Abstract
Glutaminases belong to the large superfamily of serine-dependent β-lactamases and penicillin-binding proteins, and they catalyze the hydrolytic deamidation of l-glutamine to l-glutamate. In this work, we purified and biochemically characterized four predicted glutaminases from Escherichia coli (YbaS and YneH) and Bacillus subtilis (YlaM and YbgJ). The proteins demonstrated strict specificity to l-glutamine and did not hydrolyze d-glutamine or l-asparagine. In each organism, one glutaminase showed higher affinity to glutamine (E. coli YbaS and B. subtilis YlaM; Km 7.3 and 7.6 mM, respectively) than the second glutaminase (E. coli YneH and B. subtilis YbgJ; Km 27.6 and 30.6 mM, respectively). The crystal structures of the E. coli YbaS and the B. subtilis YbgJ revealed the presence of a classical β-lactamase-like fold and conservation of several key catalytic residues of β-lactamases (Ser74, Lys77, Asn126, Lys268, and Ser269 in YbgJ). Alanine replacement mutagenesis demonstrated that most of the conserved residues located in the putative glutaminase catalytic site are essential for activity. The crystal structure of the YbgJ complex with the glutaminase inhibitor 6-diazo-5-oxo-l-norleucine revealed the presence of a covalent bond between the inhibitor and the hydroxyl oxygen of Ser74, providing evidence that Ser74 is the primary catalytic nucleophile and that the glutaminase reaction proceeds through formation of an enzyme–glutamyl intermediate. Growth experiments with the E. coli glutaminase deletion strains revealed that YneH is involved in the assimilation of l-glutamine as a sole source of carbon and nitrogen and suggested that both glutaminases (YbaS and YneH) also contribute to acid resistance in E. coli.
Glutaminases (EC 3.5.1.2) are present in most bacteria and eukaryotes and catalyze the hydrolytic deamidation of l-glutamine to l-glutamate and free NH4+(1). These enzymes are strictly specific to l-glutamine and differ from glutaminase–asparaginases (EC 3.5.1.1), which deamidate both asparagine and glutamine. Poorly characterized glutaminases belong to the large group of serine β-lactamases and penicillin-binding proteins, which have a common evolutionary origin and share the protein fold, structural motifs, and catalytic mechanism (2). This large group of enzymes includes dd-peptidases, transpeptidases, glutaminases, and three classes of well-characterized serine β-lactamases (A, C, and D) (3). β-Lactamases (EC 3.5.2.6) catalyze the hydrolysis of an amide bond (N–CO) in the β-lactam ring of antibiotics of the penicillin/cephalosporin family constituting the most common mechanism of bacterial resistance to β-lactam antibiotics, whereas penicillin-binding proteins have transpeptidase, transglycosylase, and carboxypeptidase activities and are involved in the biosynthesis of the bacterial cell wall (2, 4, 5). The representatives of all β-lactamase and dd-peptidase families have been characterized both structurally and biochemically, and the molecular mechanisms of the catalysis have been established (6–9).
In microorganisms, glutaminases have been reported from many species including Gram-positive and Gram-negative bacteria, yeasts, and fungi (1, 10). Several microbial enzymes (from Micrococcus luteus, Rhizobium etli, Bacillus pasteurii, and Lactobacillus rhamnosus) were purified and partially characterized (11–14). All of these enzymes (except for the L. rhamnosus glutaminase) were found to be soluble proteins, and they all showed low affinity to glutamine (Km from 1.5 to 9.5 mM). The recently solved crystal structure of a major fragment of the M. luteus glutaminase (PDB 2dfw) revealed the presence of a putative catalytic site in the N-terminal domain (15). Mutational analysis of this protein indicated that Ser64, Lys67, and Glu160 are essential for catalysis and suggested that the catalytic mechanism might be similar to that of the class A β-lactamases (16). Many organisms including mammals produce two glutaminases, which share 30–40% sequence identity (17–20). In Escherichia coli, the presence of two glutaminases (A and B) was experimentally demonstrated over 30 years ago (21, 22). Glutaminase A seems to be the main isoenzyme, and it was purified in apparently homogeneous form (21). Glutaminase B comprised a minute fraction of total cellular protein, and after 6000-fold purification, the purified preparation contained only about 40% of this enzyme (22). Studies on the regulation of the cellular levels of two glutaminases have revealed that glutaminase A levels are stimulated by high NH4+ and inhibited by cAMP, while glutaminase B levels were independent of the growth conditions (23–25). In the E. coli genome, as well as in the genome of the Gram-positive bacterium Bacillus subtilis, there are two genes encoding putative glutaminases, but only for the B. subtilis YbgJ has the bioinformatic prediction been recently confirmed experimentally (26). The expression of this glutaminase has been shown to be stimulated in response to glutamine in the culture medium, suggesting that this protein is involved in glutamine assimilation (26). However, the physiological functions of the other glutaminases remain unknown.
In this work, we present the results of biochemical characterization of the four predicted glutaminases from E. coli (YbaS and YneH) and B. subtilis (YbgJ and YlaM). We determined the crystal structures of YbaS and YbgJ in a free state, as well as the structure of the covalent complex of YbgJ with the glutaminase inhibitor 6-diazo-5-oxo-l-nor-leucine (DON) 1. Growth experiments with the E. coli glutaminase deletion strains suggest that YneH is involved in the assimilation of glutamine and that both YneH and YbaS also contribute to acid resistance in this organism.
EXPERIMENTAL PROCEDURES
Gene Cloning and Protein Purification
Genes (ybaS and yneH from E. coli and ylaM and ybgJ from B. subtilis) were PCR-amplified using chromosomal DNA of the E. coli W3110 strain and the B. subtilis 168 (ATCC 23857D-5). The restriction sites BamHI and NdeI were added to the PCR primers and used to directionally clone the PCR product into the modified pET15b (Novagen) as previously described (27). The recombinant plasmid was transformed into the E. coli BL21(DE3) strain for overexpression. Expression and purification of His6-tagged proteins were described previously (28, 29). All four proteins were well expressed and purified with high yield (2–5 mg/L of culture) and purity (over 95% as verified by SDS–PAGE gels and Coomassie staining). Purified proteins were stored at −80 °C, except for YbaS, which was unstable and was stored in liquid nitrogen.
Gel filtration analysis of the oligomeric state of glutaminases was performed with a Superose 12 10/300 GL column (Amersham Biosciences) equilibrated with 10 mM HEPES-K (pH 7.5) and 0.2 M NaCl using AKTA FPLC (Amersham Biosciences). Protein standards included aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa), chymotrypsinogen (25 kDa), and ribonuclease A (13.7 kDa).
Site-Directed Mutagenesis of YbaS
Site-directed mutagenesis of YbaS was performed using a protocol based on the QuikChange site-directed mutagenesis kit from Stratagene.
DNA encoding wild-type YbaS cloned into the modified pET15b was used as a template for mutagenesis. Plasmid was purified from the resulting colonies using the Qiaprep Spin Mini Prep kit (Qiagen), and all mutations were verified by DNA sequencing. Verified plasmids containing the desired mutations were transformed into the E. coli BL21(DE3) strain, and the mutant YbaS proteins were overexpressed and purified in the same manner as the wild-type YbaS.
Enzymatic Assays
Glutaminase activity was analyzed using two assays: by measurement of NH4+ production by the continuous assay with glutamate dehydrogenase (30) or by the chromogenic assay with l-glutamyl-p-nitroanilide. The reaction mixture for the continuous glutaminase assay contained (in a final volume of 1 mL) 50 mM HEPES-K buffer (pH 7.5, or pH 8.0 for YbgJ), 50 mM l-glutamine, 0.2 mM NADH, 1 mM EDTA, 0.2 mM α-ketoglutarate, 1 unit of glutamate dehydrogenase (Sigma), and 0.1–0.2 µg of glutaminase. The oxidation of NADH was followed continuously at 340 nm. Metal effects were determined using a 10 min end-point assay. pH profiles were determined using a buffer system described by Heering et al. (31) Alternatively, glutaminase activity of purified proteins was measured using the chromogenic substrate l-glutamyl-p-nitroanilide (Sigma) in a reaction mixture containing 50 mM HEPES-K buffer (pH 7.5) and 10 mM l-glutamyl-p-nitroanilide (final volume 1 mL). The activity was followed at 410 nm. For Km and Vmax determination, the glutaminase assays contained 0.1 – 150 mM l-glutamine. Kinetic parameters were determined by nonlinear curve fitting from the Lineweaver–Burk plot using the GraphPad Prism software (version 4.00 for Windows; GraphPad Software, San Diego, CA). For sigmoidal curve fitting, this program uses the equation V = (VmaxSh)/(Kh0.5 + Sh).
β-Lactamase activity of glutaminases with nitrocefin as a substrate was assayed essentially as previously described (32). Reaction mixtures (1 mL) contained 50 mM sodium phosphate buffer (pH 7.4), 50 µM nitrocefin (dissolved in DMSO), and 5–20 µg of protein. After 20 min incubation at 37 °C, the increase in absorbance at 486 nm was measured (ε = 20.5 mM−1 cm−1).
Construction of the E. coli Glutaminase Deletion Strains and Growth Experiments
The E. coli ΔybaS and ΔyneH deletion mutants were obtained from the Keio collection of E. coli deletion mutants (33). The kanamycin-resistance cassette was excised from the chromosome using the procedure described by Datsenko and Wanner (34). The double glutaminase deletion mutant (ΔybaS,ΔyneH) was prepared using reciprocal P1 transduction as described previously (35). The deletions of glutaminase genes were individually validated by colony PCR using specific primers complementary to the upstream and downstream regions of each glutaminase gene. Growth experiments with the E. coli wild-type (BW25113) and glutaminase deletion (ΔybaS, ΔyneH, and ΔybaS,ΔyneH) strains were performed essentially as previously described (36) using LB medium with various pHs (4.0, 4.5, 5.0, 5.5, 6.0, 6.5, and 7.0; adjusted after autoclaving) or minimal medium M9 containing 20 mM l-glutamine as a sole source of carbon and nitrogen (pH 7.0) or M9 medium containing 20 mM MES-K buffer (pH 4.5, 5.0, 5.5, or 6.0), 0.4% glucose, and 20 mM l-glutamine.
Protein Crystallization and Data Collection
Crystals of YbaS and YbgJ were grown at 21 °C by the hanging drop vapor diffusion method with 2 µL of protein sample mixed with an equal volume of the reservoir buffer as previously described (37). The crystals of YbaS grew after 3–5 days in the presence of 25% polyethylene glycol 3350, 0.2 M MgCl2, and 0.1 M HEPES-Na (pH 7.5). The YbgJ crystals were grown in 30% polyethylene glycol 1500, 0.2 M NaCl, 5% glycerol, 0.1 mM DTT, and 0.1 M HEPES-Na (pH 7.5), whereas the crystals of the YbgJ + DON complex were grown in 14.5% polyethylene glycol 1500, 0.5 M NaCl, 12% glycerol, 0.1 M HEPES-Na (pH 7.5), and 10 mM DON. For diffraction studies, the crystals were stabilized with the crystallization buffer supplemented with 20% ethylene glycol as a cryoprotectant and flash frozen in liquid nitrogen.
Structure Determination
All diffraction data were collected on the 19-ID beamline of the Structural Biology Center at the Advanced Photon Source (38). YbaS was crystallized in two forms, both tetragonal (space group I4) and orthorhombic (space group P212121). A MAD data set was collected for the tetragonal crystal (wavelengths of 0.97948, 0.97962, and 0.95667 Å), and a single wavelength (0.97978 Å) was collected on the orthorhombic crystal. Phases were obtained, and an initial model was built from the tetragonal data set using HKL3000 (39), but when it was realized that the orthorhombic crystal diffracted to higher resolution, the initial model from the tetragonal data set was used to obtain phases for the orthorhombic crystal by molecular replacement using AMORE (40). Subsequent model building using COOT (41) and refinement using REFMAC 5.1 (42) and TLS (43) were then performed. ARP/wARP (44) was used to generate the initial solvent model, while manual water picking was used to add additional waters. All possible waters with nearest neighbor distances to other atoms of <2.3 Å or >4.5 Å or had overly high B-factors (>80 Å2) or low electron densities (<1.0σ) were manually omitted. The final model (Rwork of 14.5% and Rfree of 17.8%) contains 10487 atoms with 4 molecules in the asymmetric unit, and each molecule of the asymmetric unit contains 309 out of 310 residues of YbaS.
For YbgJ, MAD data collection at wavelengths of 0.97910, 0.97921, and 0.954 Å was used to obtain phase information. Initial phases, initial solvent flattening, and an initial model were obtained using SOLVE/RESOLVE (45), and manual building from this initial model using COOT (41) followed by refinement using REFMAC5 (42) was used to complete the structure. Solvent molecules were added and then pruned as described for YbaS. The final model was refined to 2.0 Å resolution (with an Rcryst of 21.6% and an Rfree of 24.5%) and contains 5195 atoms including 2 molecules in the asymmetric unit, with 312 out of 327 residues of YbgJ present in each molecule of the asymmetric unit. Residues 6–12 and 36–44 of YbgJ were disordered and could not be built in the electron density map.
The structure of YbgJ plus DON was determined using SAD. Initial phases, solvent flattening, and automatic model building were performed using HKL3000 (39), with subsequent manual model building using COOT (41) and refinement using REFMAC 5.2 (42). Solvent was initially added using ARP/wARP (44), with subsequent automatic and manual water picking using COOT. Throughout refinement, picked water molecules were pruned using similar criteria as described above. However, prior to adding solvent, a long unmodeled tube of electron density originating from the hydroxyl group of Ser74 was observed (see Figure 6) and ascribed to covalently bound inhibitor. This covalently bound inhibitor fits optimally as 5-oxo-l-norleucine; a covalent bond between it and the hydroxyl group of Ser74 was made first by placing the relevant atoms within covalent-bonding distance and then using CCP4i within the CCP4 program suite (46) to form the linkage. The final model (resolution of 2.3 Å, Rcryst of 20.0% ,and Rfree of 25.2%) contains 5032 atoms with 2 molecules in the asymmetric unit. Electron density corresponding to residues 102–117 is not seen, and density around residues 8–12 and 283–286 is poor. Data collection and refinement statistics are shown in an abbreviated form in Table 2 (and in Supporting Information Table 1), which presents data collection statistics for the peak wavelength of MAD data sets. The coordinates have been deposited in the Protein Data Bank under accession codes 1u60 (YbaS), 1mki (YbgJ), and 3brm (YbgJ + DON complex).
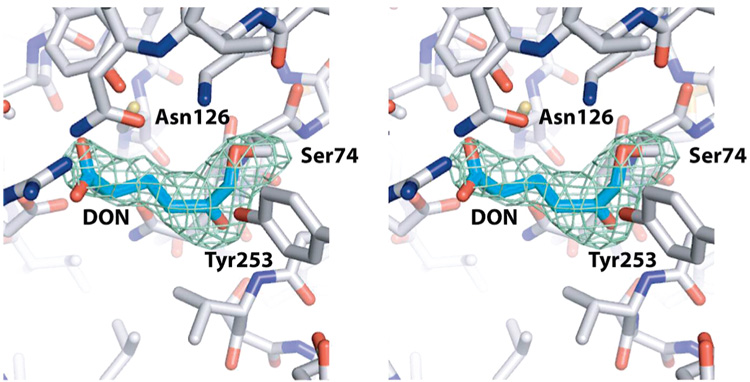
Figure 6.
Stereoview of the catalytic site of the YbgJ–DON complex. The omit map was generated by omitting both DON residues from the model and replacing both Ser74 residues by glycine. Green density represents the resulting Fo − Fc map contoured at 2.3σ. This map is shown around the model of the YbgJ/DON complex (not showing bound water molecules); protein residues (white carbon atoms) around the covalently bound DON (cyan carbon atoms) of molecule A in the asymmetric unit are shown as a stick representation, and several residues in contact with DON are labeled.
Table 2.
Crystallographic Data Collection and Model Refinement Statisticsa
| YbaSb tetragonal | YbaS (1u60) | YbgJb (1mki) | YbgJ + DON (3brm) | |
|---|---|---|---|---|
| Data Collection | ||||
| space group | I4 | P212121 | P21212 | P21212 |
| cell dimensions | ||||
| a(Å) | 239.8 | 50.5 | 71.3 | 71.2 |
| b(Å) | 155.9 | 81.5 | 184.7 | |
| c(Å) | 50.1 | 164.2 | 51.5 | 51.4 |
| wavelength | 0.97948 | 0.97978 | 0.9793 | 0.9793 |
| resolution (Å) | 50–1.8 (1.86–1.8) | 50–1.8 (1.86–1.8) | 50–2.0 (2.07–2.0) | 50–2.29 (2.38–2.29) |
| Rsym or Rmerge | 0.079 (0.388) | 0.046 (0.153) | 0.109 (0.502) | 0.138 (0.521) |
| I/σI | 27.7 (2.2) | 25.6 (5.4) | 5.4 (3.7) | 5.2 (1.9) |
| completeness (%) | 97.9 (83.4) | 93.2 (75.2) | 99.1 (98.5) | 97.6 (83.8) |
| redundancy | 6.8 (2.8) | 3.9 (3.0) | 10.2 (9.6) | 6.3 (4.0) |
| Refinement | ||||
| resolution (Å) | 40–1.80 | 40.7–2.0 | 34.5–2.29 | |
| no. of reflections | 107720/5686 | 43940/4346 | 24770/1317 | |
| Rwork/Rfree | 0.143/0.178 | 0.212/0.245 | 0.174/0.248 | |
| no. of atoms | ||||
| protein | 9258 | 4844 | 4500 | |
| major ligand | 75 | 16 | 20 | |
| solvent | 1150 | 333 | 295 | |
| average B-factors | ||||
| overall | 19.2 | 31.3 | 32.9 | |
| protein | 17.2 | 30.9 | 32.3 | |
| waters | 33.8 | 36.4 | 42.2 | |
| ligand/otherc | 39.9 | 42.6 | 42.4 | |
| Wilson B-factor | 17.1 | 15.2 | 32.4 | |
| rms deviations | ||||
| bond lengths (Å) | 0.015 | 0.006 | 0.018 | |
| bond angles (deg) | 1.4 | 1.3 | 1.7 | |
| Ramachandran plot | ||||
| % in most favored regions | 90.8 | 89.6 | 92.6 | |
| % in additionally allowed regions | 8.0 | 8.9 | 6.5 | |
| % in disallowed regions | 0.4 | 0.6 | 0.0 | |
Values in parentheses are for the highest resolution shell.
Data collection statistics are reported for data collection at the selenium peak wavelength; data were also collected for the inflection and remote wavelengths, but these are reported in the Supporting Information table.
In the YbgJ + DON covalent complex, this refers to the average temperature factors for the ligand 5-oxo-l-norleucine. In the other glutaminase structures, there was no ligand, but molecules of 1,2-ethylene glycol and formate ions were found.
RESULTS AND DISCUSSION
Sequence Analysis of Glutaminases
Glutaminases are widely distributed in bacteria and eukaryotes but seem to be absent in archaea, thermophiles, and plants (1). Many sequenced genomes contain two genes encoding predicted glutaminases of the β-lactamase superfamily. The E. coli genome encodes two predicted glutaminases, YbaS (P77454, GlsA1, 310 amino acids) and YneH (P0A6W0, GlsA2, 308 amino acids), which share 38% sequence identity. The YbaS gene is located upstream and cotranscribed with ybaT encoding an uncharacterized amino acid transporter (APC superfamily), whereas the YneH gene seems to comprise a single gene operon (EcoCyc database, http://www.ecocyc.org/). Likewise, the B. subtilis genome has two genes encoding predicted glutaminases, YbgJ (O31465, GlsA1, 327 amino acids) and YlaM (O07637, GlsA2, 309 amino acids) with 44% sequence identity to each other and 33–38% sequence identity to predicted E. coli glutaminases. The presence of glutaminase activity in YbgJ was recently reported (26). The B. subtilis ybgJ is located upstream and cotranscribed with a gene encoding the glutamine transporter YbgH (26). Sequence alignment of the predicted glutaminases from E. coli and B. subtilis with the sequences of several known glutaminases revealed the presence of over 40 absolutely conserved residues including the predicted β-lactamase motif 1 (2), a catalytic diad Ser-X-X-Lys (Ser66-X-X-Lys69 in YbaS) (Figure 1). The β-lactamase sequence motif 3 (Lys/Arg-Ser/Thr-Gly) (2) was also recognizable in glutaminases (Lys259-Ser260-Gly261 in YbaS), whereas only Ser (Ser160 in YbaS) could be identified for the Ser-Asp-Asn triad of the class A β-lactamase motif 2 (Figure 1). The class C β-lactamases contain a conserved Tyr residue (Tyr150 in AmpC from Enterobacter cloacae) instead of Ser in motif 2 (2), which also has no obvious counterpart in the glutaminase sequences. Thus, sequence analysis indicates that glutaminases retained motifs 1 and 3 of β-lactamases but differ in motif 2.
Figure 1.
Structure-based sequence alignment of β-lactamase-like glutaminases from several bacteria and humans. The secondary structure elements are shown above (YbaS) and below (YbgJ) the alignment. Highly conserved residues are shaded and boxed. Residues comprising the β-lactamase signature motifs I, II, and III are marked by asterisks. The compared glutaminases are E. coli YbaS (P77454), E. coli YneH (P0A6W0), M. luteus GlsA (Q4U1A6), H. sapiens GlsK (O94925), H. sapiens GlsL (Q9UI32), R. etli GlsA (O87405), B. subtilis YlaM (O07637), and B. subtilis YbgJ (O31465).
Protein Purification and Oligomeric State
Genes encoding the four predicted glutaminases from E. coli (YbaS and YneH) and B. subtilis (YbgJ and YlaM) were overexpressed in E. coli, and the recombinant proteins were affinity purified (see Experimental Procedures for details). All four proteins were expressed in soluble form, and a one-step purification using metal-chelate affinity chromatography on a Ni2+-NTA affinity resin produced glutaminase preparations with over 95% homogeneity as assessed by SDS–PAGE gels (not shown). Gel filtration analysis (data not shown) of the oligomeric state of purified proteins revealed that both YbaS (32.9 kDa) and YbgJ (36.2 kDa) were tetramers in solution (148.8 and 138.1 kDa, respectively), whereas YneH (33.5 kDa) and YlaM (34.0 kDa) were dimers (74.9 and 72.6 kDa, respectively).
Enzymatic Studies
All purified proteins exhibited glutaminase activity when assayed in an enzyme-coupled assay with glutamate dehydrogenase, which detects the formation of free NH4+. Glutaminase activity of YbgJ was also confirmed using the chromogenic substrate glutamate-p-nitroanilide, but the rate of hydrolysis of this substrate [0.3 µmol min−1 (mg of protein)−1] was 400–450 times lower than that of l-glutamine. With l-glutamine as a substrate, all glutaminases expressed maximal activity at pH 7.5–8.0 (Supporting Information Figure 1). However, the glutaminase activity of YbaS was more resistant to low pH than in other glutaminases, and this enzyme exhibited significant activity even at pH 4.5 (Supporting Information Figure 1A). On the basis of the pH profiles of glutaminase activity, we propose that the E. coli YbaS and YneH correspond to glutaminases A and B, respectively, that were identified in this organism over 30 years ago (21, 22, 47).
All glutaminases were highly specific to l-glutamine as a substrate and showed no activity against l-asparagine or d-glutamine. With l-glutamine, all four glutaminases demonstrated a sigmoidal saturation curve, with a Hill coefficient nH = 1.8–2.8 indicating positive cooperativity in glutamine binding. This observation is consistent with the oligomeric state of glutaminases in solution (dimers and tetramers). The E. coli YbaS and B. subtilis YlaM exhibited higher affinity to glutamine (Km 7.3 and 7.6 mM, respectively) than the E. coli YneH and B. subtilis YbgJ (Km 30.6 and 27.6 mM, respectively) (Table 1). Biochemically characterized glutami-nases from various organisms show a broad range of Km for l-glutamine (0.1–21.0 mM) and kcat/Km values (6.1 × 103 to 11.6 × 10 (5) M−1 s−1) (BRENDA database; http://www.brenda-enzymes.org/). Therefore, the glutaminases from E. coli and B. subtilis have kinetic parameters similar to those of other glutaminases. Thus, glutaminases exhibit low affinity to l-glutamine (high K m), but at the same time they are highly selective for this substrate and show no activity against d-glutamine or l-asparagine. Low substrate affinity and high substrate selectivity were also observed in glutamine synthetases (EC 6.3.1.2; Km for glutamate up to 50 mM), glucokinases (EC 2.7.1.2; Km for glucose up to 24 mM), and high-Km phosphodiesterases (EC 3.1.4.17; Km up to 3 mM) (BRENDA database; http://www.brenda-enzymes.org/).
Table 1.
Kinetic Parameters of Glutaminases from E. coli (YbaS and YneH) and B. subtilis (YbgJ and YlaM) with Glutamine as a Variable Substrate
| protein | Km (mM) | Vmax (units/mg)a | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| YbaS (wt)b | 7.3 ± 0.3 | 153.5 ± 2.9 | 91.4 ± 1.7 | 12.5 × 103 |
| YneH (wt) | 30.6 ± 2.7 | 169.7 ± 6.4 | 101.0 ± 3.8 | 3.4 × 103 |
| YlaM (wt) | 7.6 ± 0.3 | 69.9 ± 1.1 | 38.8 ± 0.6 | 5.1 × 103 |
| YbgJ (wt) | 27.6 ± 1.6 | 113.7 ± 3.2 | 67.7 ± 1.9 | 2.45 × 103 |
| YbaS (Q162A) | 8.5 ± 0.4 | 134.3 ± 2.7 | 79.9 ± 1.6 | 9.4 × 103 |
| YbaS (G261A) | 23.7 ± 1.7 | 83.9 ± 2.9 | 49.9 ± 1.7 | 2.1 × 103 |
| YbaS (wt + 1.5 mM Amp)c | 10.7 ± 0.7 | 138.6 ± 4.9 | 82.5 ± 2.9 | 7.7 × 103 |
| YbaS (wt + 3.0 mM Amp) | 12.3 ± 0.8 | 142.2 ± 4.7 | 84.6 ± 2.8 | 6.9 × 103 |
Units/mg = µmol min−1 (mg of protein)−1.
wt = wild type.
Amp = ampicillin.
In contrast to the salt-tolerant glutaminase from M. luteus (13), the rate of glutamine hydrolysis by the E. coli YbaS was not stimulated by NaCl (up to 0.2 M) and dropped two times in the presence of 0.5 M NaCl (not shown). Moreover, on the contrary to the phosphate-activated glutaminases from mammalians and B. pasteurii (12, 17), the activity of the E. coli YbaS and YneH was inhibited by low concentrations of PO43− (IC50 = 17.2–17.4 mM) (Supporting Information Figure 2). All four glutaminases were also inhibited by low concentrations of divalent metal cations, Mg 2+ and Mn2+ (Supporting Information Figure 1). YbaS was ~10 times more sensitive to these cations (IC50 for Mg2+ = 0.2 mM and IC50 for Mn2+ = 0.1 mM) than the other three glutaminases (IC50 = 1.7–5.5 mM for Mg2+ and 0.7–1.3 mM for Mn2+).
The glutamine analogue 6-diazo-5-oxo-l-norleucine (DON) has been shown to bind and irreversibly inactivate glutaminase–asparaginases from several organisms at low concentrations (60–100 µM) (48, 49). Crystal structures of three glutaminase–asparaginases complexed with DON demonstrated that this inhibitor becomes covalently attached to the catalytic threonine residue in these enzymes (49–51). Whereas glutaminases from the β-lactamase superfamily were also shown to be inhibited by DON, these enzymes exhibited much lower sensitivity to this inhibitor (21). Our work with highly purified glutaminases revealed that even high concentrations of DON (2–10 mM) produced incomplete (45–90%) inhibition of glutaminase activity of YbaS or YbgJ, suggesting that they employ a catalytic mechanism different from that of glutaminase–asparaginases.
Highly purified preparations of the E. coli YbaS showed low, but detectable β-lactamase activity [24.4–30.5 nmol min −1 (mg of protein)−1] against nitrocefin, a general β-lactamase substrate (52). This activity could not be eliminated by additional purification steps (by gel filtration or using the second Ni column after the cleavage of the His tag) but was not detectable in the catalytically inactive YbaS mutants (K69A, S160A, K259A). In addition, glutaminase activity of YbaS was inhibited by the β-lactam antibiotics penicillin (Ki = 6.4 mM) or ampicillin (Ki = 3.5 mM) (Supporting Information Figure 3A). Kinetic analysis of the YbaS glutaminase activity in the presence of ampicillin demonstrated that this antibiotic acted as a competitive inhibitor of glutamine hydrolysis (Supporting Information Figure 3B) and induced an increase in Km for glutamine without affecting its kcat (Table 1). As well, the commercial preparation of the E. cloaceae penicillinase (a class C β-lactamase) exhibited low, but detectable glutaminase activity [20.7 ± 3.4 nmol min−1 (mg of protein)−1]. Thus, the obtained results indicate that YbaS retained a low level of β-lactamase activity and the β-lactamase substrates are apparently capable of binding to the glutamine binding site and acting as competitive inhibitors of glutamine hydrolysis by glutaminases.
Crystal Structures of YbaS and YbgJ
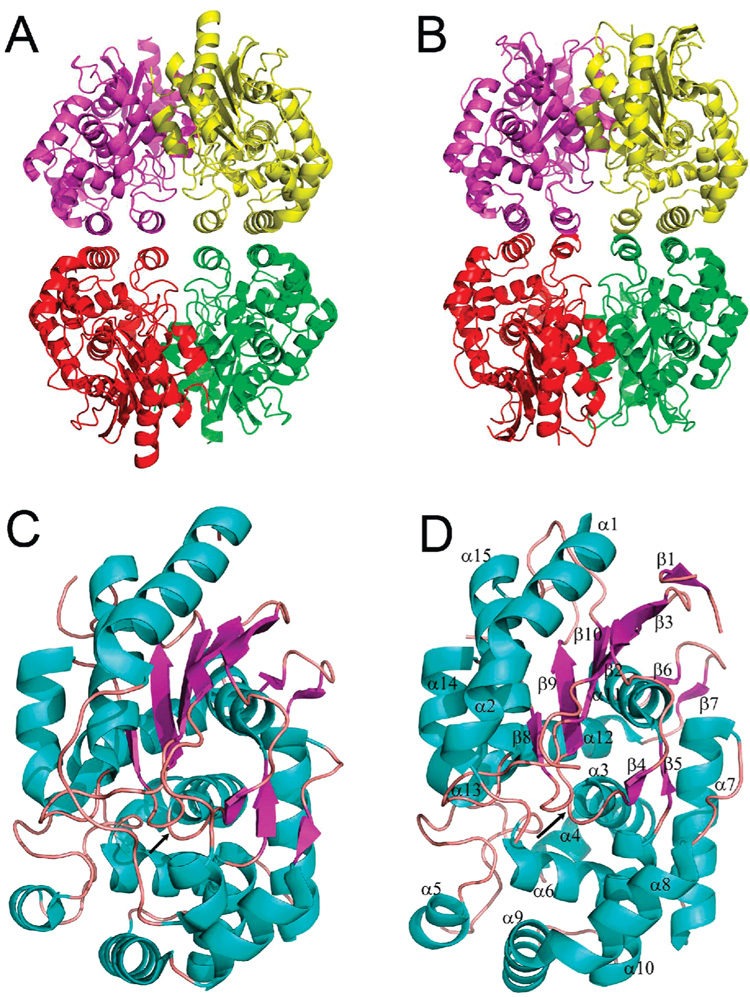
Crystal structures of unliganded YbaS (1u60) and YbgJ (1mki) were determined by MAD phasing and revealed a tetrameric protein arranged as a dimer of dimers with each subunit providing two α-helices (α6 and α10 in YbaS and α5 and α9 in YbgJ) for the dimer/dimer contact (Figure 2A,B). The monomers of both proteins have two compact domains: an α/β/α-sandwich tightly associated with a mostly α-helical domain. In the YbaS structure, five β-strands (β1, β2, β8, β9, and β10) make an antiparallel β-sheet flanked by four α-helices (α1, α15, α16, and α17) on the protein surface side and by the α-helix 12 on the opposite face (Figure 2C). YbgJ has a very similar structure, but its β-sheet contains six β-strands (Figure 2D). The recently solved crystal structure of a major fragment of the M. luteus K3 glutaminase revealed the presence of a C-terminal extension (141 amino acids) (15) representing a poorly characterized STAS domain (PROSITE entry PS50801).
Figure 2.
Crystal structures of glutaminases. Overall structure of the tetramers: YbaS (A) and YbgJ (B). Protein subunits are shown in different colors. Subunit structures of YbaS (C) and YbgJ (D) showing potential active sites. The secondary structure elements are shown in different colors (α-helices, cyan; β-strands, magenta; loops, salmon) and are numbered in the YbgJ structure (D). The potential active site is located in the area between α3, α6, and β8, and the position of the catalytic Ser (Ser66 in YbaS and Ser74 in YbgJ) is indicated by the black arrow in the center of both structures.
The search for structural homologues of YbaS using the DALI (53) (http://www.ebi.ac.uk/dali) and SSM (http://www.ebi.ac.uk/msd-srv/ssm) databases identified the B. subtilis YbgJ (1mki, this work), the M. luteus glutaminase (2dfw), and the putative glutaminase GlsA from Geobacillus kaustophilus (2pby) as the closest structural homologues (Z-score 10.9–12.3, rmsd 1.5–1.6 Å). These analyses also recognized a group of homologous structures of several β-lactamases and penicillin-binding proteins with lower structural similarity and Z-scores ranging from 3.8 to 5.8 (rmsd 2.3–2.8 Å; PDB codes 2c5w, 2bg4, 2bg3,2bg1, 2c6w, 2fff). All of these proteins have low sequence similarity to glutaminases (less than 25% sequence identity).
Putative Active Site
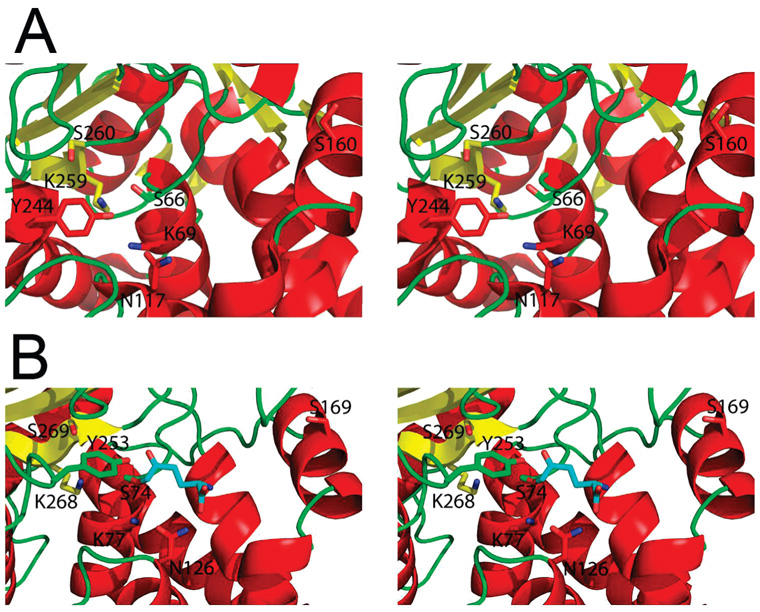
Crystal structures of YbaS and YbgJ revealed the presence of a deep cleft, whose walls are formed by the β-sheet and several α-helices of the helical domain (α3, α6, and α8) (Figure 2C,D). The YbgJ Ser74 (Ser66 in YbaS), which occurs within the β-lactamase motif 1 (Ser-X-X-Lys), is located at the bottom of this cleft and is likely to function as a catalytic nucleophile in the glutaminase reaction (Figure 3). As in the E. coli RTEM-1 β-lactamase (6), this Ser is surrounded by the side chains of three conserved residues located on the equivalent secondary structural elements: Lys77, Lys268, and Ser269 in YbgJ (Lys69, Lys259, and Ser260 in YbaS) (Figure 3). Lys77 is hydrogen bonded (2.8 Å) to the side chain of the conserved Asn126 (Asn117 in YbaS), which is equivalent to the RTEM-1 Asn132 (Figure 3). The YbgJ Asn126 (Asn117 in YbaS) is close to the conserved Asn177 (Asn168 in YbaS). Another YbgJ residue, Gly270 (Gly261 in YbaS), is conserved and located on the β7 strand in both β-lactamases and glutaminases. The absence of the side chain at this position is perhaps essential for the catalytic reaction of β-lactamases and glutaminases. The structure of YbgJ showed an additional small electron density close to the predicted catalytic nucleophile Ser74 which was interpreted as a covalently bound phosphate in the PDB submission (1mki). However, this density was not present in the structures of the YbgJ–DON complex or YbaS, and the mass spectrometric analysis of purified YbgJ produced no evidence for the presence of any covalent modification in this protein. Therefore, this additional density might actually represent a noncovalently associated solvent molecule.
Figure 3.
Close-up stereoview of the active sites of YbaS (A) and YbgJ with bound DON (B). The secondary structure elements are shown in different colors (α-helices, red; β-strands, yellow; loops, green). The side chains of the conserved catalytic residues and DON are shown as sticks (nitrogen atoms, blue; oxygens, red; the carbon atoms of DON, cyan). Note that YbgJ deamidates DON producing 5-oxo-l-norleucine (ON) covalently bound to the enzyme.
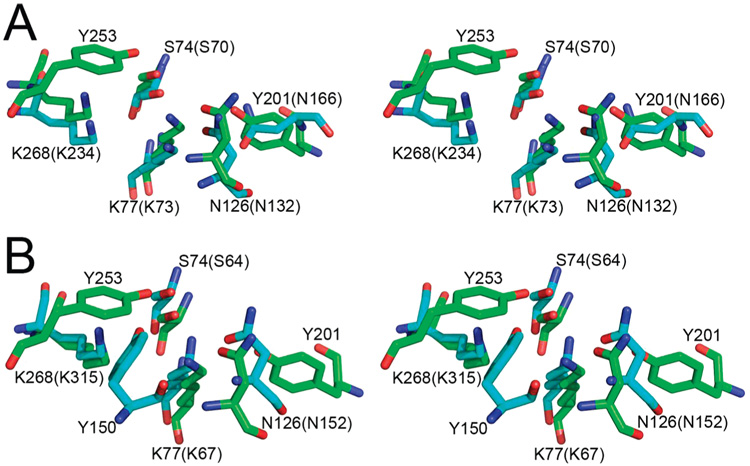
In contrast to β-lactamases, the active site of glutaminases accommodates the bulky side chains of three conserved tyrosine residues: Tyr37, Tyr201, and Tyr253 in YbgJ and Tyr29, Tyr192, and Tyr244 in YbaS. This is consistent with the smaller size of the glutaminase substrate. The YbgJ Tyr253 (Tyr244 in YbaS) occupies the position equivalent to that of the conserved Ser130 of the E. coli RTEM-1 β-lactamase (Figure 3). The latter residue is positioned close to the catalytic Ser70 and is involved in the proton transfer from Lys73 to the nitrogen atom of the substrate (6). In the class C β-lactamase from E. cloacae (AmpC), Tyr150 exists in the position occupied by Ser130 in class A enzymes (54) (Figure 4). In YbgJ, the side chain hydroxyl oxygen of Tyr253 is 3.0 Å away from the putative nucleophile Ser74 and 3.2 Å away from the side chain of conserved Lys77, which is homologous to Lys73 of RTEM-1 (Figure 4). While Tyr253 of YbgJ and Tyr150 of AmpC are not similar in orientation and located in different secondary structure elements, their oxygen atoms are only 2.14 Å apart (Figure 4), suggesting that they are functionally equivalent. Therefore, Tyr253 is likely involved in the proton relay pathway in YbgJ. Tyr201 is placed in another corner of the YbgJ catalytic cleft (4.8 Å from Ser74) and occupies the position equivalent to that of the RTEM-1 Glu166 (Asn166 is shown because the E166N mutant protein was used for crystallization) (6) (Figure 4). In class A β-lactamases, Glu166 functions as the general base of the deacylation step and coordinates the nucleophilic water molecule (6). Hence, Tyr201 might have the same role in the glutaminase reaction.
Figure 4.
Comparison of the active sites of glutaminases and β-lactamases. Stereoview of superpositions of the key catalytic residues in the active sites of YbgJ and (A) class A β-lactamase RTEM-1 from E. coli (1fqg) or (B) class C β-lactamase AmpC from E. cloacae P99 (1xx2). The side chains of the YbgJ residues are shown in green, and the β-lactamase residues are shown in cyan. The numbers of the YbgJ residues are shown, whereas those in parentheses refer to the equivalent residues of the class A β-lactamase RTEM-1 (A) or to the class C β-lactamase AmpC (B). There is no residue corresponding to Y253 in RTEM-1 or to Y201 in AmpC. The superpositions were done on the whole proteins using the combinatorial extension method (64). The YbgJ/RTEM-1 superposition had an rmsd of 3.5 Å, while the YbgJ/AmpC superposition had an rmsd of 3.3 Å.
Mutational Studies of YbaS
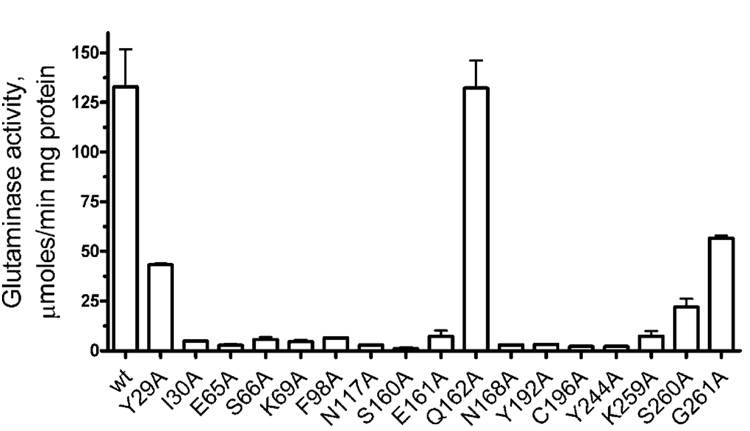
The putative catalytic cleft of glutaminases accommodates over a dozen residues absolutely conserved in these enzymes. To check the role of these residues in catalysis, we performed alanine replacement mutagenesis of YbaS using specifically designed PCR primers. Analysis of glutaminase activity of mutant proteins revealed that most mutations resulted in proteins with greatly reduced activity (Figure 5). S260A and Y29A had 16.5% and 32.5% of the wild-type activity, respectively, whereas all other mutants retained only 0.8–5.6% of activity. The G261A mutant retained 54.7% of activity (Vmax), but its affinity to glutamine dropped 3.2 times (Km increased to 23.7 mM) (Table 1). This indicates that the introduction of even the small Ala side chain to the position of the conserved Gly261 affects mainly the binding of glutamine to the enzyme active site. Glu162 is not conserved in glutaminases, and the YbaS E162A mutant showed the wild-type level of activity and substrate affinity (Figure 5 and Table 1). Thus, the putative catalytic cleft described above and conserved residues are likely to represent the glutaminase active site.
Figure 5.
Alanine replacement mutagenesis of YbaS: glutaminase activity of purified mutant proteins. The reaction mixtures contained 25 mM glutamine and 0.2 µg of YbaS, and the continuous assays were performed as described in Experimental Procedures.
Structure of the YbgJ–DON Complex and Substrate Binding in Glutaminases
YbgJ was also crystallized in the presence of the glutaminase inhibitor DON, and the structure of the YbgJ–DON complex was solved independently of the apo-YbgJ structure using SAD phasing to a resolution of 2.3 Å (3brm) (Figure 3B and Figure 6). The structure revealed continuous electron density connecting the inhibitor to the side chain of Ser74 (Ser66 in YbaS), indicating the presence of a covalent bond between these moieties (1.3 Å). Previous studies with the Pseudomonas 7A glutaminase–asparaginase demonstrated that this enzyme removes the diazo group of DON, releasing N2 and forming two covalent bonds between the inhibitor C5 (Cδ) and the hydroxyl oxygens of Thr20 and Tyr34 (50). In the structure of the YbgJ–DON complex, the Tyr253 side chain hydroxyl oxygen is positioned quite close to the inhibitor C6 (Cε) atom (2.2 Å), but the structure only shows the covalent bond between this atom and the Ser74 hydroxyl oxygen (1.3 Å) (Figure 3B and Figure 6). The formation of this covalent enzyme–inhibitor complex was previously predicted for deamidases but was never observed experimentally (50). It appears that YbgJ removes the diazo group of DON, producing N2 and 5-oxo-l-norleucine (ON) covalently bound to the Ser74 side chain through its terminal (C6) carbon atom. In the crystallization solution, this linkage cannot be further processed and thus produces a stable acylenzyme intermediate.
The YbgJ–DON (actually YbgJ–ON) complex is further stabilized by interactions between the C5 (Cδ) carbonyl oxygen of the inhibitor and the main chain NH groups of Ser74 (3.1 Å) and Val271 (2.6 Å) (oxyanione hole) (Figure 3B). The α-carboxyl oxygens of ON are coordinated by interactions with the side chains of Asn126 (3.0 Å) and Tyr201 (2.6 Å), whereas its α-amido group interacts with the side chains of Gln73 (2.8 Å) and Glu170 (2.8 Å).
Thus, we propose that the side chain Oε1 and Nε1 of l-glutamine bind to the YbgJ active site near the catalytic nucleophile Ser74 (Ser66 in YbaS) (Figure 7). The side chain δ-carbonyl oxygen of glutamine might be coordinated by the oxyanion hole formed by the main chain NH groups of Ser74 and Val271 (Ser66 and Val262 in YbaS). The side chain of Tyr253 (Tyr244 in YbaS), which is close to Ser74 (2.6 Å), is likely to be hydrogen bonded to the glutamine Nε2 (the leaving group) (Figure 7). The α-amino group of glutamine is predicted to be coordinated by hydrogen bonds with the side chains of Tyr37, Gln73, and Glu170 (Tyr29, Glu65, and Glu161 in YbaS), whereas the glutamine α-carboxyl oxygens are likely to be hydrogen bonded with the side chains of Asn126 and Asn177 of YbgJ (Asn117 and Asn168 in YbaS) (Figure 7).
Figure 7.
Proposed model of the binding of l-glutamine in the active site of YbgJ.
Implications for the Potential Catalytic Mechanism of Glutaminases
The conservation of overall β-lactamase structure and several β-lactamase catalytic residues in glutaminases, as well as the presence of low β-lactamase activity in YbaS, suggests that glutaminases use a β-lactamase-like catalytic mechanism for the deamidation of glutamine. The proposed model of the glutaminase catalytic mechanism is based on the mechanism of the E. coli RTEM-1 β-lactamase described by Strynadka et al. (6) and proposes the formation of a glutamyl-enzyme covalent intermediate at the Ser74 Oγ acting as a catalytic nucleophile (Figure 8). The important role of this residue in glutaminase catalysis is supported by the very low activity of the YbaS S66A mutant protein (Figure 5). Lys77 is likely to assist in the nucleophilic attack by acting as a general base, thereby accepting the proton from the Ser74 Oγ and transferring it to the Tyr253 side chain oxygen. This results in the tetrahedral intermediate-1 (Figure 8). The polarization of the hydrolyzable C–N bond of glutamine is enhanced by the hydrogen bonds from the oxyanion hole (the main chain NH groups of Ser74 and Val271) to the carbonyl oxygen atom Oε1. The formation of a glutamyl-enzyme intermediate is induced by the proton transfer from Tyr253 to the substrate Nε2 (the leaving group) and accompanied by the release of ammonia, the first reaction product (Figure 8). Similar to β-lactamases, the deacylation of a glutamyl-enzyme intermediate is likely accomplished by a general base-assisted nucleophilic attack of a deacylating water molecule on the ester carbonyl carbon of the intermediate resulting in tetrahedral intermediate-2 (Figure 8). In contrast to the general base Glu166 of RTEM-1 (4.3 Å from Ser70), in glutaminases this function is expected to be performed by the conserved Tyr201 (4.8 Å from Ser74). The Tyr201 side chain oxygen accepts the proton from the deacylating water molecule and transfers it to the Ser74 Oγ, thereby regenerating the active site (Figure 8).
Figure 8.
Possible reaction mechanism of the hydrolysis of l-glutamine by glutaminases. The reaction proceeds through the following steps: (1) nucleophilic attack of the Ser74 oxygen to form the tetrahedral intermediate-1; (2) decomposition of the tetrahedral intermediate-1 with the production of the glutamyl-enzyme intermediate and ammonia; (3) nucleophilic attack of water and the formation of the tetrahedral intermediate-2; (4) release of the second product (glutamate) and formation of free enzyme.
Physiological Role of Glutaminases
The main cellular function of glutaminases was proposed to be associated with the control of the intracellular pool of glutamine, which represents the central nitrogen metabolite in most organisms (17, 55, 56). Depending on the growth conditions the intracellular concentrations of glutamine in enterobacteria and Bacillus can vary in a wide range: from 0.02 to 27.0 mM (57–62). Therefore, the substrate affinities of both lower and higher affinity glutaminases from E. coli and B. subtilis fall within this range, and both forms can play a significant role in the intracellular glutamine metabolism. In B. subtilis, YbgJ was shown to be involved in the assimilation of glutamine, and the expression of its gene is activated by the GlnK-GlnL two-component system in response to glutamine (26). Moreover, increased resistance of enzymatic activity of the E. coli YbaS to low pH in vitro (Supporting Information Figure 1A) and elevated expression of its gene in response to acid shock in vivo (63) imply that this glutaminase might represent an additional component of the acid resistance system in E. coli. Our preliminary results with E. coli glutaminase deletion strains show that on M9 minimal medium with glutamine as a sole source of nitrogen and carbon the Δ yneH and Δ ybaS, Δ yneH double deletion strains grew two times slower than the wild-type or Δ ybaS strains (data not shown). In addition, both E. coli glutaminase deletion strains demonstrated a reduced growth rate during the exponential growth phase in M9 minimal medium with glucose (0.4%), glutamine (20 mM), and pH 4.5–5.0 (data not shown). These results suggest that E. coli glutaminases are involved in the assimilation of extracellular glutamine and might also contribute to acid resistance. Future work is required to further characterize the catalytic mechanism and role of glutaminases in microbial metabolism.
ACKNOWLEDGMENT
We thank all members of the Ontario Centre for Structural Proteomics at the University of Toronto and the Structural Biology Center at Argonne National Laboratory for help in conducting these experiments. James Watson (EBI) is thanked for the discussion of the glutaminase sequence motifs.
Footnotes
This work was supported by grants from Genome Canada (through the Ontario Genomics Institute), the National Institutes of Health (Grant GM074942), and the Department of Energy, Office of Biological and Environmental Research (under Contract DE-AC02-06CH11357).
Abbreviations: DON, 6-diazo-5-oxo-L-norleucine; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; MAD, multiple-wave-length anomalous diffraction; MES, 2-(N-morpholino)ethanesulfonic acid; ON, 5-oxo-l-norleucine; SAD, single anomalous diffraction; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
SUPPORTING INFORMATION AVAILABLE
Three figures showing the effect of pH, divalent metal cations, phosphate, and β-lactam antibiotics on glutaminase activity of YbaS and YbgJ, as well as one table with complete MAD data collection statistics for Ybas and YbgJ. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Nandakumar R, Yoshimune K, Wakayama M, Moriguchi M. Microbial glutaminase: biochemistry, molecular approaches and applications in the food industry. J. Mol. Catal. B: Enzym. 2003;23:87–100. [Google Scholar]
- 2.Massova I, Mobashery S. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob. Agents Chemother. 1998;42:1–17. doi: 10.1128/aac.42.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghuysen JM. Serine beta-lactamases and penicillin-binding proteins. Annu. ReV. Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 4.Fisher JF, Meroueh SO, Mobashery S. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 5.Wilke MS, Lovering AL, Strynadka NC. Beta-lactam antibiotic resistance: a current structural perspective. Curr. Opin. Microbiol. 2005;8:525–533. doi: 10.1016/j.mib.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 6.Strynadka NC, Adachi H, Jensen SE, Johns K, Sielecki A, Betzel C, Sutoh K, James MN. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 Å resolution. Nature. 1992;359:700–705. doi: 10.1038/359700a0. [DOI] [PubMed] [Google Scholar]
- 7.Gordon E, Mouz N, Duee E, Dideberg O. The crystal structure of the penicillin-binding protein 2x from Streptococcus pneumoniae and its acyl-enzyme form: implication in drug resistance. J. Mol. Biol. 2000;299:477–485. doi: 10.1006/jmbi.2000.3740. [DOI] [PubMed] [Google Scholar]
- 8.Paetzel M, Danel F, de Castro L, Mosimann SC, Page MG, Strynadka NC. Crystal structure of the class D beta-lactamase OXA-10. Nat. Struct. Biol. 2000;7:918–925. doi: 10.1038/79688. [DOI] [PubMed] [Google Scholar]
- 9.Ibuka AS, Ishii Y, Galleni M, Ishiguro M, Yamaguchi K, Frere JM, Matsuzawa H, Sakai H. Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for catalytic reaction and substrate specificity expansion. Biochemistry. 2003;42:10634–10643. doi: 10.1021/bi0342822. [DOI] [PubMed] [Google Scholar]
- 10.Nandakumar R, Wakayama M, Nagano Y, Kawamura T, Sakai K, Moriguchi M. Overexpression of salt-tolerant glutaminase from Micrococcus luteus K-3 in Escherichia coli and its purification. Protein Expression Purif. 1999;15:155–161. doi: 10.1006/prep.1998.1005. [DOI] [PubMed] [Google Scholar]
- 11.Huerta-Saquero A, Calderon J, Arreguin R, Calderon-Flores A, Duran S. Overexpression and purification of Rhizobium etli glutaminase A by recombinant and conventional procedures. Protein Expression Purif. 2001;21:432–437. doi: 10.1006/prep.2001.1394. [DOI] [PubMed] [Google Scholar]
- 12.Klein M, Kaltwasser H, Jahns T. Isolation of a novel, phosphate-activated glutaminase from Bacillus pasteurii. FEMS Microbiol Lett. 2002;206:63–67. doi: 10.1111/j.1574-6968.2002.tb10987.x. [DOI] [PubMed] [Google Scholar]
- 13.Moriguchi M, Sakai K, Tateyama R, Furuta Y, Wakayama M. Isolation and characterization of salt-tolerant glutaminases from marine Micrococcus luteus K-3. J. Ferment. Bioeng. 1994;77:621–625. [Google Scholar]
- 14.Weingand-Ziade A, Gerber-Decombaz C, Affolter M. Functional characterization of a salt- and thermotolerant glutaminase from Lactobacillus rhamnosus. Enzyme Microb. Technol. 2003;32:862–867. [Google Scholar]
- 15.Yoshimune K, Shirakihara Y, Shiratori A, Wakayama M, Chantawannakul P, Moriguchi M. Crystal structure of a major fragment of the salt-tolerant glutaminase from Micro-coccus luteus K-3. Biochem. Biophys. Res. Commun. 2006;346:1118–1124. doi: 10.1016/j.bbrc.2006.04.188. [DOI] [PubMed] [Google Scholar]
- 16.Yano S, Kamemura A, Yoshimune K, Moriguchi M, Yama-moto S, Tachiki T, Wakayama M. Analysis of essential amino acid residues for catalytic activity of glutaminase from Micrococcus luteus K-3. J. Biosci. Bioeng. 2006;102:362–364. doi: 10.1263/jbb.102.362. [DOI] [PubMed] [Google Scholar]
- 17.Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 18.Aledo JC, Gomez-Fabre PM, Olalla L, Marquez J. Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mamm. Genome. 2000;11:1107–1110. doi: 10.1007/s003350010190. [DOI] [PubMed] [Google Scholar]
- 19.Kenny J, Bao Y, Hamm B, Taylor L, Toth A, Wagers B, Curthoys NP. Bacterial expression, purification, and characterization of rat kidney-type mitochondrial glutaminase. Protein Expression Purif. 2003;31:140–148. doi: 10.1016/s1046-5928(03)00161-x. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Gomez C, Campos-Sandoval JA, Alonso FJ, Segura JA, Manzanares E, Ruiz-Sanchez P, Gonzalez ME, Marquez J, Mates JM. Co-expression of glutaminase K and L isoenzymes in human tumour cells. Biochem. J. 2005;386:535–542. doi: 10.1042/BJ20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartman SC. Glutaminase of Escherichia coli I. Purification and general catalytic properties. J. Biol. Chem. 1968;243:853–863. [PubMed] [Google Scholar]
- 22.Prusiner S, Davis JN, Stadtman ER. Regulation of glutaminase B in Escherichia coli I. Purification, properties, and cold lability. J. Biol. Chem. 1976;251:3447–3456. [PubMed] [Google Scholar]
- 23.Prusiner S, Miller RE, Valentine RC. Adenosine 3′:5′-cyclic monophosphate control of the enzymes of glutamine metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1972;69:2922–2926. doi: 10.1073/pnas.69.10.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prusiner S. Regulation of glutaminase levels in Escherichia coli. J. Bacteriol. 1975;123:992–999. doi: 10.1128/jb.123.3.992-999.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neidhardt FC, Curtiss R. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, DC: ASM Press; 1996. p. 2. [Google Scholar]
- 26.Satomura T, Shimura D, Asai K, Sadaie Y, Hirooka K, Fujita Y. Enhancement of glutamine utilization in Bacillus subtilis through the GlnK-GlnL two-component regulatory system. J. Bacteriol. 2005;187:4813–4821. doi: 10.1128/JB.187.14.4813-4821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure. 2001;9:1095–1106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez CF, Proudfoot M, Brown G, Korniyenko Y, Mori H, Savchenko AV, Yakunin AF. Molecular basis of formaldehyde detoxification. Characterization of two S-formyl-glutathione hydrolases from Escherichia coli, FrmB and YeiG. J. Biol. Chem. 2006;281:14514–14522. doi: 10.1074/jbc.M600996200. [DOI] [PubMed] [Google Scholar]
- 29.Bergmeyer HU. Enzymatic analysis of the new generation. Fresenius’ Z. Anal. Chem. 1984;319:883–889. [Google Scholar]
- 30.Neeley WE, Phillipson J. Automated enzymatic method for determining ammonia in plasma, with 14-day reagent stability. Clin. Chem. 1988;34:1868–1869. [PubMed] [Google Scholar]
- 31.Heering HA, Weiner JH, Armstrong FA. Direct detection and measurement of electron relays in a multicentered enzyme: Voltammetry of electrode-surface films of E. coli fumarate reductase, an iron-sulfur flavoprotein. J. Am. Chem. Soc. 1997;119:11628–11638. [Google Scholar]
- 32.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat. Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 33.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1984. p. 15. [Google Scholar]
- 36.Tucker DL, Tucker N, Conway T. Gene expression profiling of the pH response in Escherichia coli. J. Bacteriol. 2002;184:6551–6558. doi: 10.1128/JB.184.23.6551-6558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimber MS, Vallee F, Houston S, Necakov A, Skarina T, Evdokimova E, Beasley S, Christendat D, Savchenko A, Arrowsmith CH, Vedadi M, Gerstein M, Edwards AM. Data mining crystallization databases: knowledge-based approaches to optimize protein crystal screens. Proteins. 2003;51:562–568. doi: 10.1002/prot.10340. [DOI] [PubMed] [Google Scholar]
- 38.Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang RG, Ginell SL, Duke N, Naday I, Lazarz J, Molitsky MJ, Keefe L, Gonczy J, Rock L, Sanishvili R, Walsh MA, Westbrook E, Joachimiak A. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J. Synchrotron Radiat. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Segraves EN, Chruszcz M, Neidig ML, Ruddat V, Zhou J, Wecksler AT, Minor W, Solomon EI, Holman TR. Kinetic, spectroscopic, and structural investigations of the soybean lipoxygenase-1 first-coordination sphere mutant, Asn694Gly. Biochemistry. 2006;45:10233–10242. doi: 10.1021/bi060577e. [DOI] [PubMed] [Google Scholar]
- 40.Naaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Jones TA, Zou J-Y, Cohen SW, Kjeldgaard M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 43.Winn M, Isupov M, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 44.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 45.Terwillinger T. SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 2003;374:22–37. doi: 10.1016/S0076-6879(03)74002-6. [DOI] [PubMed] [Google Scholar]
- 46.The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Prusiner S, Stadtman ER. Regulation of glutaminase B in Escherichia coli III. Control by nucleotides and divalent cations. J. Biol. Chem. 1976;251:3463–3469. [PubMed] [Google Scholar]
- 48.Roberts J, Holcenberg JS, Dolowy WC. Isolation, crystallization, and properties of Achromobacteraceae glutaminase-asparaginase with antitumor activity. J. Biol. Chem. 1972;247:84–90. [PubMed] [Google Scholar]
- 49.Holcenberg JS, Ericsson L, Roberts J. Amino acid sequence of the diazooxonorleucine binding site of Acinetobacter and Pseudomonas 7A glutaminase-asparaginase enzymes. Biochemistry. 1978;17:411–417. doi: 10.1021/bi00596a005. [DOI] [PubMed] [Google Scholar]
- 50.Ortlund E, Lacount MW, Lewinski K, Lebioda L. Reactions of Pseudomonas 7A glutaminase-asparaginase with diazo analogues of glutamine and asparagine result in unexpected covalent inhibitions and suggests an unusual catalytic triad Thr-Tyr-Glu. Biochemistry. 2000;39:1199–1204. doi: 10.1021/bi991797d. [DOI] [PubMed] [Google Scholar]
- 51.Aghaiypour K, Wlodawer A, Lubkowski J. Do bacterial l-asparaginases utilize a catalytic triad Thr-Tyr-Glu? Biochim Biophys. Acta. 2001;1550:117–128. doi: 10.1016/s0167-4838(01)00270-9. [DOI] [PubMed] [Google Scholar]
- 52.Bebrone C, Moali C, Mahy F, Rival S, Docquier JD, Rossolini GM, Fastrez J, Pratt RF, Frere JM, Galleni M. CENTA as a chromogenic substrate for studying beta-lactamases. Antimicrob. Agents Chemother. 2001;45:1868–1871. doi: 10.1128/AAC.45.6.1868-1871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 54.Lobkovsky E, Moews PC, Liu H, Zhao H, Frere JM, Knox JR. Evolution of an enzyme activity: crystal-lographic structure at 2-Å resolution of cephalosporinase from the ampC gene of Enterobacter cloacae P99 and comparison with a class A penicillinase. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11257–11261. doi: 10.1073/pnas.90.23.11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medina MA. Glutamine and cancer. J. Nutr. 2001;131:2539S–2542S. doi: 10.1093/jn/131.9.2539S. (discussion 2550S–2551S) [DOI] [PubMed] [Google Scholar]
- 56.Prusiner SB, Stadtman ER. The enzymes of glutamine metabolism. New York: Academic Press; 1973. [Google Scholar]
- 57.Schutt H, Holzer H. Biological function of the ammonia-induced inactivation of glutamine synthetase in Escherichia coli. Eur. J. Biochem. 1972;26:68–72. doi: 10.1111/j.1432-1033.1972.tb01740.x. [DOI] [PubMed] [Google Scholar]
- 58.Fisher SH, Sonenshein AL. Bacillus subtilis glutamine synthetase mutants pleiotropically altered in glucose catabolite repression. J. Bacteriol. 1984;157:612–621. doi: 10.1128/jb.157.2.612-621.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Osorio AV, Camarena L, Salazar G, Noll-Louzada M, Bastarrachea F. Nitrogen regulation in an Escherichia coli strain with a temperature sensitive glutamyl-tRNA synthetase. Mol. Gen. Genet. 1993;239:400–408. doi: 10.1007/BF00276938. [DOI] [PubMed] [Google Scholar]
- 60.Ikeda TP, Shauger AE, Kustu S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 61.Hu P, Leighton T, Ishkhanova G, Kustu S. Sensing of nitrogen limitation by Bacillus subtilis: comparison to enteric bacteria. J. Bacteriol. 1999;181:5042–5050. doi: 10.1128/jb.181.16.5042-5050.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Javelle A, Severi E, Thornton J, Merrick M. Ammonium sensing in Escherichia coli. Role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J. Biol. Chem. 2004;279:8530–8538. doi: 10.1074/jbc.M312399200. [DOI] [PubMed] [Google Scholar]
- 63.Tucker DL, Tucker N, Ma Z, Foster JW, Miranda RL, Cohen PS, Conway T. Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 2003;185:3190–3201. doi: 10.1128/JB.185.10.3190-3201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]