Abstract
This laboratory recently identified a novel proton-coupled folate transporter (PCFT) that mediates intestinal folate absorption and transport of folates into the central nervous system. The present study focuses on the definition of the minimum transcriptional regulatory region of this gene in HeLa cells and the mechanism(s) underlying the loss of PCFT expression in the methotrexate-resistant HeLa R1–11 cell line. The PCFT transcriptional regulatory controls were localized between −42 and +96 bases from the transcriptional start site using a luciferase-reporter gene system. The promoter is a G+C rich region of 139 nucleotides contained in a CpG island. HeLa R1–11 cells have no mutations in the PCFT open-reading-frame and its promoter; the transcription/translation machinery is intact since transient transfections in HeLa R1–11 and wild-type HeLa cells produced similar luciferase activities. Hypermethylation at CpG sites within the minimal transcriptional regulatory region was demonstrated in HeLa R1–11 cells as compared to the parental PCFT-competent HeLa cells, using bisulfite conversion and sequence analysis. Treatment with 5-aza-2’-deoxycytidine resulted in a substantial restoration of transport and PCFT mRNA expression and small, but significant decreases in methylation in the promoter region. In vitro methylation of the transfected reporter plasmid inhibited luciferase gene expression. Cytogenetics/FISH indicated a loss of half the PCFT gene copies in HeLa R1–11 as compared to PCFT-competent HeLa cells. Taken together, promoter silencing via methylation and gene copy loss accounted for the loss of PCFT activity in antifolate-resistant HeLa R1–11 cells.
Keywords: PCFT, HCP1, proton-coupled folate transporter, PCFT regulation, epigenetic regulation, antifolate-resistance, methotrexate, pemetrexed
Introduction
Membrane transport of antifolates has been recognized as an important determinant of the activity of this class of agents. Traditionally, these studies have focused on methotrexate (MTX), until recently the only antifolate approved for the treatment of cancer in this country (1). Membrane transport of MTX into tumor cells is mediated almost exclusively by the reduced folate carrier (RFC), an anion exchanger and member of the superfamily of solute transporters (SLC19A1). When RFC is mutated or silenced, there is marked resistance to MTX (2). However, there had been evidence suggesting the presence of another pathway for MTX distinct from RFC in normal tissues and cancer cell lines, a folate/antifolate transport activity with a low-pH optimum (3–5). This RFC-independent mechanism was shown to have a low-pH optimum, unlike the neutral pH optimum of RFC, and a very high affinity for pemetrexed (6).
Recently, this laboratory identified the molecular basis for this low-pH transport pathway with the cloning of the proton-coupled folate transporter (PCFT – SLC46A1), that reproduces all the properties of the low-pH folate transport activity recognized in mammalian cells (7,8). From the physiological perspective, PCFT plays a critical role in the transport of folates across the apical brush-border membrane of the proximal jejunum, where there is an acid microclimate (9), and in the transport of folates into the central nervous system. This was established with the demonstration that there are loss-of-function mutations in this protein in the autosomal recessive disorder, hereditary folate malabsorption (7,10,11), in which both processes are markedly impaired (12). Hence, PCFT is required for folate homeostasis and folate sufficiency in man. From an epidemiological perspective, and from studies in mouse models, folate deficiency is a risk factor for the development of colorectal and possibly other cancers (13–15). Conversely, there is evidence that folate excess may enhance tumor growth and progression once a malignancy is established (16).
Evidence is also emerging pointing to the role PCFT plays in the pharmacology of pemetrexed and potentially other antifolates. Transfection of PCFT in cells, to levels that approximate its constitutive expression in human tumors, potentiates the pharmacological activity of pemetrexed (17). In a HeLa cell line (R5), in which RFC was deleted from the genome by chemical mutagenesis and MTX selective pressure, growth inhibition by pemetrexed was fully preserved due to transport mediated by PCFT (5). However, these cells were resistant to MTX and highly resistant to ZD1694 and PT523. When, in addition, PCFT was silenced under a second round of MTX selective pressure, the HeLa R1 cell line emerged in which there was resistance to all these antifolates (18). The antitumor effects of PCFT are likely to be greater, and have a broader spectrum of effectiveness, against solid tumors in vivo, where the drug interacts with malignant cells in a hypoxic, acidic environment (19,20) in which the activity of PCFT is enhanced, and the activity of RFC is diminished, for all antifolates.
Because of the physiological and pharmacological importance of PCFT, the basis for its regulation and the mechanisms by which it might be silenced in tumor cells is of considerable importance. The current study addresses the identification and characterization of the minimal transcriptional regulatory region of the PCFT gene and the mechanisms by which the expression of PCFT was silenced in the HeLa R1 antifolate-resistant cell line.
Materials and Methods
Materials
Tritiated methotrexate disodium salt, [3’,5’,7-3H(N)]MTX, ([3H]MTX), was obtained from Moravek Biochemicals (Brea, CA) and purified by liquid chromatography (21).
Cells and Culture Conditions
HeLa cells (cervical epitheloid carcinoma; RFC and PCFT-competent), HeLa-derived R5 cells (RFC deleted) (5), and HeLa R1–11 cells, a clonal derivative of the HeLa R1 cell line obtained from HeLa R5 cells by gene trapping mutagenesis and methotrexate selective pressure (RFC and PCFT deficient; RFC- PCFT-) (17,18), were maintained in monolayer culture at 37°C under 5% CO2 in RPMI-1640 medium containing 2.3 µM folic acid and supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin and 100 µg/ml streptomycin. HeLa R1–11 cells have a stable phenotype and retain resistance to MTX after >3 months in MTX-free medium (17).
Transient transfections and luciferase reporter gene activity
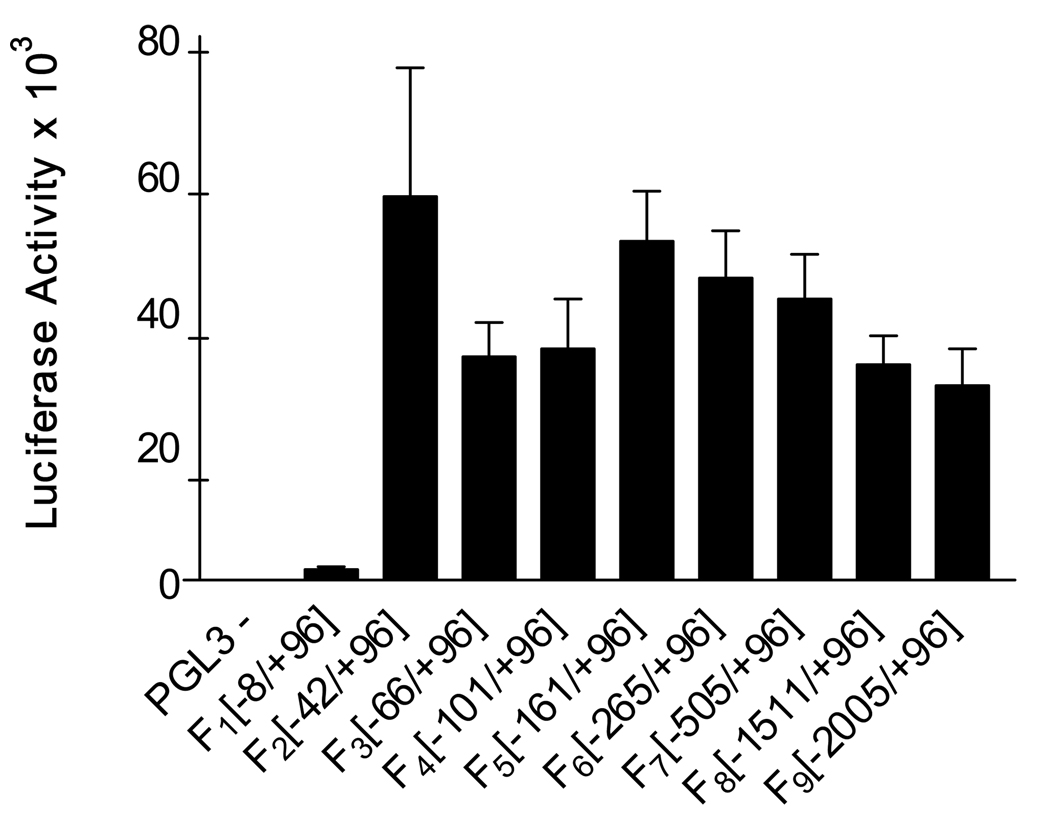
Human genomic DNA contained in the BAC Clone RP11-348E14 (Children’s Hospital Oakland Research Institute (CHORI) BACPAC Resource Center) was used as a template to amplify 9 fragments (F1–F9) of different lengths in the 5’ upstream region of PCFT. F1 contains the segment (−8/+96), F2 (−42/+96), F3 (−66/+96), F4 (−101/+96), F5 (−161/+96), F6 (−265/+96), F7 (−505/+96), F8 (−1511/+96), and F9 (−2005/+96). The numbering is relative to the transcriptional start site “A” which is based on database analysis (see Figure 2). The sequences for all the sense primers contain the Kpn I restriction site (F1–F9) and are as follows: 5’-cggggtaccaggcgcagacagcgcaagccc-3’; 5’-cggggtacccccgccggacatttaaggag-3’; 5’-cggggtaccggtggcctcaggtcacaggc-3’; 5’-cggggtacccacgcccagccaggtgcacc-3’; 5’-cggggtacctacgcacactttacaggtgag-3’; 5’-cggggtaccataccgtgcccagcacatagtaag-3’; 5’-cggggtaccatgccgaaggtagtggcagagcct-3’; 5’-cggggtacctcagctgctctgttctcagggaag-3’, 5’-cggggtacctcagctgctctgttctcagggaag-3’; and 5’-cggggtaccgagttagaaaagacctctaccctag-3’, respectively. Two antisense primers were used containing either Xho I or Bgl II restrictions sites: 5’-ccgctcgaggtgcgtgcgcggcggagctgtcg -3’ or 5’-ggaagatctgtgcgtgcgcggcggagctgtcg-3’. Amplimers were digested with Kpn I and Xho I or Bgl II, and subcloned into the multiple cloning sites of the promoter-less pGL3-Basic vector (Promega, Madison, WI) which contains the firefly luciferase gene. Clones were verified on an ABI 3730 DNA sequencer (Applied Biosystems, Foster City, CA) at the Albert Einstein Cancer Center Genomics Shared Resource using RVprimer3 and GLprimer2 (Promega). Reporter activity was assayed in HeLa cells by transient transfection using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. The pGL3-Basic (promoter-less) plasmid and the pGL3-SV40 promoter plasmid (Promega) were used as negative and positive controls, respectively. The phRG-B vector (Promega), which is identical to pGL3-Basic except that it contains the Renilla luciferase gene, was modified in this laboratory by inserting the SV40 promoter using the same flanking restriction sites as in the pGL3-promoter plasmid (Bgl II and Hind III). For normalization, the phRG-B SV40 vector was used in co-transfection experiments. Sixteen to twenty-four hours post-cotransfection, luciferase activities were measured using the Dual-Glo Luciferase kit (Promega). Firefly luciferase activities were normalized to Renilla luciferase. One-way ANOVA followed by the Tukey test were performed to determine the statistical significance of the luciferase data in Figure 1.
Figure 2.
Schematic representation of the upstream structure of the PCFT gene. The sequence [−42/+96] is shown including a predicted promoter sequence (www.fruitfly.org/seq_tools/promoter.html), BRE and TATA-like sequences (underlined), reported initiation sites (capitalized base pairs with arrows) (NM_080669, BC010691) arbitrarily positioned at +1 and +10. Conserved human and mouse nucleotides are indicated by dots; a DPE-like sequence is underlined. The GC rich nature of the human PCFT minimal transcriptional regulatory region is shown by a high G+C content (77%) and the presence of 24 CpG dinucleotides indicated in grey.
Figure 1.
Assessment of human PCFT promoter activity by luciferase gene expression reporter assays. Promoter regions of PCFT designated F1–F9 were subcloned into the pGL3-Basic vector (−). Using Lipofectamine (0.75 µl/well), HeLa cells (4000 cells seeded in a 96 well plate) were transiently co-transfected 2 days post-seeding with the pGL3 constructs (200 ng) and the phRG-B SV40 plasmid (15 ng) which contains the Renilla luciferase gene. At 16–24 hrs after transfection, luciferase activities were measured using the Dual-Glo Luciferase kit (Promega) and a POLARstar OPTIMA microplate reader (BMG LABTECH). Firefly luciferase activities were normalized with Renilla luciferase. Data represent the mean ± SE from three experiments, each performed in triplicate.
Bisulfite conversion
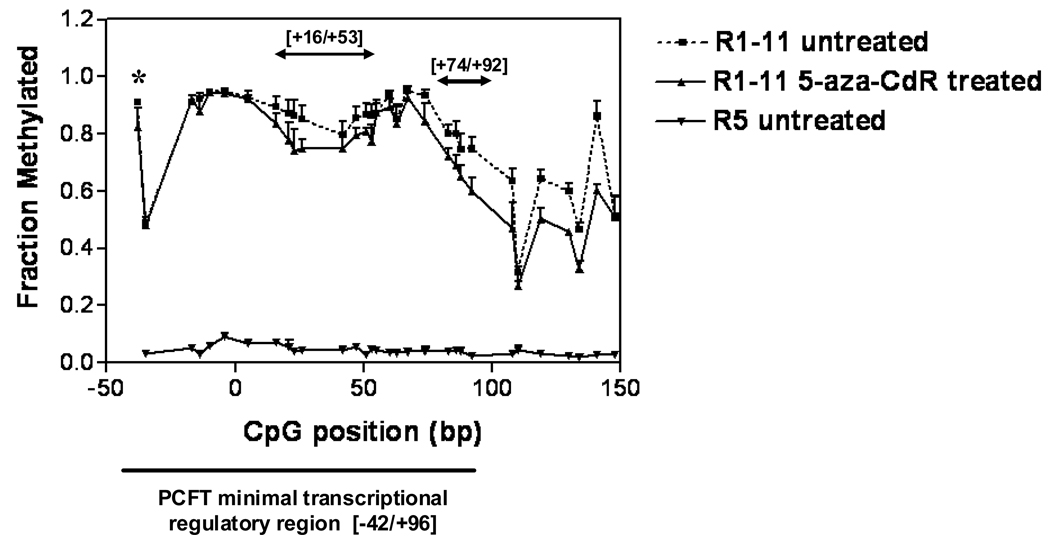
Differences in methylation patterns in the HeLa R5 and HeLa R1–11 cell lines that express moderate or no PCFT, respectively, were studied using bisulfite conversion, followed by PCR and sequencing. Genomic DNA was isolated from HeLa R5, HeLa R1–11, and HeLa R1–11 treated with 5-aza-CdR using the DNeasy Tissue kit (Qiagen Sciences, Maryland). The bisulfite reaction was performed as described previously (22) with slight modifications. Genomic DNA (~1 µg in 10 µL) was sheared five times using a 27 gauge syringe then denatured with 0.34 M NaOH for twenty minutes at room temperature. Sixty eight µL of freshly prepared bisulfite solution (2 M sodium bisulfite, 6 M urea, 0.6 mM hydroquinone at pH 5.0) was added following which PCR was performed under the following conditions: 2 min at 95 °C × 1, 2 hrs at 75 °C followed by 1 min at 95 °C × 3. The bisulfite adduct was then removed using Qiagen columns from the EpiTect Bisulfite Kit. The human PCFT minimal promoter region was amplified using the following sense and antisense primers, 5’-TAG GGT TTT TTA TTT GTT AGG TTT TT-3’ and 5’-CAC ACT TTA CAA ATA AAA TCA TCC C-3’, respectively. Amplified products were gel purified and sequenced using two degenerate inner forward and reverse primers: 5’-ATT AGY GGT TTT ATY GGG TTT YGG-3’ and 5’-CTC CCY GCY GAA CAT TTA AAA A-3’, respectively, with Y representing a C or a T. The fraction of methylated C was calculated by measuring the height of each C and T and then dividing C by the sum of the heights of C + T. Figure 4 represents the degree of methylation on the Y-axis at each CpG site plotted on the X-axis. The numbering is based on the transcriptional start site. Student paired t-tests were utilized to assess the significance of methylation changes between untreated and 5-aza-CdR-treated HeLa R1–11 cells.
Figure 4.
Assessment of DNA methylation in the PCFT minimal transcriptional regulatory region (−42 to +96) by bisulfite conversion and sequence analysis among HeLa R5, HeLa R1–11, and HeLa R1–11 cells treated with 5-aza-CdR. The fraction of methylated C was calculated by measuring the height of each C and T and then dividing C by the sum of the height of C + T shown on the Y-axis. The location of each CpG dinucleotide is indicated relative to the transcriptional start site (X-axis). The CpG sites within the minimal transcriptional regulatory region are indicated along with CpG sites within the first exon at positions beyond +100. Data are the mean ±SE from three independent experiments. The data were analyzed using the Student’s t-test. The asterix denotes statistical significance at the −38 CpG site. The regions between +16 to +53 and between +74 to +92 are also demarcated and each showed statistically significant demethylation – see text.
In vitro methylation
Construct F2 (−42/+96) was methylated in vitro by treatment with the CpG methyltransferase, M.Sss1 (New England BioLabs Inc., Beverly, MA), according to the manufacturer’s protocol. Methylation was confirmed by resistance to the methylation-sensitive restriction enzyme, BstUI.
5-aza-2’-deoxycytidine (5-aza-CdR) treatment of HeLa R1–11 and HeLa R5 cells and assessment of [3H]MTX influx
HeLa R1–11 and HeLa R5 cells (2.5 × 105) were seeded in glass vials (Research Product International Corp., Mt. Prospect, IL) and, after a day, fresh medium containing 1 µM 5-aza-CdR (Sigma-Aldrich, Saint Louis, MO) was added and changed daily. After a three day exposure to 1 µM 5-aza-CdR, cells were trypsinized, counted, and re-seeded (2.5 × 105) in drug-free medium (23). Forty-eight hours later, [3H]MTX influx was assessed over 1 min at a concentration of 0.5 µM, in MBS buffer (20 mM 4-morpholinepropane-sulfonic acid, 140 mM NaCl, 5 mM KCl, 2 mM MgCl2, and 5 mM glucose; pH 5.5) as described previously (17). HBS buffer (0°C) was added to stop uptake following which cells were washed 3 times, each for 5 min in this buffer, then lysed in 500 µl 0.2 M NaOH at 65 °C for 45 min. A portion (400 µl) of lysate was assayed for radioactivity and 10–20 µl was assayed for protein determination using the BCA protein assay kit (PIERCE, Rockford, IL). Cellular uptake is expressed in pmol/mg protein/min.
Quantitation of hPCFT mRNA levels by real time PCR
Total RNA was isolated from untreated and 5-aza-CdR-treated HeLa R5 and HeLa R1–11 cells using TRIzol® Reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. cDNA was synthesized from total RNA (5 µg) using Oligo(dT)12–18 primers and Superscript™ II reverse transcriptase (Invitrogen, Carlsbad, CA). PCFT mRNA was quantitated by real time PCR using Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA). The primers used for the PCFT gene and the housekeeping gene (β-actin) were reported previously (7). All assays were carried out in triplicate.
Fluorescence In Situ Hybridization (FISH)
BAC DNA (RP11-348E14) was labeled by nick translation using biotin-16-dUTP (Roche Diagnostics, Indianapolis, IN), hybridized to metaphase chromosomes, and detected by Alexa Fluor 647-conjugated streptavidin antibody (Invitrogen, Carlsbad, CA). A probe for human chromosome 17 (Albert Einstein Cancer Center Genome Imaging Facility) was co-hybridized. Metaphase chromosomes were derived from HeLa wild-type, R5, and R1–11 cell lines as previously described (http://www.riedlab.nci.nih.gov/protocols.asp).
Results
Definition of the PCFT minimum transcriptional regulatory region
The minimal promoter of the human PCFT gene was identified using a luciferase-reporter transfection system. Figure 1 shows the luciferase activities of constructs that map the upstream PCFT sequence. The data suggest that the −42/+96 fragment with respect to the transcriptional start site, determined by database analysis, is sufficient for maximal PCFT promoter activity. When an analysis of variance was performed, there were no significant differences among and between the F2 – F9 constructs (Figure 1). Hence, no additional enhancer or repressor elements could be identified over the region between −2005 and +96. Thus, transcriptional regulation is confined to a region that contains the core promoter and a more proximal segment (Figure 2).
Analysis of the basis for loss of PCFT activity in HeLa R1–11 cells
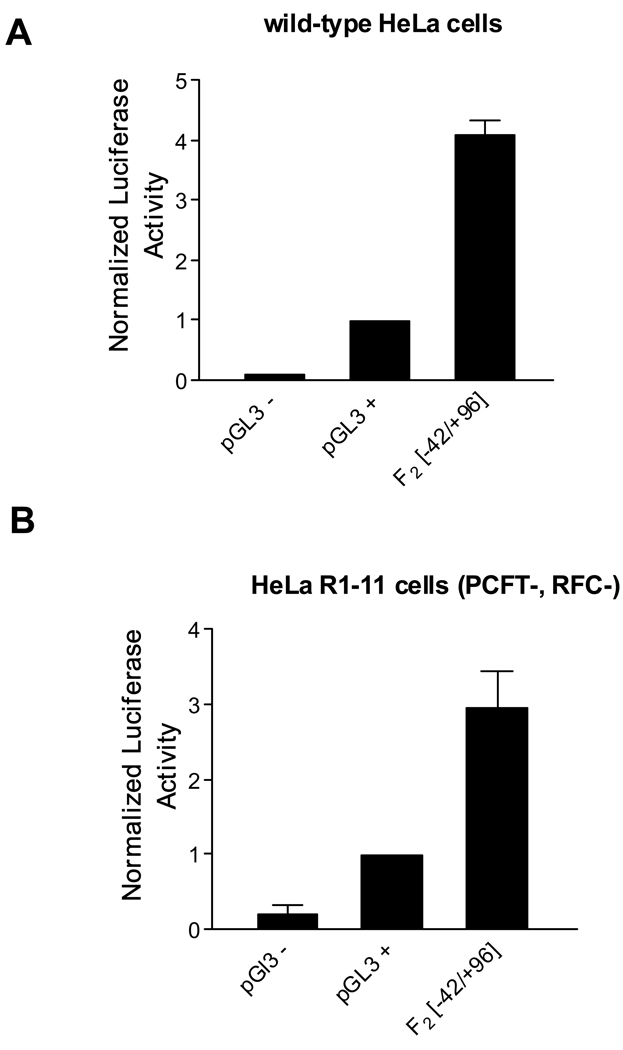
HeLa R1–11 cells were derived from HeLa R5 cells which lack genomic RFC but retain a moderate level of PCFT expression and activity. In contrast, HeLa R1–11 cells have essentially no folate transport activity and PCFT mRNA is not detected (7). Sequence analysis indicated that there are no mutations in the upstream (−1288/+96) or coding regions (data not shown). However, transient transfection of the PCFT promoter-luciferase construct [F2(−42/+96)] in HeLa R1–11 cells showed expression levels similar to those observed after transfection into wild-type HeLa cells indicative of intact transcriptional/translational mechanisms (Figures 3A and 3B). Hence, the loss of PCFT activity in HeLa R1–11 cells is consistent with a regulatory change raising the possibility of an alteration at the epigenetic level.
Figure 3.
Assessment of the human PCFT core promoter activity (F2) [−42/+96] in HeLa cells. A. Wild-type HeLa cells. B. PCFT and RFC deficient HeLa R1–11 cells (PCFT-, RFC-). The pGL3-SV40 promoter plasmid (pGL3 +) was assigned the value of 1. Data represent the mean ±SE from three experiments, each performed in triplicate.
Methylation status of the PCFT minimal transcriptional regulatory region
Studies were undertaken to explore the possibility that PCFT silencing in HeLa R1–11 cells is due to altered DNA methylation in the promoter region. The human PCFT minimal transcriptional regulatory region is G + C-rich and contained in a CpG island which comprises 584 nucleotides and includes exon 1 and 168 bp in the upstream region. The UCSC genome browser gives a CpG count of 64, a G+C percentage of 73.5 and a ratio of observed to expected CpG of 0.81 (24). Among the 139 nucleotides in the minimal regulatory region, there are 60C and 46G, thus a G+C content of 77% and 24 CpGs. Methylation analysis at those CpGs showed a striking difference between the two cell lines (Figure 4). HeLa R5 cells showed virtually no methylation; in contrast, the PCFT promoter region was highly methylated in HeLa R1–11 cells.
Impact of the methylation of the putative PCFT promoter on transcriptional activity
To directly assess the impact of methylation on transcriptional activity, the −42/+96 human PCFT construct was treated with Sss1 methyltransferase. Methylation was confirmed by the observation that this abolished cleavage of the PCFT DNA by BstUI (Figure 5A). When the methylated PCFT promoter luciferase construct was transiently expressed in HeLa cells, luciferase activity similar to a negative control plasmid was observed (Figure 5B). Hence, methylation of the PCFT promoter abolishes its activity.
Figure 5.
Impact of methylation on promoter activity and 5-aza-CdR treatment on HeLa R1–11 and HeLa R5 cells A. DNA methylation and its effect on the activity of the PCFT core promoter. Confirmation of in vitro methylation of the PCFT promoter-luciferase construct F2 (−42/+96) by protection from BstUI digestion. F2 was methylated by Sss1 methylase for 4 hrs and then treated with BstUI. Lane 1: Lambda DNA/Hind III and Phi X174 RF/Hae III molecular weight markers; sizes are shown on the left in bp; lane 2: unmethylated F2; lane 3: F2 methylated by Sss1; lane 4: unmethylated F2 treated with BstUI; lane 5: F2 methylated by Sss1 and treated with BstUI. Lanes 2–5 contain ~500 ng DNA. B. Normalized luciferase activities of unmethylated and methylated F2 (−42/+96) in wild-type HeLa cells. Data represent the mean ±SE from three experiments, each performed in triplicate. C. Reactivation of PCFT function in HeLa R1–11 and HeLa R5 cells (2.5×105 cells seeded in transport vials) treated with 1 µM 5-aza-CdR for 72 hrs. Following treatment, the same number of cells was re-seeded in drug-free media for 48 hrs following which [3H]MTX influx was assessed over 1 min at pH 5.5 and a concentration at 0.5 µM. Data are the mean ± SE from four experiments, each performed in triplicate. D. Quantitation of PCFT mRNA levels by real time PCR in HeLa R1–11 and HeLa R5 cells treated with and without 5-aza-CdR. The Y-axis represents the ratio of the relative expression levels of PCFT to β-actin mRNAs. Data are the mean ±SE from an experiment using two sets of PCFT primers and performed in triplicate.
Reactivation of PCFT gene expression in HeLa R1–11 cells treated with 5-aza-CdR
When HeLa R1–11 cells were treated for 72 hrs with the DNA methyltransferase inhibitor, 5-aza-CdR, there was a 2.6 fold increase in PCFT-mediated [3H]MTX influx compared to untreated cells (Figure 5C). Treatment of HeLa R5 cells, in which the PCFT −42/+96 region was in a demethylated state, resulted in ~ 42 % decrease in transport mediated by this carrier. This was associated with 70 ± 9.7 % inhibition of cell growth based upon cell count, consistent with the inhibitory effect of this agent on DNA synthesis (25). Likewise, there was comparable inhibition of cell growth in HeLa R1–11 cells treated with this drug (61 ± 3.2 %). Despite this growth inhibition, the level of transport activity in 5-aza-CdR-treated HeLa R1–11 cells increased to onehalf that of HeLa R5 cells (Figure 5C). These results were correlated with the effects on PCFT mRNA levels, which increased approximately 5-fold in HeLa R1–11 cells treated with 5-aza-CdR compared to untreated cells (Figure 5D). There was no difference in PCFT mRNA levels in 5-aza-CdR- treated and untreated HeLa R5 cells; hence, inhibition of transport by 5-aza-CdR was likely related to a nonspecific toxic effect of this agent associated with the inhibition of cell growth. When the level of methylation was assessed in HeLa R1–11 cells treated with 5-aza-CdR and compared to untreated cells, there was a small (7%) global (p<0.002) decrease in methylation when all the CpG sites within the minimal transcriptional regulatory region were analyzed (−38 to +92) (p<0.006) (Figure 4). Specifically, the decrease in the most upstream CpG site (−38) was significant to p= 0.03. Two other parts of the minimal transcriptional regulatory region, from +16 to +53 and +74 to +92, showed more substantial demethylation (p<0.0001). These represent potential sites for significant regulatory actions. There was, in addition, significant (p<0.003) demethylation within the coding region (+108 to +148).
Cytogenetic analysis
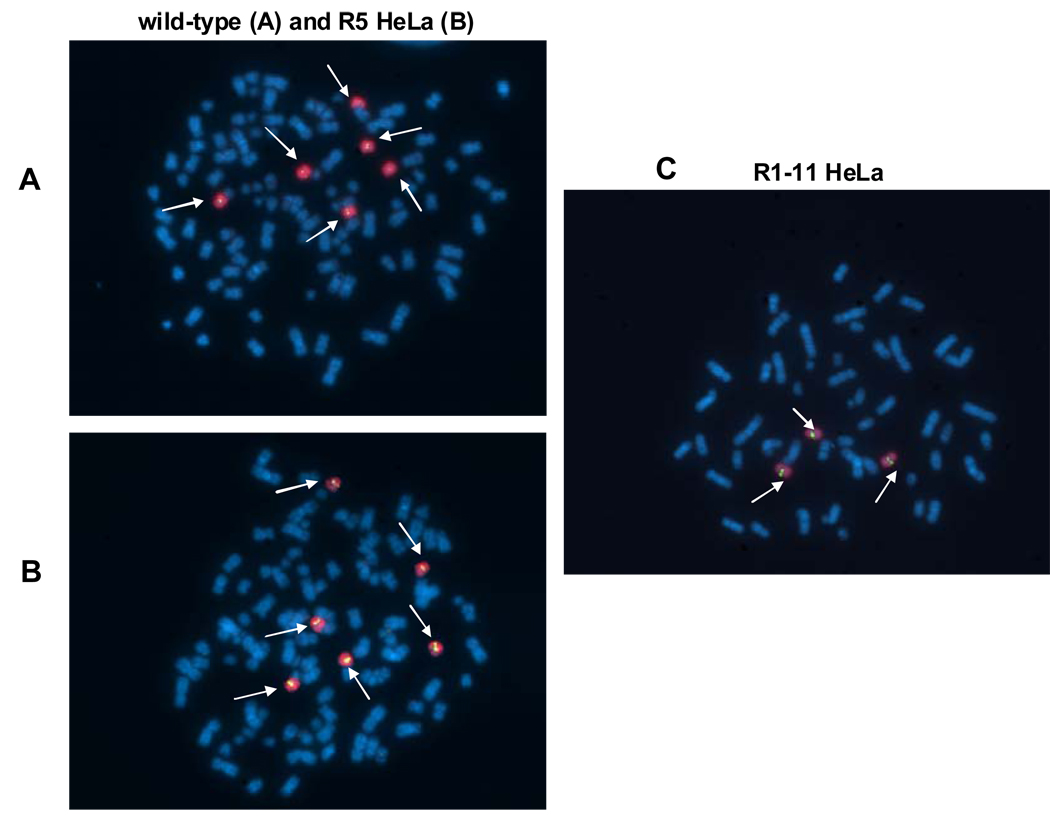
To determine whether an altered gene copy number contributed to the loss of PCFT expression and function in HeLa R1–11 cells, cytogenetic studies were undertaken using probes for chromosome 17 and the PCFT gene. As indicated in Figures 6A and 6B, wild-type and HeLa R5 cells have six copies of chromosome 17 and six copies of PCFT; HeLa R1–11 cells have three copies of chromosome 17 and three copies of PCFT (Figure 6C). Hence, HeLa R1–11 cells have lost half their PCFT DNA copies. This is consistent with the observation that treatment of HeLa R1–11 cells with 5-aza-CdR restores PCFT transport activity but only to a level ~ ½ that of HeLa R5-treated cells.
Figure 6.
Determination of PCFT gene copy number in HeLa wild-type, R5, and R1–11 cell lines by FISH analysis. A PCFT probe (labeled green) and a chromosome 17 painted probe (labeled red) were used to analyze metaphase spreads from A. wild-type HeLa cells, B. HeLa R5 cells, and C. HeLa R1–11 cells. Both signals are indicated by arrows.
Discussion
These studies provide insight into the regulation of the human PCFT gene and the mechanism by which the HeLa R1–11 cell line lost PCFT activity under MTX selective pressure. The data localized all transcriptional regulation of the PCFT gene to the interval of −42 to +96 bases, which contains the minimum transcriptional regulatory region. To explore the physiological relevance of this finding, a BLAST search was done and the databases of all expressed sequenced tags (EST) were aligned with the first 600 nucleotides of the PCFT gene (NM_080669). These clones indicated two alternative transcription start sites at +1 and +10. It is therefore possible that the entire regulatory region is contained within the interval of −42 to +96 bases upstream from the transcriptional start site. This segment includes the core promoter, but is large enough to include other cis-regulatory elements. Real time PCR showed that PCFT expression is ~1/100th that of β-actin (Figure 5D), consistent with the moderate level of PCFT protein expression in HeLa cells.
The PCFT gene was only very recently identified so there is little information on factors that alter its expression. However, a marked increase in PCFT, RFC and folate receptor mRNA expression in small intestine was observed in mice fed a folate-deficient, as compared to a folate-replete, diet (26,27). In vitro studies showed a much smaller increase in PCFT expression with folate deprivation, (2.5-fold increase in PCFT mRNA levels and 1.6-fold increase in PCFT-mediated [3H]folic acid uptake), in Caco-2 cells grown with 0.25 µM, as compared to 100 µM, folic acid (28). The present paper suggests one possible mechanism by which dietary folate availability might alter the expression of PCFT: through its effects on DNA methylation. Folate depletion has been implicated in global hypomethylation (29) which might enhance PCFT transcription resulting in enhanced intestinal folate absorption and folate repletion. Likewise, folate excess might result in PCFT hypermethylation suppressing transcriptional activity of this gene. Such a regulatory feedback mechanism could be an important factor in the maintenance of folate homeostasis. While folate status and its effects on methylation impacts on the expression of the other folate transporters (27), PCFT alone is capable of altering the level of folate absorption in the proximal small intestine thus modulating net organismal folate levels.
The HeLa cell lines utilized in the current studies played a crucial role in the identification of PCFT (5,7). The present study explored the basis for the loss of PCFT expression in the HeLa R1–11 clonal derivative of the RFC-, PCFT-, HeLa R1 line. No mutation could be detected in the coding or 5’ upstream region (−1288/+96) of this gene. Furthermore, when the PCFT promoter luciferase construct F2 (−42/+96) was transiently transfected in the MTX-resistant HeLa R1–11 and wild-type HeLa cells, the same fold increase in activity was observed with respect to the pGL3-SV40 positive control plasmid (Figure 3), consistent with intact transcription/translation machinery. The defect in HeLa R1–11 cells turned out to be due to profound hypermethylation of the minimal transcriptional regulatory region in comparison to the HeLa R5 line as established by bisulfite conversion and sequence analysis (Figure 4). PCFT mRNA expression and function in the R1–11 cells was substantially increased by treatment with 5-aza-CdR (Figures 5C and 5D). This was accompanied by small but significant changes in methylation (Figure 4). This degree of demethylation may have produced a modification in chromatin structure that was sufficient to restore a level of transcriptional activity that could account for these findings. A low level of 5-aza-CdR demethylation under these conditions is, in fact, expected since this agent only acts on newly synthesized DNA and HeLa R1–11 cells underwent less than one division over the three days of exposure to the drug. While a longer exposure to 5-aza-CdR would likely have resulted in a greater degree of demethylation, this would have been accompanied by a greater degree of cytotoxicity. It is also possible that the changes in PCFT mRNA and folate transport observed were related to a small fraction of the cell population in which there was a greater level of demethylation. These results are consistent with a recent report in which cells from patients with myelodysplastic syndrome that were treated with several cycles of low-dose 5-aza-CdR resulted in restoration of p15 protein expression after the first cycle, while demethylation of the gene occurred much more slowly over several subsequent cycles of drug treatment (30). Hence, the return of activity preceded gross demethylation in the cell population. The failure to achieve complete restoration of PCFT function is attributed to the loss of one half the PCFT gene copies in HeLa R1–11, as compared to HeLa R5 cells, as assessed by FISH (Figure 6), and a nonspecific toxic effect of 5-aza-CdR on transport unrelated to PCFT expression.
HeLa R1–11 cells were selected in the presence of MTX with folic acid as the growth source. With the loss of PCFT, there was a marked contraction of folate co-factor pools in these cells (5). However, despite the deficiency of cellular folates, the PCFT promoter was methylated and PCFT activity diminished. This suggests that the toxic effects of the antifolate produced a greater selective pressure than folate depletion.
Methylation-associated suppression of the expression of other folate transporters was reported for RFC in methotrexate-resistant MDA-MB-231 breast cancer cells (31) and folate receptor in nasopharyngeal epidermoid cancer cells, KB1BT (32). When these cells were treated with 5-aza-CdR, the RFC and folate receptor genes were re-expressed; however, the levels of expression and function in KB1BT-treated cells were far less than in wild-type cells.
In summary, the human PCFT minimal transcriptional regulatory region in HeLa cells has been defined and hypermethylation of the promoter region and gene copy loss were shown to be the basis for the silencing of the PCFT gene in a MTX-resistant cell line derived from HeLa R5. Recently, hypermethylation of a larger upstream region of this gene was shown to be associated with a low level of PCFT expression in two human leukemia cell lines. However, function was not assessed before or after treatment with 5-aza-CdR (33). Understanding the mechanisms of PCFT regulation offers the possibility of identifying approaches to modulate expression of this gene that could be used to diminish the impact of folate-deficiency states and enhance the delivery of antifolates to cancer cells.
Acknowledgments
We would like to thank Dr. John M. Greally for his advice on DNA methylation studies. This research was supported by a grant from the National Institutes of Health, CA82621, and a grant from the Mesothelioma Applied Research Foundation.
Abbreviations list
- PCFT
human Proton-Coupled Folate Tansporter
- MTX
methotrexate
- 5-aza-CdR
5-aza-2’-deoxycytidine
Footnotes
The authors disclose no conflict of interest.
References
- 1.Zhao R, Goldman ID. Resistance to antifolates. Oncogene. 2003;22:7431–7457. doi: 10.1038/sj.onc.1206946. [DOI] [PubMed] [Google Scholar]
- 2.Matherly LH, Hou Z, Deng Y. Human reduced folate carrier: translation of basic biology to cancer etiology and therapy. Cancer Metastasis Rev. 2007;26:111–128. doi: 10.1007/s10555-007-9046-2. [DOI] [PubMed] [Google Scholar]
- 3.Henderson GB, Strauss BP. Characteristics of a novel transport system for folate compounds in wild-type and methotrexate-resistant L1210 cells. Cancer Res. 1990;50:1709–1714. [PubMed] [Google Scholar]
- 4.Kuhnel JM, Chiao JH, Sirotnak FM. Contrasting effects of oncogene expression on two carrier-mediated systems internalizing folate compounds in Fisher rat 3T3 cells. J Cell Physiol. 2000;184:364–372. doi: 10.1002/1097-4652(200009)184:3<364::AID-JCP11>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 5.Zhao R, Gao F, Hanscom M, Goldman ID. A prominent low-pH methotrexate transport activity in human solid tumor cells: Contribution to the preservation of methotrexate pharmacological activity in HeLa cells lacking the reduced folate carrier. Clin Cancer Res. 2004;10:718–727. doi: 10.1158/1078-0432.ccr-1066-03. [DOI] [PubMed] [Google Scholar]
- 6.Chattopadhyay S, Moran RG, Goldman ID. Pemetrexed: biochemical and cellular pharmacology, mechanisms, and clinical applications. Mol Cancer Ther. 2007;6:404–417. doi: 10.1158/1535-7163.MCT-06-0343. [DOI] [PubMed] [Google Scholar]
- 7.Qiu A, Jansen M, Sakaris A, et al. Identification of an intestinal folate transporter and the molecular basis for hereditary folate malabsorption. Cell. 2006;127:917–928. doi: 10.1016/j.cell.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 8.Zhao R, Goldman ID. The molecular identity and characterization of a Proton-coupled Folate Transporter--PCFT; biological ramifications and impact on the activity of pemetrexed. Cancer Metastasis Rev. 2007;26:129–139. doi: 10.1007/s10555-007-9047-1. [DOI] [PubMed] [Google Scholar]
- 9.McEwan GT, Lucas ML, Denvir M, et al. A combined TDDA-PVC pH and reference electrode for use in the upper small intestine. J Med Eng Technol. 1990;14:16–20. doi: 10.3109/03091909009028758. [DOI] [PubMed] [Google Scholar]
- 10.Zhao R, Min SH, Qiu A, et al. The spectrum of mutations in the PCFT gene, coding for an intestinal folate transporter, that are the basis for hereditary folate malabsorption. Blood. 2007;110:1147–1152. doi: 10.1182/blood-2007-02-077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min SH, OH SY, Karp GI, et al. The clinical course and genetic defect in the PCFT in a 27-year-old woman with Hereditary folate malabsorption. J Pediatr. 2008;153:435–437. doi: 10.1016/j.jpeds.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geller J, Kronn D, Jayabose S, Sandoval C. Hereditary folate malabsorption: family report and review of the literature. Medicine (Baltimore) 2002;81:51–68. doi: 10.1097/00005792-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Martinez ME, Marshall JR, Giovannucci E. Diet and cancer prevention: the roles of observation and experimentation. Nat Rev Cancer. 2008;8:694–703. doi: 10.1038/nrc2441. [DOI] [PubMed] [Google Scholar]
- 14.Song J, Medline A, Mason JB, Gallinger S, Kim YI. Effects of dietary folate on intestinal tumorigenesis in the apcMin mouse. Cancer Res. 2000;60:5434–5440. [PubMed] [Google Scholar]
- 15.Ma DW, Finnell RH, Davidson LA, et al. Folate transport gene inactivation in mice increases sensitivity to colon carcinogenesis. Cancer Res. 2005;65:887–897. [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YI. Folic acid fortification and supplementation--good for some but not so good for others. Nutr Rev. 2007;65:504–511. doi: 10.1111/j.1753-4887.2007.tb00275.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhao R, Qiu A, Tsai E, et al. The proton-coupled folate transporter (PCFT): impact on pemetrexed transport and on antifolate activities as compared to the reduced folate carrier. Mol Pharmacol. 2008;74:854–862. doi: 10.1124/mol.108.045443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao R, Chattopadhyay S, Hanscom M, Goldman ID. Antifolate resistance in a HeLa cell line associated with impaired transport independent of the reduced folate carrier. Clin Cancer Res. 2004;10:8735–8742. doi: 10.1158/1078-0432.CCR-04-0932. [DOI] [PubMed] [Google Scholar]
- 19.Helmlinger G, Yuan F, Dellian M, Jain RK. Interstitial pH and pO2 gradients in solid tumors in vivo: high- resolution measurements reveal a lack of correlation. Nat Med. 1997;3:177–182. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 20.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 21.Zhao R, Babani S, Gao F, Liu L, Goldman ID. The mechanism of transport of the multitargeted antifolate, MTA- LY231514, and its cross resistance pattern in cell with impaired transport of methotrexate. Clin Cancer Res. 2000;6:3687–3695. [PubMed] [Google Scholar]
- 22.Aufsatz W, Mette MF, van der WJ, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2'-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 24.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 25.Oki Y, Aoki E, Issa JP. Decitabine--bedside to bench. Crit Rev Oncol Hematol. 2007;61:140–152. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Qiu A, Min SH, Jansen M, et al. Rodent intestinal folate transporters (SLC46A1): secondary structure, functional properties, and response to dietary folate restriction. Am J Physiol Cell Physiol. 2007;293:C1669–C1678. doi: 10.1152/ajpcell.00202.2007. [DOI] [PubMed] [Google Scholar]
- 27.Liu M, Ge Y, Cabelof DC, et al. Structure and regulation of the murine reduced folate carrier gene: identification of four noncoding exons and promoters and regulation by dietary folates. J Biol Chem. 2005;280:5588–5597. doi: 10.1074/jbc.M412662200. [DOI] [PubMed] [Google Scholar]
- 28.Ashokkumar B, Mohammed ZM, Vaziri ND, Said HM. Effect of folate oversupplementation on folate uptake by human intestinal and renal epithelial cells. Am J Clin Nutr. 2007;86:159–166. doi: 10.1093/ajcn/86.1.159. [DOI] [PubMed] [Google Scholar]
- 29.Wasson GR, McGlynn AP, McNulty H, et al. Global DNA and p53 region-specific hypomethylation in human colonic cells is induced by folate depletion and reversed by folate supplementation. J Nutr. 2006;136:2748–2753. doi: 10.1093/jn/136.11.2748. [DOI] [PubMed] [Google Scholar]
- 30.Daskalakis M, Nguyen TT, Nguyen C, et al. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2'-deoxycytidine (decitabine) treatment. Blood. 2002;100:2957–2964. doi: 10.1182/blood.V100.8.2957. [DOI] [PubMed] [Google Scholar]
- 31.Worm J, Kirkin AF, Dzhandzhugazyan KN, Guldberg P. Methylation-dependent silencing of the reduced folate carrier gene in inherently methotrexate-resistant human breast cancer cells. J Biol Chem. 2001;276:39990–40000. doi: 10.1074/jbc.M103181200. [DOI] [PubMed] [Google Scholar]
- 32.Hsueh C-T, Dolnick BJ. Regulation of folate-binding protein gene expression by DNA methylation in methotrexate-resistant KB cells. Biochem Pharmacol. 1994;47:1019–1027. doi: 10.1016/0006-2952(94)90413-8. [DOI] [PubMed] [Google Scholar]
- 33.Gonen N, Bram EE, Assaraf YG. PCFT promoter methylation and restoration of gene expression in human leukemia cells. Biochem Biophys Res Commun. 2008;376:787–792. doi: 10.1016/j.bbrc.2008.09.074. [DOI] [PubMed] [Google Scholar]