Abstract
The natural reprogramming of the mammalian egg and sperm genomes is an efficient process that takes place in less than 24 hours and gives rise to a totipotent zygote. Transfer of somatic nuclei to mammalian oocytes also leads to their reprogramming and formation of totipotent embryos, albeit very inefficiently, and requiring an activation step. Reprogramming of differentiated cells to induced-pluripotent stem (iPS) cells takes place over a period of time substantially longer than reprogramming of the egg and sperm nuclei, and is significantly less efficient. The stochastic expression of endogenous proteins during this process would imply that controlled expression of specific proteins is crucial for reprogramming to take place. The fact that OCT4, NANOG and SOX2 form the core components of the pluripotency circuitry would imply that control at the transcriptional level is important for reprogramming to iPS cells. In contradistinction, the much more efficient reprogramming of the mammalian egg and sperm genomes implies other levels of control are necessary, such as chromatin remodeling, translational regulation and efficient degradation of no-longer-needed proteins and RNAs.
Reprogramming of differentiated cells into stem cells occurs at the outset of each new generation. Sperm and egg nuclei become reprogrammed in the cytoplasm of the fertilized oocyte to form the totipotent blastomeres, the newly formed embryo. JB Gurdon was the first to report that adult Xenopus could be derived by transferring a single somatic cell nucleus into the cytoplasm of a Xenopus oocyte (Gurdon et al., 1958). It was not until almost forty years later that reprogramming of a mammalian somatic cell nucleus to totipotency in a mammalian oocyte was demonstrated by the birth of Dolly the sheep (Campbell et al., 1996). Reports of reprogramming of differentiated cells into induced pluripotent stem cells (iPS) have again challenged the existing knowledge of how reprogramming is accomplished (Jaenisch and Young, 2008). Determining the molecular circuitry of pluripotency by looking at iPS gave some insight into this problem. However, another approach would be to expand the knowledge base of how two differentiated cells, the sperm and egg, are reprogrammed to give rise to the totipotent embryo. To this day the maternal messages, and the mechanisms controlling their translation, that accomplish this natural embryonic reprogramming in mammals are only loosely understood.
Embryogenesis in metazoans
Early embryogenesis in metazoans varies at the anatomical level, but in each model organism the ooplasm contents control: oocyte maturation; reprogramming of the sperm and egg genomes; activation of the embryonic genome; and one or more mitotic divisions. Reprogramming of the sperm and egg genomes in metazoans has one purpose that is to set up totipotent stem cells to give rise to the new organism. Information gleaned from determining the regulation of maternal gene expression in the model organisms Drosophila, C. elegans, Xenopus, Danio rerio and mouse have shed some light on the molecules potentially involved in reprogramming, but by and large they are still unknown.
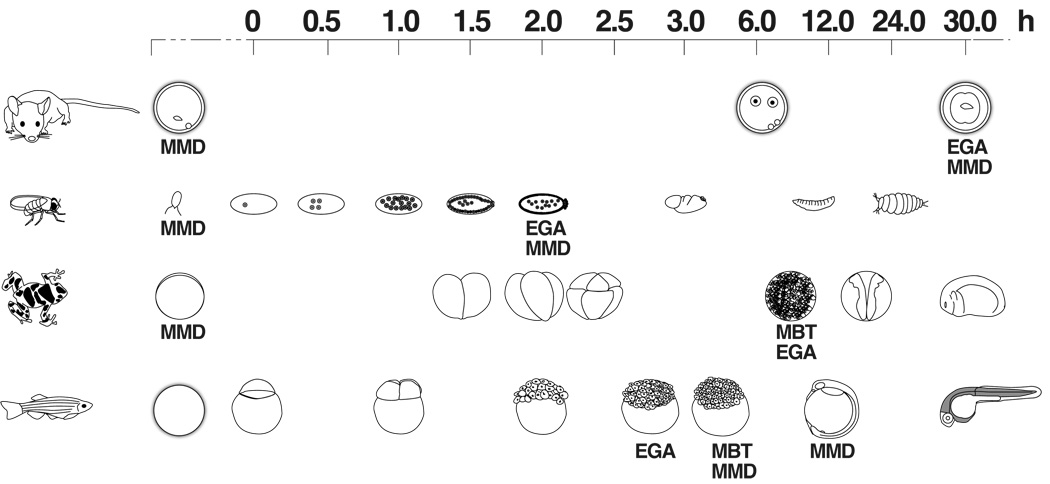
In the invertebrates, Drosophila and C. elegans, and in the vertebrates Xenopus and Danio rerio, transcription from the embryonic genome is not initiated until some number of nuclear or cell divisions have taken place (Fig. 1). Embryonic genome activation takes place in Drosophila 2 hours after egg deposition and a number of nuclear divisions, in Xenopus during the mid blastula transition (~4,000 cells) 7 hours following fertilization, and in Danio rerio at the 512 cell stage, 3 to 4 hours after fertilization. In mammals, however, the embryonic genome is activated during cleavage, at the 2-cell stage in mouse about 30 hours after fertilization, at the 4- to 8-cell stage in humans, and between the 8- and 12-cell stage in the cow (Telford et al., 1990).
Figure 1. Time line for the development of Drosophila, Xenopus and Danio rerio in relation to the oocyte-to-embryo transition in the mouse.
The time line is given in hours, with 0h fixed at fertilization. The time points when maternal message degradation (MMD), the mid-blastula transition (MBT) and embryonic genome activation (EGA) take place are indicated.
The asymmetric Drosophila, Xenopus and Danio rerio oocytes exhibit clear localization of maternal transcripts, and these localized maternal mRNAs and proteins play a crucial role in establishing the embryonic axes. In contrast, mammalian oocytes appear to be radially symmetrical and are unlikely to be patterned (Motosugi et al., 2005). Mammalian oocytes are postnatally recruited into the growth phase and at puberty the full-grown oocyte (FGO) completes the first meiotic division, progress into the second meiotic cell cycle and arrest at meiotic metaphase II until ovulation and fertilization (Eppig, 2001; Matzuk et al., 2002). Transcription decreases dramatically in the FGO and does not recommence until after the mature, ovulated oocytes are fertilized and the first cleavage division takes place (Bevilacqua et al., 1992; Worrad et al., 1995).
Programmed degradation of maternal mRNAs
A critical role in embryonic development in metazoans is fulfilled by mechanisms that degrade maternal RNAs once they are no longer required. In Drosophila, Xenopus and Danio rerio activation of the embryonic genome and degradation of maternal transcripts occur during the mid blastula transition (MBT) when the embryo is already a multi-cellular organism. In C. elegans the destruction of maternal transcripts takes place over a longer period of time. Although embryonic genome activation (EGA) takes place at the 4-cell stage in somatic cells, embryonic genome control of development doesn’t dominate until after gastrulation. In addition, transcription in the germline blastomeres is repressed until gametogenesis in the larval stage, with degradation of maternal messages taking place only at that point. In the mouse the majority of maternal transcripts are degraded during oocyte maturation (Su et al., 2006), with the remainder being degraded at the 2-cell stage upon embryonic genome activation. Although the degradation of maternal mRNAs has been well documented, the mechanisms controlling it have not. In Danio rerio, microRNA miR-430 is expressed right after the onset of MBT until gastrulation (Giraldez et al., 2006). In Drosophila SMAUG regulates destabilization of maternal mRNAs (Tadros et al., 2007) and miRNAs encoded from the miR-309 cluster are involved in degradation of a subset of maternal mRNAs (Bushati et al., 2008). Only recently has the involvement of pseudogene-derived endo-siRNAs and piRNAs in the regulation of gene expression in mouse oocytes been determined (Tam et al., 2008). These iRNAs appear solely necessary for regulation of up-regulated endogenous retroviruses.
Stabilization of maternal mRNAs
In the mouse, maternal mRNAs are degraded between the FGO and ovulated oocyte stages, but some are stabilized for utilization following fertilization and are later degraded in the embryo. Indeed, mammalian maternal transcripts must be stabilized for quite a long time to guide embryo development until embryonic genome activation. Maternal transcripts must also be protected from degradation, and activated for translation at specific times to enable successful nuclear reprogramming.
The primary knowledge base for control of maternal mRNA expression and degradation has been gleaned from study of meiotic maturation of the large and easily accessed oocytes of Xenopus. Sequences in the 3’UTR of maternal transcripts, and binding of their cognate transfactor proteins, are central to the control of maternal mRNA polyadenylation and translation. For example, the polyadenylation signal (PAS), AAUAAA, which binds the cleavage and polyadenylation specific factor, and the cytoplasmic polyadenylation element (CPE), UUUUUAU and variants thereof, which binds the cytoplasmic polyadenylation element binding protein, are known to control translation (Richter, 2007). When a CPE-binding protein is bound to a CPE-containing mRNA, a complex containing Maskin is recruited to the 3’UTR of mRNAs, which represses polyadenylation and translation by preventing entry of the mRNA into the ribosome. Phosphorylation of the CPE-binding protein by Aurora A kinase results in release of the Maskin-containing inhibitory complex from the mRNA, binding of the cleavage and polyadenylation specificity factor to the mRNA, entry into the ribosome and initiation of translation. A pumilio binding element (PBE) in the 3’UTR of mRNAs, activated/repressed by members of the PUF protein family (Nakahata et al. 2003), can also control the timing of cytoplasmic polyadenylation and translation of maternal transcripts.
Genome-wide analysis of the expression patterns and the regulatory mechanisms responsible for these patterns of expression has been undertaken in Xenopus. Piqué et al (2008), by determining the translational control of the cyclins B1-B5 during Xenopus oocyte maturation, established how a combinatorial code for the relationship between the number of, and physical distances between, the PAS, CPE and PBE in a given mRNA 3’UTR determines the time of activation of specific maternal mRNAs. The (A + U)-rich elements (AREs) are also involved in the regulation of Xenopus maternal transcripts during oocyte maturation. When bound by the protein C3H-4, which recruits the CCR4 deadenylase complex to the 3’UTR, the mRNA’s poly(A) tail is shortened, thereby repressing translation (Belloc and Mendez, 2008). An embryonic deadenylation element (EDEN) has also been described, which, upon binding of its cognate binding protein in Xenopus, prevents translation of c-Mos and Aurora A mRNA translation after fertilization (Paris and Richter, 1990; Paillard and Osborne, 2003; Graindorge et al., 2006).
Many of these mechanisms governing maternal mRNA stability and translation in Xenopus appear to be conserved in the mouse. The stability and degradation of maternal transcripts after fertilization are controlled by ARE elements characterized by AUUUA motifs repeated in an AU-rich region (Chen and Shyu, 1995; Voeltz and Steitz, 1998). These elements are bound by poly(A)-binding proteins that confer stability to mRNAs (Wagner et al., 2001). Many maternal mRNAs present in the FGO contain CPEs in their 3' UTRs and a limited number of maternal transcripts in mouse under translational control have been identified (Oh et al., 2000; Sakurai et al., 2005a, b). Maternal mRNAs containing a PBE-motif in their 3’UTR were found to be over-represented among the stable mouse maternal transcripts (Evsikov et al., 2006).
The CPE in stability and translation of mouse maternal messages
Maternal messages containing CPEs are cytoplasmically polyadenylated and translated during the oocyte-to-embryo transition (Oh et al., 2000). There is a bias toward longer 3'UTRs in stable maternal messages, i.e. in mRNAs present in both the FGO and the 2-cell stage embryo (Evsikov et al., 2006). Furthermore, a CPE and/or a PBE are also present in the majority of the stable transcripts. This suggested such motifs might contribute to regulation of polyadenylation and translational activation of messages as well as to their stability.
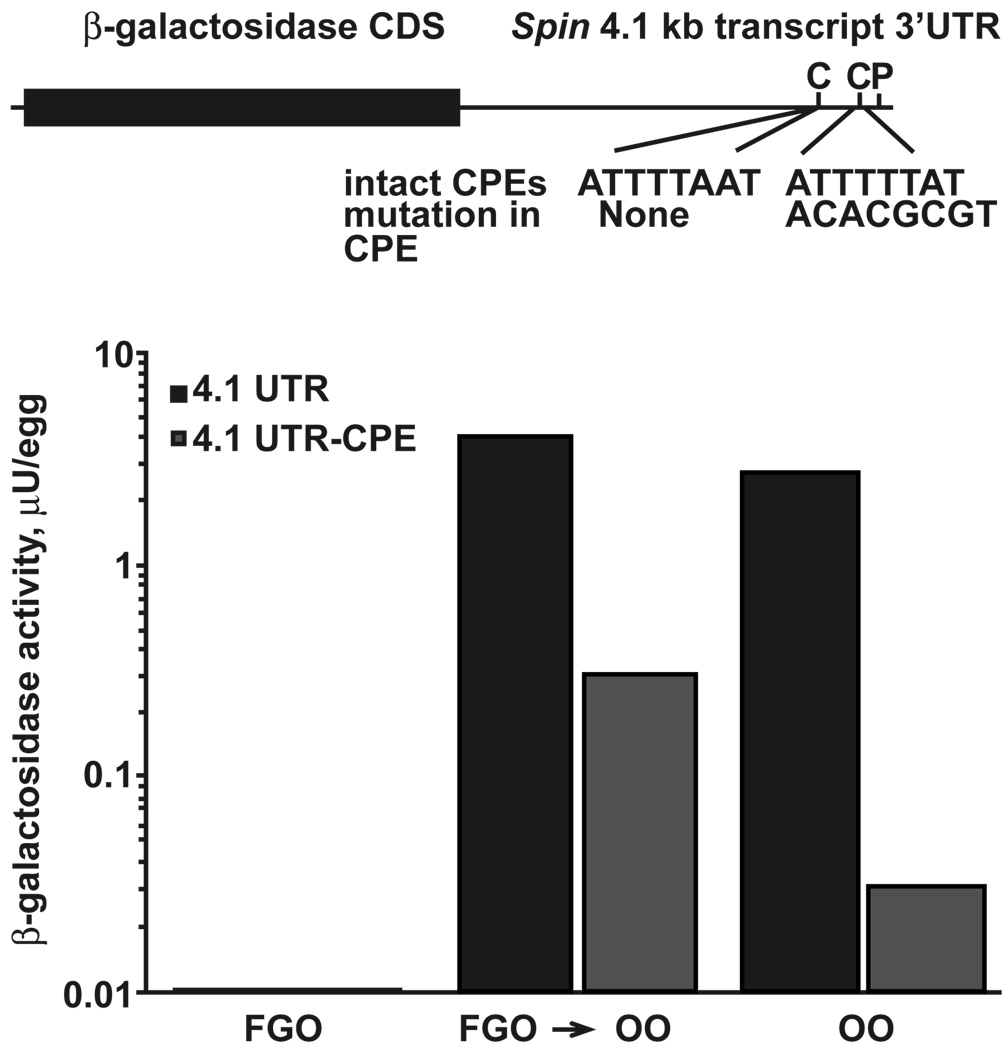
To test this hypothesis in the mouse we turned to the Spin gene, which encodes an abundant protein in the oocytes and preimplantation embryos and has three transcripts of 0.8, 1.7 and 4.1 kb (Oh et al., 2000). These three transcripts arise as a consequence of differential PAS usage resulting in different 3’UTRs, each containing the same open reading frame. While the 0.8 kb mRNA quickly degrades at the onset of maturation, the two larger transcripts are relatively stable and both are present at the 2-cell stage. The 1.7 kb transcript contains an UA-rich CPE-like sequence, whereas the 4.1 kb transcript contains two CPE sequences. The 1.7 kb transcript is polyadenylated in the ovulated oocyte and zygote, and the 4.1 kb transcript is deadenylated in the oocyte and then readenylated in the zygote. The CPE located 37 nt from the PAS of the 4.1kb Spin transcript was mutated (TTTTTAT to CACGCGT) and inserted into a previously described reporter construct pBlueβgalSpin (4.1 UTR versus 4.1 UTR -CPE; Fig. 2 upper panel). Both transcripts were in vitro transcribed, injected into full grown or ovulated oocytes and the effect of the mutation on translation of the reporter construct was measured at various times after its introduction. Neither transcript was translated in injected FGOs kept under the conditions of meiotic arrest by addition of IBMX in the culture medium. The pBlueβgalSpin4.1UTR transcript was efficiently translated in injected FGOs that were allowed to mature in vitro for 12 hours, as well as in ovulated metaphase II oocytes cultured for three hours after injection (Fig. 2, lower panel). Conversely, there was a marked decrease in the translation of the pBlueβgalSpin4.1 UTR -CPE transcript in both in vitro matured FGOs and ovulated oocytes. Thus, mutation of the CPE proximal to the PAS most likely decreases the efficiency of reporter mRNA translation, suggesting an important role of CPE in regulation of translational efficiency.
Figure 2. Mutation of the CPE in the Spin 4.1 kb 3'UTR leads to ineffective translation.
Top panel – schematic representation of the Spin 4.1 kb reporter constructs used to determine the effect of mutation of the CPE on message stability in the FGO and ovulated oocyte. The CPE sequences in the Spin 4.1 kb transcript are indicated next to “intact 3'UTR”. The 3'CPE is 37 nt from the PAS, and the 5'CPE 119 nt away from the PAS. The sequences replacing the 3'CPE sequence in the Spin 4.1 kb transcript are indicated next to “mutation in CPE”. PAS indicates the polyadenylation signal sequence. Lower panel – Graphic representation of the decrease in translation of the reporter construct when the CPE sequence in the Spin 4.1 kb transcript is mutated. FGO – full-grown oocyte; FGO -> OO – transition from FGO to ovulated oocyte; OO – ovulated oocyte.  Graph of reporter construct containing the intact CPE sequence;
Graph of reporter construct containing the intact CPE sequence;  Graph of reporter construct containing the mutated CPE sequence.
Graph of reporter construct containing the mutated CPE sequence.
Spin serves as a prototype of the kind of regulation expected for genes involved in the establishment of the totipotent embryo: it is a stable maternal transcript that undergoes translational activation at specific times during the oocyte-to-embryo transition and the message is degraded when no longer needed.
Pluripotency regulatory circuitry
Transfection of human and mouse fibroblasts and other differentiated somatic cells with viral vectors containing coding sequences for Lin28, Sox2, Klf4, Myc and Pou5f1 (Oct4) leads to reprogramming of these cells to a pluripotent state, i.e., induced pluripotent stem cells (iPS) (Meissner et al., 2007; Okita et al., 2007; Takahashi et al., 2007; Wernig et al., 2007; Yu et al., 2007; Hanna et al., 2008; Jaenisch and Young, 2008; Nakagawa et al., 2008). This reprogramming takes place in a stochastic fashion, where partially reprogrammed cells are first established, which then over time develop into fully reprogrammed iPS. Specific molecules, AP1, SSEA1, OCT4 and NANOG are sequentially expressed during the transformation of differentiated cells into iPS. Three transcription factors, OCT4, NANOG and SOX2, are responsible for activating genes involved in maintaining the ES-cell state, as well as for repressing genes that would lead to differentiation. TCF3, a transcription factor responsible for activating genes in the presence of WNT-signaling and repressing genes in the absence of WNT-signaling, also seems to play a central role in the maintenance of the stem cell state. Interestingly, TCF3 may play different roles in reprogramming in the oocyte-to embryo transition versus iPS reprogramming. During the oocyte-to-embryo transition TCF3 is expressed in an environment devoid of Wnt-signaling (De Vries et al., 2004), thus it would have a gene silencing function while during iPS reprogramming it would be expressed in the presence of Wnt-signaling and would thus have an activating function. The expression pattern of the genes used to induce iPS during the oocyte-to-embryo transition gives a clue as to the sequential expression of genes needed to trigger efficient reprogramming of differentiated genomes.
Embryonic expression of genes that effect reprogramming of differentiated cells to (iPS)
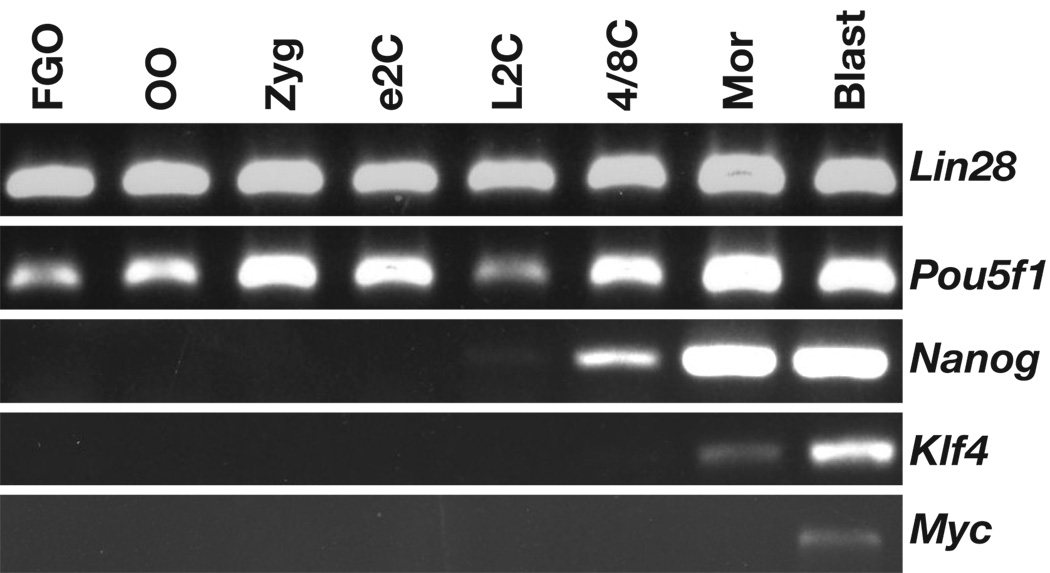
We determined the timing of expression of Nanog, Klf4, Myc, Lin28, Pou5f1 (Oct4) during the oocyte-to-embryo transition in our oocyte and 2-cell stage library databases (Evsikov et al., 2004; Evsikov et al., 2006) and also by performing RT-PCR on samples obtained throughout oocyte maturation, ovulation, fertilization and in preimplantation embryos (Fig. 3). Lin 28 and Pou5f1 (Oct4) were represented by 2 and 1 EST respectively in the oocyte and 2-cell stage cDNA libraries, i.e. at low levels, while Nanog, Klf4 and Myc were not found in either of them. RT-PCR revealed that Lin28 and Oct4 are indeed expressed throughout the oocyte-to-embryo transition and at later preimplantation stages (Fig. 3), while Nanog, Klf4 and Myc only begin to be expressed at later stages, i.e., the late 2-cell stage, the morula and the blastocyst, respectively. These data suggest that genes crucial for reprogramming to iPS cells are different than those that reprogram the egg and sperm nucleus.
Figure 3. Expression of “iPS” genes in oocytes and preimplantation embryos.
RT-PCR, using cDNA from 2 oocyte or embryo equivalents, was used to determine the expression of iPS genes during the oocyte-to-embryo transition and in later preimplantation stages. The names of the genes are indicated on the right; oocyte and embryo stages are indicated at the top. FGO - full-grown oocyte; OO - ovulated oocyte; Zyg - zygote; e2C - early 2-cell stage; L2C - late 2-cell stage; 4/8C - 4-to-8-cell stage; Mor - morula; Blast – blastocyst. Primer information, PCR conditions, and results are available in the Gene Expression Database (GXD) (http://www.informatics.jax.org; J:138685).
Oocyte to embryo transition and stem cell genes
Analysis of human embryonic stem cells and their differentiating derivatives identified a set of genes whose transcript abundance is significantly reduced upon differentiation. These are Tgf1, Gabrb3, Dnmt3b, Gdf3, Pou5f1, Fgf4, Gal, Lefty1, Ifitm1, Nodal, Tert, Utf1, Foxd3, Lin28, Grb7, Podxl, Cd9, Bxdc2, Sox2, Klf4, and Nanog (Adewumi et al., 2007). Microarray expression data ((Wang et al., 2004), obtained from GEO (http://www.ncbi.nlm.nih.gov/geo/), which were quantile normalized and corrected for background noise using the standard statistical RMA package and further analyzed using the R/MAANOVA software package (Wu et al., 2003), were searched to determine expression of these genes during the mouse oocyte-to-embryo transition and in later preimplantation stages. According to this public dataset (Wang et al., 2004) Cd9 is degraded during oocyte maturation and Tgf1, Gabrb3, Dnmt3b, Gdf3, Oct4, Fgf4, Gal, Lefty1, Ifitm1, Nodal, Tert, Utf1, Foxd3, Lin28, Grb7, Podxl, and Bxdc2 are expressed during the oocyte to embryo transition. Sox2, Klf4, and Nanog were only expressed after activation of the embryonic genome. In contrast, a search of our own full-grown oocyte EST dataset (Evsikov et al., 2006) revealed ESTs corresponding to only Oct4, Tert, Dnmt3b, Bxdc2 and Lin28, while the 2-cell stage EST database (Evsikov et al., 2004) revealed in addition ESTs corresponding to Utf1 and Sox2. The discrepancy in results obtained using these two different approaches (Tgf1, Gabrb3, Gdf3, Fgf4, Gal, Lefty1, Ifitm1, Nodal, Utf1, Foxd3, Grb7, Podxl, and Bxdc2 are present in the microarray dataset and absent in the EST databases) may reflect the different shortcomings of the two assays. Microarray analysis is very sensitive, but in some cases absence of expression could be interpreted as presence if the expression level is close to background. In contrast, ESTs in libraries represent only those genes sequenced. Since saturation sequencing is impractical, some genes expressed at a very low level can be missed. Thus the microarray approach can produce false positives and EST analysis can produce false negative results. Nevertheless, when we analyzed the expression of Nanog, Klf4 and Myc, all of which are predicted by microarray data to be expressed during the oocyte-to-embryo transition, RT-PCR confirmed our negative EST data.
To determine if there is a possibility that any of those human “stemness” genes expressed throughout the oocyte-to-embryo transition are activated for translation at specific time points during this developmental period, we searched the 3’UTR of the transcripts for known motifs. Three genes Lin28, Grb7, and Tert contain CPE sequences in their 3'UTRs. This implies that, similar to Spin, these messages could be activated for translation at different times during the oocyte-to-embryo transition.
Conclusion
Genomic reprogramming is a very complex process involving numerous and possibly alternative controlling mechanisms. Natural reprogramming, i.e. events following fertilization, is regulated by maternally inherited molecules, proteins and mRNAs synthesized and stored during oogenesis. These molecules mediate rapid reprogramming of the egg and sperm genomes likely through chromatin remodeling, differential mRNA utilization and directed mRNA and protein degradation. Reprogramming following somatic cell nuclear transfer is mechanistically similar if not identical, with the low efficiency most likely arising due to the specific state of the genome of the transferred nuclei. In contrast, artificial reprogramming to iPS cells, induced by transfection, is obviously under transcriptional control and is a much slower and less efficient process. Evolution has perfected the just-in-time supply of specific proteins to effect swift natural reprogramming, which would be very difficult to mimic in the iPS type of reprogramming. Comparing and contrasting these two models of reprogramming will help us to identify relevant molecules and mechanisms.
ACKNOWLEDGEMENTS
This work was supported by the USPHS/NIH grant 1RO1 HD37102 (B.B.K.), and 1R01 GM072706 (J.H.G.)
Contributor Information
Wilhelmine N. de Vries, The Jackson Laboratory, Bar Harbor, ME 04609
Alexei V. Evsikov, The Jackson Laboratory, Bar Harbor, ME 04609
Louise J. Brogan, The Jackson Laboratory, Bar Harbor, ME 04609
Christopher P. Anderson, The Jackson Laboratory, Bar Harbor, ME 04609
Joel H. Graber, The Jackson Laboratory, Bar Harbor, ME 04609
Barbara B. Knowles, The Jackson Laboratory, Bar Harbor, ME 04609
Davor Solter, Institute of Medical Biology, A*STAR, 8A Biomedical Grove, Immunos, Singapore 138648.
REFERENCES
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Blum B, Brooking J, Chen KG, Choo AB, Churchill GA, Corbel M, Damjanov I, Draper JS, Dvorak P, Emanuelsson K, Fleck RA, Ford A, Gertow K, Gertsenstein M, Gokhale PJ, Hamilton RS, Hampl A, Healy LE, Hovatta O, Hyllner J, Imreh MP, Itskovitz-Eldor J, Jackson J, Johnson JL, Jones M, Kee K, King BL, Knowles BB, Lako M, Lebrin F, Mallon BS, Manning D, Mayshar Y, McKay RD, Michalska AE, Mikkola M, Mileikovsky M, Minger SL, Moore HD, Mummery CL, Nagy A, Nakatsuji N, O'Brien CM, Oh SK, Olsson C, Otonkoski T, Park KY, Passier R, Patel H, Patel M, Pedersen R, Pera MF, Piekarczyk MS, Pera RA, Reubinoff BE, Robins AJ, Rossant J, Rugg-Gunn P, Schulz TC, Semb H, Sherrer ES, Siemen H, Stacey GN, Stojkovic M, Suemori H, Szatkiewicz J, Turetsky T, Tuuri T, van den Brink S, Vintersten K, Vuoristo S, Ward D, Weaver TA, Young LA, Zhang W. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Belloc E, Mendez R. A deadenylation negative feedback mechanism governs meiotic metaphase arrest. Nature. 2008;452:1017. doi: 10.1038/nature06809. [DOI] [PubMed] [Google Scholar]
- Bevilacqua A, Kinnunen LH, Mangia F. Genetic manipulation of mammalian dictyate oocytes: factors affecting transient expression of microinjected DNA templates. Mol Reprod Dev. 1992;33:124. doi: 10.1002/mrd.1080330203. [DOI] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- De Vries WN, Evsikov AV, Haac BE, Fancher KS, Holbrook AE, Kemler R, Solter D, Knowles BB. Maternal {beta}-catenin and E-cadherin in mouse development. Development. 2004;131:4435. doi: 10.1242/dev.01316. [DOI] [PubMed] [Google Scholar]
- Eppig JJ. Oocyte control of ovarian follicular development and function in mammals. Reproduction. 2001;122:829. doi: 10.1530/rep.0.1220829. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, de Vries WN, Peaston AE, Radford EE, Fancher KS, Chen FH, Blake JA, Bult CJ, Latham KE, Solter D, Knowles BB. Systems biology of the 2-cell mouse embryo. Cytogenet Genome Res. 2004;105:240. doi: 10.1159/000078195. [DOI] [PubMed] [Google Scholar]
- Evsikov AV, Graber JH, Brockman JM, Hampl A, Holbrook AE, Singh P, Eppig JJ, Solter D, Knowles BB. Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev. 2006;20:2713. doi: 10.1101/gad.1471006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Graindorge A, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of post-transcriptionally regulated Xenopus tropicalis maternal mRNAs by microarray. Nucleic Acids Res. 2006;34:986. doi: 10.1093/nar/gkj492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, Lengner CJ, Dausman JA, Jaenisch R. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science. 2002;296:2178. doi: 10.1126/science.1071965. [DOI] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Motosugi N, Bauer T, Polanski Z, Solter D, Hiiragi T. Polarity of the mouse embryo is established at blastocyst and is not prepatterned. Genes Dev. 2005;19:1081. doi: 10.1101/gad.1304805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Oh B, Hwang S, McLaughlin J, Solter D, Knowles BB. Timely translation during the mouse oocyte-to-embryo transition. Development. 2000;127:3795. doi: 10.1242/dev.127.17.3795. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Paillard L, Osborne HB. East of EDEN was a poly(A) tail. Biol Cell. 2003;95:211. doi: 10.1016/s0248-4900(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Paris J, Richter JD. Maturation-specific polyadenylation and translational control: diversity of cytoplasmic polyadenylation elements, influence of poly(A) tail size, and formation of stable polyadenylation complexes. Mol Cell Biol. 1990;10:5634. doi: 10.1128/mcb.10.11.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Sato M, Kimura M. A novel method for constructing murine cDNA library enriched with maternal mRNAs exhibiting de novo independent post-fertilization polyadenylation. Biochem Biophys Res Commun. 2005a;327:688. doi: 10.1016/j.bbrc.2004.12.057. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Sato M, Kimura M. Diverse patterns of poly(A) tail elongation and shortening of murine maternal mRNAs from fully grown oocyte to 2-cell embryo stages. Biochem Biophys Res Commun. 2005b;336:1181. doi: 10.1016/j.bbrc.2005.08.250. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;134:2593. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12:143. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford NA, Watson AJ, Schultz GA. Transition from maternal to embryonic control in early mammalian development: a comparison of several species. Mol Reprod Dev. 1990;26:90. doi: 10.1002/mrd.1080260113. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Steitz JA. AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol Cell Biol. 1998;18:7537. doi: 10.1128/mcb.18.12.7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MJ, Gogela-Spehar M, Skirrow RC, Johnston RN, Riabowol K, Helbing CC. Expression of novel ING variants is regulated by thyroid hormone in the Xenopus laevis tadpole. J Biol Chem. 2001;276:47013. doi: 10.1074/jbc.M106965200. [DOI] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6:133. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Worrad DM, Turner BM, Schultz RM. Temporally restricted spatial localization of acetylated isoforms of histone H4 and RNA polymerase II in the 2-cell mouse embryo. Development. 1995;121:2949. doi: 10.1242/dev.121.9.2949. [DOI] [PubMed] [Google Scholar]
- Wu H, Kerr MK, Cui X, Churchill GA. MAANOVA: A software package for the analysis of spotted cDNA experiments. In: Parmigiani G, Garett ES, Irizarri RA, Zeger SL, editors. The analysis of gene expression data: methods and software. New York: Springer-Verlag; 2003. p. 313. [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]