracy of clinical diagnosis alone, ApoE genotyping alone and their combination in a clinicopathologic series of 2188 patients from 26 AD centers, 1833 of whom had clinical and 1770 of whom had pathologic diagnosis of AD, with 418 patients who had pathologic diagnosis of other causes of dementia [108]. The sensitivity and specificity of clinical diagnosis alone were 93% and 55% respectively, whereas ApoE genotyping alone had 65% and 68% sensitivity and specificity, respectively. The addition of ApoE genotyping to the clinical diagnosis of AD decreased the sensitivity to 61%, but increased specificity to 84%, thus reducing false positive rate. Tsuang et al. found similar findings in a community-based longitudinal series of 132 patients with cognitive complaints and subsequent autopsy [109]. The sensitivity and specificity of clinical diagnosis alone were 84% and 50% in this study; whereas those for ApoE alone were 59% and 71%, respectively. In the presence of clinical diagnosis of AD, ApoE decreased sensitivity to 49% and increased specificity to 84%. In the absence of clinical diagnosis of AD, ApoE increased sensitivity to 94% and decreased specificity to 37%, indicating high false positive rate of diagnosing AD in individuals with an ApoE ε4 allele but no pathologic evidence of AD. Although ApoE genotyping increased specificity (decreased false positive rate) when a clinical diagnosis of AD is made, this association is not absolute, such that there are individuals who lack the ApoE ε4 allele and have pathologically confirmed AD, as well as those with ApoE ε4 and cognitive impairment but non-AD pathology. Thus, ApoE genotyping is not advocated as part of the diagnostic work-up for AD [106].
Given the importance of early detection for development of AD from prodromal cognitive impairment states, multiple studies focused on the use of ApoE genotyping as a predictor for the development of AD. Most population or clinic-based longitudinal prospective studies which utilized mild cognitive impairment (MCI) or other clinical criteria for pre-dementia cognitive impairment found ApoE ε4 to be a significant predictor for the development of AD [111–114]. In two longitudinal studies assessing cognitive testing, ApoE ε4 was found to be a predictor for development of AD when used independently but not in combination with cognitive assessment [115–116]. This is in contrast to the findings by Petersen et al. where ApoE ε4 was a strong predictor for conversion from MCI to AD, even in the presence of cognitive testing [111]. The discrepancy may arise from the definition of cognitive state in the starting populations in these studies. Cervilla et al. assessed conversion from cognitively normal state [115]; and Lee et al. studied subjects with questionable dementia [116]. Both of these populations are likely more heterogeneous than the MCI subjects studied by Petersen et al. [111]. In conclusion although ApoE ε4 appears to be an important predictor of conversion to AD, especially from well-defined cognitive impairment states, its use in the clinical setting is not substantiated. Nonetheless, future studies of other potential premorbid biomarkers of AD need to include this genetic factor for increased accuracy.
Whole genome linkage and association studies
1) Study characteristics
The extensive data from prior twin, segregation, family aggregation, EOFAD autosomal dominant families and ApoE association studies discussed above have led to the conclusion that AD has a substantial genetic component not explained by the known EOFAD mutations or ApoE. This, along with the development of high-throughput techniques for genotyping an ever-expanding number of markers (microsatellites and single nucleotide polymorphisms-SNPs), statistical methodologies geared towards identification of genetic loci in the absence of known Mendelian patterns of inheritance and collective efforts to obtain collections of LOAD families, relative pairs and case-control series led to the pursuit of a number of whole genome linkage and association studies in the literature [117–136]. A summary of the key characteristics of these studies are presented in Table 2. As seen here, there are essentially three groups of studies: “A” denotes those whole genome linkage studies on AD families or sibpairs [117, 121, 126, 127, 129, 132, 135, 136]. These studies are independent in that they were either performed entirely on ethnically distinct datasets of Caribbean Hispanic [126, 132], Amish [135] or Swedish [136] subjects; or even if they did have overlapping data [NIMH or NIA datasets], there was considerable non-overlapping data as well (117, 121, 127, 129]. The initial linkage analysis by Pericak-Vance et al. [117] performed on a total of 54 LOAD families may be a subset of their subsequent work on a significantly larger dataset [121] based on the similar sources of the study subjects, although the extent of overlap is not explicitly mentioned in their latter publication. Nonetheless, the results from their initial study is presented here separately (Tables 3–8), because this was the first whole genome linkage analysis conducted in LOAD and also because it led to the identification of the chromosome 12 locus, subsequently replicated in other studies [126, 127, 130]. Although the extent of overlap is not clearly evident between the three predominantly US sample-based studies utilizing the NIMH and NIA datasets [121, 127, 129], Blacker et al. [129] tried to address this issue by reporting results from both their entire dataset, as well as the subset of subjects non-overlapping with the first [120] and second stages [127] of the Washington University study. Here, their results from the non-overlapping (non-Kehoe) subset are depicted.
TABLE 2.
Characteristics of the whole genome linkage and association studies in AD.
| Study Ref. |
Authors | Study Type |
Sample Source | Sample Type |
Age at Onset |
Sample Size | Analytic Approach |
# Markers | Marker Density |
Subset analysis |
|---|---|---|---|---|---|---|---|---|---|---|
| 117 | Pericak-Vance et al. | A | US (NIA, Duke, UCLA, MGH) | multiplex families | ≥60 | 16 families (52 ADs vs. 83 controls) | ARP, ASP, LOD3 | 280 microsatellites | 10–15 cM | E4+ (all ADs with ≥ 1 E4 = 27 families) vs. E4− (≥1 AD member without E4 =27 families)5 |
| 117 | ibid. follow-up | A | 38 families (89 ADs vs. 216 controls) | |||||||
| 118 | Pericak-Vance et al.6 | D | US (NIA, Duke, UCLA, MGH) | multiplex families | >60 | 16 families (52 ADs vs. 83 controls) | ARP, ASP, LOD | 280 microsatellites | 10–15 cM | |
| 118 | ibid. follow-up | D | 38 families (89 ADs vs. 216 controls) | |||||||
| 119 | Zubenko et al. | B | US (Belmont, Pittsburgh) | case-control | >60 | 100 AD cases (autopsied) vs. 100 controls (50 autopsied, 50 90+) | Chi-square | 391 microsatellites | 10 cM | |
| 120 | Kehoe et al. | D | US (NIMH) | sibpairs | ≥65 | 292 sibpairs (230families) | ASP | 237 microsatellites | 16.3 cM | |
| 121 | Pericak-Vance et al.7 | A | US (NIMH, NIA, Duke, UCLA, Vanderbilt) | families | >60 | 466 families (1051 ADs vs. 591 controls; 199 families autopsied) | ARP, ASP | 326 microsatellite | 10 cM | Autopsied (N=199) vs. Not (N=267) |
| 122 | Curtis et al.8 | C | US (NIMH) | families | most ≥65 | 241 families | Conditional non-parametric linkage | 237 microsatellites | 16.3 cM | |
| 123 | Hiltunen et al. | B | Finland | case-control | >60 | 47 ADs vs. 51 controls | Chi-square | 366 microsatellites | 10 cM | |
| 124 | Bacanu et al.9 | C | US (NIMH) | families1 | ≥65 | 65 families (42 with E4+) | ARP (LOAD+psychosis) | 237 microsatellites | 16.3 cM | E4+ (≥2 members with E4+) |
| 125 | Li et al.10,11 | C | US (NIMH, NIA, Duke, UCLA, Vanderbilt) | families | ≥49 | 449 families (1121 ADs vs. 746 controls) | QTL (AAO)3 | 323 microsatellite | 10 cM | |
| 126 | Mayeux et al.12 | A | Caribbean Hispanics | families | mixed (>65 and ≤65) | 79 families (320 subjects) | ARP | 35 markers (Ch12) | 3.4 cM | Late-onset (≥ 65) vs. Early/Mixed onset (<65); E4+ (≥75% of ADs with ≥1 E4) vs. E4− (<75% of ADs with ≥ 1 E4) |
| 127 | Myers et al.2 | A | US (NIMH, NIA)UK | sibpairs | ≥65 | 451 sibpairs (174 new from ref. 120) | ASP | 328 microsatellites (91 additional to ref. 120) | 5 cM (select regions) | E4+ (each sib with ≥ 1 E4= 280 ASP) vs. E4− (all E4−/E4− = 76 ASP) |
| 128 | Olson et al.9 | C | US (NIMH) | sibpairs | ≥60 | 272 sibpairs | ASP with covariates (age, ApoE) | (see 4) | (see 4) | |
| 129 | Blacker et al.8 | A | US (NIMH) | families | ≥50 | 437 families (994 AD vs. 445 controls; 238non-Kehoe families) | ARP, LOD | 381 microsatellites | 9 cM | Late-onset (≥65) vs. Early/Mixed onset (<65) |
| 130 | Farrer et al.13 | B | Inbred Arab community | case-control | ≥60 | 5 ADs vs. 5 controls | Chi-square | 375 microsatellites | 10 cM | |
| 130 | ibid. follow-up | B | 100 ADs vs. 110controls | |||||||
| 131 | Scott et al.14 | C | US (NIMH, NIA, Duke, UCLA, Vanderbilt) | families | ≥49 | 437 multiplex families (1014 ADs vs. 238controls) | Ordered subsets analysis | 336 microsatellites | 10 cM | |
| 132 | Lee et al. | A | Caribbean Hispanics | families | mixed (>65 and ≤65) | 96 families (490subjects; 237 ADs vs.157 controls, others unknown); 104 families (2nd stage, 521 subjects) | ASP, ARP, Affected only dominant model | 340 microsatellites | 10.2 cM | Late-onset (≥65) vs. Early/Mixed onset (<65) |
| 132 | ibid. follow-up | A | 104 families (521subjects) | additional markers on Ch 10, 18, 12 | ||||||
| 133 | Avramopoulos et al.15 | C | US (NIMH) | families | >50 | 148 families (348 ADs vs. 153 controls) | ASP with covariates (delusions, hallucinations) | 381 microsatellites | 9 cM | Late-onset (>65) vs. Early onset (<65) |
| 134 | Holmans et al.16 | C | US (NIMH, NIA) UK | sibpairs | ≥65 | 453 sibpairs (600–789 subjects depending on covariate) | ASP with covariates (mean AAO, difference AAO, mean ROD, difference ROD)3 | 328 microsatellites (91 additional to ref. 120) | 5 cM (select regions) | |
| 135 | Hahs et al. | A | US (Amish) | extended families | ≥58 | 5 families (115 subjects; 40 AD, 9 MCI, 66 controls) | LOD | 407 microsatellites | 7 cM | |
| 136 | Sillen et al. | A | Sweden | multiplex and sibpair families | ≥65 | 71 families (188subjects) | ARP | 365 microsatellites | 9 cM | E4+ = all ADs with ≥ 1 E4 vs. E4− = ≥ 1 AD without any E4 (in E4− families, the E4+ individuals were excluded from analysis) |
Explanation of study types:
A: Linkage analysis of families, relative pairs, independent studies
B: Association analysis of case-controls, independent studies
C: Linkage analysis of families, using alternative analytical methods on largely overlapping datasets with previously published results D: First stage linkage results or results on subsets of other datasets
Explanation of superscripts:
Families with ≥ 2 members with LOAD and psychosis and ≥ 2 members with ApoE e4+
Follow-up study of 120; 80% sample overlap with 121
ARP=affected relative pair; ASP=affected sibpair; LOD=parametric linkage; QTL=quantitative trait loci; AAO=age at onset; ROD=rate of decline
Likely same markers as ref. 120, but not mentioned explicitly in text.
E4 = ApoE e4
Appears to be same study as ref. 117
Sample overlap significantly with ref. 120
Subset of ref. 120
Only results for AD are used herein
Subset of ref. 132
Ch12 results are not follow-up from stage I, but based on previous studies
Subset of 121
Sample overlaps with other NIMH data
Same dataset as ref. 127
TABLE 3.
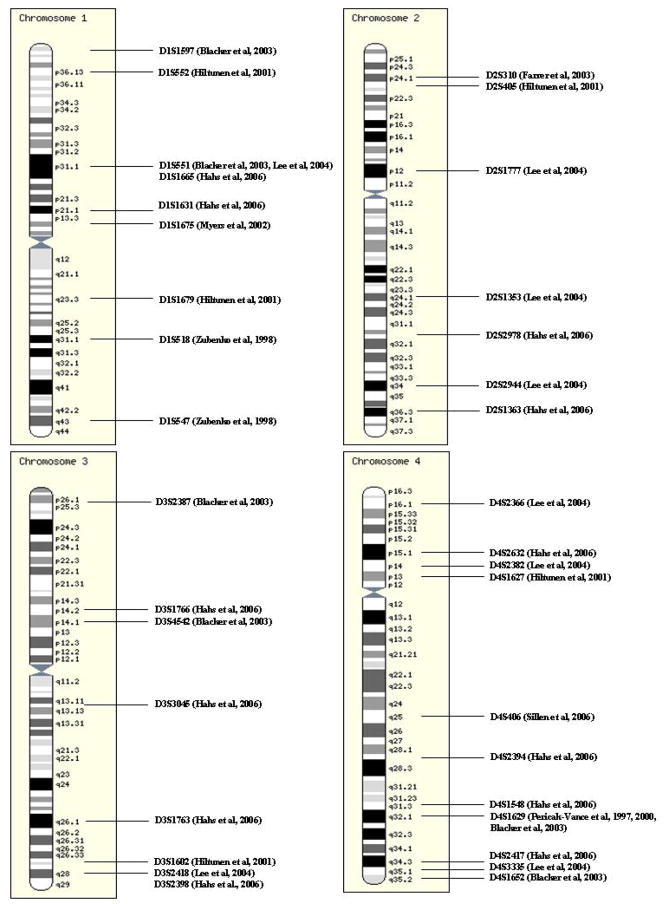
Results of the whole genome linkage and association studies in AD-Chromosomes 1–4.
| Ch | 1 | 2 | 3 | 4 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study Ref. |
Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result |
| 117 | D4S1629 | 4q32.1 | >1.0 | |||||||||
|
| ||||||||||||
| 121 | D4S1629 | 4q32.1 | 1.32 | |||||||||
|
| ||||||||||||
| 127 | D1S1675 | 1p13.2 | 1.7 (E4+) | |||||||||
|
| ||||||||||||
| 129 | D1S1597 | 1p36.21 | 1.3 (late) | D3S2387 | 3p26.3 | 2.9 (early/mixed) | D4S1629 | 4q32.1 | 1.1 (total) | |||
| D1S551 | 1p31.1 | 1.7 (early/mixed) | D3S4542 | 3p14.1 | 1.5 (total) | D4S1652 | 4q35.2 | 1.1 (late) | ||||
|
| ||||||||||||
| 126,132 | D1S551 | 1p31.1 | 1.12 (late) | D2S1777 | 2p12 | 1.51 (all) | D3S2418 | 3q28 | 1.07 (total) | D4S2366 | 4p16.1 | 1.59 (all) |
| D2S1353 | 2q24.1 | 1.59 (late) | D4S2382 | 4p14 | 1.26 (late) | |||||||
| D2S2944 | 2q34 | 1.31 (all and late) | D4S3335 | 4q35.1 | 1.25 (late) | |||||||
|
| ||||||||||||
| 135 | D1S1665 | 1p31.1 | 1.07 | D2S2978 | 2q31.3 | 1.94 | DS31766 | 3p14.2 | 1.78 | D4S2632 | 4p15.1 | 1.33 |
| D1S1631 | 1p21.1 | 1.83 | D2S1363 | 2q36.3 | 1.17 | D3S3045 | 3q13.12 | 1 | D4S2394 | 4q28.2 | 2.12 | |
| D3S1763 | 3q26.1 | 1.69 | D4S1548 | 4q31.3 | 3.01 | |||||||
| D3S2398 | 3q28 | 2.16 | D4S2417 | 4q34.3 | 1.21 | |||||||
|
| ||||||||||||
| 136 | D4S406 | 4q25 | 2 (E4+) | |||||||||
|
| ||||||||||||
| 119 | D1S518 | 1q31.1 | 0.003 | |||||||||
| D1S547 | 1q43 | 0.001 | ||||||||||
|
| ||||||||||||
| 123 | D1S552 | 1p36.13 | 0.006 | D2S405 | 2p23.2 | 0.009 | D3S1602 | 3q27.2-27.3 | 0.007 | D4S1627 | 4p13 | 0.004 |
| D1S1679 | 1q23.3 | 0.015 | ||||||||||
|
| ||||||||||||
| 130 | D2S310 | 2p24.1 | 0.005 | |||||||||
References (descriptors in superscripts):
117 Pericak-Vance et al.1
121 Pericak-Vance et al.2
127 Myers et al.3
129 Blacker et al.4
135 Hahs et al.2
136 Sillen et al.3
119 Zubenko et al.6
130 Farrer et al.6
Descriptors:
1 twopoint (38 families, except Ch 12 = 54 families; Ch 19 results are significant but due to ApoE and therefore not shown in this paper).
2 twopoint
3 multipoint
4 multipoint (non-Kehoe dataset)
5 Ch 10, 12, 18 second stage multipoint linkage, others single stage twopoint
6 chi-square p
TABLE 8.
Results of the whole genome linkage and association studies in AD-Chromosomes 21-X.
The second type of study denoted by “B” identifies whole genome association studies conducted on case-control series [119, 123, 130]. These three studies are entirely independent and assess subjects within the US [119], Finland [123] or an inbred Arab community [130]. Their results are shown in Tables 3–8. Studies in group “C” are performed on datasets which have complete or major overlaps with other published genome scans [122, 124, 125, 128, 131, 133, 134]. Group C studies re-assess these datasets either using alternative analytical strategies, such as conditional linkage analysis [122], ordered subsets analyses [131], linkage analysis with covariates [128, 133, 134]; employing alternative phenotypes such as age at onset [125] or defined subsets within the data such as LOAD with psychosis [124]. The summary results from these alternative approaches are depicted in the text rather than Tables 3–8. Finally, the first stage of the Washington University genome scan, which is a subset of their subsequent study [127] and the Pericak-Vance et al. study from 1998 [118], which appears to be the same study as ref. 117, are denoted as a separate category “D”. Their results are not discussed separately.
As seen in Table 2, most of the studies are based on AD families or sibpairs with age at onset greater than 60 [117, 118, 119, 120, 121, 123, 124, 127, 128, 130, 134, 136]. Most studies with both late and early/mixed age at onset datasets showed separate results for these different age groups [126, 129, 132, 133]. Where applicable, the age subsets for the results are shown in Tables 3–8. Many of the studies performed subset analyses based on ApoE genotypes [117, 124, 126, 127, 136], though their definitions of E4+ (ApoE ε4 +) or E4− subsets varied between the studies. Again, results for the various ApoE subsets are shown in Tables 3–8.
2) Results of whole genome linkage and association studies
Tables 3–8 summarize the results of the whole genome linkage (group A studies from Table 2) and association (group B studies from Table 2) studies in the literature. Different analytical approaches have been used between and within the studies. Where there is more than one analytical approach for a given study, findings from the approach yielding the most significant result is depicted. Likewise, when there are subset analyses, the results of the subset with the most significant result are used; and this is depicted in the table. For multi-stage genome scans, the results from the second stage or follow-up study are used where available. When both twopoint and multipoint linkage results are available, the latter result is shown. LOD scores (parametric or non-parametric linkage results) above 1.0 or p values below 0.05 are shown, unless study authors determined a more significant cutoff in their paper. When ≥2 “interesting” results exist on a single chromosome separated by an arbitrary distance cutoff of >15 cM, each one is reported separately. Otherwise, the most significant result is reported. When a multipoint linkage curve spans >15 cM with ≥1 peaks separated only by sharp “dips” (as opposed to a plateau of “no suggestive linkage” with LOD scores consistently <1 in-between), the highest multipoint result is reported. Otherwise, each multipoint result is reported separately. For locations of the multipoint LOD score results, unless given explicitly in the paper, the nearest marker location is utilized.
Figure 1 is a compilation of all chromosomes where >1 suggestive/significant result exists in >1 study from Tables 3–8. The plain karyotype figures (not showing the study results) are taken from the Ensemble website (http://www.ensembl.org/info/software/website/installation/build.html), [138]. The chromosomal locations for the markers are as per the March 2006 human genome assembly on the UCSC Genome Browser (http://genome.ucsc.edu/) [139].
FIGURE 1.
FIGURE 1A–E. Chromosomal map summaries of whole genome linkage and association studies in AD. The plain karyotype figures were taken from the Ensemble website (http://www.ensembl.org/info/software/website/installation/build.html). The markers’ chromosomal locations were determined according to the March 2006 assembly on the UCSC Genome Browser (http://genome.ucsc.edu/). Chromosomes 16, 17, 21 and 22 are not shown since there is only a single suggestive/significant result from a single study for these chromosomes.
Despite the differences in the study subjects, study designs, and analytical approaches between the various studies, inspection of Tables 3–8 and Figure 1 reveals a number of chromosomal regions where evidence for linkage or association exists based on a number of studies. This is evident for chromosome 19, where all but two studies [130, 132] showed significant/suggestive evidence of linkage or association. One of these studies was performed on a specific ethnic group of an inbred Arab community where ApoE frequency is low both in controls and AD cases [130]. The other study was on a collection of Caribbean-Hispanic families where there was evidence of association but no significant linkage [132]. These results strongly suggest the existence of genetic risk factors for AD other than ApoE, especially in ethnic groups of non-European or non-North American origin. Furthermore, there are three studies where one or more locus yields greater significance for linkage or association compared with the ApoE locus on chromosome 19 [123, 127, 135]. Hiltunen et al. identified loci on chromosomes 1–6, 10, 13, and 18 in their study of a Finnish case-control series where evidence for association was greater than that for ApoE [123]. They reported simulated p values. Myers et al. found strongest evidence of linkage on chromosome 10q [127] with LOD score of 3.9 in their whole dataset vs. 1.3 for chromosome 19q [127]. Finally Hahs et al., in their study of extended Amish families from the US, where ApoE was previously shown not to be an important risk factor for AD, identified both novel (Ch 4–7, 11, 18–20) and previously reported (Ch 4–7, 11, 18–20) regions with evidence for linkage [135]. Thus, there is ample evidence from these whole genome linkage and association studies for the existence of novel LOAD risk loci.
Given the plethora of positive findings, it is important to be able to discern the true positive vs. false positive results. Evidence of linkage or association from multiple independent studies is an important indicator for a result as being a true positive. Although studies with overlapping datasets [117, 121, 127, 129] render such a comparison somewhat difficult, attempts are being made to approach this analytical difficulty. Blacker et al. [129], reported results of their linkage analyses on both overlapping and non-overlapping (non-Kehoe) datasets separately, where latter results can be seen as an independent assessment of findings by Pericak-Vance et al. [117, 121] and Myers et al. [127]. Despite these confounders, several chromosomal loci emerge as potential risk loci for AD. Chromosomal loci 1p31.1 [129, 132, 135], 2p24.1-23.2 [123, 130], 3p14.1-2 [129, 135], 3q27.2-28 [123, 132, 135], 4p13-15.1 [123, 132, 135], 4q31.3-32.1 [117, 121, 129, 135], 4q34.3-35.2 [129, 132, 135], 13q12.12 [121, 123], 14q22.2 [129, 132], 14q24.1-2 [123, 135], and 18q22.3-23 [121, 132] have been implicated in at least two completely independent whole genome linkage and/or association studies. Of the chromosomal loci with independent replication those on chromosomes 6 [121, 123, 127, 129, 132], 9 [121, 127, 129, 130], 10 [119, 121, 123, 127, 130, 135] and 12 [117, 126, 127] received the greatest interest due to the large number of confirmatory studies, strength of these results and the existence of multiple strong candidate genes in these regions with positive association results from one or more studies.
a) Chromosome 6
Chromosome 6p21.2-12.3 (117, 123, 127, 132) and 6q27 (121, 129) regions have been implicated in whole genome linkage and association studies. Initially, Pericak-Vance et al. [117] identified linkage to chromosome 6p21.2 and 6q15 in a follow-up analyses of 38 LOAD families, where evidence for linkage was also replicated for chromosomes 4, 12 and 20. In their larger subsequent analyses of 466 LOAD families, continued linkage signal was observed for the 6q27 region [121]. Chromosome 6p21.1-12.1 region showed significant association in a whole genome association study from Finland [123]. Myers et al., in a sample with “as much as 80%” overlap with Pericak-Vance et al. [121] also detected linkage to chromosome 6p21.1-12.3 region in their ApoE ε4+ sample [127]. Interestingly, their maximum multipoint LOD score is near marker D6S1017, which showed evidence of association in the Finnish association study [123]. Blacker et al. [129] confirmed the linkage in the 6p21 and 6q27 regions, which is perhaps not surprising given their sample overlap with the two prior linkage studies [121, 127]. Nonetheless, the D6S1017 marker also yielded a twopoint LOD score of 1.2 in their non-Kehoe families, which do not overlap with the Myers et al. [127] study. Furthermore, this subset continued to have suggestive multipoint linkage to the 6q27 region. Finally, in a Caribbean-Hispanic series evidence for linkage was identified to chromosome 6p22.3 and 12.3 regions [132]. These studies provide independent evidence for the existence of two AD risk regions of interest on chromosome 6 on the p and q arms. A number of candidate genes have been analyzed in these regions which are depicted graphically on the AlzGene website (http://www.alzforum.org/res/com/gen/alzgene/) [140] and reviewed previously [141, 142]. In a recent meta-analysis of the available candidate gene studies compiled by the AlzGene database, tumor necrosis factor (TNF) emerged as a promising candidate gene. The rs4647198 (-1031) SNP in a transcriptional regulatory site for TNF showed significant association with AD in a meta-analysis of three studies [140].
b) Chromosome 10
Several chromosome 10 regions were identified as candidate AD risk loci in numerous linkage and association studies [119, 121, 123, 127, 129, 130, 132, 135]. Zubenko et al. identified significant AD association with D10S1423 on 10p12.33 in their 10 cM whole genome scan of 100 AD cases and 100 controls [119]. Chromosome 10p11.23, ~13 cM downstream of this region showed suggestive linkage in 199 autopsy-confirmed LOAD families [121], studied by Pericak-Vance et al. Two studies implicated the more upstream chromosome 10p region [123, 129], while Farrer et al. confirmed the findings in the 10p11.23 region [130]. The 10p13 region implicated in the Finnish association study [123], 10p14 identified with linkage in early/mixed onset non-Kehore LOAD families [129] along with the initial association region by Zubenko et al. [119] span ~17 cM on the short arm of chromosome 10. Whether this region and the more downstream 10p11.23 region identified independently in a US-based linkage [121] and an inbred Arab-based association study [130] represent the same locus is not as yet known.
The most significant linkage evidence for chromosome 10 identified 10q21.3 as a potential AD risk region, with a maximum multipoint LOD score of 3.9 at ~82.2 cM, which is significant at the whole genome level [127]. In this study of 451 affected sibpairs by Myers et al., the ApoE ε4+ group yielded the strongest linkage signal. Importantly, in an independent study of 10 extended LOAD families, using plasma Aβ42 levels as a quantitative phenotype, Ertekin-Taner et al. found evidence of linkage to the same region on chromosome 10 with greatest evidence of linkage in those 5 extended LOAD families whose AD proband had extremely high plasma Aβ levels [143]. This study was the first one in AD genetics using a quantitative biologically relevant phenotype for mapping an AD risk locus. Jointly, these two independent studies provided evidence for a novel AD risk locus on 10q21.3 which acts via the Aβ pathway [143, 144]. The linkage peak in the Myers et al. study covers a broad region (59 cM–103 cM). In a study of Amish pedigrees, Hahs et al. found linkage to ~101 cM [135] on 10q22.3. Similarly, Farrer et al. found evidence of association in their inbred Arab sample to a number of chromosome 10 markers spanning 81–115 cM, with greatest single marker association at 10q22.3 (97 cM) and four contiguous markers showing association between 105–115 cM [130]. Not surprisingly, Blacker et al. found linkage in their overlapping NIMH sample at 10q22 and 10q24, but not in the non-Kehoe sample. In another analysis of the same dataset using a different set of markers, this group reported significant linkage and association at 115–127 cM near the IDE gene [145]. Finally, using age at onset as a quantitative phenotype Li et al. [125] identified linkage to 10q25.3 at 139 cM. These studies provide evidence for the existence of one or more AD risk loci on chromosome 10, some/all of which appear to affect both Aβ levels and age at onset. According to the AlzGene website (www.alzgene.org), 69 candidate genes have been studied on chromosome 10 [140]. Some of these candidate gene studies will be discussed briefly below.
c) Chromosome 12
The first evidence of linkage to chromosome 12 came from Pericak-Vance et al. who detected the strongest linkage signal with a maximum multipoint lod score of 3.5 at 12p11.23 in their 54 LOAD families [117]. The evidence for linkage was strongest in their subset of ApoE4-families, defined as those families with at least one patient who lacked the ApoE ε4 allele. This group subsequently genotyped additional markers and re-analyzed the same dataset using different subgroups by their ApoE genotypes and underlying pathology [146]. Their analyses of all affected sibpairs confirmed the initial linkage findings ~12p11.23 though identified a second, smaller peak ~18 cM more downstream at 12q13.13. When the analyses were weighted by ApoE ε4− individuals, the peak linkage occurred at 12q13.2 and was significantly stronger than the ApoE ε4+ weighted linkage. Furthermore, subset analyses of the 8 families with Lewy body pathology yielded stronger linkage than that in the remaining 46 families. The families with Lewy body pathology had linkage to the 12p region whereas the other families linked to the 12q region. These results suggest the presence of one or more loci in the ~30cM region encompassing the linkage peaks in the various subgroups at 12p and 12q. Myers et al. found a suggestive linkage to chromosome 12 at 12p13.2, also with stronger results in the ApoE ε4-subset [127]. Finally, Rogaeva et al. [147] found strongest evidence of linkage near marker D12S96 at 12q13.13, though their linkage region extended to 12p13.2 spanning ~42 cM region (according to the marker locations from the Marshfield map). An independent analysis of Caribbean-Hispanic LOAD families [126] found evidence of linkage at the 12p13.31 region, as well as the more downstream 12p12.1 regions, close to the Myers et al. [127] and Pericak-Vance et al. linkage regions [117], respectively. The evidence of linkage in this study was stronger in the ApoE ε4- subset, as well. Two independent association studies also found significant results on chromosome 12 [119, 130], however the loci identified in these reports were ~15 cM [130] to 93 cM [119] distal to the nearest linkage region.
Thus, similar to the findings on chromosomes 10 and 6, multiple potential LOAD risk regions have been identified on chromosome 12. More than 20 candidate genes have been analyzed on this chromosome of which α2-macroglobulin (A2M) mapping to 12p13.31 and low-density lipoprotein-like protein (LRP1) at 12q13.3 received the greatest interest due to their potential involvement in the Aβ clearance pathway [reviewed in 148]. Both genes are excellent functional and positional candidates, however their role as Aβ-susceptibility genes have not yet been established unequivocally due to inconsistent results from association studies [reviewed in 149]. Recently, LOAD association has been detected for glyceraldehyde-3 phosphate dehydrogenase (GAPD) variants in multiple series [150, 151].
d) Chromosome 9
Although, chromosome 9 has received interest due to the evidence of linkage or association from several studies [121, 127, 129, 130], it should be noted that there is overlap between datasets assessed by three of these studies [121, 127, 129] and they, therefore, cannot be considered as providers for independent evidence of linkage. Chromosome 9 is therefore considered last in this section. In a whole genome scan of 466 LOAD families, Pericak-Vance et al. identified a region on chromosome 9p to have strong evidence for linkage with a multipoint lod score of 2.97 [121]. The evidence was even stronger in the autopsy-confirmed subset of 199 families with a multipoint lod score (MLS=4.31) even higher than that found in the ApoE region on chromosome 19 (MLS=3.42). The autopsy confirmed subset also had another downstream region on chromosome 9q with evidence of linkage. In the substantially overlapping dataset analyzed by Myers et al., evidence for linkage was identified to regions on 9p and 9q [127]. The 9p linkage coincides with that in the Pericak-Vance et al. study, although the 9q linkage findings for the two studies are ~46 cM apart. Blacker et al. also identified two suggestive linkage peaks on 9p21.1 and 9q22.2 in their overall dataset, however could not detect a suggestive signal in their non-overlapping, non-Kehoe families [129]. Finally, Farrer et al., in an association analysis of an independent series of Arab case-controls determined four markers on chromosome 9 spanning 32–47 cM (9p22.2-21.2) with significant LOAD association [130]. Their most significant marker (AFM220XF2, p=2.3×10−7) resides in a region coinciding with two other linkage signals [121, 127] from overlapping datasets.
Several functional and positional candidate genes reside on chromosome 9, including DAPK1 (death-associated protein kinase) [151], ABCA1-2 (ATP binding cassette A1-2 transporters) [153, 154], VLDLR (very low density lipoprotein receptor) [155], and UBQLN1 (ubiquilin 1) [156], with roles in apoptosis [151], lipid metabolism [153–155] and protein degradation [156]. Recent meta-analyses of genetic association studies on LOAD from the AlzGene database determined that variants in DAPK1 showed significant association in meta-analyses of 3–6 independent series [140] and also significantly altered expression of this gene [151].
3) Alternative analytical methods to detect risk loci for LOAD
In addition to the 9 whole genome linkage or association analyses in predominantly non-overlapping samples [117, 119, 121, 123, 126, 127, 129, 130, 132, 135, 136], seven linkage studies are presented here, which utilized alternative analytical strategies to re-assess the data in overlapping datasets [122, 124, 125, 128, 131, 133, 134]. We will briefly discuss their approaches and results, as they bring potential new understanding and evidence to the LOAD risk loci. Curtis et al. [122] re-analyzed the NIMH families, which have significant overlap with the genome-wide scans by Kehoe et al. [120] and Myers et al. [127], using ApoE genotypes as liability classes, thus introducing a novel method for simultaneous analysis of any loci conditional on a known risk locus. They were able to confirm the 10q21.3 linkage at D10S1211 and 12p13.2-1 at D12S358. Scott et al. [131] utilized “ordered subsets analysis” developed by Hauser et al. [157] to identify subsets of families ordered by their age-at-onset and which yielded stronger evidence of linkage to certain loci, compared to the analyses using the whole dataset. This approach was applied to 437 families with complete age-at-onset information, from the original set of 466 families in the Pericak-Vance et al. whole genome screen [121]. Two novel loci were detected at chromosomes 2q34 in early onset AD (50–60 years) and 15q22 in very late onset AD (≥79 years), in addition to increasing the previously detected linkage signal at 9p from 3.0 to 4.6 (n=334 families, 60–75 years). The chromosome 2q34 locus also showed evidence of linkage in Caribbean Hispanics [132], along with two other regions encompassing 2q34 detected in Amish families [135]. The 15q region is at least 17 cM away from the nearest risk region identified in another study [123].
Several groups included a number of variables as covariates in linkage analyses [128, 133, 134]. Olson et al. studied 272 sibpairs from the NIMH Genetics Initiative with complete covariate information on age-at-onset, current age and ApoE genotype, to determine the effects of these covariates on linkage [128]. They were able to obtain stronger evidence of linkage to regions on chromosomes 5, 6, 9 and 21, previously identified in genome scans using the NIMH data [121, 127]. More interestingly, they detected signals at chromosomes 14 and 20 in regions not previously identified by inclusion of covariates. The most significant evidence came from chromosomes 20p and 21p near the APP region, where linkage was strongest in the oldest age group, lacking the ApoE ε4 allele. There was also significant epistasis between these two regions suggesting a biological link. Avramopoulos et al. found linkage to chromosome 14q24 region near the PSEN1 locus using presence of psychotic features as a covariate [133] in a subset of the NIMH sibpairs. Their strongest linkage was detected in earlier onset AD families without comorbid hallucinations. Two other studies identified 14q24 as a risk region [123, 135]. Recent meta-analyses of an intronic SNP in PSEN1 was found to have significant association with AD in 34 different studies, drawing attention to the potential involvement of this established EOAD gene for LOAD, as well [140]. Bacanu et al. analyzed the subset of the NIMH families with psychotic symptoms and ApoE ε4 allele and determined linkage to chromosomes 2p, 6q and 21q [124]. These regions do not overlap with those from the Avrampoulos et al. study [133]. Although both studies showed linkage to 2p in LOAD with psychotic features, their locations are ~40 cM apart. Holmans et al. [134] analyzed the effects of multiple covariates including age-at-onset, rate of decline and ApoE on the linkage of LOAD series with major overlap with the Myers et al. study [127]. Their most significant finding was on chromosome 21 in the NIMH dataset, where linkage was strongest in the sibpairs with high age-at-onset, thus confirming the results of Olson et al. [128]. Their approach led to improved linkage findings in several other chromosomal loci.
Quantitative trait analyses in LOAD were first performed by Ertekin-Taner et al. using plasma Aβ levels as the phenotype [143] where evidence of linkage to chromosome 10 was determined in a locus overlapping with that from the Myers et al. study [127]. Li et al. used age-at-onset as a quantitative phenotype in AD and PD families [125]. Their strongest evidence for linkage in AD families came from chromosome 10q at a locus >50 cM downstream that of the Myers et al. [127] and Ertekin-Taner et al. [143] linkage.
These linkage studies with alternative analytical approaches have several important uses. They can: 1) lead to identification of novel loci by reducing heterogeneity and increasing power; 2) help confirm prior findings; 3) identify the subset of individuals where effect of the risk locus is strongest, thus potentially help guide future mapping/association studies; 4) bring some potential understanding to the pathophysiologic effects of the risk locus (e.g. decreases age at onset, acts via the amyloid pathway etc.).
Candidate gene studies
There have been 968 association studies on 398 AD candidate genes to date according to the AlzGene database with update on February 8, 2007 (http://www.alzforum.org/res/com/gen/alzgene/) [140]. It is neither possible nor effectively informative to provide a thorough review of these studies. Instead, this section will focus on some general pitfalls and guidelines on the association studies for complex diseases, with highlight on a few AD candidate genes as examples. The lack of a universally accepted genetic risk factor(s) for LOAD besides ApoE, despite the substantial genetic component underlying this disorder and the large numbers of genes studied, is testimony in part to the complexity of the genetic underpinnings of AD as well as the inadequacy of some of the approaches utilized. Several reviews have highlighted potential reasons for the failure to identify the “next ApoE” gene for AD [141, 158–160]. These include 1) initial false positive results followed by lack of replication; 2) lack of power due to small sample size (false negatives); 3) lack of informative markers; 4) genetic heterogeneity (different sets of genes underlying the same AD phenotype); 5) clinical heterogeneity (multiple clinical subtypes with different sets of susceptibility genes);.
Although, initial false positive associations could account for some of the lack of replication in LOAD association studies, it is unlikely to account for all cases of failure to replicate. Lohmueller et al. assessed 301 published studies on 25 single variant associations and determined a highly substantial excess of studies with significant replication of the original association results [161]. They determined that of the 301 follow-up studies to an original significant association, 59, 26 and 10 studies showed significant associations with p<0.05, <0.01 and <0.001, respectively. These numbers were significantly higher than the numbers of significant association studies expected by chance alone (15, 3 and <1, respectively). Ioannidis et al. systematically assessed by meta-analysis 370 published studies on 36 genetic associations [162]. Both studies concluded that the initial estimate of the genetic effect size from the original positive association study tended to be inflated [161, 162], with modest true effect sizes with ORs of 0.5–2.0 in most cases. Given this, many association studies assessed by these authors and as well from the LOAD literature will be sorely underpowered to detect true but modest effects. Two groups systematically analyzed AD association studies published within a given period of time [141, 160]. Bertram and Tanzi [141] assessed 105 (38 positive, 67 negative) AD association studies published in 2003, whereas Blomqvist et al. [160] analyzed 138 (86 positive, 52 negative) articles published between January 2004 to April 2005. Both studies excluded reports on ApoE. They both observed insufficient sample sizes. Blomqvist et al. found that 59% of the studies (82/138) had <500 subjects, whereas Bertram and Tanzi have determined that ~20% of the studies had <200 individuals. Since sample sizes on the order of 1000–10,000 will often be required to achieve sufficient power to detect the expected modest effect sizes for most complex disease susceptibility genes, a large number of the AD association studies are underpowered. It should also be noted that small sample size is not only problematic for lack of power for follow-up studies, but also an important determinant for an initial false positive association. Initial sample size of <150 was significantly less likely to be replicated compared with larger studies, suggesting a higher false positive rate for initial studies of smaller size [162].
Association of a disease with a genetic variant does not imply causation and may simply be due to linkage disequilibrium (LD) between the tested marker and an untested susceptibility variant. Furthermore, more than one susceptibility variant could be present in a single gene, acting in an additive or multiplicative fashion. Both of these issues complicate association studies and could potentially lead to false negative results due to a) different extent of LD between tested marker and untested susceptibility variant between different populations or b) incomplete assessment of the true susceptibility conferred by a genic region. To address these issues, choosing potentially functional variants (e.g. missense SNPs, insertions/deletions, variants in highly conserved regions) and assessing multiple variants and haplotypes have been advocated [141, 163]. The two recent surveys of AD association studies have revealed that the majority (>60%) have been performed on a single variant [141, 160] and less than 20% pursued haplotype analysis [141].
Genetic heterogeneity due to ethnic or environmental differences leading to false negative results, as well as false positive results secondary to population substructure have been raised as potential problems affecting association studies [reviewed in 163], though their true impact on AD genetics is yet to be determined. Finally, use of biologically relevant, quantitative phenotypes have been proposed as a means to overcome potential clinical heterogeneity, as such quantitative traits may a) be a less heterogeneous and more direct correlate for the genetic variation(s); b) be less prone to the imprecisions of a clinical phenotype; c) be amenable to inclusion of subjects with pre-clinical disease; and d) allow for downstream functional testing of putative susceptibility variants [158, 160, 163]. More recently, the use of quantitative traits such as Aβ levels, mini-mental state examination scores, CSF tau levels, age-at-onset, pathological disease burden and CSF cholesterol levels [164–168] have emerged as novel approaches to study candidate AD genes.
Despite the pitfalls and shortcomings of association studies on AD, a recent meta-analysis of 127 polymorphisms in 69 AD candidate genes, identified 20 polymorphisms in 13 genes (other than ApoE and related genes) with significant results [140]. It should be noted that the effects of these variants were modest with average ORs of 0.82–1.25. Additionally, several recent reports on AD candidate genes have applied many of the suggested guidelines for complex disease association studies and found significant results [164–167, 174, 176, 178, 179]. While the role of these genes for AD remains to be established with further confirmatory studies and despite presence of negative results, these findings are nonetheless encouraging.
Chromosome 10 has been an area of great interest in AD due to the convergence of multiple linkage and association signals as well as the presence of multiple excellent positional and functional candidate genes. We will thus give examples of several AD candidate gene association studies from this chromosome highlighting study designs as well as briefly summarizing the conclusions.
Ertekin-Taner et al. [164] analyzed alpha-T-catenin (VR22) as a candidate gene on chromosome 10 due to its location and indirect interaction with presenilin 1 via β-catenin. They have identified two intronic SNPs which showed association with plasma Aβ42 levels in two independent sets of families, showed that these SNPs accounted for their plasma Aβ linkage on chromosome 10 and bounded the association within the large VR22 gene by analyzing an additional 49 SNPs within and around this gene, none of which showed as strong association. The strengths of this study were the use of a quantitative phenotype, multiple variants, internal replication, bounding of the association within a gene and assessment of linkage conditional on association. Subsequently three affirmative [169–171] and four negative [172–175] association studies were published on this genic region.
In 2003, Prince et al. [167] identified a haplotype block spanning insulin degrading enzyme (IDE), which showed significant association with AD and/or its quantitative traits (mini-mental state examination scores, CSF tau levels, age at onset) in an analysis of 5 independent case-control series. The strengths of this study were the use of multiple variants, haplotype analysis, multiple independent series from different populations and both clinical and quantitative phenotypes. Assessing the same haplotypes in two case-control and one family series, Ertekin-Taner et al. [165] found evidence of association with AD and plasma Aβ42 levels with directions of effect similar to those from the Prince et al. study. Sixteen additional association studies exist on the IDE gene with five confirmatory reports, all of which are summarized on the AlzGene website [140].
Li et al. [176] screened 22,000 genes by an expression assay and identified 52 genes with significant expression difference between ADs and controls, four of which resided on chromosome 10. They assessed these candidate genes in their AD and PD series, and determined variants in glutathione S-transferase omega-1 and -2 genes (GSTO1, GSTO2) to be significantly associated with age-at-onset of AD and PD. They subsequently determined that this association contributed significantly to their linkage results on chromosome 10 [177]. The main strength of this study is the selection of candidate genes based on a functional screen, in addition to use of multiple markers, multiple series, quantitative phenotypes and assessing effect of association on linkage. Of the four follow-up studies on GSTO1-2, two show a positive trend [summarized in AlzGene, 140].
Grupe et al. [174] genotyped ~1,400 gene-based SNPs in the chromosome 10q region to identify 69 with significant association in their exploratory series. Of these one SNP in LOC439999 showed significant association in four of their six case-control series. The strengths of this study were the use of a high-throughput screening strategy to survey a large number of genes in a non-hypothesis driven fashion and the presence of internal replication using >1000 cases and controls. Their association did not account for the linkage on chromosome 10. Only two negative follow-up studies exist for this association to date [140]. Using a similar screening approach with 1206 chromosome 10 SNPs in Japanese case-control series with no ApoE ε4 allele, Kuwano et al. [178] identified six SNPs in a two-stage association study, three of which resided in the dynamin binding protein gene (DNMBP). The strengths of this study are similar to those of Grupe et al. [174]. Kuwano et al. also determined decreased expression levels for DNMBP in the AD vs. control brains, thus providing some functional correlate for this gene.
Finally, though not on chromosome 10, recent findings on neuronal sortilin related receptor SORL1 on chromosome 11 will be briefly discussed [179] to highlight the study approach and challenges of complex disease association studies. Rogaeva et al. assessed SNPs in several genes from the vacuolar protein sorting family and identified SORL1 as a candidate for further analyses. Utilizing a two-stage approach with a total of 6 series from different ethnic groups, with two family-based series in the exploratory set and four combined case-control and family series in the replication set, they assessed SORL1 variants in >1500 cases and controls. In addition, they analyzed three more independent series of 1405 ADs and 2124 controls. The authors found significant association with SORL1 variants in multiple independent data sets, though there was no single variant with significant association across all data sets. There was also considerable functional data demonstrating reduced expression of SORL1 in subjects who are carriers of a SORL1 risk haplotype. Finally, the authors showed experimental results depicting the role of SORL1 in differential sorting of the APP holoprotein. The strengths of this study are the use of large numbers of independent series of both family and case-control type, with multiple different ethnic groups, hypothesis-driven assessment of candidate genes in a specific pathway, multiple single-SNP and haplotype variants assessed, and the presence of expression data in support of the association findings.
Synopsis
Alzheimer’s disease is one of the most challenging disorders of the century due to its personal and public impact. Discovering the underlying genetics of this common, complex disorder harbors the hope for its early detection, prevention and treatment. With the identification of exceedingly rare, early-onset autosomal dominant familial AD gene mutations over the past two decades, substantial progress has been made in understanding the disease pathophysiology. The most common, late-onset form of AD has a substantial genetic component which remains unexplained despite the identification of the susceptibility allele ApoE ε4. Evidence from whole genome linkage, association and candidate gene studies strongly implicate multiple different genetic loci responsible for susceptibility to AD. The ever-growing numbers of variants in the human genome with systematic cataloguing efforts, development of high-throughput genotyping and phenotyping assays, generation of well-characterized and large study series, and advent of novel analytical strategies will provide the tools essential for studying AD genetics. Though, significant progress is as yet lacking in this area, recent studies hold promise for illuminating important genetic aspects of this multifaceted disorder.
TABLE 4.
Results of the whole genome linkage and association studies in AD-Chromosomes 5–8.
| Ch | 5 | 6 | 7 | 8 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study Ref. |
Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result |
| 117 | D6S1004 | 6q15 | >1.5 | |||||||||
| D6S1019 | 6p21.2 | >1.0 | ||||||||||
|
| ||||||||||||
| 121 | D5S1470 | 5p13.3 | 2.23 (autopsy) | D6S1027 | 6q27 | 1.20 (autopsy) | D7S2847 | 7q31.31 | 2.18 (autopsy) | |||
|
| ||||||||||||
| 127 | D5S1470 | 5p13.3 | 1.0/1.6 (all, E4+) | D6S1017–D6S1018 | 6p21.1-p12.3 | 1.3 (E4+) | ||||||
|
| ||||||||||||
| 129 | D6S1027 | 6q27 | 2.7 (total) | |||||||||
|
| ||||||||||||
| 126,132 | D5S2488 | 5p15.33 | 1.76 (late) | GAAT3A06 | 6p22.3 | 1.65 (all) | D8S1136 | 8q13.1 | 1.08 (late) | |||
| D6S1280 | 6p12.3 | 1.58 (late=all) | ||||||||||
|
| ||||||||||||
| 135 | D5S1725 | 5q14.3 | 1.47 | D6S1040 | 6q23.1 | 1.94 | D7S2204 | 7q21.11 | 1.14 | |||
| D7S3070 | 7q36.1 | 1.74 | ||||||||||
|
| ||||||||||||
| 136 | D5S498 | 5q35.2 | 2.4 | |||||||||
|
| ||||||||||||
| 119 | ||||||||||||
|
| ||||||||||||
| 123 | D5S807 | 5p15.2 | 0.001 | D6S1017 | 6p21.1 | 0.003 | GAAT1A4 | 8q22.2 | 0.028 | |||
|
| ||||||||||||
| 130 | ||||||||||||
TABLE 5.
Results of the whole genome linkage and association studies in AD-Chromosomes 9–12.
| Ch | 9 | 10 | 11 | 12 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study Ref. |
Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result |
| 117 | D12S1042 | 12p11.23 | 3.2 (multi all=3.5; multi E4−=3.9) | |||||||||
| D12S390 | 12q13.13 | 2.3 | ||||||||||
|
| ||||||||||||
| 121 | D9S741 | 9p21.3 | 4.31 (autopsy) | D10S1426 | 10p11.23 | 1.22 (autopsy) | ||||||
| D9S1818 | 9q34.2 | 2.05 (autopsy) | ||||||||||
|
| ||||||||||||
| 127 | D9S741 | 9p21.3 | 1.8 (all) | D10S1211 | 10q21.3 | 3.9/3.2 (all/E4+) | LOX-1 | 12p13.2 | 1.3/1.4 (whole/E4−) | |||
| D9S176 | 9q22.33 | 1.0/1.8 (all/E4+) | ||||||||||
|
| ||||||||||||
| 129 | D10S189 | 10p14 | 1.2 (early/mixed) | D11S968 | 11q25 | 1.1 (total) | ||||||
|
| ||||||||||||
| 126,132 | D10S190 | 10q26.11 | 2.02 (all) | D12S1623 | 12p13.31 | 2.01 (late) | ||||||
| D12S1057 | 12p12.1 | 1.63 (late) | ||||||||||
|
| ||||||||||||
| 135 | D10S2327 | 10q22.3 | 2.42 | D11S1392 | 11p13 | 2.14 | ||||||
|
| ||||||||||||
| 136 | ||||||||||||
|
| ||||||||||||
| 119 | D10S1423 | 10p12.33 | 0.001 | D12S1045 | 12q24.33 | 0.03 | ||||||
|
| ||||||||||||
| 123 | D10S1664 | 10p13 | 0.007 | |||||||||
|
| ||||||||||||
| 130 | AFM220XF2 | 9p21.3 | 2.3×10^−7 | D10S580 | 10q22.3 | 0.001 | D12S1722 | 12q15 | 1.3×10^^−6 | |||
| D10S1426 | 10p11.23 | <0.01 | ||||||||||
TABLE 6.
Results of the whole genome linkage and association studies in AD-Chromosomes 13–16.
| Ch | 13 | 14 | 15 | 16 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study Ref. |
Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result |
| 117 | ||||||||||||
| 121 | D13S787 | 13q12.12 | 1.05 | |||||||||
| 127 | ||||||||||||
| 129 | D13S793 | 13q32.1 | 1.4 (late) | D14S587 | 14q22.2 | 2.9 (early/mixed) | D15S642 | 15q26.3 | 1.2 (early/mixed) | |||
| 126,132 | D14S587 | 14q22.2 | 2.0 (total) | |||||||||
| 135 | D14S588 | 14q24.1 | 1.54 | |||||||||
| 136 | ||||||||||||
| 119 | ||||||||||||
| 123 | D13S292 | 13q12.12 | 0.002 | D14S77 | 14q24.2 | 0.048 | D15S659 | 15q21.1 | 0.036 | D16S403 | 16p12.1 | 0.012 |
| 130 | ||||||||||||
TABLE 7.
Results of the whole genome linkage and association studies in AD-Chromosomes 17–20.
| Ch | 17 | 18 | 19 | 20 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study Ref. |
Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result | Marker | Locus | Result |
| 117 | D20S94 | 20q13.32 | <1.0 | |||||||||
|
| ||||||||||||
| 121 | D18S878 | 18q22.1 | 1.02 (autopsy) | D19S246 | 19q13.33 | 3.64 (autopsy) | ||||||
| D18S1371 | 18q22.3-23 | 1.14 (autopsy) | APOE | 19q13.32 | 11.85 | |||||||
|
| ||||||||||||
| 127 | D19S412 | 19q13.32 | 1.3 | |||||||||
|
| ||||||||||||
| 129 | D19S178 | 19q13.31 | 7.6 (early/mixed) | |||||||||
|
| ||||||||||||
| 126,132 | D18S1106 | 18q22.3 | 3.65 (all) | |||||||||
|
| ||||||||||||
| 135 | D18S858 | 18q21.31 | 1.88 | D19S586 | 19p13.2 | 2.06 | D20S477 | 20p11.21 | 1.05 | |||
| D19S245 | 19q13.11 | 1.41 | ||||||||||
| D19S246 | 19q13.33 | 1.57 | ||||||||||
|
| ||||||||||||
| 136 | D19S902 | 19q13.32 | 4.84 | |||||||||
|
| ||||||||||||
| 119 | APOE | 19q13.32 | <0.0001 | |||||||||
|
| ||||||||||||
| 123 | D17S1293 | 17q12 | 0.025 | D18S66 | 18q11.2 | 0.025 | D19S1034 | 19p13.3 | 0.013 | D20S604 | 20p12.1 | 0.036 |
| D18S59 | 18p11.32 | 0.009 | D19S433 | 19q12 | 0.021 | |||||||
|
| ||||||||||||
| 130 | ||||||||||||
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mayeux R. Epidemiology of neurodegeneration. Annu Rev Neurosci. 2003;26:81–104. doi: 10.1146/annurev.neuro.26.043002.094919. [DOI] [PubMed] [Google Scholar]
- 2.Rice DP, Fillit HM, Max W, Knopman DS, Lloyd JR, Duttagupta S. Prevalence, costs, and treatment of Alzheimer’s disease and related dementia: a managed care perspective. Am J Manag Care. 2001 Aug;7(8):809–818. [PubMed] [Google Scholar]
- 3.Brookmeyer R, Gray S, Kawas C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am J Public Health. 1998 Sep;88(9):1337–1342. doi: 10.2105/ajph.88.9.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borenstein AR, Copenhaver CI, Mortimer JA. Early-life risk factors for Alzheimer disease. Alzheimer Dis Assoc Disord. 2006 Jan–Mar;20(1):63–72. doi: 10.1097/01.wad.0000201854.62116.d7. [DOI] [PubMed] [Google Scholar]
- 5.Whalley LJ, Dick FD, McNeill G. A life-course approach to the aetiology of late-onset dementias. Lancet Neurol. 2006 Jan;5(1):87–96. doi: 10.1016/S1474-4422(05)70286-6. [DOI] [PubMed] [Google Scholar]
- 6.Lazarov O, Robinson J, Tang YP, et al. Environmental enrichment reduces Abeta levels and amyloid deposition in transgenic mice. Cell. 2005 Mar 11;120(5):701–713. doi: 10.1016/j.cell.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Jankowsky JL, Melnikova T, Fadale DJ, et al. Environmental enrichment mitigates cognitive deficits in a mouse model of Alzheimer’s disease. J Neurosci. 2005 May 25;25(21):5217–5224. doi: 10.1523/JNEUROSCI.5080-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alzheimer A. A new disease of the cortex (Ger) Allg Z Psychiatr. 1907;64:146–148. [Google Scholar]
- 9.Harris R. Genetics of Alzheimer’s disease. Br Med J (Clin Res Ed) 1982 Apr 10;284(6322):1065–1066. doi: 10.1136/bmj.284.6322.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitner JC, Silverman JM, Mohs RC, Davis KL. Familial aggregation in Alzheimer’s disease: comparison of risk among relatives of early-and late-onset cases, and among male and female relatives in successive generations. Neurology. 1988;38(2):207–212. doi: 10.1212/wnl.38.2.207. [DOI] [PubMed] [Google Scholar]
- 11.Farrer LA, Myers RH, Cupples LA, et al. Transmission and age-at-onset patterns in familial Alzheimer’s disease: evidence for heterogeneity. Neurology. 1990;40(3 Pt 1):395–403. doi: 10.1212/wnl.40.3_part_1.395. [DOI] [PubMed] [Google Scholar]
- 12.Lautenschlager NT, Cupples LA, Rao VS, et al. Risk of dementia among relatives of Alzheimer’s disease patients in the MIRAGE study: What is in store for the oldest old? Neurology. 1996;46(3):641–650. doi: 10.1212/wnl.46.3.641. [DOI] [PubMed] [Google Scholar]
- 13.Green RC, Cupples LA, Go R, et al. Risk of dementia among white and African American relatives of patients with Alzheimer disease. JAMA. 2002;287(3):329–336. doi: 10.1001/jama.287.3.329. [DOI] [PubMed] [Google Scholar]
- 14.Farrer LA, Cupples LA, van Duijn CM, et al. Apolipoprotein E genotype in patients with Alzheimer’s disease: implications for the risk of dementia among relatives. Ann Neurol. 1995 Nov;38(5):797–808. doi: 10.1002/ana.410380515. [DOI] [PubMed] [Google Scholar]
- 15.Martinez M, Campion D, Brice A, et al. Apolipoprotein E epsilon4 allele and familial aggregation of Alzheimer disease. Arch Neurol. 1998 Jun;55(6):810–816. doi: 10.1001/archneur.55.6.810. [DOI] [PubMed] [Google Scholar]
- 16.Devi G, Ottman R, Tang M, et al. Influence of APOE genotype on familial aggregation of AD in an urban population. Neurology. 1999 Sep 11;53(4):789–794. doi: 10.1212/wnl.53.4.789. [DOI] [PubMed] [Google Scholar]
- 17.Farrer LA, Myers RH, Connor L, Cupples LA, Growdon JH. Segregation analysis reveals evidence of a major gene for Alzheimer disease. Am J Hum Genet. 1991 Jun;48(6):1026–1033. [PMC free article] [PubMed] [Google Scholar]
- 18.van Duijn CM, Farrer LA, Cupples LA, Hofman A. Genetic transmission of Alzheimer’s disease among families in a Dutch population based study. J Med Genet. 1993 Aug;30(8):640–646. doi: 10.1136/jmg.30.8.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao VS, van Duijn CM, Connor-Lacke L, Cupples LA, Growdon JH, LA F. Multiple etiologies for Alzheimer disease are revealed by segregation analysis. Am J Hum Genet. 1994;55(5) [PMC free article] [PubMed] [Google Scholar]
- 20.Warwick Daw E, Payami H, Nemens EJ, et al. The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet. 2000;66(1):196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raiha I, Kaprio J, Koskenvuo M, Rajala T, Sourander L. Alzheimer’s disease in Finnish twins. Lancet. 1996;347(9001):573–578. doi: 10.1016/s0140-6736(96)91272-6. [DOI] [PubMed] [Google Scholar]
- 22.Bergem AL, Engedal K, Kringlen E. The role of heredity in late-onset Alzheimer disease and vascular dementia. A twin study. Arch Gen Psychiatry. 1997;54(3):264–270. doi: 10.1001/archpsyc.1997.01830150090013. [DOI] [PubMed] [Google Scholar]
- 23.Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006 Feb;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 24.Breitner JC, Gatz M, Bergem AL, et al. Use of twin cohorts for research in Alzheimer’s disease. Neurology. 1993 Feb;43(2):261–267. doi: 10.1212/wnl.43.2.261. [DOI] [PubMed] [Google Scholar]
- 25.Raiha I, Kaprio J, Koskenvuo M, Rajala T, Sourander L. Alzheimer’s disease in twins. Biomed Pharmacother. 1997;51(3):101–104. doi: 10.1016/s0753-3322(97)86906-5. [DOI] [PubMed] [Google Scholar]
- 26.Steffens DC, Plassman BL, Helms MJ, Welsh-Bohmer KA, Newman TT, Breitner JC. APOE and AD concordance in twin pairs as predictors of AD in first-degree relatives. Neurology. 2000 Feb 8;54(3):593–598. doi: 10.1212/wnl.54.3.593. [DOI] [PubMed] [Google Scholar]
- 27.Bergem AL, Lannfelt L. Apolipoprotein E type epsilon4 allele, heritability and age at onset in twins with Alzheimer disease and vascular dementia. Clin Genet. 1997 Nov;52(5):408–413. doi: 10.1111/j.1399-0004.1997.tb04362.x. [DOI] [PubMed] [Google Scholar]
- 28.Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349(6311):704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 29.Fraser J, Mitchell A. Kalmuc idiocy: report of a case with autopsy, with notes on sixty-two cases. J Ment Sci. 1876;22:161–179. [Google Scholar]
- 30.Struwe F. Histopathologische Untersuchungen uber Entstehung und Wesen der senilen Plaques. Z Neurol Psychiatrie. 1929;122:291–307. [Google Scholar]
- 31.Jervis G. Early senile dementia in mongoloid idiocy. Am J Psychiatry. 1948;105:102–106. doi: 10.1176/ajp.105.2.102. [DOI] [PubMed] [Google Scholar]
- 32.Pary RJ, Stonecipher GR. A Overview of Alzheimer’s disease in Down syndrome. In: Prasher VP, editor. DOWN SYNDROME AND ALZHEIMER’S DISEASE biological correlates. Radcliffe Publishing; 2006. [Google Scholar]
- 33.Glenner GG, Wong CW. Alzheimer’s disease and Down’s syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem Biophys Res Commun. 1984;122(3):1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- 34.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82(12):4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.St George-Hyslop PH, Tanzi RE, Polinsky RJ, et al. The genetic defect causing familial Alzheimer’s disease maps on chromosome 21. Science. 1987;235(4791):885–890. doi: 10.1126/science.2880399. [DOI] [PubMed] [Google Scholar]
- 36.Goldgaber D, Lerman MI, McBride OW, Saffiotti U, Gajdusek DC. Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer’s disease. Science. 1987;235(4791):877–880. doi: 10.1126/science.3810169. [DOI] [PubMed] [Google Scholar]
- 37.Tanzi RE, Gusella JF, Watkins PC, et al. Amyloid beta protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science. 1987;235(4791):880–884. doi: 10.1126/science.2949367. [DOI] [PubMed] [Google Scholar]
- 38.Tanzi RE, St George-Hyslop PH, Haines JL, et al. The genetic defect in familial Alzheimer’s disease is not tightly linked to the amyloid beta-protein gene. Nature. 1987;329(6135):156–157. doi: 10.1038/329156a0. [DOI] [PubMed] [Google Scholar]
- 39.Van Broeckhoven C, Genthe AM, Vandenberghe A, et al. Failure of familial Alzheimer’s disease to segregate with the A4-amyloid gene in several European families. Nature. 1987;329(6135):153–155. doi: 10.1038/329153a0. [DOI] [PubMed] [Google Scholar]
- 40.Schellenberg GD, Bird TD, Wijsman EM, et al. Absence of linkage of chromosome 21q21 markers to familial Alzheimer’s disease. Science. 1988;241(4872):1507–1510. doi: 10.1126/science.3420406. [DOI] [PubMed] [Google Scholar]
- 41.Goate AM, Haynes AR, Owen MJ, et al. Predisposing locus for Alzheimer’s disease on chromosome 21. Lancet. 1989;1(8634):352–355. doi: 10.1016/s0140-6736(89)91725-x. [DOI] [PubMed] [Google Scholar]
- 42.Theuns J, Marjaux E, Vandenbulcke M, et al. Alzheimer dementia caused by a novel mutation located in the APP C-terminal intracytosolic fragment. Hum Mutat. 2006 Sep;27(9):888–896. doi: 10.1002/humu.20402. [DOI] [PubMed] [Google Scholar]
- 43.Eckman C. This issue. [Google Scholar]
- 44.Schellenberg GD, Bird TD, Wijsman EM, et al. Genetic linkage evidence for a familial Alzheimer’s disease locus on chromosome 14. Science. 1992;258(5082):668–671. doi: 10.1126/science.1411576. [DOI] [PubMed] [Google Scholar]
- 45.Mullan M, Houlden H, Windelspecht M, et al. A locus for familial early-onset Alzheimer’s disease on the long arm of chromosome 14, proximal to the alpha 1-antichymotrypsin gene. Nat Genet. 1992;2(4):340–342. doi: 10.1038/ng1292-340. [DOI] [PubMed] [Google Scholar]
- 46.St George-Hyslop P, Haines J, Rogaev E, et al. Genetic evidence for a novel familial Alzheimer’s disease locus on chromosome 14. Nat Genet. 1992;2(4):330–334. doi: 10.1038/ng1292-330. [DOI] [PubMed] [Google Scholar]
- 47.Van Broeckhoven C, Backhovens H, Cruts M, et al. Mapping of a gene predisposing to early-onset Alzheimer’s disease to chromosome 14q24.3. Nat Genet. 1992;2(4):335–339. doi: 10.1038/ng1292-335. [DOI] [PubMed] [Google Scholar]
- 48.Sherrington R, Rogaev EI, Liang Y, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375(6534):754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- 49.Alzheimer’s Disease Collaborative Group. The structure of the presenilin 1 (S182) gene and identification of six novel mutations in early onset AD families. Nat Genet. 1995;11(2):219–222. doi: 10.1038/ng1095-219. [DOI] [PubMed] [Google Scholar]
- 50.Hutton M, Busfield F, Wragg M, et al. Complete analysis of the presenilin 1 gene in early onset Alzheimer’s disease. Neuroreport. 1996;7(3):801–805. doi: 10.1097/00001756-199602290-00029. [DOI] [PubMed] [Google Scholar]
- 51.Cruts M, Van Broeckhoven C. Presenilin mutations in Alzheimer’s disease. Hum Mutat. 1998;11(3):183–190. doi: 10.1002/(SICI)1098-1004(1998)11:3<183::AID-HUMU1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 52.Levy-Lahad E, Wijsman EM, Nemens E, et al. A familial Alzheimer’s disease locus on chromosome 1. Science. 1995;269(5226):970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 53.Levy-Lahad E, Wasco W, Poorkaj P, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269(5226):973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- 54.Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, Chi H, Lin C, Holman K, Tsuda T, et al. Familial Alzheimer’s disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer’s disease type 3 gene. Nature. 1995;376:775–778. doi: 10.1038/376775a0. [DOI] [PubMed] [Google Scholar]
- 55.Sherrington R, Froelich S, Sorbi S, et al. Alzheimer’s disease associated with mutations in presenilin 2 is rare and variably penetrant. Hum Mol Genet. 1996;5(7):985–988. doi: 10.1093/hmg/5.7.985. [DOI] [PubMed] [Google Scholar]
- 56.Finckh U, Kuschel C, Anagnostouli M, et al. Novel mutations and repeated findings of mutations in familial Alzheimer disease. Neurogenetics. 2005 May;6(2):85–89. doi: 10.1007/s10048-005-0211-x. [DOI] [PubMed] [Google Scholar]
- 57.Campion D, Dumanchin C, Hannequin D, et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet. 1999;65(3):664–670. doi: 10.1086/302553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schoenberg BS, Anderson DW, Haerer AF. Severe dementia. Prevalence and clinical features in a biracial US population. Arch Neurol. 1985;42(8):740–743. doi: 10.1001/archneur.1985.04210090004002. [DOI] [PubMed] [Google Scholar]
- 59.Sulkava R, Wikstrom J, Aromaa A, et al. Prevalence of severe dementia in Finland. Neurology. 1985;35(7):1025–1029. doi: 10.1212/wnl.35.7.1025. [DOI] [PubMed] [Google Scholar]
- 60.Kokmen E, Beard CM, Offord KP, Kurland LT. Prevalence of medically diagnosed dementia in a defined United States population: Rochester, Minnesota, January 1, 1975. Neurology. 1989;39(6):773–776. doi: 10.1212/wnl.39.6.773. [DOI] [PubMed] [Google Scholar]
- 61.Croes EA, Dermaut B, van Der Cammen TJ, van Broeckhoven C, van Duijn CM. Genetic testing should not be advocated as a diagnostic tool in familial forms of dementia. Am J Hum Genet. 2000;67(4):1033–1035. doi: 10.1086/303074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cruts M, van Duijn CM, Backhovens H, et al. Estimation of the genetic contribution of presenilin-1 and -2 mutations in a population-based study of presenile Alzheimer disease. Hum Mol Genet. 1998;7(1):43–51. doi: 10.1093/hmg/7.1.43. [DOI] [PubMed] [Google Scholar]
- 63.Janssen JC, Beck JA, Campbell TA, et al. Early onset familial Alzheimer’s disease: Mutation frequency in 31 families. Neurology. 2003 Jan 28;60(2):235–239. doi: 10.1212/01.wnl.0000042088.22694.e3. [DOI] [PubMed] [Google Scholar]
- 64.Nussbaum RL, Ellis CE. Alzheimer’s disease and Parkinson’s disease. N Engl J Med. 2003 Apr 3;348(14):1356–1364. doi: 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- 65.Sleegers K, Roks G, Theuns J, et al. Familial clustering and genetic risk for dementia in a genetically isolated Dutch population. Brain. 2004 Jul;127(Pt 7):1641–1649. doi: 10.1093/brain/awh179. [DOI] [PubMed] [Google Scholar]
- 66.Jimenez-Escrig A, Gomez-Tortosa E, Baron M, et al. A multigenerational pedigree of late-onset Alzheimer’s disease implies new genetic causes. Brain. 2005 Jul;128(Pt 7):1707–1715. doi: 10.1093/brain/awh501. [DOI] [PubMed] [Google Scholar]
- 67.Giedraitis V, Hedlund M, Skoglund L, et al. New Alzheimer’s disease locus on chromosome 8. J Med Genet. 2006 Dec;43(12):931–935. doi: 10.1136/jmg.2006.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ashley-Koch AE, Shao Y, Rimmler JB, et al. An autosomal genomic screen for dementia in an extended Amish family. Neurosci Lett. 2005 May 13;379(3):199–204. doi: 10.1016/j.neulet.2004.12.065. [DOI] [PubMed] [Google Scholar]
- 69.Rademakers R, Cruts M, Sleegers K, et al. Linkage and association studies identify a novel locus for Alzheimer disease at 7q36 in a Dutch population-based sample. Am J Hum Genet. 2005 Oct;77(4):643–652. doi: 10.1086/491749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinha S, Anderson JP, Barbour R, et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402(6761):537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 71.Vassar R, Bennett BD, Babu-Khan S, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 72.Yan R, Bienkowski MJ, Shuck ME, et al. Membrane-anchored aspartyl protease with Alzheimer’s disease beta-secretase activity. Nature. 1999;402(402):533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 73.Vassar R, Citron M. Abeta-generating enzymes: recent advances in beta- and gamma-secretase research. Neuron. 2000;27(3):419–422. doi: 10.1016/s0896-6273(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 74.Lammich S, Kojro E, Postina R, et al. Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A. 1999;96(7):3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hardy J. Amyloid, the presenilins and Alzheimer’s disease. Trends Neurosci. 1997;20(4):154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- 76.Scheuner D, Eckman C, Jensen M, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2(8):864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 77.Citron M, Oltersdorf T, Haass C, et al. Mutation of the beta-amyloid precursor protein in familial Alzheimer’s disease increases beta-protein production. Nature. 1992;360(6405):672–674. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- 78.Suzuki N, Cheung TT, Cai XD, et al. An increased percentage of long amyloid beta protein secreted by familial amyloid beta protein precursor (beta APP717) mutants. Science. 1994;264(5163):1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 79.Borchelt DR, Thinakaran G, Eckman CB, et al. Familial Alzheimer’s disease-linked presenilin 1 variants elevate Abeta1-42/1-40 ratio in vitro and in vivo. Neuron. 1996;17(5):1005–1013. doi: 10.1016/s0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 80.Eckman CB, Mehta ND, Crook R, et al. A new pathogenic mutation in the APP gene (I716V) increases the relative proportion of A beta 42(43) Hum Mol Genet. 1997;6(12):2087–2089. doi: 10.1093/hmg/6.12.2087. [DOI] [PubMed] [Google Scholar]
- 81.Hsiao K, Chapman P, Nilsen S, et al. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274(5284):99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 82.Duff K, Eckman C, Zehr C, et al. Increased amyloid-beta42(43) in brains of mice expressing mutant presenilin1. Nature. 1996;383(6602):710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Z, Nadeau P, Song W, et al. Presenilins are required for gamma-secretase cleavage of beta-APP and transmembrane cleavage of Notch-1. Nat Cell Biol. 2000;2(7):463–465. doi: 10.1038/35017108. [DOI] [PubMed] [Google Scholar]
- 84.Golde TE, Younkin SG. Presenilins as therapeutic targets for the treatment of Alzheimer’s disease. Trends Mol Med. 2001;7(6):264–269. doi: 10.1016/s1471-4914(01)02064-0. [DOI] [PubMed] [Google Scholar]
- 85.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006 Oct 19;443(7113):774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 86.Klafki HW, Staufenbiel M, Kornhuber J, Wiltfang J. Therapeutic approaches to Alzheimer’s disease. Brain. 2006 Nov;129(Pt 11):2840–2855. doi: 10.1093/brain/awl280. [DOI] [PubMed] [Google Scholar]
- 87.Masters CL, Beyreuther K. Alzheimer’s centennial legacy: prospects for rational therapeutic intervention targeting the Abeta amyloid pathway. Brain. 2006 Nov;129(Pt 11):2823–2839. doi: 10.1093/brain/awl251. [DOI] [PubMed] [Google Scholar]
- 88.Pericak-Vance MA, Yamaoka LH, Haynes CS, et al. Genetic linkage studies in Alzheimer’s disease families. Exp Neurol. 1988;102(3):271–279. doi: 10.1016/0014-4886(88)90220-8. [DOI] [PubMed] [Google Scholar]
- 89.Pericak-Vance MA, Bebout JL, Gaskell PC, et al. Linkage studies in familial Alzheimer disease: evidence for chromosome 19 linkage. Am J Hum Genet. 1991;48(6):1034–1050. [PMC free article] [PubMed] [Google Scholar]
- 90.Schellenberg GD, Deeb SS, Boehnke M, et al. Association of an apolipoprotein CII allele with familial dementia of the Alzheimer type. J Neurogenet. 1987 Apr;4(2–3):97–108. [PubMed] [Google Scholar]
- 91.Namba Y, Tomonaga M, Kawasaki H, Otomo E, Ikeda K. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer’s disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 1991;541(1):163–166. doi: 10.1016/0006-8993(91)91092-f. [DOI] [PubMed] [Google Scholar]
- 92.Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 94.Myers RH, Schaefer EJ, Wilson PW, et al. Apolipoprotein E epsilon4 association with dementia in a population-based study: The Framingham study. Neurology. 1996;46(3):673–677. doi: 10.1212/wnl.46.3.673. [DOI] [PubMed] [Google Scholar]
- 95.Farrer LA, Cupples LA, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278(16):1349–1356. [PubMed] [Google Scholar]
- 96.Bickeboller H, Campion D, Brice A, et al. Apolipoprotein E and Alzheimer disease: genotype-specific risks by age and sex. Am J Hum Genet. 1997;60(2):439–446. [PMC free article] [PubMed] [Google Scholar]
- 97.Slooter AJ, Cruts M, Kalmijn S, et al. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam Study. Arch Neurol. 1998 Jul;55(7):964–968. doi: 10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- 98.Tang MX, Stern Y, Marder K, et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998 Mar 11;279(10):751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 99.Breitner JC, Wyse BW, Anthony JC, et al. APOE-epsilon4 count predicts age when prevalence of AD increases, then declines: the Cache County Study. Neurology. 1999;53(2):321–331. doi: 10.1212/wnl.53.2.321. [DOI] [PubMed] [Google Scholar]
- 100.Graff-Radford NR, Green RC, Go RC, et al. Association between apolipoprotein E genotype and Alzheimer disease in African American subjects. Arch Neurol. 2002;59(4):594–600. doi: 10.1001/archneur.59.4.594. [DOI] [PubMed] [Google Scholar]
- 101.Evans DA, Bennett DA, Wilson RS, et al. Incidence of Alzheimer disease in a biracial urban community: relation to apolipoprotein E allele status. Arch Neurol. 2003 Feb;60(2):185–189. doi: 10.1001/archneur.60.2.185. [DOI] [PubMed] [Google Scholar]
- 102.Murrell JR, Price B, Lane KA, et al. Association of apolipoprotein E genotype and Alzheimer disease in African Americans. Arch Neurol. 2006 Mar;63(3):431–434. doi: 10.1001/archneur.63.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hendrie HC, Murrell J, Gao S, Unverzagt FW, Ogunniyi A, Hall KS. International studies in dementia with particular emphasis on populations of African origin. Alzheimer Dis Assoc Disord. 2006 Jul–Sep;20(3 Suppl 2):S42–46. doi: 10.1097/00002093-200607001-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gureje O, Ogunniyi A, Baiyewu O, et al. APOE epsilon4 is not associated with Alzheimer’s disease in elderly Nigerians. Ann Neurol. 2006 Jan;59(1):182–185. doi: 10.1002/ana.20694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Romas SN, Santana V, Williamson J, et al. Familial Alzheimer disease among Caribbean Hispanics: a reexamination of its association with APOE. Arch Neurol. 2002 Jan;59(1):87–91. doi: 10.1001/archneur.59.1.87. [DOI] [PubMed] [Google Scholar]
- 106.Olarte L, Schupf N, Lee JH, et al. Apolipoprotein E epsilon4 and age at onset of sporadic and familial Alzheimer disease in Caribbean Hispanics. Arch Neurol. 2006 Nov;63(11):1586–1590. doi: 10.1001/archneur.63.11.1586. [DOI] [PubMed] [Google Scholar]
- 107.Arboleda GH, Yunis JJ, Pardo R, et al. Apolipoprotein E genotyping in a sample of Colombian patients with Alzheimer’s disease. Neurosci Lett. 2001 Jun 8;305(2):135–138. doi: 10.1016/s0304-3940(01)01829-8. [DOI] [PubMed] [Google Scholar]
- 108.Mayeux R, Saunders AM, Shea S, et al. Utility of the apolipoprotein E genotype in the diagnosis of Alzheimer’s disease. Alzheimer’s Disease Centers Consortium on Apolipoprotein E and Alzheimer’s Disease. N Engl J Med. 1998 Feb 19;338(8):506–511. doi: 10.1056/NEJM199802193380804. [DOI] [PubMed] [Google Scholar]
- 109.Tsuang D, Larson EB, Bowen J, et al. The utility of apolipoprotein E genotyping in the diagnosis of Alzheimer disease in a community-based case series. Arch Neurol. 1999 Dec;56(12):1489–1495. doi: 10.1001/archneur.56.12.1489. [DOI] [PubMed] [Google Scholar]
- 110.Waldemar G, Dubois B, Emre M, et al. Recommendations for the diagnosis and management of Alzheimer’s disease and other disorders associated with dementia: EFNS guideline. Eur J Neurol. 2007 Jan;14(1):e1–26. doi: 10.1111/j.1468-1331.2006.01605.x. [DOI] [PubMed] [Google Scholar]
- 111.Petersen RC, Smith GE, Ivnik RJ, et al. Apolipoprotein E status as a predictor of the development of Alzheimer’s disease in memory-impaired individuals. JAMA. 1995 Apr 26;273(16):1274–1278. [PubMed] [Google Scholar]
- 112.Hsiung GY, Sadovnick AD, Feldman H. Apolipoprotein E epsilon4 genotype as a risk factor for cognitive decline and dementia: data from the Canadian Study of Health and Aging. Cmaj. 2004 Oct 12;171(8):863–867. doi: 10.1503/cmaj.1031789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Devanand DP, Pelton GH, Zamora D, et al. Predictive utility of apolipoprotein E genotype for Alzheimer disease in outpatients with mild cognitive impairment. Arch Neurol. 2005 Jun;62(6):975–980. doi: 10.1001/archneur.62.6.975. [DOI] [PubMed] [Google Scholar]
- 114.Tschanz JT, Welsh-Bohmer KA, Lyketsos CG, et al. Conversion to dementia from mild cognitive disorder: the Cache County Study. Neurology. 2006 Jul 25;67(2):229–234. doi: 10.1212/01.wnl.0000224748.48011.84. [DOI] [PubMed] [Google Scholar]
- 115.Cervilla J, Prince M, Joels S, Lovestone S, Mann A. Premorbid cognitive testing predicts the onset of dementia and Alzheimer’s disease better than and independently of APOE genotype. J Neurol Neurosurg Psychiatry. 2004 Aug;75(8):1100–1106. doi: 10.1136/jnnp.2003.028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee DY, Youn JC, Choo IH, et al. Combination of clinical and neuropsychologic information as a better predictor of the progression to Alzheimer disease in questionable dementia individuals. Am J Geriatr Psychiatry. 2006 Feb;14(2):130–138. doi: 10.1097/01.JGP.0000192487.58426.d2. [DOI] [PubMed] [Google Scholar]
- 117.Pericak-Vance MA, Bass MP, Yamaoka LH, et al. Complete genomic screen in late-onset familial Alzheimer disease. Evidence for a new locus on chromosome 12. JAMA. 1997;278(15):1237–1241. [PubMed] [Google Scholar]
- 118.Pericak-Vance MA, Bass ML, Yamaoka LH, et al. Complete genomic screen in late-onset familial Alzheimer’s disease. Neurobiol Aging. 1998 Jan–Feb;19(1 Suppl):S39–42. doi: 10.1016/s0197-4580(98)00037-2. [DOI] [PubMed] [Google Scholar]
- 119.Zubenko GS, Hughes HB, Stiffler JS, Hurtt MR, BBK A genome survey for novel Alzheimer disease risk loci: results at 10-cM resolution. Genomics. 1998;50(2):121–128. doi: 10.1006/geno.1998.5306. [DOI] [PubMed] [Google Scholar]
- 120.Kehoe P, Wavrant-De Vrieze F, Crook R, et al. A full genome scan for late onset Alzheimer’s disease. Hum Mol Genet. 1999;8(2):237–245. doi: 10.1093/hmg/8.2.237. [DOI] [PubMed] [Google Scholar]
- 121.Pericak-Vance MA, Grubber J, Bailey LR, et al. Identification of novel genes in late-onset Alzheimer’s disease. Exp Gerontol. 2000;35(9–10):1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 122.Curtis D, North BV, Sham PC. A novel method of two-locus linkage analysis applied to a genome scan for late onset Alzheimer’s disease. Ann Hum Genet. 2001 Sep;65(Pt 5):473–481. doi: 10.1017/S0003480001008776. [DOI] [PubMed] [Google Scholar]
- 123.Hiltunen M, Mannermaa A, Thompson D, et al. Genome-wide linkage disequilibrium mapping of late-onset Alzheimer’s disease in Finland. Neurology. 2001;57(9):1663–1668. doi: 10.1212/wnl.57.9.1663. [DOI] [PubMed] [Google Scholar]
- 124.Bacanu SA, Devlin B, Chowdari KV, DeKosky ST, Nimgaonkar VL, Sweet RA. Linkage analysis of Alzheimer disease with psychosis. Neurology. 2002 Jul 9;59(1):118–120. doi: 10.1212/wnl.59.1.118. [DOI] [PubMed] [Google Scholar]
- 125.Li YJ, Scott WK, Hedges DJ, et al. Age at Onset in Two Common Neurodegenerative Diseases Is Genetically Controlled. Am J Hum Genet. 2002;70(4):985–993. doi: 10.1086/339815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mayeux R, Lee JH, Romas SN, et al. Chromosome-12 mapping of late-onset Alzheimer disease among Caribbean Hispanics. Am J Hum Genet. 2002 Jan;70(1):237–243. doi: 10.1086/324773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Myers A, Wavrant De-Vrieze F, Holmans P, et al. Full genome screen for Alzheimer disease: stage II analysis. Am J Med Genet. 2002;114(2):235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- 128.Olson JM, Goddard KA, Dudek DM. A second locus for very-late-onset Alzheimer disease: a genome scan reveals linkage to 20p and epistasis between 20p and the amyloid precursor protein region. Am J Hum Genet. 2002 Jul;71(1):154–161. doi: 10.1086/341034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Blacker D, Bertram L, Saunders AJ, et al. Results of a high-resolution genome screen of 437 Alzheimer’s Disease families. Hum Mol Genet. 2003;12(1):23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 130.Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, Baldwin CT. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12(4):415–422. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- 131.Scott WK, Hauser ER, Schmechel DE, et al. Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. Am J Hum Genet. 2003 Nov;73(5):1041–1051. doi: 10.1086/379083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee JH, Mayeux R, Mayo D, et al. Fine mapping of 10q and 18q for familial Alzheimer’s disease in Caribbean Hispanics. Mol Psychiatry. 2004 Nov;9(11):1042–1051. doi: 10.1038/sj.mp.4001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Avramopoulos D, Fallin MD, Bassett SS. Linkage to chromosome 14q in Alzheimer’s disease (AD) patients without psychotic symptoms. Am J Med Genet B Neuropsychiatr Genet. 2005 Jan 5;132(1):9–13. doi: 10.1002/ajmg.b.30074. [DOI] [PubMed] [Google Scholar]
- 134.Holmans P, Hamshere M, Hollingworth P, et al. Genome screen for loci influencing age at onset and rate of decline in late onset Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2005 May 5;135(1):24–32. doi: 10.1002/ajmg.b.30114. [DOI] [PubMed] [Google Scholar]
- 135.Hahs DW, McCauley JL, Crunk AE, et al. A genome-wide linkage analysis of dementia in the Amish. Am J Med Genet B Neuropsychiatr Genet. 2006 Mar 5;141(2):160–166. doi: 10.1002/ajmg.b.30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sillen A, Forsell C, Lilius L, et al. Genome scan on Swedish Alzheimer’s disease families. Mol Psychiatry. 2006 Feb;11(2):182–186. doi: 10.1038/sj.mp.4001772. [DOI] [PubMed] [Google Scholar]
- 137.Hiltunen M, Mannermaa A, Koivisto AM, Lehtovirta M, Helisalmi S, Ryynanen M, et al. Linkage disequilibrium in the 13q12 region in Finnish late onset Alzheimer’s disease patients. Eur J Hum Genet. 1999;7(6):652–658. doi: 10.1038/sj.ejhg.5200342. [DOI] [PubMed] [Google Scholar]
- 138.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, et al. Ensembl 2007. Nucleic Acids Res. 2007;35(Database issue):D610–617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007 Jan;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 141.Bertram L, Tanzi RE. Alzheimer’s disease: one disorder, too many genes? Hum Mol Genet. 2004 Apr 1;13(Spec No 1):R135–141. doi: 10.1093/hmg/ddh077. [DOI] [PubMed] [Google Scholar]
- 142.Kamboh MI. Molecular genetics of late-onset Alzheimer’s disease. Ann Hum Genet. 2004 Jul;68(Pt 4):381–404. doi: 10.1046/j.1529-8817.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- 143.Ertekin-Taner N, Graff-Radford N, Younkin LH, et al. Linkage of Plasma Aβ42 to a Quantitative Locus on Chromosome 10 in Late-Onset Alzheimer’s Disease Pedigrees. Science. 2000;290(5500):2303–2304. doi: 10.1126/science.290.5500.2303. [DOI] [PubMed] [Google Scholar]
- 144.Myers A, Holmans P, Marshall H, et al. Susceptibility locus for Alzheimer’s disease on chromosome 10. Science. 2000;290(5500):2304–2305. doi: 10.1126/science.290.5500.2304. [DOI] [PubMed] [Google Scholar]
- 145.Bertram L, Blacker D, Mullin K, et al. Evidence for genetic linkage of Alzheimer’s disease to chromosome 10q. Science. 2000;290(5500):2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- 146.Scott WK, Grubber JM, Conneally PM, et al. Fine mapping of the chromosome 12 late-onset Alzheimer disease locus: potential genetic and phenotypic heterogeneity. Am J Hum Genet. 2000;66(3):922–932. doi: 10.1086/302828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rogaeva E, Premkumar S, Song Y, et al. Evidence for an Alzheimer disease susceptibility locus on chromosome 12 and for further locus heterogeneity. JAMA. 1998;280(7):614–618. doi: 10.1001/jama.280.7.614. [DOI] [PubMed] [Google Scholar]
- 148.Kovacs DM. alpha2-macroglobulin in late-onset Alzheimer’s disease. Exp Gerontol. 2000 Jul;35(4):473–479. doi: 10.1016/s0531-5565(00)00113-3. [DOI] [PubMed] [Google Scholar]
- 149.D’Introno A, Solfrizzi V, Colacicco AM, et al. Current knowledge of chromosome 12 susceptibility genes for late-onset Alzheimer’s disease. Neurobiol Aging. 2006 Nov;27(11):1537–1553. doi: 10.1016/j.neurobiolaging.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 150.Li Y, Nowotny P, Holmans P, et al. Association of late-onset Alzheimer’s disease with genetic variation in multiple members of the GAPD gene family. Proc Natl Acad Sci U S A. 2004 Nov 2;101(44):15688–15693. doi: 10.1073/pnas.0403535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lin PI, Martin ER, Bronson PG, et al. Exploring the association of glyceraldehyde-3-phosphate dehydrogenase gene and Alzheimer disease. Neurology. 2006 Jul 11;67(1):64–68. doi: 10.1212/01.wnl.0000223438.90113.4e. [DOI] [PubMed] [Google Scholar]
- 152.Li Y, Grupe A, Rowland C, et al. DAPK1 variants are associated with Alzheimer’s disease and allele-specific expression. Hum Mol Genet. 2006 Sep 1;15(17):2560–2568. doi: 10.1093/hmg/ddl178. [DOI] [PubMed] [Google Scholar]
- 153.Katzov H, Chalmers K, Palmgren J, et al. Genetic variants of ABCA1 modify Alzheimer disease risk and quantitative traits related to beta-amyloid metabolism. Hum Mutat. 2004 Apr;23(4):358–367. doi: 10.1002/humu.20012. [DOI] [PubMed] [Google Scholar]
- 154.Mace S, Cousin E, Ricard S, et al. ABCA2 is a strong genetic risk factor for early-onset Alzheimer’s disease. Neurobiol Dis. 2005 Feb;18(1):119–125. doi: 10.1016/j.nbd.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 155.Okuizumi K, Onodera O, Namba Y, et al. Genetic association of the very low density lipoprotein (VLDL) receptor gene with sporadic Alzheimer’s disease. Nat Genet. 1995 Oct;11(2):207–209. doi: 10.1038/ng1095-207. [DOI] [PubMed] [Google Scholar]
- 156.Bertram L, Hiltunen M, Parkinson M, et al. Family-based association between Alzheimer’s disease and variants in UBQLN1. N Engl J Med. 2005 Mar 3;352(9):884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 157.Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M. Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol. 2004 Jul;27(1):53–63. doi: 10.1002/gepi.20000. [DOI] [PubMed] [Google Scholar]
- 158.Emahazion T, Feuk L, Jobs M, et al. SNP association studies in Alzheimer’s disease highlight problems for complex disease analysis. Trends Genet. 2001;17(7):407–413. doi: 10.1016/s0168-9525(01)02342-3. [DOI] [PubMed] [Google Scholar]
- 159.Finckh U. The future of genetic association studies in Alzheimer disease. J Neural Transm. 2003;110(3):253–266. doi: 10.1007/s00702-002-0775-7. [DOI] [PubMed] [Google Scholar]
- 160.Blomqvist ME, Reynolds C, Katzov H, et al. Towards compendia of negative genetic association studies: an example for Alzheimer disease. Hum Genet. 2006 Mar;119(1–2):29–37. doi: 10.1007/s00439-005-0078-9. [DOI] [PubMed] [Google Scholar]
- 161.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet. 2003;33(2):177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 162.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29(3):306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 163.Newton-Cheh C, Hirschhorn JN. Genetic association studies of complex traits: design and analysis issues. Mutat Res. 2005 Jun 3;573(1–2):54–69. doi: 10.1016/j.mrfmmm.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 164.Ertekin-Taner N, Ronald J, Asahara H, et al. Fine mapping of the alpha-T catenin gene to a quantitative trait locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Hum Mol Genet. 2003 Dec 1;12(23):3133–3143. doi: 10.1093/hmg/ddg343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ertekin-Taner N, Allen M, Fadale D, et al. Genetic variants in a haplotype block spanning IDE are significantly associated with plasma Abeta42 levels and risk for Alzheimer disease. Hum Mutat. 2004 Apr;23(4):334–342. doi: 10.1002/humu.20016. [DOI] [PubMed] [Google Scholar]
- 166.Ertekin-Taner N, Ronald J, Feuk L, et al. Elevated amyloid beta protein (Abeta42) and late onset Alzheimer’s disease are associated with single nucleotide polymorphisms in the urokinase-type plasminogen activator gene. Hum Mol Genet. 2005 Feb 1;14(3):447–460. doi: 10.1093/hmg/ddi041. [DOI] [PubMed] [Google Scholar]
- 167.Prince JA, Feuk L, Gu HF, et al. Genetic variation in a haplotype block spanning IDE influences Alzheimer disease. Hum Mutat. 2003;22(5):363–371. doi: 10.1002/humu.10282. [DOI] [PubMed] [Google Scholar]
- 168.Wollmer MA, Streffer JR, Tsolaki M, et al. Genetic association of acyl-coenzyme A: cholesterol acyltransferase with cerebrospinal fluid cholesterol levels, brain amyloid load, and risk for Alzheimer’s disease. Mol Psychiatry. 2003 Jun;8(6):635–638. doi: 10.1038/sj.mp.4001296. [DOI] [PubMed] [Google Scholar]
- 169.Martin ER, Bronson PG, Li YJ, et al. Interaction between the alpha-T catenin gene (VR22) and APOE in Alzheimer’s disease. J Med Genet. 2005 Oct;42(10):787–792. doi: 10.1136/jmg.2004.029553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cellini E, Bagnoli S, Tedde A, Nacmias B, Piacentini S, Sorbi S. Insulin degrading enzyme and alpha-3 catenin polymorphisms in Italian patients with Alzheimer disease. Alzheimer Dis Assoc Disord. 2005 Oct–Dec;19(4):246–247. doi: 10.1097/01.wad.0000189030.50826.86. [DOI] [PubMed] [Google Scholar]
- 171.Bertram L, Mullin K, Parkinson M, et al. Is alpha-T catenin (VR22) an Alzheimer’s disease risk gene? J Med Genet. 2007 Jan;44(1):e63. doi: 10.1136/jmg.2005.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Blomqvist ME, Andreasen N, Bogdanovic N, Blennow K, Brookes AJ, Prince JA. Genetic variation in CTNNA3 encoding alpha-3 catenin and Alzheimer’s disease. Neurosci Lett. 2004 Apr 1;358(3):220–222. doi: 10.1016/j.neulet.2004.01.032. [DOI] [PubMed] [Google Scholar]
- 173.Busby V, Goossens S, Nowotny P, et al. Alpha-T-catenin is expressed in human brain and interacts with the Wnt signaling pathway but is not responsible for linkage to chromosome 10 in Alzheimer’s disease. Neuromolecular Med. 2004;5(2):133–146. doi: 10.1385/NMM:5:2:133. [DOI] [PubMed] [Google Scholar]
- 174.Grupe A, Li Y, Rowland C, et al. A scan of chromosome 10 identifies a novel locus showing strong association with late-onset Alzheimer disease. Am J Hum Genet. 2006 Jan;78(1):78–88. doi: 10.1086/498851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kuwano R, Miyashita A, Arai H, et al. Dynamin-binding protein gene on chromosome 10q is associated with late-onset Alzheimer’s disease. Hum Mol Genet. 2006 Jul 1;15(13):2170–2182. doi: 10.1093/hmg/ddl142. [DOI] [PubMed] [Google Scholar]
- 176.Li YJ, Oliveira SA, Xu P, et al. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum Mol Genet. 2003;12(24):3259–3267. doi: 10.1093/hmg/ddg357. [DOI] [PubMed] [Google Scholar]
- 177.Li YJ, Scott WK, Zhang L, et al. Revealing the role of glutathione S-transferase omega in age-at-onset of Alzheimer and Parkinson diseases. Neurobiol Aging. 2006 Aug;27(8):1087–1093. doi: 10.1016/j.neurobiolaging.2005.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kuwano R, Miyashita A, Arai H, et al. Dynamin-binding protein gene on chromosome 10q is associated with late-onset Alzheimer’s disease. Hum Mol Genet. 2006 Jul 1;15(13):2170–2182. doi: 10.1093/hmg/ddl142. [DOI] [PubMed] [Google Scholar]
- 179.Rogaeva E, Meng Y, Lee JH, et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007 Feb;39(2):168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]