Abstract
NRAGE (also known as Maged1, dlxin) is a member of the MAGE gene family that may play a role in the neuronal apoptosis that is regulated by the p75 neurotrophin receptor (p75NTR). To test this hypothesis in vivo, we generated NRAGE knockout mice and found that NRAGE deletion caused a defect in developmental apoptosis of sympathetic neurons of the superior cervical ganglia, similar to that observed in p75NTR knockout mice. Primary sympathetic neurons derived from NRAGE knockout mice were resistant to apoptosis induced by brain-derived neurotrophic factor (BDNF), a pro-apoptotic p75NTR ligand, and NRAGE deficient sympathetic neurons show attenuated BDNF-dependent JNK activation. Hair follicle catagen is an apoptosis-like process that is dependent on p75NTR signaling; we show that NRAGE and p75NTR show regulated co-expression in the hair follicle and that identical defects in hair follicle catagen are present in NRAGE and p75NTR knockout mice. Interestingly, NRAGE knockout mice have severe defects in motoneuron apoptosis that are not observed in p75NTR knockout animals, raising the possibility that NRAGE may facilitate apoptosis induced by receptors other than p75NTR. Together, these studies demonstrate that NRAGE plays an important role in apoptotic signaling in vivo.
Keywords: neurotrophin, BDNF, motoneuron, sympathethic neuron, hair follicle, Mage, Necdin, Dlxin, apoptosis
Introduction
The p75 neurotrophin receptor (p75NTR) is a TNF receptor superfamily member that is expressed at high levels during embryogenesis and functions as a pro-apoptotic receptor during development of neuronal and non-neuronal tissues 1–3. The ligand requirements and downstream signaling cascades activated by p75NTR are beginning to emerge and mounting data indicates that p75NTR induces cell death through mechanisms that are distinct from those activated by other pro-apoptotic TNFR superfamily members. p75NTR-dependent apoptosis does not involve caspase-8 activation but instead requires activation of Jun kinase (JNK), phosphorylation of BH3-domain-only family members, release of mitochondrial contents, and activation of caspase-9 4,5,6,7.
Independent studies have shown that several members of the MAGE protein family bind to the p75NTR cytosolic region 8,9,10 and one of them, termed NRAGE (also known as Maged1 and dlxin) is a potent activator of JNK and inducer of apoptosis5,11. NRAGE is broadly expressed during development and in vitro studies have implicated NRAGE in cellular functions ranging from cell cycle regulation and cell adhesion to transcriptional regulation 12,13,14,15. We have previously shown that NRAGE is recruited to p75NTR in a neurotrophin dependent manner and proposed that NRAGE functions as an adaptor protein that links p75NTR to JNK activation.
In this study, we generated NRAGE knockout mice to assess the physiological role of NRAGE and to specifically test the hypothesis that NRAGE functions as a pro-apoptotic p75NTR adaptor protein in vivo. We demonstrate that NRAGE plays an essential non-redundant role in developmental apoptosis of sympathetic neurons in vivo. In primary sympathetic neurons derived from NRAGE knockout mice, apoptosis induced by brain-derived neurotrophic factor (BDNF), a p75NTR ligand, is sharply attenuated. We confirmed these results in rat sympathetic neurons in which NRAGE levels were reduced by RNA interference. We also report that NRAGE knockout mice exhibit defects in hair follicle catagen that mimic those previously reported for p75NTR knockouts. Together, these findings establish that NRAGE functions as a pro-apoptotic adaptor protein that mediates p75NTR apoptotic signaling events in vivo. In addition, we show that NRAGE knockout mice, but not p75NTR knockout mice, have a severe defect in motoneuron apoptosis, suggesting that NRAGE may function as an adaptor protein for pro-apoptotic receptors distinct from p75NTR. Taken together, these data demonstrate that NRAGE is a versatile pro-apoptotic protein required for developmental apoptosis.
Results
NRAGE Knockouts are Viable and Fertile
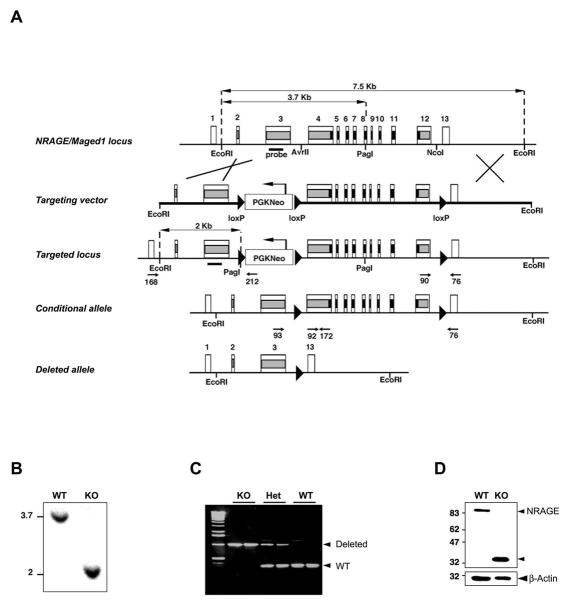
To determine if NRAGE is required for p75NTR-induced apoptosis in vivo, we generated mice in which exons 4 – 12 of the NRAGE gene were deleted (Figure 1A). The deleted region contains the conserved Mage Homology Domain (MHD) as well as the NRAGE segment that binds p75NTR. Southern blotting and PCR confirmed that the NRAGE gene was successfully targeted (Figure 1B and 1C) and immunoblotting of brain lysates demonstrated that full-length NRAGE was absent in the deleted animals. Exons 1–3 of the NRAGE gene remained intact in the deleted animals and we observed the presence of a truncated NRAGE fragment on immunoblots that presumably contained the initiator codon in exon 2 and the subsequent coding sequence in exons 2–3 (Figure 1D). This portion of the NRAGE protein has no obvious protein motifs and is poorly conserved and our analyses assumed that the mice lacking exons 4 – 12 of the NRAGE gene represent functional knockouts.
Figure 1. Targeted mutation of NRAGE/Maged1.
(A) Genomic organization of the NRAGE/Maged1 locus and schematic representation of the inactivation procedure. The conditional allele (Maged1tm1Urfm) and the deleted allele (Maged1tm1.1Urfm), generated after Cre-mediated recombination, are shown. Filled boxes represent the coding region of NRAGE, the segment encoding the p75NTR binding domain is in black. The loxP sites are represented by solid triangles. (B) Southern blot analysis of DNA isolated from wild-type ES cells and from ES cells that have undergone gene replacement (EcoRI/PagI double digestion). (C) PCR analysis of DNA isolated from tail biopsies of NRAGE knockout, heterozygous and wild-type mice. PCR amplification with primers 93 and 76 gives rise to a 940-bp product specific of the deleted allele (Maged1tm1.1Urfm) while amplification with primers 92 and 172 produces a 270-bp fragment specific of the wild-type allele. (D) Immunoblot blot analysis of protein lysates from NRAGE knockout and wild-type E13.5 brains.
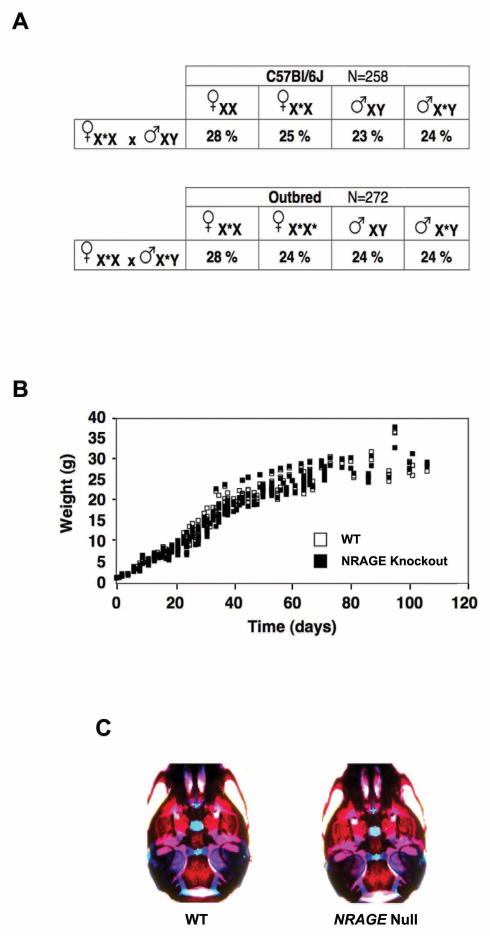
NRAGE knockout mice were born with normal Mendelian frequencies when maintained as an inbred line on the C57Bl6 strain or when outbred to CD1 animals (Figure 2A). Gross morphological and histopathological features of NRAGE knockout mice were normal (data not shown) and weight gain of NRAGE knockout mice was not different from wild-type littermates (Figure 2B). In vitro studies have raised the possibility that NRAGE may play a role in skeletal development or in cell cycle control yet whole mount alcian blue/alizarin red staining did not reveal abnormalities in skeletal development (Figure 2C) and cell cycle control of NRAGE knockout dermal fibroblasts, analyzed by FACS analysis, was indistinguishable from that observed in dermal fibroblasts from wild-type animals (Supplemental Figure 1). We therefore focused our efforts to determine if NRAGE plays a role in developmental apoptosis that is regulated by p75NTR.
Figure 2. NRAGE deficiency has no impact on viability.
(A) The fraction of each genotype generated by crossing heterozygous females (X*X) with either wild-type (XY) or hemizygous (X*Y) males do not differ from Mendelian predictions on either a pure C57Bl/6 background or in outbred CD1 strains. (B) NRAGE knockout and wild-type littermates display similar weight gain. (C) Alcian blue blue/alizarin red staining of P0 mice did not reveal abnormalities in skeletal development.
Hair Follicle Catagen is Defective in NRAGE Knockouts
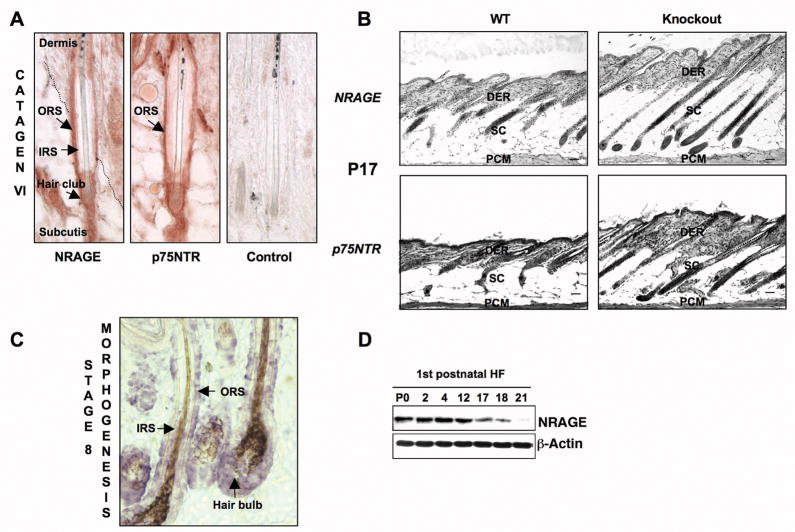
After morphogenesis, hair follicles undergo alternating periods of cell growth (anagen), apoptosis-driven regression (catagen) and resting (telogen). HF morphogenesis and the first HF catagen and telogen occur with stereotypic developmental timing; in C57BL/6 genetic background, HF morphogenesis is complete by postnatal day 9 (P9), the catagen phase is detected at P17 and telogen starts three days later 16. The apoptosis that occurs in the catagen phase relies on neurotrophin-driven activation of p75NTR 17,18. During catagen, most cells expressing p75NTR lie in the outer root sheath (ORS) and loss of p75NTR is associated with a decreased apoptosis in those cells. We used immunostaining to analyze the expression of NRAGE and p75NTR proteins in HF undergoing catagen at P18 in C57BL/6 mice. Figure 3A shows that both proteins co-localize in the ORS, the site of p75NTR-dependent apoptosis. To compare NRAGE and p75NTR knockout mice for defects in catagen, we examined the anagen to catagen transition, during the first postnatal HF cycle and after depilation, using the criteria established by Paus and colleagues 16. Back skin samples were collected at P17 for the first HF cycle (WT N=6, KO N=6) and 19 days post depilation (WT N=13, KO N=13). NRAGE deficient mice showed a profound delay in catagen-dependent dermal thinning, reduction in the hair follicle bulb volume and hair follicle shortening during the first hair follicle cycle (Figure 3B) and after depilation (data not shown). This defect observed in NRAGE nulls did not reflect a delay of HF maturation since the dynamic of HF morphogenesis and growth, analyzed at P1-P2 (WT N=11, KO N=11) and P14 (WT N=4, KO N=4), was undistinguishable between NRAGE wild-type and knockout littermates (Supplemental Figure 2 and data not shown). At P21 (WT N=3, KO N=3), a light delay in catagen completion was still observed in NRAGE knockout animals when compare to wild-type littermates (Supplemental Figure 2).
Figure 3. Hair follicle catagen is retarded in NRAGE knockout mice.
(A) Immunocytochemistry performed on serial back skin sections of a P18 C57BL/6 mouse showing expression of NRAGE and p75NTR protein in the outer root sheath of a hair follicle on catagen VI. (B) Retardation of hair follicle regression in P17 NRAGE knockout mice resembles that observed in P17 p75NTR knockout mice. Note the thickening of the dermis in each of the null strains. (C) In situ hybridization using an NRAGE probe labeled with digoxygenin 45 showing NRAGE RNA expression in the inner root sheath, the outer root sheath and hair bulb within a hair follicle of a P4 C57BL/6 mouse. (D) NRAGE protein expression at different stages of the first hair follicle cycle. Scale bar in B is 50 μm. IRS, inner root sheath; ORS, outer root sheath; DER, dermis; SC, subcutis; PCM, panniculus carnosus muscle.
p75NTR levels vary during the hair follicle cycle, with highest levels observed in the anagen phase, just before the hair follicle initiates regression 18. To establish if NRAGE mRNA and protein levels are regulated in a similar manner, we performed in situ hybridization and immunoblot analyses on dermal sections and lysates. We found abundant NRAGE mRNA expression during all stages of morphogenesis, moderate expression during catagen and no expression during telogen (Figure 3C and data not shown). Consistent with this, expression levels of NRAGE protein are low in telogen, intermediate in catagen and high during morphogenesis, similar to those previously described for p75NTR (Figure 3D). The tight co-expression and co-regulation of p75NTR and NRAGE levels, together with the precise phenocopying of catagen defects in p75NTR and NRAGE knockout mice, support the hypothesis that a p75NTR-NRAGE signaling pathway regulates hair follicle regression in vivo.
NRAGE is Required for p75NTR-Dependent Apoptosis of Sympathetic Neurons
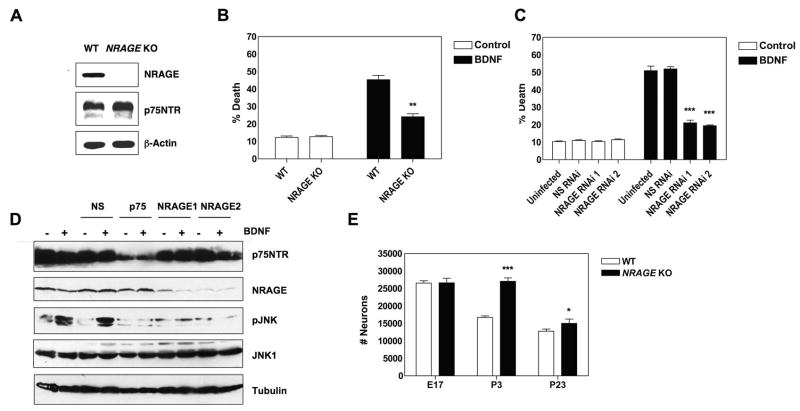
NGF and BDNF induce neuronal cell death in vivo by binding p75NTR and activating apoptotic signaling cascades. This can be replicated in vitro, where BDNF induces p75NTR-dependent apoptosis of sympathetic neurons derived from the superior cervical ganglia (SCG) 19,20. To determine if NRAGE could play a role in BDNF-induced death of sympathetic neurons, we first examined NRAGE and p75NTR expression levels in SCG during the period of naturally occurring cell death. Figure 4A shows that NRAGE protein is expressed in postnatal day 4 SCGs and that p75NTR protein expression levels do not change in NRAGE knockout mice. To determine if NRAGE lies on the p75NTR apoptotic cascade, we produced primary cultures of SCG sympathetic neurons from NRAGE knockout mice and wild-type littermates and analyzed the induction of apoptosis in these cells following BDNF exposure. Figure 4B shows that in wild-type neurons, BDNF treatment increased sympathetic neuron cell death by approximately 4-fold (from 11% to 43%). In NRAGE knockout neurons, the effect of BDNF was significantly reduced, with exposure to the ligand causing only 2-fold increase in SCG cell death (from 12% to 22%). Therefore, we conclude that NRAGE plays an important, but not exclusive, role in p75NTR-dependent apoptosis activated by BDNF exposure.
Figure 4. NRAGE knockout SCG sympathetic neurons are deficient in p75NTR-induced apoptosis.
(A) NRAGE and p75NTR expression in lysates from P4 SCGs of NRAGE knockout and wild-type control pups was determined by immunoblot. (B) Sympathetic neurons derived from NRAGE null animals or from wild-type littermates were exposed to BDNF and apoptotic nuclei were quantified 48 hours later. Approximately 50–70 neurons were counted for each condition. The experiment was performed in triplicate. **: P=0.0022. (C) Rat sympathetic neurons were infected with lentivirus expressing miRNA that targets NRAGE or were infected with a negative control lentivirus expressing miRNA that does not target a mammalian protein (NS RNAi), then cultured for 5 days. Neurons were then exposed to BDNF for 72 hours and apoptotic nuclei were quantified. ***: P<0.0001. (D) Rat sympathetic neurons were infected as in (C) but were also infected with a lentivirus expressing miRNA that targets p75NTR. 6 days later, neurons were exposed to BDNF for 1 hour, lysed and analyzed by immunoblot, as indicated. (E) Number of neurons within the superior cervical ganglia were quantified in NRAGE null and wild-type littermates at E17 (n=3), at P3 (n=5) and at P23 (NRAGE knockout: n=8, wild-type controls: n=10). ***: P<0.0001, *: P=0.05. For B, C and E, significance values were calculated using unpaired student t tests. Error bars represent SEM.
To confirm these results, we produced lentivirus expressing miRNA that targets NRAGE mRNA for destruction and asked whether depletion of NRAGE reduces BDNF-induced killing in primary rat sympathetic neurons. When sympathetic neurons infected with lentivirus that target NRAGE mRNA were exposed to BDNF, cell death was dramatically reduced, both at 48 and 72 hours after BDNF exposure was initiated (Figure 4C and Supplemental Figure 3). Identical results were obtained using two distinct lentivirus that target distinct NRAGE regions whereas a lentivirus expressing a control miRNA had no effect on BDNF-induced killing.
Activation of Jun kinase (JNK) is required for sympathetic neurons apoptosis induced by p75NTR and we therefore assessed if loss of p75NTR or NRAGE caused a reduction in BDNF-induced JNK activation in primary sympathetic neurons. Primary rat sympathetic neurons were infected with lentivirus targeting p75NTR or NRAGE and 6 days later, these cells were examined for BDNF-induced JNK activation. Figure 4D shows that lentivirus targeting p75NTR dramatically reduced levels of the receptor (>90%) and that, as expected, BDNF-induced activation of JNK was strongly reduced in these cells. Lentivirus targeting NRAGE sharply reduced NRAGE protein levels and, importantly, loss of NRAGE resulted in an almost complete loss of BDNF-induced JNK activation. We conclude that NRAGE is a key element linking p75NTR to JNK activation in sympathetic neurons. As above, identical results were obtained using lentivirus that target different NRAGE regions, and the control miRNA had no effect on BDNF-induced JNK activation.
p75NTR plays an important role in the developmental apoptosis of sympathetic neurons within the superior cervical ganglia (SCG). In mice, the pool of SCG neurons normally declines by more than 50% over the first three weeks of life and previous studies have shown that this decrease is significantly attenuated in p75NTR knockout animals19,21. To determine if NRAGE plays a role in the developmental death of sympathetic neurons in vivo, we performed neuronal counts on SCGs from wild-type and NRAGE knockout mice. At E17, before programmed cell death takes place in this ganglia, the number of sympathetic neurons was essentially identical in wild-type and NRAGE knockout littermates. The peak of cell death in the SCG occurs in early postnatal stages and at P3, we found that the number of surviving sympathetic neurons was much higher in NRAGE knockout animals. Between P3 and P23, when most programmed cell death has normally ended, the number of sympathetic neurons in the NRAGE null animals had dropped substantially but they were still significantly higher than in wild-type littermates (Figure 4E). We conclude that NRAGE is required for normal developmental cell death of SCG sympathetic neurons.
Neuronal Growth is Normal in NRAGE Knockout Mice
Mice rendered null for p75NTR have defects in neuronal growth that are manifest early in development. To assess whether NRAGE contributes to p75NTR-dependent neuronal growth, we compared wild-type, p75NTR knockout and NRAGE knockout embryos for neuronal growth defects at E11.5, E12.5 and E13.5, using whole-mount immunostaining for β3-tubulin to mark neuronal projections. As previously reported22–24, p75NTR knockout embryos displayed profound defects in several neuronal projections at these stages; as an example, Figure 5 shows that the ophthalmic branch of the trigeminal ganglion is severely truncated in p75NTR knockout animals at E12.5. In contrast, neuronal projections in NRAGE knockout embryos were indistinguishable from wild-type littermates at all stages examined (Figure 5 and data not shown). Therefore, although NRAGE is involved in p75NTR-dependent apoptosis, it is dispensable for effects on neuronal growth.
Figure 5. Abnormal outgrowth of the ophthalmic branch of the trigeminal ganglion in p75NTR knockout mice but not in NRAGE knockout mice.
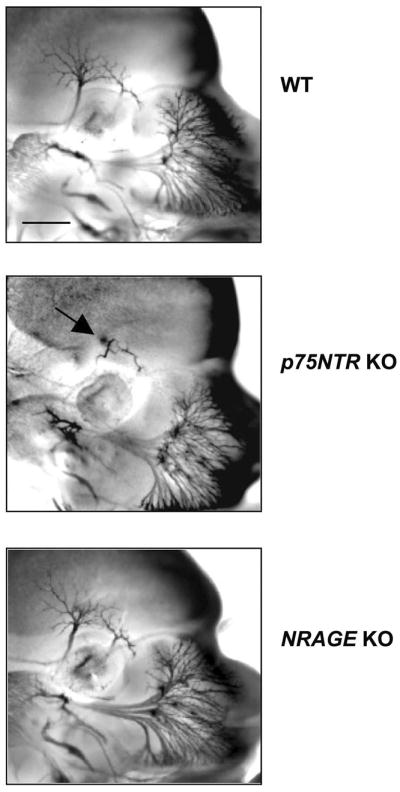
Whole-mount immunostaining of wild-type, p75NTR knockout and NRAGE knockout embryos with TuJ1, a monoclonal antibody raised against the neuron specific marker β3 tubulin. Arrow indicates growth defect in the ophthalmic branch of the trigeminal ganglion in p75NTR knockout embryos. Scale bar: 0.35 mm.
NRAGE Knockout Mice Display Defects in Developmental Apoptosis of Motoneurons
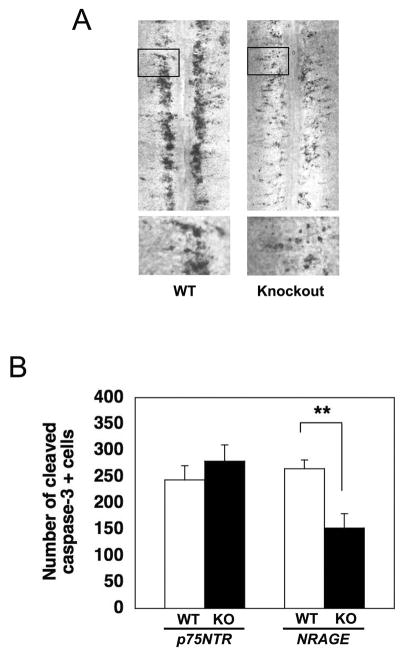
p75NTR isexpressed at high levels in developing motoneurons25 and some studies have suggested that NGF facilitates p75NTR-dependent developmental apoptosis in spinal motoneurons 26,27. However, the spinal cord motoneuron pool present in wild-type and p75NTR knockouts is identical at P7, suggesting that the receptor is not involved in the programmed cell death of these cells in vivo 28. Whether p75NTR plays a role in motoneuron apoptosis earlier in development has not been examined and we therefore used the whole mount spread method of Yamamoto and Henderson (1999) to ask whether p75NTR or NRAGE deletion alters developmental death of motoneurons. We found that mice lacking p75NTR do not have a defect in developmental motoneuron death but surprisingly, motoneuron apoptosis was sharply reduced in NRAGE null embryos (Figure 6A). Quantification of motoneuron apoptosis in the lumbar region of E13.5 embryos revealed that developmental apoptosis of motoneurons was reduced by 42% in NRAGE knockout mice compared to wild-type littermates, a highly significant effect (Figure 6B; p = 0.008). These data indicate that NRAGE functions in a p75NTR-independent manner to facilitate motoneuron cell death in vivo.
Figure 6. NRAGE, but not p75NTR, is required for motoneuron apoptosis in vivo.
(A) Whole-mount cleaved caspase-3 immunostaining of the lumbar region of E13.5 spinal cords from wild-type and NRAGE knockout embryos. Bottom panels represent magnifications of the inset regions. (B) Quantification of lumbar spinal motoneuron apoptosis. NRAGE knockout: n=4, wild-type littermates: n=5; p75NTR knockout: n=8, wild-type littermates: n=12. **: P=0.0081. P value was calculated using unpaired, two-tailed student t test. Error bars represent s.e.m
Discussion
Over 25 MAGE genes are expressed in the human genome but their functions remain poorly understood 15,29. MAGE proteins are characterized by a ~200 amino acid region, of unknown function, termed the MAGE homology domain 15. On the basis of their expression pattern, MAGE genes fall into two categories: Type I MAGE genes are activated in a wide variety of tumors but are silent in normal tissues, with the exception of testis and, in some case, placenta. Type II MAGE genes are broadly expressed in embryonic and adult somatic tissues. Most MAGE genes are single exon-encoding genes, likely formed by retrotransposition events, and are poorly conserved during mammalian evolution. NRAGE (also known as Maged1 and Dlxin) is a Type II MAGE gene that shows strong phylogenetic conservation with the unique MAGE gene found in Drosophila and other lower species 30,31, both with regard to its amino acid sequence and its genomic structure. Thus, NRAGE is likely to perform a fundamental cellular function which has been phylogenetically conserved.
NRAGE has been implicated in a large number of cellular processes. In vitro studies have indicated that NRAGE binds Dlx and Msx transcription factors and modulates their transcriptional activity 13,14,32 and that NRAGE regulates p53 -dependent transcription 33. NRAGE has also been shown to alter cell cycle progression 34, to disrupt E-cadherin dependent adhesion 12 and to facilitate p38 MAPK activation in response to bone morphogenetic proteins 35. Finally, NRAGE binds to the p75NTR intracellular domain and facilitates apoptosis following neurotrophin exposure, suggesting that NRAGE is an adaptor protein that links p75NTR to the apoptotic cascade 5,11,36. Our goal in this study was to test the hypothesis that NRAGE functions as an adaptor protein that is required for apoptosis induced by the p75 neurotrophin receptor (p75NTR) in vivo.
NRAGE knockout mice are fertile, grossly normal and show no obvious defects at birth or in adulthood. Although NRAGE has been implicated in cell proliferation and in transcriptional events that regulate skeletal development, our analyses did not reveal obvious defects in cell cycle control or in the skeletal system. Given the high degree of homology between NRAGE and the other MAGED genes, it is possible that compensatory effects from other alleles dampen an NRAGE loss of function phenotype. The targeted allele produced in this study lacked exons 4–12 (which encodes the MHD as well as the portion of NRAGE that binds p75NTR) but left exons 1–3 intact. The fragment encoded by these exons contains no identifiable protein motifs and does not bind to p75NTR but we cannot rule out the possibility that this fragment has cellular consequences that may ameliorate or exacerbate the consequences of NRAGE gene deletion.
p75NTR plays an important role in the developmental cell death of sympathetic neurons. SCGs in mice lacking p75NTR have considerably more sympathetic neurons during and shortly after the period of naturally occurring cell death than their wild-type counterparts19. We performed neuronal counts of the SCG in NRAGE knockouts and in wild-type littermates and found that NRAGE null mice exhibit a defect in sympathetic neuron apoptosis very similar to that observed in the p75NTR mice. For example, recent results from Ginty and collaborators have indicated that at P3, wild-type mice have about 15500 SCG neurons whereas p75NTR nulls have approximately 24000; we report that P3 wild-type animals have 17000 SCG neurons whereas the NRAGE null mice approximately 27000. At P21, Deppmann et al report that wild-type SCGs have about 12000 neurons whereas p75NTR nulls have about 15000 and we report that wild-type animals possess 12500 neurons versus 15000 in NRAGE nulls. Therefore, SCG survival defects are precisely phenocopied in p75NTR and NRAGE animals, consistent with the hypothesis that a p75NTR-NRAGE signaling cascade is required for normal SCG neuronal neuron death in vivo.
To confirm that NRAGE functions in a p75NTR apoptotic cascade that is activated by neurotrophin, we generated primary sympathetic neuron cultures from NRAGE knockout animals and showed that apoptosis induced by the pro-apoptotic ligand, BDNF, was severely attenuated in these cells. Because NRAGE knockout cells harbour a fragment of the NRAGE gene that may skew these results, we confirmed these findings in primary rat sympathetic neurons, using lentivirus driving miRNA targeting NRAGE to knockdown NRAGE gene products. Results obtained using this approach were entirely consistent with data derived from the NRAGE null neurons and we conclude that NRAGE normally plays an important role in BDNF-induced apoptosis of sympathetic neurons. It is noteworthy that NRAGE reduces but does not completely block p75NTR-dependent cell death in vitro and therefore independent pro-apoptotic pathways may lie downstream of this receptor. NRIF, has been shown to play an important role in BDNF-induced sympathetic neuronal death in vitro and BDNF may therefore activate p75NTR-dependent apoptosis in sympathetic neurons through the actions of least two adaptor proteins, NRAGE and NRIF. We had previously shown that NRAGE overexpression can activate JNK11 and we therefore tested whether NRAGE knockdown attenuated BDNF-induced JNK activation in sympathetic neurons. We found that knockdown of either NRAGE or p75NTR strongly inhibited JNK activation induced by BDNF, indicating that a p75NTR-NRAGE signaling cascade is required for BDNF-dependent JNK activation in SCG neurons.
We have shown that NRAGE knockout animals show delays in hair follicle catagen that are remarkably similar to those observed in mice lacking p75NTR18,37, suggesting that NRAGE lies on the p75NTR catagen cascade in this organ. Expression analyses showed that NRAGE and p75NTR are co-expressed in the hair follicle compartment where p75NTR-dependent apoptosis occurs and that NRAGE and p75NTR are similarly regulated in this tissue, with increased expression just prior to catagen. Given that a recent study has shown that proNGF expression peaks during catagen 38, it seems likely that the defect in catagen observed in NRAGE knockouts reflects loss of a proNGF-p75NTR-NRAGE signaling cascade.
Approximately one-half of spinal motoneurons initially produced are removed by programmed cell death during development 39. p75NTR is highly expressed in these cells at this stage and several studies have addressed its ability to induce motoneuron cell death. Sedel et al (1999) found that motoneurons in dissected neural tubes underwent NGF apoptosis that was blocked by antibodies to p75NTR and Wiese and colleagues (1999) showed that administration of exogenous NGF increases p75NTR-dependent death of motoneurons, particularly in injured facial motoneurons 26,27. More recently, Pehar et al (2004) have shown that NGF can induce apoptosis of normal spinal motoneurons but not those derived from p75NTR knockout mice 40. Despite these intriguing in vitro findings, Murray and colleagues (1999) found that the pool of spinal cord motoneurons present at P7 is completely normal in p75NTR knockout animals. We examined motoneuron apoptosis during the period of naturally occurring cell death and similarly, found no evidence for a defect in motoneuron apoptosis in p75NTR nulls.
Surprisingly, we found that NRAGE knockout animals show a major defect in the developmental death of spinal motoneurons. The mechanism by which NRAGE participates in programmed cell death in motoneurons are unknown and, while we cannot rule out the possibility that the NRAGE fragment present in knockout animals results in a toxic gain-of-function in motoneurons, a more interesting possibility is that NRAGE functions as an adaptor protein for receptors other than p75NTR to mediate this effect. One interesting candidate for this role is the UNC5A netrin receptor. UNC5A directly binds to NRAGE (through a domain lacking in the NRAGE knockout) and UNC5A induces apoptosis in cultured cells through an NRAGE-dependent pathway 41. Intriguingly, UNC5A knockout mice have defects in the developmental apoptosis of motoneurons42 similar to those reported here for the NRAGE knockout. The defects in motoneuron apoptosis reported in UNC5A knockouts42 are not phenocopied in Netrin-1 knockouts and the ligand responsible for this effect has not been identified. Identifying the ligand(s) required Unc5A-induced motoneuron apoptosis and assessing the role of NRAGE in this cascade are important future challenges.
In closing, we have shown that NRAGE is required for p75NTR-dependent apoptosis in sympathetic neurons and required for normal hair follicle catagen, an apoptotic event that is dependent on p75NTR. In addition, we show that NRAGE induces developmental apoptosis of motoneurons, perhaps through functional interactions with other pro-apoptotic receptors. Taken together, these studies demonstrate that NRAGE is a p75NTR adaptor protein that facilitates programmed cell death in vivo.
Materials and Methods
Animal strains
NRAGE/Maged1 knockout mice are described below. p75NTR knockout mice are exon 3 targeted mice originally generated by Dr. Kuo Fee Lee and colleagues 43. The morning on which a vaginal plug was observed was considered as E0.5; the day of birth was considered P0. All animal procedures were performed under guidelines of institutional animal ethics committees of the University of Louvain (Belgium), University of Namur (Belgium), IBDM (France) and McGill University (Canada).
Generation of NRAGE/Maged1-deficient mice
A conditional allele of NRAGE was generated by homologous recombination in ES cells using standard gene targeting techniques. A 7.5-kb EcoRI genomic fragment served as template for the targeting vector. A loxP site was introduced into a unique NcoI site located in intron 12 by inserting a floxed PGK-Neomycin resistance cassette in Cre-expressing E. coli cells. The same PGK-Neo cassette was then inserted into an AvrII site located in intron 3 to generate the targeting construct. The vector was linearized using NotI, electroporated in E14 ES cells and gene replacement was analysed by PCR (primer 168: 5′-GTCCCTTCACCCTGGATTCGG-3′ and primer 212: 5′-CGAAGTTATACGCTCTCCTGAG-3′ for the 5′ side; and primer 90: 5′-TGACTGGACTGCACAGTT-3′ and primer 76: 5′-CATGCCACTCTCAGTCAACAGG-3′ for the 3′ side) and confirmed by Southern blot analysis (EcoRI/PagI double digestion) using a probe covering nucleotides 70–642 (AF319975). The Neomycin gene was removed by transient expression of Cre after electroporation of a plasmid containing a PGK-Cre gene and a puromycin-resistance cassette. After a short selection (24 hours) with puromycin (2μg/ml) and cloning by dilution, ES clones containing the conditional allele (Maged1tm1Urfm) were identified by PCR (primer 93: 5′-GACCAGCGCAGGTATCTC-3′ and primer 172: 5′-TCTCAGATTAGGTGAGGGTCG-3′). Chimeric males were obtained from three independent Maged1tm1Urfm ES clones after injection into CD1 blastocysts (Eurogentec, Belgium). Heterozygous Maged1tm1Urfm females were bred with Cre deleter males to generate females heterozygous for the knockout mutation. Homozygous males and females were obtained by subsequent crossings. Absence of NRAGE was confirmed by Western blot analysis (anti-NRAGE, 1:1000 36). All phenotypic analysis was performed on mice that had been backcrossed to the C57BL/6 genetic background for a minimum of 6 generations. Genotyping was performed by PCR using primers 93 and 76 to detect the knockout allele; and primer 92: 5′-CAGTGATCTGGCCAAACC-3′ and primer 172 to detect the wild-type allele.
Histopathology and necropsy
Standard phenotyping was carried out at the “Institut Clinique de la Souris” (ICS, Strasbourg), on adult NRAGE knockout males with age-matched wild-type controls. The phenotyping battery included macroscopic inspection of organs, organ weights (liver, kidney, spleen, heart, visceral fat), total body weight and histopathological evaluation of organs (integument, cardiovascular, respiratory, immune/heamatopoietic, digestive tract, digestive organs, urogenital, musculoskeletal, endocrine system, central nervous system, peripheral nervous system and sensory organs). Analysis of skeletons was performed on newborn mice following standard alcian blue and alizarin red staining.
Cell cycle analysis
Mouse embryonic fibroblasts were isolated from E13.5 NRAGE knockout and wild-type littermates. Cells were synchronized to G0 phase by serum starvation for 48h, then allow to enter the cell cycle by adding serum to the medium. Analysis of relative cellular DNA content was analyzed using propidium iodide and flow cytomety following standard protocols.
Hair follicle analysis
Expression of NRAGE during the first hair follicle cycle was analyzed by in situ hybridization and immunoblot on dermal cryosections and lysates from C57BL/6 mice at P0, P2, P4, P8, P12, P17, P18 and P21 following standard protocols. For the immunostaining analysis, P18 back skin samples were immersion fixed in 4% paraformaldehyde (PFA) in phosphate buffer (PBS) for 4 h, left overnight in 30 % PBS-sucrose, and embedded in OCT. 15-μm-thick sections were serially cut and mounted on superfrost®/Plus slides (Fisherbrand). Primary antibodies against NRAGE and p75NTR36 were used at a dilution of 1/100 following standard protocols. For the analysis of the first hair follicle cycle, back skins from NRAGE knockout and wild-type littermates were harvested at P1-P2, P14, P17 (2 p75NTR knockout and 2 wild-type littermates used as control) and P21. For the anagen-catagen transition after HF cycle induction, 6- to 8-wk-old male mice were depilated with a wax and rosin mixture and skin samples were harvested 19 days post depilation. All skin samples were fixed either in 4% PFA or Bouin’s fixative, embedded in paraffin and sectioned parallel to the vertebral line in order to obtain longitudinal HF sections.
Sympathetic Neurons Analysis
Apoptosis assays
Primary sympathetic neurons were isolated from P6 SCG of NRAGE knockout and wild-type mice and cultured as previously described 44. Neurons were maintained in 20 ng/ml of NGF (Harlan) for 2 days, then NGF was removed, the cells rinsed and switched to media containing anti-NGF (0.1 μg/ml, Chemicon International) together with 12.5 mM KCl, to promote survival, with or without the addition of 200 ng/ml of BDNF for 48 hr. Then cells were fixed in 4% paraformaldehyde and stained with DAPI (Vector Labs) to score the nuclei as apoptotic or non-apoptotic. Approximately 50–70 neurons were counted for each condition. The experiment was performed in triplicate.
Lentiviral infection
p75NTR and NRAGE knockdowns were achieved by infecting rat sympathetic neurons with lentivirus encoding miRNA with sequences directed against rat p75NTR or NRAGE mRNA. The miR targeting sequences were designed using the Invitrogen miR prediction algorithm and cloned following manufacturer’s instructions into a variant of the pcDNA6.2/GW-EmGFP-miR vector (Invitrogen) in which the EmGFP has been replaced with mRFP. VSV-G pseudotyped viral particles were produced in 293-T cells and particles were purified by ultracentrifugation, resuspended in DMEM, and the amount of active viral particles was determined by titration on HEK293T cells. For miR-mediated knock-down, sympathetic neurons were transduced at an MOI of 10 and analyzed 5–6 days post-transduction. Apoptosis assays were realized by treating the rat cells with BDNF for 48h and 72h following the protocol described above. JNK activation was analyzed by stimulating the cell with BDNF (200 ng/ml) for one hour. The cells were then lysed in NP-40 buffer and immunoblot for p-JNK, JNK1, NRAGE, p75NTR and Tubulin.
Neuronal counts of SCGs
SCGs from E17, P3 and P23 NRAGE knockout and wild-type littermates were immersion fixed overnight in formalin and embedded in paraffin. 6-μm-thick sections were serially cut and mounted on superfrost®/Plus slides (Fisherbrand). Slides were stained with cresyl violet, scanned with Miraxscan scanner (Zeiss) and neuronal numbers were determined by counting nucleoli present in neuronal profiles on every third section for P3 and P23 and every sixth for E17.
Developmental motoneuron death
Apoptosis in spinal cords was analyzed by whole-mount immunohistochemistry using an antibody directed against cleaved caspase-3 (Asp175, Cell Signaling Technologies; 1/200) following a previously described protocol 39. For quantification, spinal cords were divided into 5 regions (cervical, brachial, thoracic, lumbar and sacral) and the number of apoptotic cells was determined in each region. 4 NRAGE knockout and 5 wild-type littermates and 8 p75NTR knockout and 12 wild-type littermates were analyzed.
Nerve outgrowth
Peripheral innervation was visualized at E11.5, E12.5 and E13.5 by whole mount immunohistochemistry using a monoclonal antibody (TuJ1) recognizing the neuron-specific b-tubulin III protein. A minimum of 3 knockout and 3 wild-type littermates were observed for each genotype at all stages of development.
Supplementary Material
Supplemental Figure 1. Cell cycle analysis of NRAGE knockout fibroblasts: Mouse embryonic fibroblasts were synchronized to G0 phase by serum starvation for 48h, then allow to re-enter cell cycle by adding serum to the medium. The cell cycle entry and progression were followed by measuring the relative DNA content in the cells using propidium iodide staining and flow cytometry. M1: G0/G1 phase, M2: S phase, M3: G2/M phase.
Supplemental Figure 2. Hair Follicle Development at P14 and P21 in NRAGE Null Mice: Comparison of back skin section between NRAGE knockout and wild-type littermates. At P14, prior to catagen, no difference is observed in the dynamic of hair follicle growth between the two genotypes. At P21, after catagen, a slight delay in catagen completion is detected in the knockout skin, as observed by the presence of hair follicles in the subcutis.
Supplemental Figure 3: NRAGE knockdown blocks BDNF-induced apoptosis of sympathetic neurons. Rat sympathetic neurons were infected with lentivirus expressing miRNA that targets NRAGE or were infected with a negative control lentivirus expressing miRNA that does not target a mammalian protein (NS RNAi), then cultured for 5 days. Neurons were then exposed to BDNF for 48 hours and apoptotic nuclei were quantified. ***: P<0.0001. P value was calculated using unpaired, two-tailed student t test. Error bars represent SEM.
Acknowledgments
We are grateful to Prof. M. Herin for help with hair follicle analysis and to P. Auquier, D. Desnoeck, C. Dernoncourt and R. Déom for technical assistance. Dr. Barry Bedell provided valuable help with sympathetic neuron counts and Drs. Peter McPherson and Brigitte Ritter provided the protocol for producing miRNA-expressing lentivirus. This work was supported by Action de Recherche Concertée n°99/03-248 of the Communauté Française de Belgique (to ODB), by the Fonds de la Recherche Scientifique et Médicale (grant n° 3.4527.04) of the Fonds National de la Recherche Scientifique of Belgium (to ODB), and by operating funds from the Canadian Institutes of Health Research (to PAB). MB was supported by the Fonds pour la formation à la Recherche dans l’Industrie et l’Agriculture (FNRS of Belgium), DA was supported by the French Association pour la Recherche sur le Cancer (ARC) and PAB is an Investigator of the Canadian Institutes of Health Research.
Abbreviations
- MAGE
Melanoma AntiGEn encoding gene
- NRAGE
neurotrophin-interacting MAGE homolog
- p75NTR
p75 neurotrophin receptor
- BDNF
brain-derived neurotrophic factor
- JNK
jun kinase
- MHD
mage homology domain
- HF
hair follicle
- SCG
superior cervical ganglion
References
- 1.Roux PP, Barker PA. Neurotrophin signaling through the p75 neurotrophin receptor. Prog Neurobiol. 2002;67:203–33. doi: 10.1016/s0301-0082(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 2.Nykjaer A, Willnow TE, Petersen CM. p75NTR--live or let die. Curr Opin Neurobiol. 2005;15:49–57. doi: 10.1016/j.conb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Teng KK, Hempstead BL. Neurotrophins and their receptors: signaling trios in complex biological systems. Cell Mol Life Sci. 2004;61:35–48. doi: 10.1007/s00018-003-3099-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beattie MS, Harrington AW, Lee R, Kim JY, Boyce SL, Longo FM, et al. ProNGF induces p75-mediated death of oligodendrocytes following spinal cord injury. Neuron. 2002;36:375–86. doi: 10.1016/s0896-6273(02)01005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhakar AL, Howell JL, Paul CE, Salehi AH, Becker EB, Said F, et al. Apoptosis induced by p75NTR overexpression requires Jun kinase-dependent phosphorylation of Bad. J Neurosci. 2003;23:11373–81. doi: 10.1523/JNEUROSCI.23-36-11373.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrington AW, Leiner B, Blechschmitt C, Arevalo JC, Lee R, Morl K, et al. Secreted proNGF is a pathophysiological death-inducing ligand after adult CNS injury. Proc Natl Acad Sci U S A. 2004 doi: 10.1073/pnas.0305755101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Bauer JH, Li Y, Shao Z, Zetoune FS, Cattaneo E, et al. Characterization of a p75(NTR) apoptotic signaling pathway using a novel cellular model. J Biol Chem. 2001;276:33812–20. doi: 10.1074/jbc.M010548200. [DOI] [PubMed] [Google Scholar]
- 8.Kuwako K, Taniura H, Yoshikawa K. Necdin-related MAGE proteins differentially interact with the E2F1 transcription factor and the p75 neurotrophin receptor. J Biol Chem. 2004;279:1703–12. doi: 10.1074/jbc.M308454200. [DOI] [PubMed] [Google Scholar]
- 9.Salehi AH, Roux PP, Kubu CJ, Zeindler C, Bhakar A, Tannis LL, et al. NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor-dependent apoptosis. Neuron. 2000;27:279–88. doi: 10.1016/s0896-6273(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 10.Tcherpakov M, Bronfman FC, Conticello SG, Vaskovsky A, Levy Z, Niinobe M, et al. The p75 neurotrophin receptor interacts with multiple MAGE proteins. J Biol Chem. 2002;277:49101–4. doi: 10.1074/jbc.C200533200. [DOI] [PubMed] [Google Scholar]
- 11.Salehi AH, Xanthoudakis S, Barker PA. NRAGE, a p75 neurotrophin receptor-interacting protein, induces caspase activation and cell death through a JNK-dependent mitochondrial pathway. J Biol Chem. 2002;277:48043–50. doi: 10.1074/jbc.M205324200. [DOI] [PubMed] [Google Scholar]
- 12.Xue B, Wen C, Shi Y, Zhao D, Li C. Human NRAGE disrupts E-cadherin/beta-catenin regulated homotypic cell-cell adhesion. Biochem Biophys Res Commun. 2005;336:247–51. doi: 10.1016/j.bbrc.2005.08.069. [DOI] [PubMed] [Google Scholar]
- 13.Masuda Y, Sasaki A, Shibuya H, Ueno N, Ikeda K, Watanabe K. Dlxin-1, a novel protein that binds Dlx5 and regulates its transcriptional function. J Biol Chem. 2001;276:5331–8. doi: 10.1074/jbc.M008590200. [DOI] [PubMed] [Google Scholar]
- 14.Kuwajima T, Taniura H, Nishimura I, Yoshikawa K. Necdin interacts with the Msx2 homeodomain protein via MAGE-D1 to promote myogenic differentiation of C2C12 cells. J Biol Chem. 2004;279:40484–93. doi: 10.1074/jbc.M404143200. [DOI] [PubMed] [Google Scholar]
- 15.Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61:5544–51. [PubMed] [Google Scholar]
- 16.Muller-Rover S, Handjiski B, van der Veen C, Eichmuller S, Foitzik K, McKay IA, et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 17.Peters EM, Stieglitz MG, Liezman C, Overall RW, Nakamura M, Hagen E, et al. p75 Neurotrophin Receptor-Mediated Signaling Promotes Human Hair Follicle Regression (Catagen) Am J Pathol. 2006;168:221–34. doi: 10.2353/ajpath.2006.050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Botchkarev VA, Botchkareva NV, Albers KM, Chen LH, Welker P, Paus R. A role for p75 neurotrophin receptor in the control of apoptosis-driven hair follicle regression. Faseb J. 2000;14:1931–42. doi: 10.1096/fj.99-0930com. [DOI] [PubMed] [Google Scholar]
- 19.Bamji SX, Majdan M, Pozniak CD, Belliveau DJ, Aloyz R, Kohn J, et al. The p75 neurotrophin receptor mediates neuronal apoptosis and is essential for naturally occurring sympathetic neuron death. J Cell Biol. 1998;140:911–23. doi: 10.1083/jcb.140.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenchappa RS, Zampieri N, Chao MV, Barker PA, Teng HK, Hempstead BL, et al. Ligand-dependent cleavage of the P75 neurotrophin receptor is necessary for NRIF nuclear translocation and apoptosis in sympathetic neurons. Neuron. 2006;50:219–32. doi: 10.1016/j.neuron.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 21.Deppmann CD, Mihalas S, Sharma N, Lonze BE, Niebur E, Ginty DD. A model for neuronal competition during development. Science. 2008;320:369–73. doi: 10.1126/science.1152677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergmann I, Priestley JV, McMahon SB, Brocker EB, Toyka KV, Koltzenburg M. Analysis of cutaneous sensory neurons in transgenic mice lacking the low affinity neurotrophin receptor p75. Eur J Neurosci. 1997;9:18–28. doi: 10.1111/j.1460-9568.1997.tb01349.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita T, Tucker KL, Barde YA. Neurotrophin binding to the p75 receptor modulates Rho activity and axonal outgrowth. Neuron. 1999;24:585–93. doi: 10.1016/s0896-6273(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 24.Lee K-L, Bachman K, Landis S, Jaenisch R. Dependence on p75 for innervation of some sympathetic targets. Science. 1994;263:1447–1449. doi: 10.1126/science.8128229. [DOI] [PubMed] [Google Scholar]
- 25.Ernfors P, Henschen A, Olson L, Persson H. Expression of nerve growth factor receptor mRNA is developmentally regulated and increased after axotomy in rat spinal cord motoneurons. Neuron. 1989;2:1605–13. doi: 10.1016/0896-6273(89)90049-4. [DOI] [PubMed] [Google Scholar]
- 26.Wiese S, Metzger F, Holtmann B, Sendtner M. The role of p75NTR in modulating neurotrophin survival effects in developing motoneurons. Eur J Neurosci. 1999;11:1668–76. doi: 10.1046/j.1460-9568.1999.00585.x. [DOI] [PubMed] [Google Scholar]
- 27.Sedel F, Bechade C, Triller A. Nerve growth factor (NGF) induces motoneuron apoptosis in rat embryonic spinal cord in vitro. Eur J Neurosci. 1999;11:3904–12. doi: 10.1046/j.1460-9568.1999.00814.x. [DOI] [PubMed] [Google Scholar]
- 28.Murray SS, Bartlett PF, Cheema SS. Differential loss of spinal sensory but not motor neurons in the p75NTR knockout mouse. Neurosci Lett. 1999;267:45–8. doi: 10.1016/s0304-3940(99)00330-4. [DOI] [PubMed] [Google Scholar]
- 29.Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res. 2002;67:705–12. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- 30.Bischof JM, Ekker M, Wevrick R. A MAGE/NDN-like gene in zebrafish. Dev Dyn. 2003;228:475–9. doi: 10.1002/dvdy.10398. [DOI] [PubMed] [Google Scholar]
- 31.Lopez-Sanchez N, Gonzalez-Fernandez Z, Niinobe M, Yoshikawa K, Frade JM. Single mage gene in the chicken genome encodes CMage, a protein with functional similarities to mammalian type II Mage proteins. Physiol Genomics. 2007;30:156–71. doi: 10.1152/physiolgenomics.00249.2006. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki A, Masuda Y, Iwai K, Ikeda K, Watanabe K. A RING finger protein Praja1 regulates Dlx5-dependent transcription through its ubiquitin ligase activity for the Dlx/Msx-interacting MAGE/Necdin family protein, Dlxin-1. J Biol Chem. 2002;277:22541–6. doi: 10.1074/jbc.M109728200. [DOI] [PubMed] [Google Scholar]
- 33.Wen CJ, Xue B, Qin WX, Yu M, Zhang MY, Zhao DH, et al. hNRAGE, a human neurotrophin receptor interacting MAGE homologue, regulates p53 transcriptional activity and inhibits cell proliferation. FEBS Lett. 2004;564:171–6. doi: 10.1016/S0014-5793(04)00353-9. [DOI] [PubMed] [Google Scholar]
- 34.Kendall SE, Goldhawk DE, Kubu C, Barker PA, Verdi JM. Expression analysis of a novel p75(NTR) signaling protein, which regulates cell cycle progression and apoptosis. Mech Dev. 2002;117:187–200. doi: 10.1016/s0925-4773(02)00204-6. [DOI] [PubMed] [Google Scholar]
- 35.Kendall SE, Battelli C, Irwin S, Mitchell JG, Glackin CA, Verdi JM. NRAGE mediates p38 activation and neural progenitor apoptosis via the bone morphogenetic protein signaling cascade. Mol Cell Biol. 2005;25:7711–24. doi: 10.1128/MCB.25.17.7711-7724.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salehi AH, Roux PP, Kubu CJ, Zeindler C, Bhakar A, Tannis LL, et al. NRAGE, a novel MAGE protein, interacts with the p75 neurotrophin receptor and facilitates nerve growth factor–dependent apoptosis. Neuron. 2000:279–288. doi: 10.1016/s0896-6273(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 37.Botchkareva NV, Botchkarev VA, Chen LH, Lindner G, Paus R. A role for p75 neurotrophin receptor in the control of hair follicle morphogenesis. Dev Biol. 1999;216:135–53. doi: 10.1006/dbio.1999.9464. [DOI] [PubMed] [Google Scholar]
- 38.Peters EM, Hendrix S, Golz G, Klapp BF, Arck PC, Paus R. Nerve growth factor and its precursor differentially regulate hair cycle progression in mice. J Histochem Cytochem. 2006;54:275–88. doi: 10.1369/jhc.4A6585.2005. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto W, Henderson CE. Patterns of programmed cell death in populations of developing spinal motoneurons in chicken, mouse, and rat. Dev Biol. 1999;1999:60–71. doi: 10.1006/dbio.1999.9413. [DOI] [PubMed] [Google Scholar]
- 40.Pehar M, Cassina P, Vargas MR, Castellanos R, Viera L, Beckman JS, et al. Astrocytic production of nerve growth factor in motor neuron apoptosis: implications for amyotrophic lateral sclerosis. J Neurochem. 2004;89:464–73. doi: 10.1111/j.1471-4159.2004.02357.x. [DOI] [PubMed] [Google Scholar]
- 41.Williams ME, Strickland P, Watanabe K, Hinck L. UNC5H1 induces apoptosis via its juxtamembrane region through an interaction with NRAGE. J Biol Chem. 2003;278:17483–90. doi: 10.1074/jbc.M300415200. [DOI] [PubMed] [Google Scholar]
- 42.Williams ME, Lu X, McKenna WL, Washington R, Boyette A, Strickland P, et al. UNC5A promotes neuronal apoptosis during spinal cord development independent of netrin-1. Nat Neurosci. 2006;9:996–8. doi: 10.1038/nn1736. [DOI] [PubMed] [Google Scholar]
- 43.Lee K-F, Li E, Huber J, Landis SC, Sharpe AH, Chao MV, et al. Targeted mutation of the gene encoding the low affinity NGF receptor leads to deficits in the peripheral sensory nervous system. Cell. 1992;69:737–749. doi: 10.1016/0092-8674(92)90286-l. [DOI] [PubMed] [Google Scholar]
- 44.Palmada M, Kanwal S, Rutkoski NJ, Gustafson-Brown C, Johnson RS, Wisdom R, et al. c-jun is essential for sympathetic neuronal death induced by NGF withdrawal but not by p75 activation. J Cell Biol. 2002;158:453–61. doi: 10.1083/jcb.200112129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bertrand M, Huijbers I, Chomez P, De Backer O. Comparative expression analysis of the MAGED genes during embryogenesis and brain development. Dev Dyn. 2004;230:325–34. doi: 10.1002/dvdy.20026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Cell cycle analysis of NRAGE knockout fibroblasts: Mouse embryonic fibroblasts were synchronized to G0 phase by serum starvation for 48h, then allow to re-enter cell cycle by adding serum to the medium. The cell cycle entry and progression were followed by measuring the relative DNA content in the cells using propidium iodide staining and flow cytometry. M1: G0/G1 phase, M2: S phase, M3: G2/M phase.
Supplemental Figure 2. Hair Follicle Development at P14 and P21 in NRAGE Null Mice: Comparison of back skin section between NRAGE knockout and wild-type littermates. At P14, prior to catagen, no difference is observed in the dynamic of hair follicle growth between the two genotypes. At P21, after catagen, a slight delay in catagen completion is detected in the knockout skin, as observed by the presence of hair follicles in the subcutis.
Supplemental Figure 3: NRAGE knockdown blocks BDNF-induced apoptosis of sympathetic neurons. Rat sympathetic neurons were infected with lentivirus expressing miRNA that targets NRAGE or were infected with a negative control lentivirus expressing miRNA that does not target a mammalian protein (NS RNAi), then cultured for 5 days. Neurons were then exposed to BDNF for 48 hours and apoptotic nuclei were quantified. ***: P<0.0001. P value was calculated using unpaired, two-tailed student t test. Error bars represent SEM.