Abstract
Recent work has shown that the incorporation of an all-hydrocarbon “staple” into peptides can greatly increase their α-helix propensity, leading to an improvement in pharmaceutical properties such as proteolytic stability, receptor affinity and cell-permeability. Stapled peptides thus show promise as a new class of drugs capable of accessing intractable targets such as those that engage in intracellular protein-protein interactions. The extent of α-helix stabilization provided by stapling has proven to be substantially context dependent, requiring cumbersome screening to identify the optimal site for staple incorporation. In certain cases, a staple encompassing one turn of the helix (attached at residues i and i+4) furnishes greater helix stabilization than one encompassing two turns (i,i+7 staple), which runs counter to expectation based on polymer theory. These findings highlight the need for a more thorough understanding of the forces that underlie helix stabilization by hydrocarbon staples. Here we report all-atom Monte Carlo folding simulations comparing unmodified peptides derived from RNAse A and BID BH3 with various i,i+4 and i,i+7 stapled versions thereof. The results of these simulations were found to be in quantitative agreement with experimentally determined helix propensities. We also discovered that staples can stabilize quasi-stable decoy conformations, and that the removal of these states plays a major role in determining the helix stability of stapled peptides. Finally, we critically investigate why our method works, exposing the underlying physical forces that stabilize stapled peptides.
Keywords: Stapled Peptides, Monte-Carlo simulations, Drug Discovery, Folding Traps, Entropic Stabilization
INTRODUCTION
In principle, an appealing approach to the modulation of protein-protein interactions is the use of “dominant-negative” peptides representing only the fragment of a protein that engages a target. In practice, however, such dominant-negative peptide fragments rarely exhibit potent biologic activity, because their removal from the folded context of the parent protein tends to deprive them of conformational stability, leading to poor binding affinity, proteolytic instability, cell impermeability, and rapid clearance in vivo by renal filtration. One approach toward circumventing this problem involves the introduction of synthetic crosslinking functionality into the peptide, so as to restore and enforce its bioactive conformation 1–5. Given that the α-helix is the most common secondary structural motif in proteins, it is not surprising that this particular folding motif has received the greatest deal of attention with respect to crosslink-induced stabilization.6–14 Experience has shown that the extent of α-helix stabilization conferred by introduction of a crosslink is highly context dependent. For example, helix induction by lactam crosslinks has been found to be dependent on the length of the lactam linker, the placement and orientation of the amide moiety along the crosslink, and the position of the linking residues in a peptide sequence.6 Similar context-dependence has been reported for an all-hydrocarbon, α-methylated system referred to as a “staple”.15,16 Furthermore, staples have been described that encompass one or two helical turns (i,i+4 and i,i+7, respectively), and in certain cases, one or the other type has been found to confer greater helix stability.15,16
Polymer theory has long held that crosslink-induced stabilization of folded structure in polypeptide chains results primarily from the reduction in configurational entropy of the unfolded state.17–19 Should helix stabilization result exclusively from such entropic contributions, polymer theory would predict that the extent of stabilization would vary as a function of the logarithm of the number of residues that lie in a loop closed by a crosslink, with longer loops being more stable.17–19 Models based on this theoretical construct have had some success in predicting the stability of proteins stabilized by crosslinks,20–22 and we found qualitative agreement with these models when simulating disulfide crosslinked lattice proteins.23 However, these models have failed in other cases,24 suggesting that other factors may sometimes make even greater contributions to the overall stability of crosslinked polypeptide chains. In some regards it is not surprising that these models might not apply to crosslinked peptides as they assume a Gaussian distribution of crosslinking residue end to end distances, and while this might be true for a long polypeptide, it has been demonstrated that this is not the case for short peptides.25 Furthermore, in the case of the stapled peptides, the linking residues are connected by relatively long flexible chains (8- and 11-carbon chains in the i,i+4 and i,i+7 staples, respectively), resulting in a system that might be described as a heteropolymer (peptide) and homopolymer (hydrocarbon) chain connected with two crosslinks.
Our current work aims to understand, at atomic resolution, how staples affect α-helicity, with the ultimate aim being to develop a tool that can be used a priori to predict which crosslinking configurations will be most stabilizing for specific peptide sequences. Such a computational tool would be of great practical utility, as stapled α-helical peptides have proven capable of targeting therapeutically relevant macromolecules that are difficult to target using other types of molecules,16,26–32 and computational screening could eliminate the cumbersome process of synthesizing and screening multiple stapled versions of a candidate peptide.
Knowledge-based (KB) potentials have proven useful in the prediction of protein33 and peptide34 conformational stability. With this in mind, we used as a starting point the Monte Carlo (MC) approach to all-atom protein folding with a KB potential we have previously described.35 Our KB potential was able to accurately predict the structure of a diverse set of proteins,35 and a recent study employing our KB potential to study the folding trajectory as well as Φ-values of three helix bundles was in excellent agreement with experimental evidence.36 Furthermore, a critical assessment of our potential revealed that it recapitulates physically meaningful quantities.37 We anticipated that it would also be able to predict conformational stabilities of peptides in the present study. It was necessary to modify the method to accommodate the atomic representation of the crosslink, and we also added a KB side-chain torsional energy term (Supporting Information). The MC move set used to study thermodynamics also required modification in order to appropriately sample the macrocyclic conformations introduced by the crosslink (Supporting Information).
METHODS
The all-atom energy function H in our previous study35 has been further developed to model cross-linked peptides and consists of the following energy terms:
| (1) |
where Econ is the pairwise atom-atom contact potential, Ehb is the geometrically sensitive hydrogen bonding potential, Ebbtor is the sequence-dependent backbone torsional potential based on the statistics of sequential amino acid triplets, Esct is the side-chain torsional angle potential, and Elinktor is the torsional potential for hydrocarbon linkers. The first three energy terms are described in detail in our previous publication,35 while the last two are described in the Supporting Information. Each of our potential terms are trained on a database of crystal structures,35 and they are in no way trained or fitted based on the experimental helical propensities of the peptides used in the present study. Details of the MC ensemble simulations and analysis procedures are provided in the Supporting Information.
RESULTS
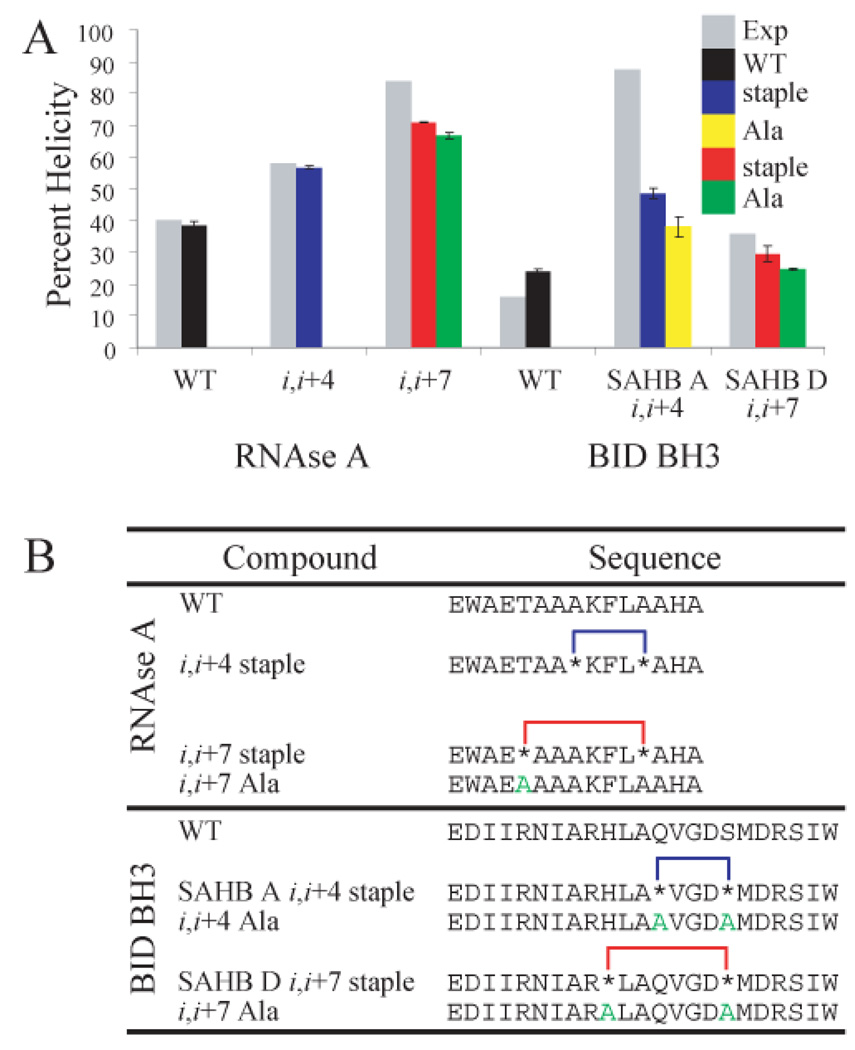
We have tested our new potential and move set on peptides that have been shown experimentally to be either preferentially stabilized by the i,i+4 staple (stapled alpha helical BID BH3 (SAHB) peptides, Fig. 1B),16 or by the i,i+7 staple (RNAse A peptides, Fig. 1B).15 Conformations that were obtained during MC simulations were analyzed for percent helicity using DSSP (Supporting Information).38 When we compared the average helicities obtained in such a manner with experimentally determined ones, we found that our potential was able to predict which staple would be more stabilizing for each peptide (Fig. 1A). It should be noted that the experimental conditions used to determine helical content for RNAse A and BID BH3 differed,39 and as such we sought to predict the best stapling system in each case, without comparing the relative helicities of the two peptide systems.
Figure 1.
(A) Experimental (grey) and simulation (black, blue, and red, T=0.72) percent helicities for WT and stapled peptides. Simulation results for alanine mutants (yellow or green, T=0.72) are also depicted. (B) Peptide sequences used in this study. Astericks (*) denote unnatural amino acids connecting the staple to the peptide. Residues mutated to alanine in control peptides are colored green.
The helical propensity of the unnatural crosslinking amino acids is expected to be enhanced as they are α-methylated, which reduces backbone flexibility.40 In order to tease apart energetic contributions such as backbone torsions from the geometric constraint of the staple, we also included mutant versions in which alanine is inserted in the place of the stapling residues as controls (Fig. 1). Alanine has a similar helical propensity to aminoisobutyric acid (AIB),41 and we used its backbone torsional energy for the unnatural amino acids as well. In all cases, the stapled peptides are more helical than the corresponding alanine-substituted versions, suggesting that the physical constraint conferred by macrocyclic ring formation contributes to increased helical propensity. In some cases the alanine mutants show increased helical propensity as compared with the corresponding wild-type peptide, suggesting that the overall helical propensity observed in the stapled peptides is due to both the torsional preferences of the backbone as well as the physical constraint imposed by the hydrocarbon crosslink.
In stark contrast to what we might expect from polymer theory predictions, the i,i+7 staple is not always more stabilizing than the i,i+4 staple. This unexpected experimental finding was reproduced in our work: the RNAse A peptide is more stabilized by the i,i+7 staple, while the BID BH3 peptide is more stabilized by the i,i+4 staple. We sought to understand the physical basis for the discrepancy between these peptides. By carefully studying these peptides with microscopic resolution afforded by our equilibrium simulations, we have been able lay down a physically sound, albeit unexpected, explanation that follows below.
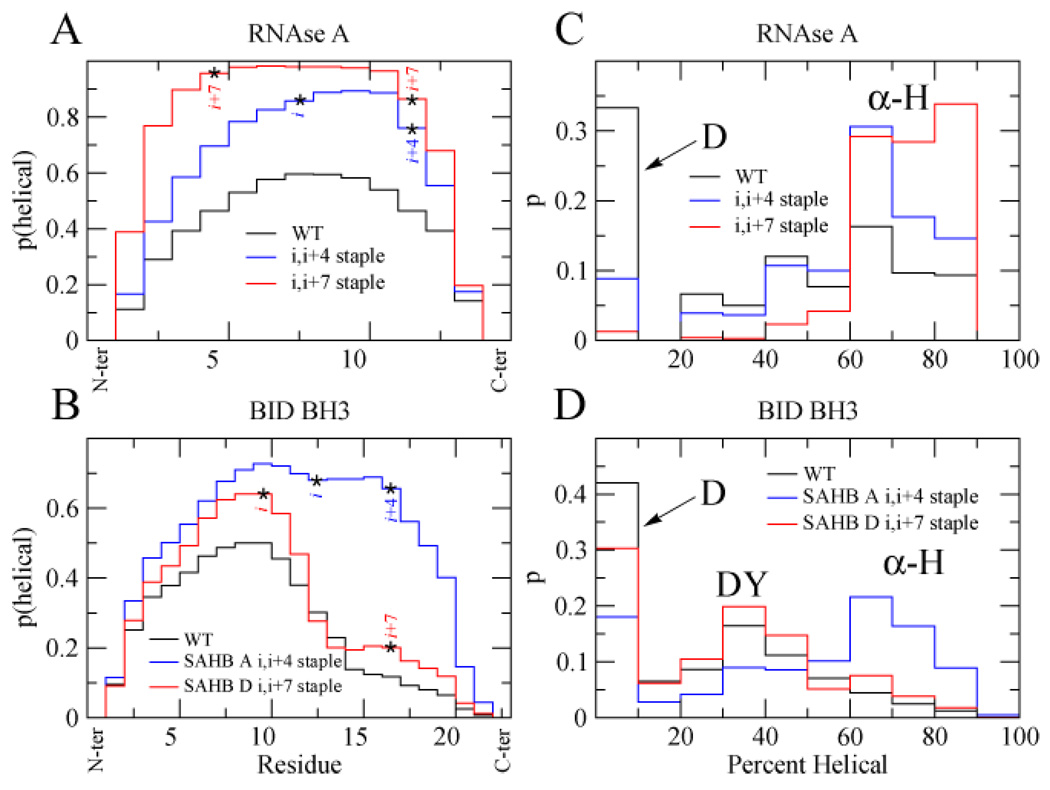
To understand how localized the effect of the staple was, the likelihood of each residue to be in a helical conformation was examined. In the case of RNAse A, the probability that a residue exists in an α-helix increases for residues encompassed by either the i,i+4 or i,i+7 staples (Fig. 2A). Residues lying outside the stapled region are also more likely to reside in helices, although this increased helical propensity drops off moving away from the staple. This may suggest that the staple increases the local helical propensity, and this templates further elongation of the helix.
Figure 2.
Probability of each residue in RNAse A (A) or BID BH3 (B) peptides to reside in a helical region during simulations (T=0.72). Probabilities of finding RNAse A (C) or BID BH3 (D) peptides in α-helical (α-H), denatured (D), or decoy (DY) state during simulations (T=0.72).
In the case of BID BH3, we observed a very different profile (Fig. 2B). Although the i,i+4 staple seemed to increase the helical propensity of all residues (especially residues 10–20), the i,i+7 staple increased the helix propensity of residues towards the N-terminus of the staple but failed to significantly increase the helical propensity of residues encompassed by the staple, especially the residues in position i+3 (Q) and i+4 (V), and only slightly increased those near the C-terminus of the staple.
To further understand the unexpected behavior of the BID BH3 peptide relative to the other stapled peptides, we looked at what fraction and to what extent the obtained conformations were helical. In the case of RNAse A, we observed largely a two-state distribution, comprised of either highly helical conformations or random coil conformations (Fig. 2C). In the case of the BID BH3 peptides, however, we noticed that there was apparently a third partially helical state present which we term a decoy state (Fig. 2D). We regard this as a decoy state because although it is partially folded, it is not the fully folded α-helical state that we expected to be stabilized by the staple. The population of this state is increased when the i,i+7 staple is present, while the population of the α-helical state is increased when the i,i+4 staple is present.
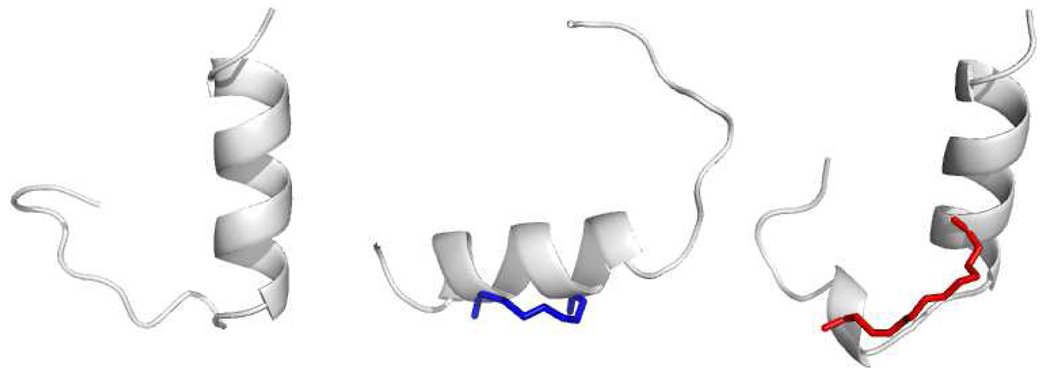
We next employed graph theory36,42,43 to examine the nature of the unexpected decoy state that was stabilized by the BID BH3 i,i+7 staple. In an attempt to identify the most representative conformations of this population, BID BH3 peptide structures with 9–11 helical residues were selected from thermodynamic runs for clustering analysis. Single linkage clustering was employed using Cα RMSD between each conformation as the clustering metric (Supporting Information). The size of the largest cluster, the giant component (GC), was monitored versus RMSD cutoff. Cutoffs were selected at the GC transition, when the GC contains half of the total structures. Representative structures that characterize the decoy state were selected from the GC by choosing the structure with the greatest connectivity k (Fig. 3).44
Figure 3.
Representative decoy structures for the BID BH3 WT (left), i,i+4 stapled (middle), and i,i+7 stapled (right) peptides. The N-termini are at the top of the image and the staples are rendered blue (i,i+4) or red (i,i+7).
As might be expected from our residue helicities (Fig. 2B) the N-terminus was folded in an α-helix for the wild-type (Fig. 3, left) and i,i+7 stapled (Fig. 3, right) peptides. In contrast, this conformation is not stabilized by the i,i+4 staple. Instead we see a helical fold that is centered on the staple with unfolded ends (Fig. 3, middle). These findings demonstrate that adding crosslinks to certain peptides may preferentially stabilize a decoy state over the native fold that is present in the context of a full protein.
In all stapled peptides, however, we observed at least some stabilization of the α-helical fold, and we were interested in understanding what energetic factors contribute to this stabilization. In a preliminary effort we looked at the Cα distance distribution of the i,i+4 and i,i+7 residues when either an i,i+4 or i,i+7 staple is present (Fig. S4). We used a temperature (T=1.0) well above the melting temperature (Fig. S5) of each peptide. Even so, although the peptides are largely unfolded, we still see that the average distance of the i and i+4 α-carbons is less when stapled in an i,i+4 fashion, and similarly the average distance of the i and i+7 α-carbons is less when stapled in an i,i+7 fashion. Furthermore, in both cases there are distances that the unstapled peptide can access that the stapled peptide cannot. This demonstrates that specific unfolded conformations present in the wild type peptide are geometrically not allowed in the stapled peptide – pointing to the importance of the entropic component in helical stabilization of stapled peptides.
We were curious to see whether the reduction of configurational entropy in the unfolded state of the stapled peptides was the dominant thermodynamic feature that increased the overall helical propensity. Using the number of helical residues h as an order parameter the average energy (enthalpy, H(h)) and entropy S(h) of peptides with specified helical residues were ascertained from long equilibrium simulations (Supporting Information), and the free energy G(h) was calculated from these values (Fig. S9). When we refer to entropy in this text, we specifically mean the configurational entropy of the peptide and staple. Our enthalpy refers to the total score of our KB potential. It is important to note that KB potentials are intended to correlate with free energies, rather than enthalpies, and thus specific terms may reflect both enthalpic and entropic contributions. For example, the atom-atom pairwise interaction energy Econ is a potential of mean force where the solvent degrees of freedom are averaged out, and entropic contributions from solvent (such as from the desolvation of hydrophobic residues) might be included. Furthermore, our backbone torsional term based on the geometric arrangement of amino acid triplets Ebbtor will score certain conformations as favorable because they tend to be observed frequently in our training database, but these conformations are likely physically stabilized by a mixture of enthalpic and entropic contributions which we refrain from attempting to deconvolute. Thus, we cannot easily interpret what we term “enthalpic” contributions from our potential, as these may include entropic contributions as well, and when we refer to enthalpy it is important to keep these types of considerations in mind.
In good agreement with expectations, helix formation is driven by our enthalpy while being disfavored by configurational entropy (Table 1). In order to more deeply understand what contributions each of our potential terms were making to the overall stability of helix formation, we divided the total enthalpy into individual components (Table S2). The most significant terms in helix formation were the hydrogen bonding ΔH(hb) and the backbone torsion ΔH(bbtor) terms. When we calculated the change in thermodynamic terms relative to the WT peptide (Table 1), it is clear that for all stapled peptides ΔΔH makes a larger contribution to ΔΔG than -TΔΔS. We also observed that although the stapled peptides were not always stabilized entropically compared to the WT peptide, they were always more entropically stabilized relative to the Ala mutant controls, except for the i,i+7 stapled BID BH3. Since ΔΔH was the most significant stabilizing contribution, especially for the most stabilized peptides of each series (i,i+7 stapled RNAse A and i,i+4 stapled BID BH3), we computed the individual potential terms to understand what factors were most important. Once again, the most significant changes that took place in the stapled peptides relative to the WT peptides were in the hydrogen bonding ΔΔH(hb) and backbone torsional ΔΔH(bbtor) terms (Table S2).
Table 1.
Thermodynamic analysis of RNAse A and BID BH3 simulations using the number of helical residues as the order parameter. Thermodynamic values for folding to the helical state (ΔH, ΔS, ΔG) are given, as well as these values relative to the WT peptide (ΔΔH, ΔΔS, ΔΔG).
| ΔH | −TΔS | ΔG | ΔΔH | −TΔΔS | ΔΔG | ||
|---|---|---|---|---|---|---|---|
| WT | −29.20 ± 0.60 | 32.50 ± 0.60 | 3.30 ± 0.10 | ||||
| i,i+4 staple | −30.00 ± 0.50 | 31.90 ± 0.40 | 1.90 ± 0.10 | −0.8 ± 0.5 | −0.6 ± 0.5 | −1.37 ± 0.07 | |
| i,i+7 staple | −33.25 ± 0.06 | 33.40 ± 0.10 | 0.12 ± 0.09 | −4.0 ± 0.6 | 0.9 ± 0.5 | −3.14 ± 0.07 | |
| i,i+7 Ala | −32.59 ± 0.09 | 34.16 ± 0.09 | 1.57 ± 0.04 | −3.4 ± 0.5 | 1.7 ± 0.5 | −1.70 ± 0.20 | |
| WT | −47.00 ± 0.40 | 48.20 ± 0.40 | 1.2_ ± 0.10 | ||||
| i,i+4 staple | −48.30 ± 0.30 | 47.70 ± 0.30 | −0.63 ± 0.03 | −1.4 ± 0.5 | −0.5 ± 0.4 | −1.80 ± 0.10 | |
| i,i+4 Ala | −49.20 ± 0.40 | 49.00 ± 0.50 | −0.16 ± 0.04 | −2.2 ± 0.3 | 0.8 ± 0.4 | −1.40 ± 0.10 | |
| i,i+7 staple | −47.50 ± 0.20 | 48.00 ± 0.10 | 0.56 ± 0.05 | −0.5 ± 0.5 | −0.1 ± 0.5 | −0.70 ± 0.10 | |
| i,i+7 Ala | −46.50 ± 0.40 | 47.90 ± 0.30 | 1.38 ± 0.04 | 0.4 ± 0.5 | −0.2 ± 0.4 | 0.20 ± 0.10 | |
The temperatures used for RNAse A and BID BH3 were 0.78 and 0.70, respectively. Errors are reported as standard deviations of averages of 3 groups of 50 runs (RNAse A) or 200 runs (BID BH3).
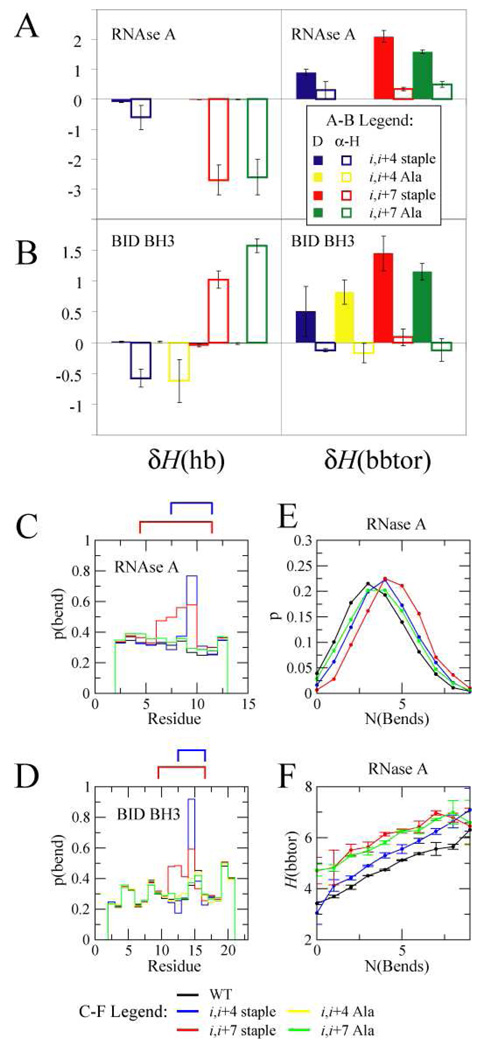
Changes in enthalpy could result from either destabilizing the denatured state or by stabilizing the folded state. By defining the change in enthalpy of a specific helical state as the difference in energy of of a stapled peptide in a specific folded state relative to the WT peptide in the same state, we were able to elucidate whether the overall changes in hydrogen bonding and backbone torsional energy arose from changes that occurred in the folded state δHα-H or unfolded state δHD (Fig S10, 4A, and 4B). In the RNAse A stapled peptides, and the i,i+4 stapled BID BH3 peptide, the hydrogen bonding term is stabilizing by lowering the energy of the folded state. Interestingly for the i,i+7 stapled BID BH3 peptide, this term is destabilizing by increasing the energy of the folded state. The backbone torsional term, on the other hand, stabilizes all of the stapled peptides by increasing the energy of the denatured state. To understand why this was the case, we statistically analyzed the secondary structure (Fig S11) and tertiary structure (Fig S12) that is present in the denatured state, and noticed that the most significant difference when a staple is present is in the bend propensity (local curvature)38,45 of residues lying between the stapling residues (Fig. 4C and 4D). In the i,i+4 stapled peptides there is a dramatic increase in the bend propensity of the residue in position i+2. In the case of the i,i+7 stapled peptides, the bend propensity increases for residues in the i+2 through i+5 positions. As a result, the distribution of peptides with residues in bends shifts towards peptides with higher bend content when stapled in an i,i+4 fashion, and this shift is even more drastic when stapled in the i,i+7 positions (Fig. 4E and S13A). We also observed that the average backbone torsional energy H(bbtor) of conformations with a given number of bends is higher in stapled and Ala control peptides compared to WT peptides, and that for all peptides, the backbone energy increases with more bends (Fig. 4F and S13B). Thus staples destabilize the denatured state in our model by increasing the backbone torsional energy of bent conformations, and by increasing the bend content in the denatured ensemble.
Figure 4.
RNAse A (A) and BID BH3 (B) changes in hydrogen bonding δH(hb) and sequence-dependent backbone torsions δH(bbtor) of denatured (D) and helical (α-H) states of stapled and Ala mutant peptides relative to the corresponding WT peptide. Probability that a residue is classified as a bend for RNAse A (C) and BID BH3 (D). A cartoon depicting the staple is above the plot. E: Probability of finding conformations with a certain number of “bend” residues in the denatured state for RNAse A peptides. F: The average backbone triplet energy H(bbtor) of conformations in the denatured state versus number of “bend” residues. RNAse A and BID BH3 simulations were carried out at T=0.78 and T=0.70, respectively. Colors of peptides in C, D, E, and F are as follows: WT (black), i,i+4 stapled (blue), i,i+7 stapled (red), i,i+4 Ala (yellow), i,i+7 stapled (green).
DISCUSSION
Our initial goal was to understand why in some cases an i,i+7 staple can be more stabilizing than an i,i+4 staple, while in other cases the opposite is true. We then sought to uncover the physical forces that underlie stapled peptide stabilization. Our findings suggest that there are three primary modes of stabilization of helical content in stapled peptides. By adding a staple, all peptides except one were stabilized compared to Ala controls by the reduction of configurational entropy in the denatured state, although they were not all stabilized compared to the WT peptide. Staples also stabilize or destabilize the α-helical fold by modulating the hydrogen bonding energy in the folded state. We also found that they destabilize the denatured state by making regions of high curvature more energetically demanding while concomitantly increasing the likelihood of these unfavourable residue “bends.” Stabilizing the native state of proteins by disrupting favourable interactions in the denatured ensemble has garnered much attention experimentally and theoretically.46,47 We present an interesting example of stabilizing the native state of peptides by disrupting favourable backbone conformations in the denatured state, and replacing them with energetically demanding ones.
The increase in bend propensity induced by staples can also help explain why in the case of the BID BH3 peptide, the i,i+7 staple stabilizes a decoy conformation over the all helical state. A partially folded conformation with some bend propensity present in the WT ensemble is stabilized by increasing the intrinsic bend propensity with a i,i+7 staple that spans the bend. The idea that the denatured state of peptides is composed of a heterogeneous ensemble of energetically trapped states as well as entropically stabilized random coils has been experimentally and theoretically supported.48–52 We did not classify the “decoy” state as part of the denatured state, but using a different theoretical framework for thermodynamic evaluation this could have been the case, and we consider it similar to “trapped” states reported by other groups. Thus, in the case of BID BH3, the i,i+7 staple stabilizes what we termed a decoy state over the native state, resulting in the i,i+4 staple being more stabilizing in this case.
It is important to point out that these results would not be realized with a simpler model. For example, we might assume predictions based on incorporating a helix-promoting amino acid such as alanine in the stapling positions might correlate with stapled peptide helicities. Using such sequences (identical to our Ala controls), computations employing AGADIR,53–56 a state of the art helix prediction tool, were unable to predict the correct helical trends (Fig. S14). Models such as the Gō model do not include non-native interactions, and would be unable to detect the crucial decoy state in the BID BH3 system which led to the i,i+4 staple being more stabilizing than the i,i+7 staple. While a previous study employing the Gō model and treating amino acids as beads found that the BID BH3 i,i+4 stapled peptide was more stable than the WT peptide, the BID BH3 i,i+7 stapled peptide was not included in this work.57 Furthermore the Gō potential is non-transferable and therefore cannot be used for predictive purposes.57–59 It is worthwhile to note that when we employed the Gō potential with an all-atom representation of the peptide and crosslink, we were unable to reproduce experimental helical propensity trends (Fig. S15 and S16).
In conclusion, we have developed an all-atom model that predicts helical propensities of stapled peptides in excellent agreement with experimental values. In addition, the current study revealed that in certain peptides specific staples may stabilize a decoy state rather than the all helical state. Prior to the present study, it has been tempting to attribute the overall α-helical stabilization of staples to a decrease in the configurational entropy of the unfolded state. By carefully examining our ensemble simulations, we were able to elucidate a more complex mixture of energetic factors that act on both the denatured and folded states. Our model yields accurate results with microscopic resolution, and we are currently using it in the design of therapeutically relevant stapled peptides.
Supplementary Material
Details of computational and data analysis procedures. Tables S1-S3, Figures S1-S16.
ACKNOWLEDGEMENTS
This work was supported by an NSF fellowship (to P.S.K.) and NIH grant GM52126 (to E.I.S.). We thank Stefan Wallin for helpful discussions.
REFERENCES
- 1.Ghalit N, Poot AJ, Furstner A, Rijkers DTS, Liskamp RM. J. Org. Lett. 2005;7:2961–2964. doi: 10.1021/ol0508781. [DOI] [PubMed] [Google Scholar]
- 2.Ghalit N, Rijkers DTS, Kemmink J, Versluis C, Liskamp RM. J. Chem. Commun. 2005:192–194. doi: 10.1039/b415555f. [DOI] [PubMed] [Google Scholar]
- 3.Angell Y, Burgess K. J. Org. Chem. 2005;70:9595–9598. doi: 10.1021/jo0516180. [DOI] [PubMed] [Google Scholar]
- 4.Miller SJ, Blackwell HE, Grubbs RH. J. Am. Chem. Soc. 1996;118:9606–9614. [Google Scholar]
- 5.Fink BE, Kym PR, Katzenellenbogen JA. J. Am. Chem. Soc. 1998;120:4334–4344. [Google Scholar]
- 6.Andrews MJI, Tabor AB. Tetrahedron. 1999;55:11711–11743. [Google Scholar]
- 7.Guerrero L, Smart OS, Woolley GA, Allemann RK. J Am Chem Soc. 2005;127:15624–15629. doi: 10.1021/ja0550428. [DOI] [PubMed] [Google Scholar]
- 8.Chi L, Sadovski O, Woolley GA. Bioconjug. Chem. 2006;17:670–676. doi: 10.1021/bc050363u. [DOI] [PubMed] [Google Scholar]
- 9.Jackson DY, King DS, Chmielewski J, Singh S, Schultz PG. J. Am. Chem. Soc. 1991;113:9391–9392. [Google Scholar]
- 10.Bracken C, Gulyas J, Taylor JW, Baum J. J. Am. Chem. Soc. 1994;116:6431–6432. [Google Scholar]
- 11.Phelan JC, Skelton NJ, Braisted AC, McDowell RS. J. Am. Chem. Soc. 1997;119:455–460. [Google Scholar]
- 12.Blackwell HE, Grubbs RH. Angew. Chem. Int. Ed. 1998;37:3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 13.Yang B, Liu D, Huang Z. Bioorg Med Chem Lett. 2004;14:1403–1406. doi: 10.1016/j.bmcl.2003.09.101. [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto K, Oimoto N, Katsuno K, Inouye M. Chem Commun (Camb) 2004:1280–1281. doi: 10.1039/b403615h. [DOI] [PubMed] [Google Scholar]
- 15.Schafmeister CE, Po J, Verdine GL. J. Am. Chem. Soc. 2000;122:5891–5892. [Google Scholar]
- 16.Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schellman JA. C R Trav Lab Carlsberg [Chim] 1955;29:230–259. [PubMed] [Google Scholar]
- 18.Flory JP. J. Am. Chem. Soc. 1956;78:5222–5235. [Google Scholar]
- 19.Poland DC, Scheraga HA. Biopolymers. 1965;3:379–399. [Google Scholar]
- 20.Pace CN, Grimsley GR, Thomson JA, Barnett BJ. J. Biol. Chem. 1988;263:11820–11825. [PubMed] [Google Scholar]
- 21.Johnson RE, Adams P, Rupley JA. Biochemistry. 1978;17:1479–1484. doi: 10.1021/bi00601a019. [DOI] [PubMed] [Google Scholar]
- 22.Lin SH, Konishi Y, Denton ME, Scheraga HA. Biochemistry. 1984;23:5504–5512. doi: 10.1021/bi00318a019. [DOI] [PubMed] [Google Scholar]
- 23.Abkevich VI, Shakhnovich EI. J. Mol. Biol. 2000;300:975–985. doi: 10.1006/jmbi.2000.3893. [DOI] [PubMed] [Google Scholar]
- 24.Johnson CM, Oliveberg M, Clarke J, Fersht AR. J. Mol. Biol. 1997;268:198–208. doi: 10.1006/jmbi.1997.0928. [DOI] [PubMed] [Google Scholar]
- 25.Mutter M. J. Am. Chem. Soc. 1977;99:8307–8314. [Google Scholar]
- 26.Sia SK, Carr PA, Cochran AG, Malashkevich VN, Kim PS. Proc. Natl. Acad. Sci. USA. 2002;99:14664–14669. doi: 10.1073/pnas.232566599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leduc AM, Trent JO, Wittliff JL, Bramlett KS, Briggs SL, Chirgadze NY, Wang Y, Burris TP, Spatola AF. Proc. Natl. Acad. Sci. USA. 2003;100:11273–11278. doi: 10.1073/pnas.1934759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Liao W, Arora PS. Angew. Chem. Int. Ed. Engl. 2005;44:6525–6529. doi: 10.1002/anie.200501603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. J. Am. Chem. Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, Datta SR, Pitter KL, Bird GH, Wikstrom JD, Deeney JT, Robertson K, Morash J, Kulkarni A, Neschen S, Kim S, Greenberg ME, Corkey BE, Shirihai OS, Shulman GI, Lowell BB, Korsmeyer S. J. Nat Med. 2008;14:144–153. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Zhao Q, Bhattacharya S, Waheed AA, Tong X, Hong A, Heck S, Curreli F, Goger M, Cowburn D, Freed EO, Debnath AK. J Mol Biol. 2008;378:565–580. doi: 10.1016/j.jmb.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhattacharya S, Zhang H, Debnath AK, Cowburn D. J Biol Chem. 2008;283:16274–16278. doi: 10.1074/jbc.C800048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoppe C, Schomburg D. Protein Sci. 2005;14:2682–2692. doi: 10.1110/ps.04940705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noy K, Kalisman N, Keasar C. BMC Struct Biol. 2008;8:27. doi: 10.1186/1472-6807-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang JS, Chen WW, Skolnick J, Shakhnovich EI. Structure. 2007;15:53–63. doi: 10.1016/j.str.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 36.Yang JS, Wallin S, Shakhnovich EI. Proc. Natl. Acad. Sci. USA. 2008;105:895–900. doi: 10.1073/pnas.0707284105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen WW, Shakhnovich EI. Protein Sci. 2005;14:1741–1752. doi: 10.1110/ps.051440705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 39.RNAse A: 4 C, 0.1% TFA; BID BH3: 20 C, buffer.
- 40.Prasad BV, Balaram P. CRC Crit. Rev. Biochem. 1984;16:307–348. doi: 10.3109/10409238409108718. [DOI] [PubMed] [Google Scholar]
- 41.O'Neil KT, DeGrado WF. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 42.Hubner IA, Deeds EJ, Shakhnovich EI. Proc. Natl. Acad. Sci. USA. 2005;102:18914–18919. doi: 10.1073/pnas.0502181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hubner IA, Deeds EJ, Shakhnovich EI. Proc. Natl. Acad. Sci. USA. 2006;103:17747–17752. doi: 10.1073/pnas.0605580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shortle D, Simons KT, Baker D. Proc Natl Acad Sci U S A. 1998;95:11158–11162. doi: 10.1073/pnas.95.19.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rose GD, Seltzer JP. J Mol Biol. 1977;113:153–164. doi: 10.1016/0022-2836(77)90046-8. [DOI] [PubMed] [Google Scholar]
- 46.Shortle D. FASEB J. 1996;10:27–34. doi: 10.1096/fasebj.10.1.8566543. [DOI] [PubMed] [Google Scholar]
- 47.Shortle D. Current Opinion in Structural Biology. 1993;3:66–74. [Google Scholar]
- 48.Ihalainen JA, Bredenbeck J, Pfister R, Helbing J, Chi L, van Stokkum IH, Woolley GA, Hamm P. Proc. Natl. Acad. Sci. USA. 2007;104:5383–5388. doi: 10.1073/pnas.0607748104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caflisch A. Curr. Opin. Struct. Biol. 2006;16:71–78. doi: 10.1016/j.sbi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 50.Rao F, Caflisch A. J. Mol. Biol. 2004;342:299–306. doi: 10.1016/j.jmb.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 51.Krivov SV, Karplus M. Proc. Natl. Acad. Sci. USA. 2004;101:14766–14770. doi: 10.1073/pnas.0406234101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bredenbeck J, Helbing J, Kumita JR, Woolley GA, Hamm P. Proc. Natl. Acad. Sci. USA. 2005;102:2379–2384. doi: 10.1073/pnas.0406948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munoz V, Serrano L. Journal of Molecular Biology. 1995;245:275–296. doi: 10.1006/jmbi.1994.0023. [DOI] [PubMed] [Google Scholar]
- 54.Munoz V, Serrano L. Journal of Molecular Biology. 1995;245:297–308. doi: 10.1006/jmbi.1994.0024. [DOI] [PubMed] [Google Scholar]
- 55.Munoz V, Serrano L. Biopolymers. 1997;41:495–509. doi: 10.1002/(SICI)1097-0282(19970415)41:5<495::AID-BIP2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 56.Lacroix E, Viguera AR, Serrano L. Journal of Molecular Biology. 1998;284:173–191. doi: 10.1006/jmbi.1998.2145. [DOI] [PubMed] [Google Scholar]
- 57.Hamacher K, Hubsch A, McCammon JA. J. Chem. Phys. 2006;124:164907. doi: 10.1063/1.2185645. [DOI] [PubMed] [Google Scholar]
- 58.Karanicolas J, Charles L, Brooks I. Protein Science. 2002;11:2351–2361. doi: 10.1110/ps.0205402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Go N, Abe H. Biopolymers. 1981;20:991–1011. doi: 10.1002/bip.1981.360200511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of computational and data analysis procedures. Tables S1-S3, Figures S1-S16.