SUMMARY
Rats have important advantages over mice as an experimental system for physiological and pharmacological investigations. The lack of rat embryonic stem (ES) cells has restricted the availability of transgenic technologies to create genetic models in this species. Here, we show that rat ES cells can be efficiently derived, propagated, and genetically manipulated in the presence of small molecules that specifically inhibit GSK3, MEK, and FGF receptor tyrosine kinases. These rat ES cells express pluripotency markers and retain the capacity to differentiate into derivatives of all three germ layers. Most importantly, they can produce high rates of chimerism when reintroduced into early stage embryos and can transmit through the germline. Establishment of authentic rat ES cells will make possible sophisticated genetic manipulation to create models for the study of human diseases.
INTRODUCTION
Embryonic stem (ES) cells are derived from the inner cell mass (ICM) of preimplantation blastocysts (Brook and Gardner, 1997). They can be maintained in culture indefinitely while retaining the capacity to generate nearly any type of cell in the body (Keller, 2005). The pluripotency of ES cells, combined with the ease with which they can be manipulated genetically, has provided a powerful means to elucidate gene function and create disease models via the generation of transgenic, chimeric, and knock-out animals. Although ES cells have been routinely derived from mice since 1981 (Evans and Kaufman, 1981; Martin, 1981), authentic rat ES cells have never been established.
In general, rats are more relevant to humans, both physiologically and pharmacologically, than mice, providing an important experimental model system for the study of human diseases (Jacob and Kwitek, 2001). For example, rats have been used extensively in studies of hypertension (Rapp, 2000). Because of the lack of rat ES cells, the generation of novel rat models for studying specific aspects of human diseases largely depends on selection for specific traits using existing rat strains. Although strategies based on chemical mutagenesis using the supermutagen N-ethyl-N-nitrosourea (ENU) or mutagenesis using the L1 retrotransposon have been developed to introduce random mutations into rats (Ostertag et al., 2007; Smits et al., 2006), germline-competent ES cells will be required to achieve robust, facile, and precise genetic modification in this species.
Derivation and maintenance of the undifferentiated state of mouse ES cells originally relied on cocultivation with feeder cells, usually mitotically inactivated mouse embryonic fibroblasts (MEFs), and the presence of serum. Later, it was shown that leukemia inhibitory factor (LIF) is the key cytokine secreted by feeders in supporting mouse ES cell self-renewal (Smith et al., 1988; Williams et al., 1988). We recently demonstrated that bone morphogenetic proteins (BMPs) can replace serum and act together with LIF to maintain mouse ES cell self-renewal (Ying et al., 2003). Several groups have attempted to derive ES cells from rats under similar culture conditions developed for mouse ES cells; however, no authentic rat ES cell lines have ever been established (Brenin et al., 1997; Buehr et al., 2003; Demers et al., 2007; Fandrich et al., 2002; Ueda et al., 2008; Vassilieva et al., 2000). Pluripotent EpiSCs (postimplantation epiblast-derived stem cells) have been derived from rat embryos at 7.5 days postcoitus (dpc) (Brons et al., 2007). However, EpiSCs do not contribute to chimeras, seriously limiting their potential use.
Although rat and mouse take much the same course of embryogenesis during the early stages of development, the early embryos differ significantly in their differentiation potential in vitro or in vivo when they are transplanted to an ectopic site. For instance, the isolated mouse epiblast can no longer regenerate parietal endoderm (Gardner, 1985), whereas the rat epiblast predominantly differentiates into parietal endoderm cells in culture (Nichols et al., 1998). Mouse egg cylinders form teratocarcinomas containing pluripotent embryonic carcinoma stem cells after being implanted to ectopic sites (Solter et al., 1970; Stevens, 1970). When the same procedure is carried out in the rat, only a yolk sac carcinoma develops (Damjanov and Sell, 1977). These differences may account for the failure of rat ES cell derivation using conditions developed for mouse ES cell cultures. Although the derivation of putative ES-like cells from other species has been reported, only ES cells from mice have proven to be able to efficiently contribute to chimeras and re-enter the germline, which is the defining feature of true ES cells.
Extrinsic stimuli are thought to be necessary for the maintenance of ES cell self-renewal. These stimuli may be provided in an integrated manner by a cellular microenvironment or by administration of cocktails of growth factors and cytokines in vitro (Smith, 2001). Recently, we have made a striking discovery in understanding mouse ES cell self-renewal (Ying et al., 2008). We found that, contrary to dogma, mouse ES cell self-renewal does not require activating signals from the LIF/STAT3 and BMP/SMAD pathways, but only that ES cells be shielded from inductive differentiation cues. On the basis of these findings, we developed a culture medium containing three inhibitors (3i): CHIR99021, PD184352, and SU5402. CHIR99021 is a well-characterized highly selective small molecule inhibitor of glycogen synthase kinase 3 (GSK3) (Murray et al., 2004). PD184352 and SU5402 are selective pharmacological inhibitors of mitogen-activated protein kinase kinase (MEK) and fibroblast growth factor (FGF) receptor tyrosine kinase, respectively (Davies et al., 2000; Mohammadi et al., 1997). 3i can support efficient derivation and maintenance of ES cells from different strains of mice (Ying et al., 2008).
Here, we examine the effect of 3i on rat ES cell derivation to determine whether this principle may be more broadly applicable. We report for the first time that germline-competent rat ES cells can also be established under 3i conditions.
RESULTS
3i Maintains the Expression of Oct4 in the Outgrowths of Rat Blastocysts
To examine the effect of 3i on sustaining the undifferentiated state of rat pluripotent stem cells, we plated E4.5 Dark Agouti (DA) rat blastocysts on MEFs in serum-free N2B27 medium with or without 3i. The expression of the pluripotency marker Oct4 was used as an assay for the presence of undifferentiated stem cells in the primary outgrowths of rat blastocysts. In the presence of 3i, Oct4 expression was maintained in the majority of the outgrowths (six out of nine) 10 days after plating (Figure 1A). In contrast, Oct4 was rapidly extinguished in all outgrowths of rat blastocysts placed in N2B27 medium alone (zero out of eight) or in cultures supplemented with LIF and serum (zero out of eight) conditions permissive for mouse ES cell derivation (Figures 1B and 1C). These data suggest that the uncommitted state in the primary culture of rat blastocysts can also be sustained in 3i conditions, as we have previously demonstrated in different strains of mice (Ying et al., 2008).
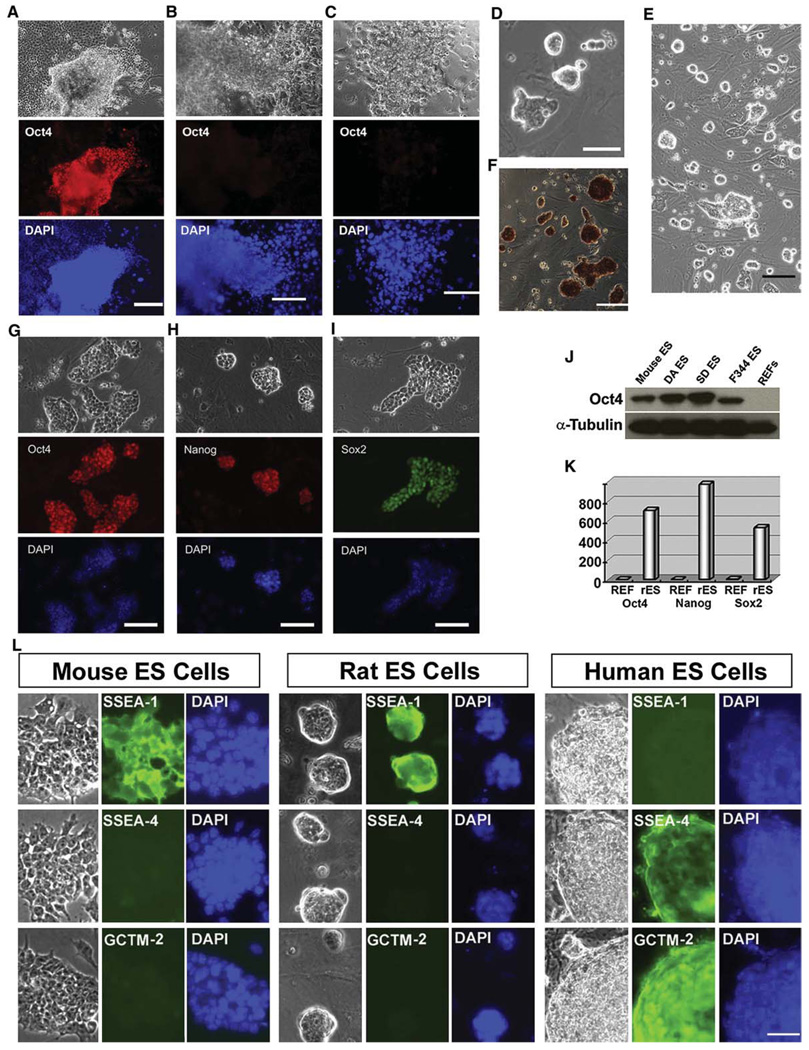
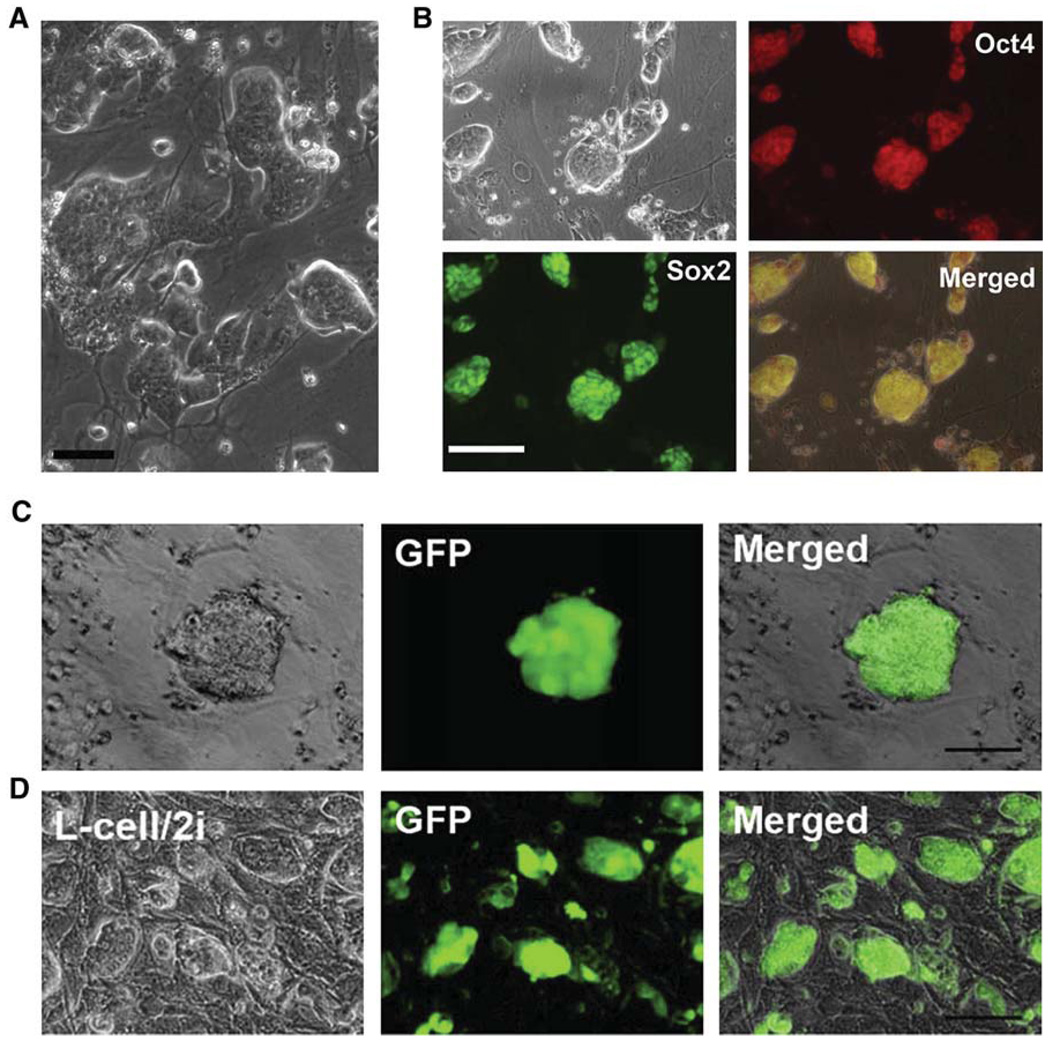
Figure 1. Rat ES Cells Derived and Maintained in 3i Conditions Express Pluripotency Markers.
(A–C) Immunofluorescence staining for Oct4 in the outgrowths of DA rat blastocysts. E4.5 DA rat blastocysts were plated onto mitotically inactivated MEFs with different culture medium: serum-free N2B27 medium supplemented with 3i (A), N2B27 medium only (B), or GMEM/10% FBS supplemented with 10 ng/ml LIF (C). Ten days after plating, the cultures were fixed and stained for Oct4. Scale bars represent 50 µm.
(D) Five days after plating, the outgrowths of DA rat blastocysts were disaggregated and replated onto new wells of 4-well plates. This phase contrast image shows colonies of DA rat cells formed from a single outgrowth of blastocyst 4 days after the first disaggregation. The scale bar represents 50 µm.
(E) Phase contrast image of DA rat ES cells at passage 25. The scale bar represents 100 µm.
(F) Alkaline phosphatase staining of DA rat ES cells at passage 15. The scale bar represents 100 µm.
(G–I) Immunofluorescence staining for Oct4 (G), Nanog (H), and Sox2 (I) in DA rat ES cells after 15 passages in 3i conditions. Scale bars represent 50 µm.
(J) Western blot analysis of Oct4 expression in mouse and rat ES cells. Rat embryonic fibroblasts (REFs) were used as control.
(K) qRT-PCR analysis of Oct4, Nanog, and Sox2 expression in undifferentiated rat ES cells (rES) and REFs. Data were average of triplicate samples and represent relative expression levels of indicated genes in rES and REFs.
(L) Expression of cell surface markers in mouse, rat, and human ES cells. Mouse ES cells were grown on gelatin in GMEM/10% FBS medium supplemented with LIF. Rat and human ES cells were maintained in MEF/3i and human ES cell culture conditions, respectively. The scale bar represents 50 µm.
3i Enables Efficient Derivation and Maintenance of Rat ES Cells
To investigate whether 3i conditions can support the long-term expansion of these rat blastocyst-derived undifferentiated cells, we dissociated the outgrowths of DA rat blastocysts 5–7 days after in 3i culture and replated them into wells of 4-well plates under the same MEF/3i condition. Cells loosely attached to the feeders and proliferated to form morphologically undifferentiated colonies (Figure 1D). These cells could be passaged repeatedly, resulting in the establishment of stable cell lines (Figure 1E). We established 11 cell lines from 32 plated DA blastocysts. These established lines exhibited positive staining for alkaline phosphatase (AP) (Figure 1F) and expressed pluripotency markers Oct4, Nanog, and Sox2 (Figures 1G–1J). The expression of Oct4, Nanog, and Sox2 was also verified by quantitative RT-PCR (qRT-PCR) (Figure 1K). We routinely passage these cells every 3–4 days by dissociating them into single cells using trypsin and replating onto wells preseeded with MEFs in 3i medium. Cultures have been expanded for over 60 passages without overt differentiation. We also found that these 3i rat ES cells, like mouse ES cells, can be cryopreserved and recovered with high efficiency by standard techniques. We tested the MEF/3i condition for the derivation of ES cells from other strains of rats. We have so far established six Sprague Dawley (SD) rat ES cell lines from 20 blastocysts and five Fischer 344 (F344) rat ES cell lines from 18 blastocysts.
To further confirm the identity of these cells, we examined genome-wide gene expression using an Affymetrix GeneChip Rat gene 1.0 ST array. These 3i rat cells express most of the pluripotency-related genes at high levels (Table S1 available online). We also performed hierarchical clustering analysis to compare with the global gene expression patterns of rat embryonic fibroblasts (REFs) and mouse ES cells. This microarray analysis shows that the gene expression profile of 3i rat cells resembles mouse ES cells, and is distinct from that of REFs derived from the same strain (Figure S1). All of these data together suggest that these rat cells derived and maintained in 3i conditions remain undifferentiated.
Rat ES Cells Have a Similar Expression Pattern of Cell Surface Markers with Mouse, but Not Human ES Cells
Mouse and human ES cells have a distinct expression pattern of cell surface markers that characterize the undifferentiated state (Ginis et al., 2004). SSEA-1 is expressed in mouse ES cells but is absent in human ES cells, whereas SSEA-4, TRA-1-60, and TRA-1-81 are expressed in human ES cells but not in mouse ES cells. To test whether rat ES cells are similar to mouse or human ES cells in terms of cell surface marker expression, we performed immunofluorescence staining for cell-surface antigens SSEA-1, SSEA-4, and GCTM-2. Antibody GCTM-2 recognizes a 200 kD keratan-sulfate proteoglycan that also bears the TRA-1-60 antigen. We found that, similar to mouse ES cells, rat ES cells expressed SSEA-1, but not SSEA-4 or GCTM-2 (Figure 1L).
Epigenetic Status of Rat ES Cells
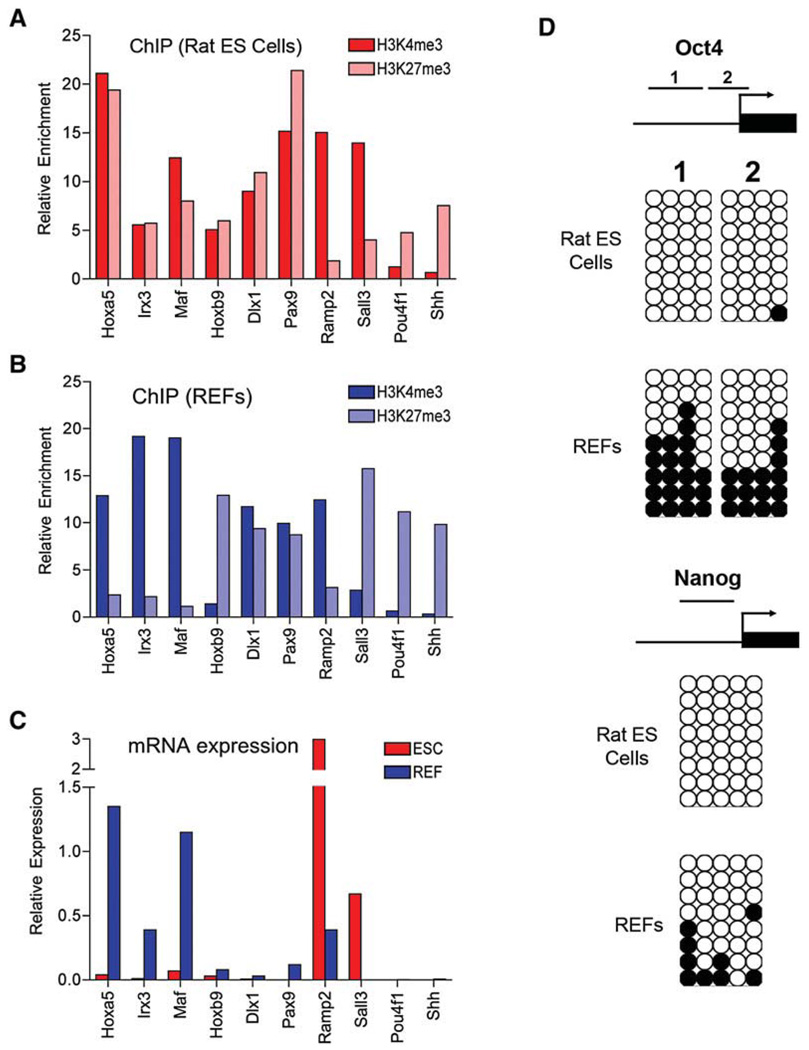
ES cells have some epigenetic features that are distinguishable from those of differentiated cells. One such feature is that a set of promoters in ES cells carry both trimethylated histone H3 lysine 4 (H3K4me3) and H3K27me3 chromatin marks, which are associated with gene activation and repression, respectively. These so-called “bivalent domains” preferentially occur at transcription start sites (TSSs) of key developmental genes in mouse ES cells and function to silence these genes in ES cells while keeping them poised for lineage-specific activation and repression (Bernstein et al., 2006; Mikkelsen et al., 2007). To determine whether rat ES cells also have this epigenetic feature, we analyzed H3K4 and K27 trimethylation patterns of ten selected genes that have been shown to contain bivalent promoters in mouse ES cells and resolve to monovalent state (H3K4me3 or H3K27me3) or remain bivalent in MEFs (Mikkelsen et al., 2007). Using chromatin immunoprecipitation (ChIP) and qPCR, we found that six genes (Hoxa5, Irx3, Maf, Hoxa9, Dlx1, and Pax9) were associated with bivalent promoters in rat ES cells, with four resolving into either Lys4 or Lys27 trimethylation and two remaining bivalent in REFs (Figures 2A and 2B). Interestingly, the other four genes (Ramp2, Sall3, Pou4f1, and Shh) were marked predominantly by either Lys4 or Lys27 trimethylation in rat ES cells. They remained the same modification in REFs except one (Sall3) (Figures 2A and 2B). Using qRT-PCR, we confirmed that genes marked by H3K4me3 were expressed at high levels, whereas those associated with H3k27me3 or bivalent domains were barely detectable (Figure 2C). These results suggest that some developmental genes are also marked by a bivalent chromatin structure in rat ES cells, although divergences of chromatin modification may exist between mouse and rat ES cells. Generation of a genome-wide map of chromatin states is needed to better understand epigenetic mechanisms of gene regulation in rat ES cells.
Figure 2. Gene-Specific Histone and DNA Methylation Profiles in Rat ES Cells and REFs.
(A and B) The enrichment of H3K4me3 and H3k27me3 at the TSSs of indicated genes was determined by ChIP qPCR in DAc2 rat ES cells (A) and in REFs derived from E14.5 DA rat embryos (B). Data are the average of triplicate qPCR results.
(C) Expression levels of indicated genes in rat ES cells (ESC) and REFs were determined by qRT-PCR and were normalized to Gapdh. Data are the average of triplicate samples.
(D) Bisulfite sequencing of DNA methylation of the Oct4 and Nanog promoter regions in rat ES cells and REFs. Arrows indicate the transcriptional start sites. Unmethylated and methylated CpGs are shown with open and filled circles, respectively.
Another important epigenetic feature of ES cells is that the promoter regions of key pluripotency genes are unmethylated (Meissner et al., 2008). We used bisulfite sequencing to examine the DNA methylation status of the Oct4 and Nanog promoters in rat ES cells and REFs. As shown in Figure 2D, both Oct4 and Nanog promoter elements were fully unmethylated in rat ES cells. However, the Oct4 promoter became hypermethylated in REFs, and the Nanog promoter was also methylated, but to a lesser extent.
Rat ES Cells Derived and Maintained in 3i Retain the Capacity to Differentiate into Derivatives of All Three Germ Layers
The classical method to induce ES cell differentiation is to allow ES cells to grow in suspension and form three-dimensional aggregates known as embryoid bodies (EBs) (Keller, 1995). Within the EBs, ES cell differentiation proceeds on a schedule similar to that in the embryo. We performed rat ES cell differentiation using this EB method. Dissociated rat ES cells were plated into noncoated dishes in the presence of serum. Instead of forming EBs, the majority of rat ES cells died 2 days after plating. The presence of 3i and feeders seems to be critical for the survival of rat ES cells. We made two modifications to the EB protocol: (1) 3i was still added at half of the original concentration for the first 2 days of differentiation, and (2) GMEM/10% fetal bovine serum (FBS)-conditioned medium collected from feeders was then substituted for 3i to instigate EB formation. Under these conditions, rat ES cells formed EBs (Figure 3A), although at much lower efficiency compared with EB formation from mouse ES cells.
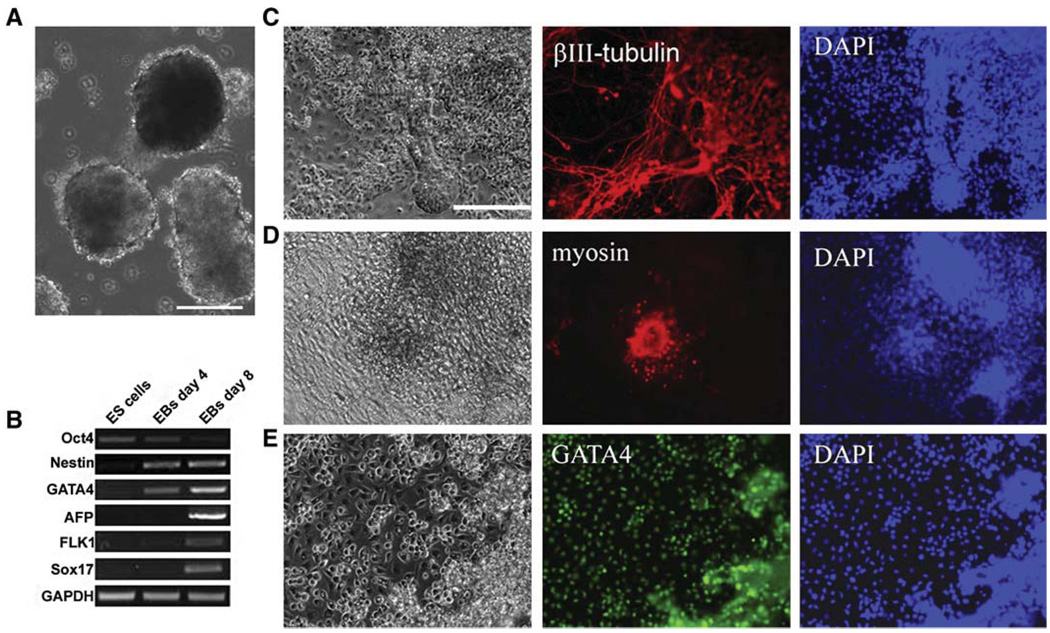
Figure 3. In Vitro Differentiation of Rat ES Cells.
(A) Phase contrast image of Day 4 EBs formed from DA rat ES cells. The scale bar represents 100 µm.
(B) RT-PCR analysis of gene expression in undifferentiated rat ES cells and EBs formed from rat ES cells.
(C) Day 4 rat ES cell-derived EBs were plated onto matrigel-coated dishes and cultured in N2B27 medium. Nine days after plating, cells were fixed and stained for neuronal marker βIII-tubulin. The scale bar represents 50 µm.
(D) Fourteen days after replating of DA rat ES cell-derived EBs into GMEM medium plus 10% FBS, spontaneously beating areas appeared. The cultures were then fixed and stained for cardiomyocyte marker myosin.
(E) GATA4 immunofluorescence staining of differentiated cells derived from DA rat ES cells.
We examined the expression of markers of pluripotency and lineage commitment by RT-PCR during the process of EB differentiation. Oct4 was significantly downregulated, whereas the expression of markers for ectoderm (nestin), primitive endoderm (Gata4 and Sox17), endoderm (AFP), and mesoderm (Flk1) was induced in EBs (Figure 3B). We plated day 4 EBs onto matrigel-coated dishes with either serum-free N2B27 medium (Figure 3C) or serum-containing medium (Figures 3D and 3E). In N2B27 medium, a large proportion of cells differentiated into βIII-tubulin- positive neurons (Figure 3C). We observed spontaneously beating areas in the cultures with serum (Movie S1). The cells within the beating areas were positive for myosin staining (Figure 3D). In the outgrowth of EBs, large proportions of cells displayed morphology typical of parietal endoderm cells and were positive for Gata4 staining (Figure 3E). These results suggest that the rat ES cells derived and maintained in 3i conditions are pluripotent.
Rat ES Cells Derived in 3i Can Contribute to High Rates of Chimerism and Transmit through the Germline
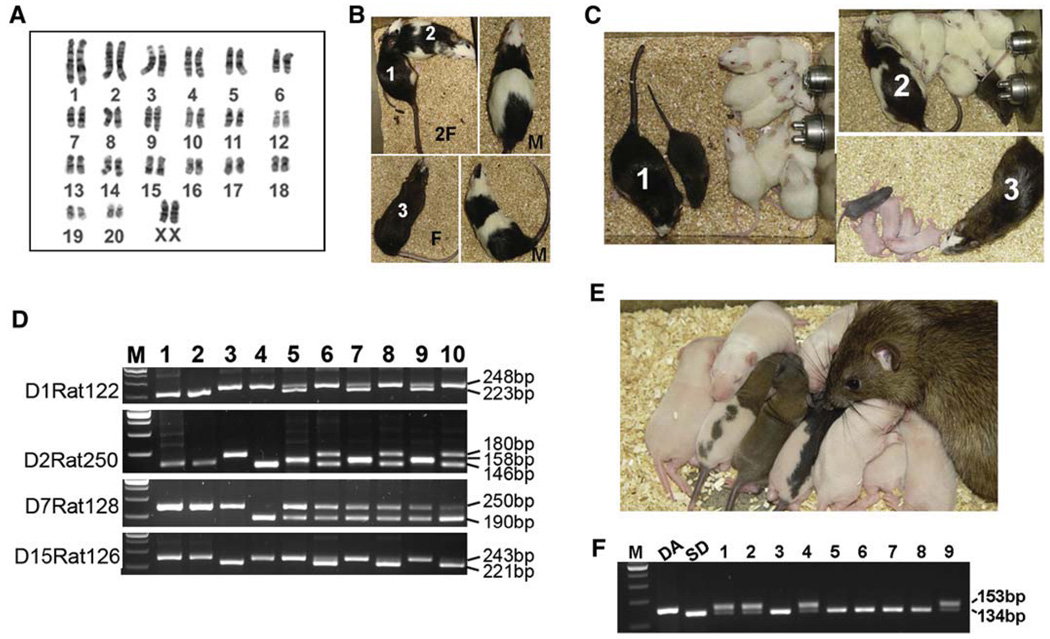
The defining feature of authentic ES cells is their capacity to incorporate into the developing embryo and transmit through the germline (Smith, 2001). We injected rat ES cells into blastocysts to determine their potential to form germline chimeras. A total of 43 F344 blastocysts were injected with DAc2 cells (female, Figure 4A) and transferred to five pseudopregnant SD rats. Seven pups were born, among which two male and three female exhibited coat coloring indicative of the presence of DA rat cells (Figure 4B and Table 1). DA rat ES cells have an agouti (A/A) genetic background, whereas F344 and SD rats are both albino (c/c). The albino coat color mutation is recessive to agouti; thus, when only one allele is present, there should be no partial expression. As a result, when a DA/F344 or DA/SD chimeric rat is mated with a SD rat, the germline transmission of DA rat ES cells can be easily identified by the presence of agouti coat color. These DA/F344 chimeras were mated with SD rats, and all three female chimeras have produced offspring with agouti coat color, indicating the germline transmission of the DA rat ES cell genome (Figure 4C). Microsatellite analysis further confirmed the presence of the DA rat ES cell genome in the resulting germline offspring (Figure 4D and Table S2). One of the female germline offspring has developed into a fertile adult and produced a total of nine pups; six had an albino coat color, one was agouti/berkshire, one was agouti/hooded, and one was black/hooded (Figure 4E). The two agouti pups and two out of the six albino pups carried both the agouti gene from DA genetic background and the nonagouti gene from the SD rat, whereas the black/hooded and other four albino pups only had the nonagouti gene, as expected (Figure 4F and Table S3) (Kuramoto et al., 2001). Four more DA/F344 chimeras (three male, one female) were generated with a male DA rat ES cell line DAc3 (Figure S2A). Two male chimeras were infertile, whereas the female and the other male chimeras produced all white pups. DAc3 cells had an abnormal karyotype, which may account for their inability to transmit through the germline (Table 1). We have also generated one highly pigmented male DA/F344 chimera using karyotypically normal DAc8 cells (Table 1 and Figure S2B). This chimera will be ready for mating in January, 2009.
Figure 4. Chimeras and Germline Offspring Produced from Rat ES Cells.
(A) Cytogenetic analysis of DAc2 rat ES cells used for the production of germline chimeras.
(B) Five chimeras generated by injection of DAc2 rat ES cells into F344 rat blastocysts. The agouti coat color denotes the presence of introduced DA ES cells in albino F344 hosts. Chimera numbers 1, 2, and 3 were female. The other two chimeras were male. F, female; M, male.
(C) All three DA/F344 female chimeras mated with SD males have produced pigmented offspring, indicating the transmission of the DA rat ES cell genome.
(D) DNA microsatellite analysis of DA rat ES cells, the three germline offspring, and their littermates. M, 100 bp DNA marker; 1, DAc2 rat ES cells; 2, DA rat; 3, F344 rat; 4, SD rat; 5, 7, and 9, the three germline offspring; 6, 8, and 10, their littermates. The PCR primers for different microsatellite markers are listed in Table S5, and the expected sizes of PCR products for different strains of rats are listed in Table S2.
(E) The first germline offspring (female) produced by chimera number 1 was mated with an SD male and had produced a litter of nine pups with different coat color patterns.
(F) Genotyping analysis of the above nine pups for the agouti gene. The agouti gene (A/A) is present in DA rats, but not in SD rats. Instead, SD rats have a nonagouti gene (a/a) resulting from a loss-of-function mutation of the agouti gene (Kuramoto et al., 2001). The sizes of PCR products for agouti and nonagouti genes are 153 bp and 134 bp, respectively. M, 100bp DNA marker; 1, the agouti/berkshire pup; 2, the agouti/hooded pup; 3, the black/hooded pup; 4–9, the six albino pups.
Table 1.
Rat ES Cell Karyotype and Chimera Production
| Cell Line | DAc2 (42,XX) |
DAc5 (42,XY) |
DAc3 (43,XXY) |
DAc8 (42,XY) |
F1 (42,XY) |
||
|---|---|---|---|---|---|---|---|
| Karyotype | |||||||
| Background | DA | DA | DA | DA | DA | DA | F344 |
| Passage Number | 25 | 39 | 25 | 26 | 12 | 20 | N/A |
| Diploid Cells, Normal Count | 19 | 22 | 18 | 0 | 20 | 16 | N/A |
| Diploid Cells, Cells with Gain or Loss | 2 | 0 | 3 | 20a | 0 | 4 | N/A |
| Polyploid: Diploid | 17:83 | 15:85 | 22:78 | 21:79 | 5:95 | 6:94 | N/A |
| Blasotocyst Injection | |||||||
| Passage Number | 15 | 12 | 12 | 15 | 11 | 12 | |
| Host Blastocyst | F344 | SD | SD | F344 | F344 | DA | |
| Foster Female | SD | SD | SD | SD | SD | SD | |
| Number of Blastocysts Injected | 43 | 104 | 9 | 28 | 24 | 29 | |
| Number Born | 7 | 18 | 1 | 4 | 2 | 3 | |
| Number of Chimeras | 2M, 3F | 8M, 7F | 1F | 3M, 1F | 1M | 0 | |
| Number of Germline Chimeras | 3F | 0 | 0 | 0 | N/Ab | 0 | |
M, male; F, female.
Of the 20 diploid cells examined, seven had an extra copy of the X chromosome (43,XXY). Nine cells had a translocation involving a chromosome 12, and an X chromosome in addition to a normal X and a normal Y chromosome [42, XY, −12, +Rb(12.X)]. Two cells showed random loss of the Y chromosome, one other cell showed random loss of an X chromosome, and another cell showed random loss of a chromosome 4.
This chimera (Figure S2B) will be ready for mating in January, 2009.
The genetic backgrounds of the host embryos and the donor ES cells are critical for germline transmission of the resulting chimeras in mice (Schwartzberg et al., 1989). This is also likely to be true in the rat. We have generated a total of 16 DA/SD chimeras by injection of DA ES cells into SD blastocysts (Table 1 and Figure S2C). All the 16 DA/SD chimeras developed into fertile adults and produced over 200 offspring. However, none of these offspring, as judged by coat color, were products of the transmission of the DA ES cell genome through the germline. This result suggests that SD blastocysts may be not suitable as a host embryo for DA ES cells. We also injected F344 rat ES cells into a total of 29 DA blastocysts and transferred them to three pseudopregnant SD rats. Three pups were born, and none of them were chimeras (Table 1).
L Cell/2i Conditions Support Robust Propagation and Genetic Manipulation of Rat ES Cells
Germline competent rat ES cells can be derived at relatively high efficiency and can be maintained in long-term culture under the MEF/3i condition, as shown above. However, we found that rat ES cells adhere poorly and tend to form small clumps that easily detach from feeders under this MEF/3i condition. Cell adhesion is mainly mediated by the integrin receptor family. The majority of integrins are expressed at a low level or are absent in rat ES cells, as suggested from microarray analysis and qRT-PCR results (Table S4 and Figure S3). Interestingly, integrin α7 and integrin α6 are highly expressed in rat ES cells, suggesting that rat ES cells may attach well if the right extracellular matrix (ECM) is provided. We have tested over 20 different types of cells as feeder layers for the growth of rat ES cells. Although rat ES cells did not adhere, or adhered poorly, to most of the feeders tested, we found that L cells, which were derived from subcutaneous connective tissues of adult male C3H/An mice, can efficiently support adherent growth of undifferentiated rat ES cells. We also found that the viability and growth of rat ES cells can be further improved if we use a more potent MEK inhibitor PD0325901 to replace both PD184352 and SU5402 (Ying et al., 2008). Rat ES cells readily attached to mitotically inactivated L cells after plating under the 2i (CHIR99021+PD0325901) condition and did not detach even after forming large colonies (Figure 5A). More importantly, rat ES cells maintained on this L cell/2i condition remained morphologically undifferentiated and expressed pluripotency markers Oct4 and Sox2 after long-term culture (Figure 5B).
Figure 5. Propagation and Genetic Manipulation of Rat ES Cells in L Cells/2i Conditions.
(A) DAc2 rat ES cells, 4 days after transfer from the MEF/3i condition to the L cell/2i condition. The scale bar represents 25 µm.
(B) Immunofluorescence staining for Oct4 and Sox2 in DAc2 rat ES cells after 15 passages in the L cell/2i condition. The scale bar represents 50 µm.
(C) One of the GFP-positive DAc2 rat ES cell colonies formed after transfection with GFP plasmids by nucleofection and selection with puromycin in L cell/2i conditions. The scale bar represents 50 µm.
(D) DAc2-GFP rat ES cells maintained in L cell/2i conditions for six passages. The scale bar represents 50 µm.
We next used the CAG-eGFP-IRES-pac plasmid to assess the feasibility of performing genetic manipulation in rat ES cells under L cell/2i conditions. The eGFP transgene is driven by a robust CAG promoter. GFP-positive cells should also be resistant to puromycin because of the IRES-pac cassette. Initially, eGFP plasmids were transfected into rat ES cells via the conventional electroporation method. Cell viability was low after transfection, and very few colonies grew up 1 week after selection in puromycin. We then tested the nucleofection method with which high efficiency of gene transfer can be achieved (Hohenstein et al., 2008). About 40% of rat ES cells were GFP positive a day after transfection by nucleofection in L cell/2i conditions. We then added 0.4 µg/ml puromycin to select for stable transfectants. Approximately 200 GFP-positive colonies were formed out of 10,000 rat ES cells plated 7 days after selection (Figure 5C). GFP-positive colonies were picked and expanded to establish stable GFP-positive rat ES cell lines (Figure 5D).
Elevated LIF/STAT3 Signaling Supports Rat ES Cell Self-Renewal
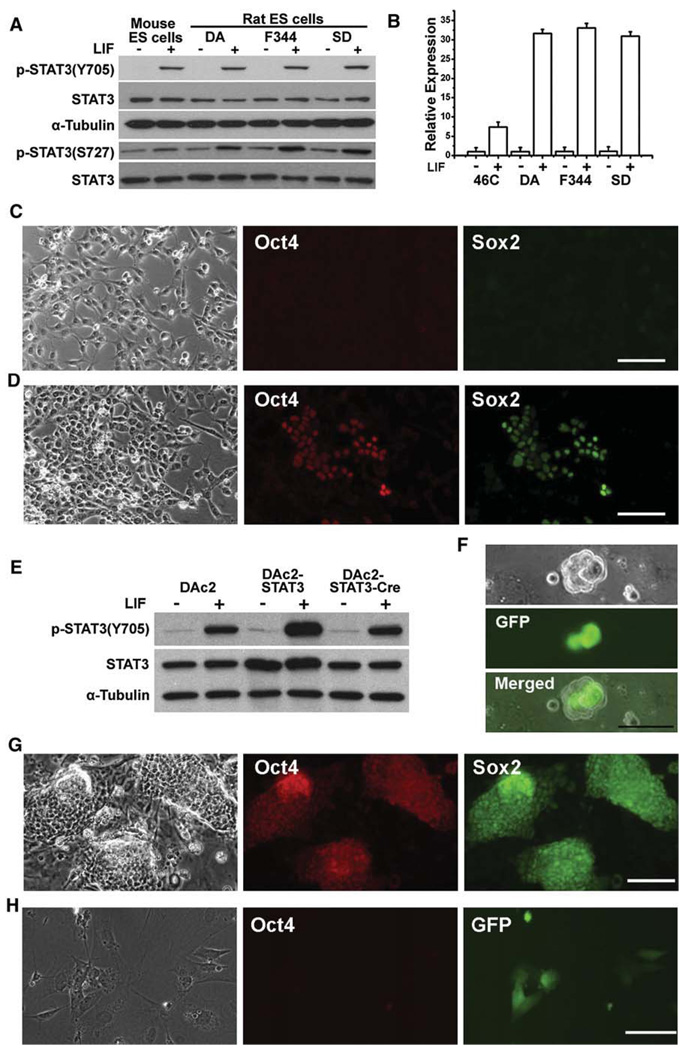
Signal transducer and activator of transcription 3 (STAT3) is activated by tyrosine phosphorylation at a single site Tyr705, as well as by serine phosphorylation at Ser727. Activation of STAT3 is essential for mouse ES cell self-renewal mediated by signaling through LIF/gp130 receptors (Niwa et al., 1998). Interestingly, LIF/STAT3 signaling fails to maintain self-renewal of mouse EpiScs and human ES cells (Brons et al., 2007; Daheron et al., 2004; Humphrey et al., 2004; Tesar et al., 2007). To investigate the potential role of LIF/STAT3 signaling in rat ES cells, we first examined whether STAT3 is functional in rat ES cells. LIF strongly induced phosphorylation of STAT3 at Tyr705 and Ser727 in mouse and rat ES cells (Figure 6A). The expression of suppressor of cytokine signaling 3 (Socs3), which is one of STAT3’s direct target genes, was highly induced after stimulation with LIF (Figure 6B). These data suggest that the LIF/STAT3 pathway is functional in rat ES cells. We next examined whether LIF has any beneficial effects on rat ES cells. Feeders produce LIF and may mask any effects of exogenous addition of the factor, so we plated rat ES cells onto laminin-coated dishes in N2B27 medium because rat ES cells do not attach to gelatin. All the cells differentiated rapidly under this condition (Figure 6C). In the presence of LIF, approximately 10%–20% cells remained undifferentiated after 7 days in culture (Figure 6D), suggesting that LIF has a positive effect on rat ES cell self-renewal. However, the undifferentiated cells could not be maintained beyond two passages. Serum or BMP4 had no beneficial effect. In fact, we found that addition of serum rapidly induced cell death and differentiation even in 3i or 2i conditions. LIF alone was also insufficient in retaining the undifferentiated state of rat ES cells grown on feeders, which may account for the previous failures in establishing rat ES cells.
Figure 6. Rat ES Cells Are Responsive to LIF/STAT3 Signaling for Self-Renewal.
(A) Analysis of STAT3 activation by western blot in mouse and rat ES cells stimulated with 10 ng/ml LIF for 30 min.
(B) qRT-PCR analysis of Socs3 induction by LIF treatment in mouse 46C ES cells and rat ES cells. Data represent mean ± SD of triplicate samples from two independent experiments.
(C and D) Immunofluorescence staining for Oct4 and Sox2 in DAc2 rat ES cells 7 days after they were cultured in laminin/N2B27 (C) or laminin/N2B27+LIF (D) conditions. Scale bars represent 50 µm.
(E) Western blot analysis of STAT3 activation in DAc2, DAc2-STAT3, and DAc2-STAT3-Cre rat ES cells after treatment with 10 ng/ml LIF for 30 min.
(F) DAc2-STAT3 cells, 1 day after transient transfection with Cre to remove the STAT3 transgene and simultaneously activate GFP. The scale bar represents 50 µm.
(G) Immunofluorescence staining for Oct4 and Sox2 in DAc2-STAT3 cells after nine passages in L cell/LIF conditions. The scale bar represents 50 µm.
(H) Immunofluorescence staining for Oct4 in DAc2-STAT3-Cre cells at second passage in L cell/LIF conditions. The presence of GFP denotes the removal of the STAT3 transgene. The scale bar represents 50 µm.
Feeder-dependent mouse ES cells from strains other than 129 undergo apoptosis and differentiation after the removal of feeders even in the presence of LIF. We found that this phenotype can be rescued simply by increasing STAT3 activation, suggesting that STAT3’s function is dose-dependent (E.N.S. and Q.-L.Y., unpublished data). To examine the effect of elevated STAT3 activation in rat ES cells, we introduced a STAT3 transgene into DAc2 cells. Overexpression of STAT3 in DAc2-STAT3 cells was confirmed by Western blot (Figure 6E). The STAT3 transgene was flanked by loxP sites. Treatment with Cre removed the STAT3 transgene and simultaneously activated GFP (Figures 6E and 6F) (Ying et al., 2008). DAc2-STAT3 cells have been maintained in L cell/LIF conditions for 11 passages without overt differentiation (Figure 6G), whereas the removal of LIF resulted in rapid cell death and differentiation (data not shown). Cre revertants (GFP positive) also died or differentiated after just one passage, despite the presence of LIF (Figure 6H), which is similar to wild-type DAc2 cells. Undifferentiated DAc2-STAT3 cells could also be maintained in laminin/N2B27+LIF conditions for up to five passages, after which they gradually died or differentiated. These results suggest that rat ES cells, like mouse ES cells, are also responsive to LIF/STAT3 signaling for self-renewal, although factor(s) provided by feeders are still required for long-term culture.
DISCUSSION
Our results demonstrate that the use of 3i enables us for the first time to efficiently derive and maintain cells from rat blastocysts with all the key features of ES cells: expression of pluripotency markers Oct4, Nanog, and Sox2; long-term self-renewal; the capacity to differentiate into derivatives of all three germ layers; and most importantly, the ability to produce high rates of chimerism and to transmit through the germline. We have also developed methods that enable us to robustly propagate and genetically manipulate rat ES cells, paving the way for the application of gene targeting and related genome engineering technologies in the rat.
In the past two decades, ES cells have been routinely used to create loss of function (knockout) or gene replacement (knockin) mutations by homologous recombination in the mouse, providing an invaluable tool for the functional characterization of genes (Capecchi, 2005). Now, the availability of true rat ES cells provides an opportunity to adapt the technology developed in the mouse to the rat. However, we still need to overcome several obstacles before rat ES cells can be used routinely to produce transgenic and gene knockout rat models. We need to optimize culture conditions so that we can efficiently perform genetic manipulation in rat ES cells while their germline competency is retained. In addition, we need to find the optimal host, donor, and recipient combination for efficient germline transmission of rat ES cells.
We found that it is technically challenging to genetically manipulate rat ES cells in the MEF/3i condition. This is mainly because rat ES cells are deficient in cell adhesion and are very sensitive to drug selection under this condition. In contrast, L cell/2i conditions allow us to robustly propagate and genetically manipulate rat ES cells. We are currently investigating whether germline competence of rat ES cells is retained after long-term culture in L cell/2i conditions. ES cells are subjected to selection pressures and are likely to acquire genetic and epigenetic changes that favor self-renewal over differentiation. Chromosomal abnormality is one the major causes for the loss of germline competence of mouse ES cells (Liu et al., 1997). This is likely also the case in rat ES cells. We found that the karyotype of rat ES cells is reasonably stable at early passage numbers, but chromosomal abnormalities increase with higher passages (data not shown). Developing optimal culture conditions for the maintenance of chromosome stability in rat ES cells is clearly important for their future broad applications. We routinely use trypsin to passage rat ES cells. Several studies indicate that the use of trypsin selects for aneuploid human ES cells in culture (Chan et al., 2008). It will be of interest to find out whether nonenzymatic passaging methods will improve the rat ES cell quality after long-term expansion. CHIR99021 is the key component in the 3i or 2i formulations. A recent report suggests that CHIR99021 can delay chromosome alignment and induce chromosome instability in cultured HeLa cells (Tighe et al., 2007). If the compound has the same effect on rat ES cells, we then need to test other factors that can replace CHIR99021, such as Wnt3a (Willert et al., 2003).
Because of the advantage of the dominant agouti coat color, we chose DA rat ES cells to perform most of the experiments described in this paper. A relatively high efficiency of germline transmission was achieved by injection of DA ES cells into F344 blastocysts and the subsequent transfer of the embryos to the recipient pseudopregnant SD rats. However, we still do not know the homologous recombination efficiency in DA rat ES cells and whether they can still efficiently transmit through the germline after extensive genetic manipulation and long-term culture. These properties are essential if we are to exploit all the genetic manipulation now only possible in mice. We may need to test more different strains of rat ES cell lines and different strain blastocysts as host embryos before we can identify one combination that can yield desirable efficiency for both homologous recombination and germline transmission.
Germline-competent mouse and rat ES cells can both be derived and maintained in the same 3i condition. They also share similar molecular and epigenetic signatures and are responsive to LIF/STAT3 signaling for self-renewal. We hypothesize that the fundamental mechanism underlying self-renewal of authentic ES cells might be conserved among different species. It will be of interest to investigate whether ES cells from other mammals, such as cows and pigs, can also be established using culture formulations on the basis of the 3i principle.
EXPERIMENTAL PROCEDURES
Media, Feeders, Animals, and Primers
Serum-free N2B27 medium was prepared as described (Nichols and Ying, 2006). CHIR99021, PD184352, PD0325901, and SU5402 were provided by Stem Cell Sciences plc. 3i medium was prepared by the addition of 3 µM CHIR99021, 0.8 µM PD184352, and 2 µM SU5402 to N2B27 medium. In 2i medium, 0.4 µM PD0325901 was used to replace PD184352 and SU5402. γ-irradiated MEFs were used as feeders and maintained in GMEM (Sigma)/ 10% FBS (HyClone) medium. REFs were derived from E14.5 DA rat embryos. Timed pregnant DA, F344, and SD rats were purchased from Harlan. Animal experiments were performed according to the investigator’s protocols approved by the USC Institutional Animal Care and Use Committee. All the primer sequences are listed in Table S5.
Derivation, Propagation, and Gene Transfection of Rat ES Cells
Rat blastocysts were gently flushed out from the uteruses of E4.5 timed-pregnant rats with N2B27 medium. After the removal of the zona with acid tyrodes solution (Sigma), whole blastocysts were transferred into 4-well plates and cultured on MEFs with 3i medium. After 5–7 days, the outgrowths of blastocysts were disaggregated and replated in the same MEF/3i conditions. Emerging ES cell colonies were then trypsinized and expanded. Established rat ES cell lines were routinely maintained in MEF/3i conditions. Medium was changed every other day, and cells were split with 0.05% trypsin every 3–4 days. In L cell/2i conditions, feeders were prepared by mixing of γ-irradiated L cells (ATCC Number: CCL-1) with either MEFs or DR4 feeders at a ratio of 1–2:1 and plating onto gelatin coated dishes at a density of 5 × 104/cm2. DR4 feeders were prepared from E13.5 mouse embryos that have been engineered to be resistant to hygromycin, puromycin, 6-thioguanine, and neomycin. For nucleofection, 10 µg linearized pCAG-eGFP-IP or pPyFloxedMT-STAT3-IPgfp plasmids were transfected into 2,000,000 rat ES cells with Mouse ES Cell Nucleofector Kit (amaxa Inc.) according to the manufacturer’s instruction. The pPyFloxedMTIPgfp vector has been described previously (Ying et al., 2008). The full-length mouse STAT3 transgene was obtained from addgene. There is only one amino acid difference between mouse and rat STAT3 proteins. After nucleofection, cells were cultured in L cell/2i conditions (mixed with DR4 feeders), and 0.4 µg/ml puromycin was applied to select for stable transfectants 2 days after transfection. Individual stably transfected colonies were picked and expanded. For removal of the STAT3 transgene, DAc2-STAT3 cells were transfected with pCAG-Cre-IP plasmids using Lipofectin LTX (Invitrogen) and the resulting GFP-positive cells were separated by FACS sorting.
Rat Blastocyst Injection
Blastocyst injection was performed as described (Nagy et al., 2003) with some modification. Blastocysts from E4.5 timed-pregnant rats were placed into a droplet of M2 medium and incubated in M16 medium (Sigma) for 2–3 hr, and the well-expanded blastocysts were used for microinjection. Eight to twelve rat ES cells (passage numbers between 12 and 15) were injected into each blastocyst and incubated at 37° C for 1 hr in M16 medium to allow the recovery of embryos. Eight to ten embryos were then transferred into the uterine horn of each E3.5 pseudopregnant female SD rat. Chimeric rats were identified by coat color. Germline transmission was tested by mating chimeras with SD rats.
In Vitro Differentiation of Rat ES Cells
Differentiation of rat ES cells was induced by the formation of EBs. Rat ES cells were plated into noncoated Petri dishes in REF-conditioned medium containing GMEM/10% FBS plus 1 mM glutamine. Cells grew in suspension and formed EBs, which were then plated onto matrigel coated dishes in either N2B27 or GMEM/10% FBS medium. The expression of markers for the three germ layers was examined by RT-PCR or immunostaining.
Immunostaining and AP Staining
Immunostaining was performed via standard protocols. Primary antibodies used include the following: Oct4 (C-10, Santa Cruz, 1:200), Sox2 (Y-17, Santa Cruz, 1:200), SSEA-1 (480, Santa Cruz, 1:200), SSEA-4 (813-70, Santa Cruz, 1:200), GATA-4 (G-4, Santa Cruz, 1:200), Nanog (Abcam, 1:200), βIII-tubulin (Sigma, 1:200), and Myosin (MF-20, 1:5). GCTM-2 antibody was provided by Martin Pera’s lab. Alexa Fluor fluorescent secondary antibodies (Invitrogen) were used at 1:1000 dilution. Nuclei were visualized with DAPI staining. AP staining was performed with an alkaline phosphatase kit (Sigma) according to the manufacturer’s instructions.
RT-PCR and qRT-PCR
Total RNA was extracted with RNA Easy Mini Kit (QIAGEN). cDNA was synthesized with 1 µg of total RNA using Cloned AMV First-Strand cDNA Synthesis Kit. AMV and Oligo (dT) 20 primers were used in a 20 µl reaction according to the manufacturer’s instructions. PCR reaction mixtures were prepared with Taq DNA polymerase (Invitrogen) and 1/20 of the above reaction as template. qRT-PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) according to manufacturer’s instructions. Signals were detected with an ABI7900HT Real-Time PCR System (Applied Biosystems). The relative expression level was determine by the 2−ΔDCT method and normalized against GAPDH.
Western Blot
Western blotting was performed according to a standard protocol. For the detection of phosphorylated STAT3, cells were stimulated with 10 ng/ml human LIF (Sigma) for 30 min. The primary antibodies used include the following: anti-STAT3 (BD, 1:2000), anti-phospho-STAT3 (Tyr705) (Cell signaling, 1:2000), and anti-phospho-STAT3 (S727) (Santa Cruz, 1:2000).
Genotyping and Karyotyping
Genotyping of animals was carried out by PCR on tail DNA. Primer sequences for microsatellite analysis were obtained from rat genome database website (http://rgd.mcw.edu/). For karyotyping, rat ES cells were treated with colcemid (Sigma) at a final concentration of 100 ng/ml for 2 hr before being harvested for metaphase preparation by standard methods. The GTW banding method was used for chromosome analysis as described (Hsieh, 1997).
Bisulfite Sequencing
Genomic DNA was extracted with a genomic DNA purification kit (QIAGEN). DNA (500 ng) from each sample was treated with EZ DNA methylation kit (ZYMO) to convert the unmethylated C to U. The promoter regions of Oct4 and Nanog were amplified by PCR and cloned into pCR-Blunt II-TOPO vector (Invitrogen) and sequenced with T7 and Sp6 primers.
Chromatin Immunoprecipitation
ChIP assays of cultured rat ES cells and REFs were performed as described previously (Jia et al., 2006). In brief, chromatin from 1 × 107 fixed cells was sonicated to a size range of 200–1000 bp as analyzed on agarose gels. Solublilized chromatin was subjected to immunoprecipitation with antibody against H3K4me3 (Abcam #8580) or H3K27me3 (Upstate #07-449). DNA samples from ChIP preparation were analyzed by qPCR with SYBR Green PCR Master Mix. The relative enrichment of each site was determined by the 2−ΔCT method and normalized against input DNA.
DNA Microarray Analysis
Total RNA was extracted form rat ES cells or REFs with Trizol (Invitrogen) and purified by RNeasy Clean up column (QIAGEN). RNA was amplified, labeled, and hybridized to the Rat Affymetrix GeneChip Gene 1.0 ST Array according to standard Affymetrix protocols.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jing Du, Yu Su, Robin Wesselschmidt, Hongjun Zhang, and Shelley Hough for technical assistance; Gerald Chester for ordering animals; Ramiro Montano, Gloria Martinez, and colleagues for rat husbandry; and Dr. Wange Lu for sharing research materials. This work was supported by USC Startup fund (Q.-L.Y.) and in part by California Institute for Regenerative Medicine (CIRM) Scientific Excellence through Exploration and Development (SEED) grants RS1-00327-1 (Q.-L.Y.).
Footnotes
ACCESSION NUMBERS
Microarray data have been deposited in the Gene Expression Omnibus Database with the accession number GSE13681.
SUPPLEMENTAL DATA
Supplemental Data include three figures, five tables, and one movie and can be found with this article online at http://www.cell.com/supplemental/S0092-8674(08)01567-5.
REFERENCES
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Brenin D, Look J, Bader M, Hubner N, Levan G, Iannaccone P. Rat embryonic stem cells: a progress report. Transplant. Proc. 1997;29:1761–1765. doi: 10.1016/s0041-1345(97)00046-8. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Brook FA, Gardner RL. The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA. 1997;94:5709–5712. doi: 10.1073/pnas.94.11.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M, Nichols J, Stenhouse F, Mountford P, Greenhalgh CJ, Kantachuvesiri S, Brooker G, Mullins J, Smith AG. Rapid loss of Oct-4 and pluripotency in cultured rodent blastocysts and derivative cell lines. Biol. Reprod. 2003;68:222–229. doi: 10.1095/biolreprod.102.006197. [DOI] [PubMed] [Google Scholar]
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005;6:507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- Chan EM, Yates F, Boyer LF, Schlaeger TM, Daley GQ. Enhanced plating efficiency of trypsin-adapted human embryonic stem cells is reversible and independent of trisomy 12/17. Cloning Stem Cells. 2008;10:107–118. doi: 10.1089/clo.2007.0064. [DOI] [PubMed] [Google Scholar]
- Daheron L, Opitz SL, Zaehres H, Lensch WM, Andrews PW, Itskovitz-Eldor J, Daley GQ. LIF/STAT3 signaling fails to maintain self-renewal of human embryonic stem cells. Stem Cells. 2004;22:770–778. doi: 10.1634/stemcells.22-5-770. [DOI] [PubMed] [Google Scholar]
- Damjanov I, Sell S. Yolk sac carcinoma grown from rat egg cylinders. J. Natl. Cancer Inst. 1977;58:1523–1525. doi: 10.1093/jnci/58.5.1523. [DOI] [PubMed] [Google Scholar]
- Davies SP, Reddy H, Caivano M, Cohen P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers SP, Yoo JG, Lian L, Therrien J, Smith LC. Rat embryonic stem-like (ES-like) cells can contribute to extraembryonic tissues in vivo. Cloning Stem Cells. 2007;9:512–522. doi: 10.1089/clo.2007.0029. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fandrich F, Lin X, Chai GX, Schulze M, Ganten D, Bader M, Holle J, Huang DS, Parwaresch R, Zavazava N, et al. Preimplantation-stage stem cells induce long-term allogeneic graft acceptance without supplementary host conditioning. Nat. Med. 2002;8:171–178. doi: 10.1038/nm0202-171. [DOI] [PubMed] [Google Scholar]
- Gardner RL. Regeneration of endoderm from primitive ectoderm in the mouse embryo: fact or artifact? J. Embryol. Exp. Morphol. 1985;88:303–326. [PubMed] [Google Scholar]
- Ginis I, Luo Y, Miura T, Thies S, Brandenberger R, Gerecht-Nir S, Amit M, Hoke A, Carpenter MK, Itskovitz-Eldor J, et al. Differences between human and mouse embryonic stem cells. Dev. Biol. 2004;269:360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- Hohenstein KA, Pyle AD, Chern JY, Lock LF, Donovan PJ. Nucleofection mediates high-efficiency stable gene knockdown and trans-gene expression in human embryonic stem cells. Stem Cells. 2008;26:1436–1443. doi: 10.1634/stemcells.2007-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CL. Basic cytogenetic techniques: culture, slidemaking and banding. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. Second Edition. New York: Academic Press; 1997. pp. 391–396. [Google Scholar]
- Humphrey RK, Beattie GM, Lopez AD, Bucay N, King CC, Firpo MT, Rose-John S, Hayek A. Maintenance of pluripotency in human embryonic stem cells is STAT3 independent. Stem Cells. 2004;22:522–530. doi: 10.1634/stemcells.22-4-522. [DOI] [PubMed] [Google Scholar]
- Jacob HJ, Kwitek AE. Rat genetics: attaching physiology and pharmacology to the genome. Nat. Rev. Genet. 2001;3:33–42. doi: 10.1038/nrg702. [DOI] [PubMed] [Google Scholar]
- Jia L, Shen HC, Wantroba M, Khalid O, Liang G, Wang Q, Gentzschein E, Pinski JK, Stanczyk FZ, Jones PA, et al. Locus-wide chromatin remodeling and enhanced androgen receptor-mediated transcription in recurrent prostate tumor cells. Mol. Cell. Biol. 2006;26:7331–7341. doi: 10.1128/MCB.00581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Keller GM. In vitro differentiation of embryonic stem cells. Curr. Opin. Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- Kuramoto T, Nomoto T, Sugimura T, Ushijima T. Cloning of the rat agouti gene and identification of the rat nonagouti mutation. Mamm. Genome. 2001;12:469–471. doi: 10.1007/s003350020010. [DOI] [PubMed] [Google Scholar]
- Liu X, Wu H, Loring J, Hormuzdi S, Disteche CM, Bornstein P, Jaenisch R. Trisomy eight in ES cells is a common potential problem in gene targeting and interferes with germ line transmission. Dev. Dyn. 1997;209:85–91. doi: 10.1002/(SICI)1097-0177(199705)209:1<85::AID-AJA8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi M, McMahon G, Sun L, Tang C, Hirth P, Yeh BK, Hubbard SR, Schlessinger J. Structures of the tyrosine kinase domain of fibroblast growth factor receptor in complex with inhibitors. Science. 1997;276:955–960. doi: 10.1126/science.276.5314.955. [DOI] [PubMed] [Google Scholar]
- Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, Peggie M, Bain J, Bloomberg GB, Grahammer F, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem. J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo, A Laboratory Manual. Third Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- Nichols J, Smith A, Buehr M. Rat and mouse epiblasts differ in their capacity to generate extraembryonic endoderm. Reprod. Fertil. Dev. 1998;10:517–525. doi: 10.1071/rd98075. [DOI] [PubMed] [Google Scholar]
- Nichols J, Ying QL. Derivation and propagation of embryonic stem cells in serum- and feeder-free culture. Methods Mol. Biol. 2006;329:91–98. doi: 10.1385/1-59745-037-5:91. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Madison BB, Kano H. Mutagenesis in rodents using the L1 retrotransposon. Genome Biol. 2007;8 Suppl 1:S16. doi: 10.1186/gb-2007-8-s1-s16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp JP. Genetic analysis of inherited hypertension in the rat. Physiol. Rev. 2000;80:135–172. doi: 10.1152/physrev.2000.80.1.135. [DOI] [PubMed] [Google Scholar]
- Schwartzberg PL, Goff SP, Robertson EJ. Germ-line transmission of a c-abl mutation produced by targeted gene disruption in ES cells. Science. 1989;246:799–803. doi: 10.1126/science.2554496. [DOI] [PubMed] [Google Scholar]
- Smith AG. Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Smits BM, Mudde JB, van de Belt J, Verheul M. Generation of gene knockouts and mutant models in the laboratory rat by ENU-driven target-selected mutagenesis. Pharmacogenet. Genomics. 2006;16:159–169. doi: 10.1097/01.fpc.0000184960.82903.8f. [DOI] [PubMed] [Google Scholar]
- Solter D, Skreb N, Damjanov I. Extrauterine growth of mouse egg-cylinders results in malignant teratoma. Nature. 1970;227:503–504. doi: 10.1038/227503a0. [DOI] [PubMed] [Google Scholar]
- Stevens LC. The development of transplantable teratocarcinomas from intratesticular grafts of pre- and postimplantation mouse embryos. Dev. Biol. 1970;21:364–382. doi: 10.1016/0012-1606(70)90130-2. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Tighe A, Ray-Sinha A, Staples OD, Taylor SS. GSK-3 inhibitors induce chromosome instability. BMC Cell Biol. 2007;8:34. doi: 10.1186/1471-2121-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda S, Kawamata M, Teratani T, Shimizu T, Tamai Y, Ogawa H, Hayashi K, Tsuda H, Ochiya T. Establishment of rat embryonic stem cells and making of chimera rats. PLoS ONE. 2008;3:e2800. doi: 10.1371/journal.pone.0002800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilieva S, Guan K, Pich U, Wobus AM. Establishment of SSEA-1- and Oct-4-expressing rat embryonic stem-like cell lines and effects of cytokines of the IL-6 family on clonal growth. Exp. Cell Res. 2000;258:361–373. doi: 10.1006/excr.2000.4940. [DOI] [PubMed] [Google Scholar]
- Willert K, Brown JD, Danenberg E, Duncan AW, Weissman IL, Reya T, Yates JR, 3rd, Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.