Abstract
The localization of genes within the nuclear space is of paramount importance for proper genome functions. However, very little is known on the cis-acting elements determining subnuclear positioning of chromosome segments. We show here that the D4Z4 human subtelomeric repeat localizes a telomere at the nuclear periphery. This perinuclear activity lies within an 80 bp sequence included within a region known to interact with CTCF and A-type Lamins. We further show that a reduced level of either CTCF or A-type Lamins suppresses the perinuclear activities of D4Z4 and that an array of multimerized D4Z4 sequence, which has lost its ability to bind CTCF and A-type Lamins, is not localized at the periphery. Overall, these findings reveal the existence of an 80 bp D4Z4 sequence that is sufficient to position an adjacent telomere to the nuclear periphery in a CTCF and A-type lamins-dependent manner. Strikingly, this sequence includes a 30 bp GA-rich motif, which binds CTCF and is present at several locations in the human genome.
Keywords: CTCF, D4Z4, FSHD, nuclear periphery, telomere
Introduction
The spatial distribution of chromosomal domains within the nucleoplasm has an important function in the regulation of eukaryotic genomes (Csink and Henikoff, 1996; Maillet et al, 1996; Brown et al, 1997; Andrulis et al, 1998; Taddei et al, 2006; Nagai et al, 2008). Very little is known on the cis-acting elements governing the subnuclear localization of genes or groups of genes, especially in mammals. However, these elements are expected to contribute to accurate genetic and epigenetic regulations. As examples, the localization of chromosome regions at the periphery of the nucleus seems to have a central function in gene regulation and DNA repair (Therizols et al, 2006; Finlan et al, 2008; Kumaran and Spector, 2008; Reddy et al, 2008). Nevertheless, the precise regulation of genes located at the nuclear rim remains poorly understood as the expression of some genes appears unaffected by their proximity to the nuclear periphery (Finlan et al, 2008; Kumaran and Spector, 2008).
In metazoan, the lamina, which coats the inner surface of the nuclear envelope, has an important function in the perinuclear localization of chromosome segments. For instance, in mice, lamin B1 is involved in the perinuclear localization of chromosome 18 (Malhas et al, 2007) and recent large-scale mappings of chromosomal domains attached to B-type lamins performed in Drosophila and human cells, defined the Lamin-associated domains or LADs (Pickersgill et al, 2006; Guelen et al, 2008), which correlate with silenced regions. In human cells, some of the LADs are flanked by binding sites for CTCF, suggesting that the insulator protein allows a functional partitioning of the human genome by separating active and inactive domains.
The telomeres are not randomly localized within the nucleoplasm and can be found at the nuclear periphery (Gilson et al, 1993). This positioning varies greatly among organisms, cell types, cell cycle stages and individual telomeres. At the bouquet stage for instance, the clustering of all telomeres at the edge of the nucleus, is a nearly universal feature of meiosis (Scherthan, 2007). In budding yeast, the 32 telomeres gather into 4–6 foci, associated with the nuclear envelope (Gotta et al, 1996) that sequesters heterochromatic factors (Gotta et al, 1996; Maillet et al, 1996). The peripheral positioning of yeast telomere is mediated by at least two complexes bound to the telomeric chromatin, suggesting that, in this model organism, the telomere by itself behaves as a perinuclear positioning element (Taddei et al, 2005). Consistent with this hypothesis, de novo telomeres formed in yeast, devoid of natural subtelomeric repeats adopt a perinuclear localization (Tham et al, 2001). A perinuclear positioning of telomeres is also observed in Plasmodium, where it favours subtelomeric gene conversion (Freitas-Junior et al, 2000), whereas in plants, telomeres are observed either close to the nuclear periphery (Rawlins and Shaw, 1990) or around the nucleolus (Fransz et al, 2002). In mammalian nuclei, telomeres adopt different locations (Luderus et al, 1996). Although human telomeres are clustered at the nuclear periphery in sperm (Zalenskaya et al, 2000), most telomeres in lymphocyte nuclei are located in the interior of the nucleoplasm (Weierich et al, 2003). Thus, it seems that by default, human telomeres are localized internally in most cell types. However, some subtelomeric elements may antagonize this internal localization and target their proximal telomere to the nuclear envelope as suggested by the presence of LADs at different subtelomeres (Guelen et al, 2008). Such an example of localization at the periphery of the nucleus is the positioning of the 4q35 subtelomeric locus, involved in the facio-scapulo-humeral dystrophy (FSHD) (Masny et al, 2004; Tam et al, 2004).
The existence in metazoan of cis-acting DNA elements targeting chromosomal regions to the nuclear envelope remains elusive and a key question regarding the function of perinuclear localization is to know whether it is the cause or consequence of differential gene expression or genomic nature.
To identify and characterize such elements, we investigated the ability of D4Z4, a 3.3 kb macrosatellite repeat present at the 4q35 locus (Hewitt et al, 1994; Lyle et al, 1995), to direct the localization of an associated telomere towards nuclear periphery. D4Z4 is repeated in tandem at several chromosomal loci including the short arm of acrocentric chromosomes, the pericentric region of chromosome 1q and the telomeric regions of the long arms of chromosomes 4 and 10 (Hewitt et al, 1994; Lyle et al, 1995). Normal 4q35 ends carry from 11 up to 150 copies of this element, whereas this number is reduced to 1–10 repeats in patients affected with the autosomal dominant FSHD (van Deutekom et al, 1993; Winokur et al, 1994). We report here that D4Z4, which we previously characterized as a CTCF- and A-type Lamins-dependent insulator (Ottaviani et al, 2009), behaves as a potent perinuclear cis-acting positioning element. Impressively, this property depends on CTCF and A-type Lamins binding and is lost upon multimerization of the repeats, uncovering a striking link between perinuclear localization, CTCF and A-type Lamins and transcriptional insulation.
Results and discussion
D4Z4 behaves as a perinuclear positioning element
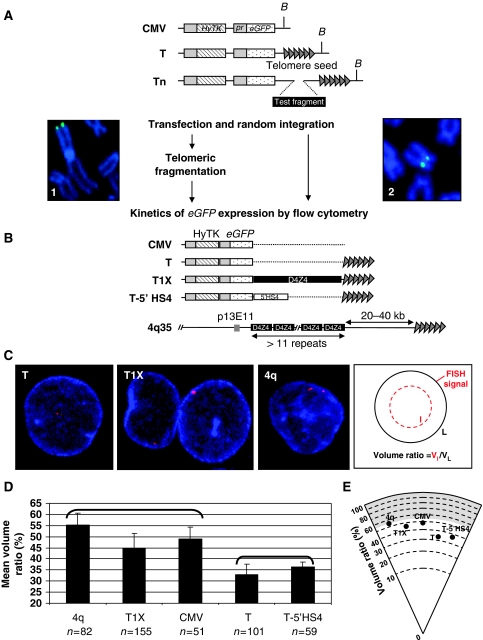
To evaluate the putative role of D4Z4 in the subnuclear localization of telomeres, we artificially tagged different telomeres by transfecting human cells with DNA molecules carrying D4Z4 sequences inserted between a seed of (TTAGGG)n telomeric repeats and a hygromycin-eGFP cassette (Figure 1A and B). Telomere fragmentation is based on the non-targeted introduction of cloned telomeres into mammalian cells and de novo telomere formation after telomeric healing (Farr et al, 1992). Successful de novo formation of hygromycin-tagged telomeres was confirmed in individual clones by detection of a diffuse hybridization signal in Southern blot or in the population of transfected cells by fluorescence in situ hybridization (FISH) of metaphase spreads (data not shown and Figure 1A). In agreement with earlier data (Koering et al, 2002), the rate of de novo telomere formation in stably transfected C33A cells is very high reaching 85–94% of the hygromycin-resistant cells (Supplementary Table S1). Therefore, polyclonal populations of stably transfected cells are representative of pools of independent clones of hygromycin-tagged telomeres and further experiments were performed on populations.
Figure 1.
D4Z4 relocates eGFP-tagged telomeres towards the nuclear periphery. (A) The constructs are derived from the pCMV vector, which carries a Hygromycin resistance gene fused to the herpes simplex virus type 1 thymidine kinase suicide gene (HyTK) and an eGFP reporter gene both driven by CMV promoters, and carry a telomere seed (depicted by arrows) that allows telomeric fragmentation. We inserted different sequences between the reporter and the telomere to investigate their respective effect on telomere positioning. Successful de novo formation of eGFP-tagged telomeres or internal integration of the CMV construct were confirmed by fluorescence in situ hybridization (FISH) on metaphase spreads (photographs 1, 2, respectively, Supplementary Table S1). M-FISH analysis performed on cells containing the pCMV Telo vector (T) or a vector with D4Z4 (T1X) confirmed that constructs carrying the D4Z4 sequence do not integrate preferentially at certain chromosomes (Supplementary Figure S2). (B) CMV: empty vector; T: empty telomeric vector; T1X: insertion of a 3.3 kb D4Z4 element between eGFP and the telomere in T; T-5′ HS4: insertion of the 1.2 kb chicken β-globin insulator (Chung et al, 1993). Schematic representation of the normal 4q35 allele. (C) Confocal section of T, T1X cells stained with an eGFP probe (single red dot) or endogenous 4q with a 4qtel probe staining both alleles (two red dots). Representation of the analysis of a nucleus after two-colour 3D-FISH (right panel). We considered the outer limit of the Lamin B signal (blue) as the edge of the nucleus (100%). Distribution of the FISH signals within the nuclear volume was calculated from the centre (0%) to the outer edge of the sphere after reduction of the outer signal (VL=nuclear volume=100%) until it overlaps with the FISH signal (Vl=x% of VL) (Supplementary Figure S1). Experiments were performed on three to four independent populations of cells (Supplementary Table S1). (D) Histogram displaying the mean positioning of natural and fragmented telomeres as their mean values of volume ratio (Vl/VL)±s.d. shown by error bars (y-axis). Data sets were compared with the Kruskal–Wallis statistic test (P<2.4 × 10−9, n=number of interphase nuclei). Brackets identify two groups where all conditions are significantly different from the other group, based on FDR determination. (E) Distribution of the FISH signal from the centre to the outer rim of the nucleus. Lamin B signal occupies the outermost 18% of the nuclear radius considered (grey shadow).
The tri-dimensional distribution of the FISH signal corresponding to the hygromycin-eGFP cassette was determined in different cell populations as a function of the nuclear volume after delineation of the nuclear periphery by simultaneous immunolocalization of B-type Lamins (Figure 1C; Supplementary Figure S1). Using this 3D-immuno-FISH method, we showed that de novo formed telomeres, devoid of natural subtelomeric sequences, have the tendency to localize in the innermost nuclear volume (T construct in Figure 1C–E). By contrast, the eGFP-hygromycin construct but devoid of telomere seed appears randomly distributed between the interior and the periphery of the nucleoplasm (Figure 1D and E). These results are in agreement with several reports suggesting that human telomeres have the intrinsic property to be anchored to internal sites (Luderus et al, 1996; Weierich et al, 2003; Tam et al, 2004).
As expected from earlier observations, the 4q35 locus is enriched in a peripheral zone of the nucleus (Figure 1C–E) (Masny et al, 2004; Tam et al, 2004). Strikingly, telomeres associated to a single copy of D4Z4 have a more peripheral distribution (T1X construct) than D4Z4-less telomeres (T construct) (Figure 1C–E), suggesting that this element might be specialized in the subnuclear positioning of telomeres. The perinuclear localization of D4Z4-associated telomeres unlikely results from biases in telomere seeding or in chromosome choice as we observed a similar rate of telomere formation (Supplementary Table S1) and no preferential insertion between populations of cells transfected with seeding constructs with or without D4Z4 (Supplementary Figure S2).
We recently showed that D4Z4 act as a CTCF-dependent insulator protecting from position effect variegation and enhancer-promoter communication (Ottaviani et al, 2009). The protection against silencing is mediated in part by CTCF and A-type Lamins, which bind in vivo to D4Z4 as shown by chromatin immunoprecipitation. Upon multimerization of D4Z4, both CTCF binding and insulator activity are lost suggesting that FSHD is associated with a gain of function of CTCF (Ottaviani et al, 2009). Interestingly, insulators are often anchored at fixed nuclear substructures including the nuclear periphery in eukaryotes (Gerasimova and Corces, 2001; Gaszner and Felsenfeld, 2006; Valenzuela and Kamakaka, 2006). Therefore, to test whether peripheral tethering of telomeres is a common property of CTCF-dependent vertebrate insulators, we compared D4Z4 with the 5′ HS4 chicken β-globin CTCF-dependent insulator inserted near telomeres (Figure 1B). We showed that 5′HS4-tagged telomeres are localized in the innermost nuclear volume as observed for the T construct, suggesting that the peripheral positioning activity is an intrinsic property of D4Z4 (Figure 1C–E).
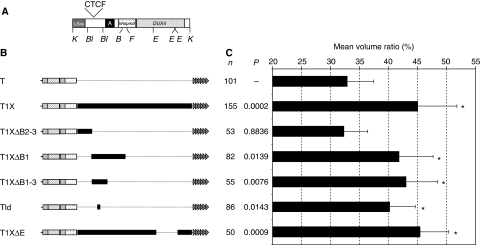
We have previously identified a shorter fragment with anti-silencing activity within D4Z4 and we mapped by deletion the region of D4Z4 involved in this perinuclear positioning activity. We found three truncated forms of D4Z4 that recapitulate both the insulator activity and the peripheral localization of telomeres of the full-length repeat (Figure 2, T1XΔB1, T1XΔB1-3, T1XΔE), whereas the very proximal 382 bp at the 5′ end of D4Z4 (Figure 2, T1XΔB2-3) harbours the same distribution than a D4Z4-less telomere. Impressively, a short sequence of 80 bp that harbours anti-silencing activity (Supplementary Figure S3) is sufficient to localize a telomere closer to the periphery (Figure 2, Tld). Therefore, we conclude that D4Z4 has the ability to direct a subtelomeric region at or near the nuclear envelope and that this property is mediated, at least in part, by a segment present at the 5′ end of the D4Z4 repeat.
Figure 2.
Identification of the tethering element within D4Z4. (A) Schematic representation of this element from position 1–3303 relative to the two flanking KpnI sites (K) (to scale), the different regions within D4Z4 are shown. Each repeat contains two classes of repetitive DNA, LSau, a repetitive element associated with heterochromatin regions, a GC-rich low copy repeat, hhspm3 displaying sperm-specific DNA hypomethylation and a region named region A. DUX4 corresponds to an ORF with a double homeobox encoding the DUX4 protein putatively involved in the disease. The position of the CTCF site is shown (Ottaviani et al, 2009). The restriction sites used for the cloning of D4Z4 subfragments are indicated (B: BamHI; Bl: BlpI; E: EheI) and the different constructs used are depicted (B). (C) Subnuclear positioning of eGFP-tagged telomeres in cells containing different fragments of D4Z4. The number of nuclei analysed per construct is indicated (n) together with the P-values determined with the Mann–Whitney tests using the T construct as the reference. Significant conditions after false discovery rate (FDR, α=0.05) correction for multiple comparisons are marked by asterisk. These results reveal that constructs containing the proximal insulator of D4Z4 displace a telomere towards the nuclear periphery, whereas T1XΔ2-3 occupies the same positioning as the T constructs.
CTCF and A-Lamins contribute to the localization of D4Z4 at the nuclear periphery
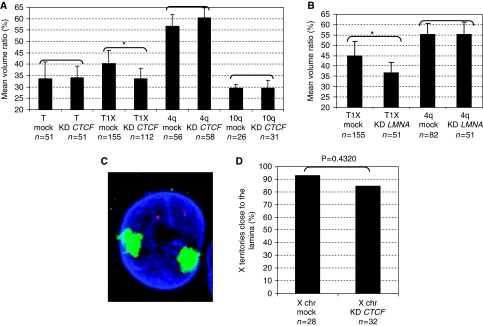
As the different fragments involved in the perinuclear positioning possess an anti-silencing activity mediated in vivo by CTCF and A-type Lamins (Ottaviani et al, 2009; Supplementary Figure S3), we wondered whether these proteins are involved in the positioning activity of D4Z4 as well. When the expression of CTCF or LMNA is significantly reduced by RNA interference (Supplementary Figure S4), the distribution of D4Z4-tagged telomeres is more internal (Figure 3A and B) revealing an essential role for these proteins in the perinuclear distribution of D4Z4. Importantly, the effect of CTCF depletion is specific for D4Z4 as our RNA interference conditions are (i) not accompanied by a modification of the localization of D4Z4-less telomeres (ii) nor by any global disturbance of the nuclear architecture as shown by the distribution of the 10q locus and X chromosome (Figure 3A, C and D) and (iii) the CTCF-dependent 5′HS4 insulator of the chicken β-globin locus artificially integrated at chromosome ends (T-5′ HS4) remains in the inner nuclear space (data not shown). We conclude that D4Z4 positions its associated seeded telomere to the nuclear periphery through an interaction with CTCF and A-type Lamins, whereas in the absence of CTCF, the wild-type 4q35 locus remains at the nuclear periphery (Figure 3A), suggesting additional pathways for the anchoring of this region.
Figure 3.
CTCF and A-type Lamins contribute to the positioning of D4Z4-tagged telomeres. (A) Cells containing the T1X or the T transgene transiently transfected with CTCF (KD CTCF) or negative (mock) siRNA were compared using the Mann–Whitney test. Depletion of CTCF correlates with the relocation of D4Z4-tagged telomere to the interior of the nucleus (P=0.0198) whereas the T construct (P=0.597) and the 4q (P=0.536) are not affected. (B) LMNA depletion significantly displaces the T1X construct (P=0.0376) but does not affect the distribution of the 4q telomere (P=0.6973) (Mann–Whitney). (C) To ascertain the global integrity of the nucleus architecture after the transient knock-down of CTCF, we analysed the distribution of the 4q or 10q telomeres (red signal) and the whole X chromosome (green signal) in T1X cells after transfection of CTCF siRNA by 3D-FISH as described earlier (the blue signal corresponds to Lamin B). We do not observe a redistribution of the FISH signal for the 4q and 10q probes (panel A) or X chromosome territories (D) in cells transfected with siRNA compared with the control, suggesting that the reduced CTCF expression does not globally perturb the distribution of these chromosomes inside the nuclear space as observed for the D4Z4-tagged telomeres. For 4q and 10q distributions, we compared mock and knock-down conditions with the Mann–Whitney statistical test. The numbers of X territories associated to the Lamin B signal in these conditions were compared with the Fisher exact test.
The multimerization of D4Z4 suppresses peripheral localization
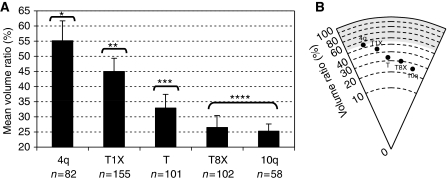
D4Z4 is repeated in tandem at several chromosomal loci including the 4q35 region linked to FSHD. Moreover, we demonstrated earlier that the binding of CTCF to D4Z4 is lost on multimerization of the repeat (Ottaviani et al, 2009) and that the positioning activity of D4Z4 is CTCF-dependent. Hence, we hypothesized that localization of a telomere-containing multiple D4Z4 repeats would be more internal and explored whether the multimerization alters D4Z4 perinuclear positioning activity. For this purpose, we transfected linearized DNA containing eight copies of the D4Z4 sequence abutting a telomere seed (T8X construct). As observed with the other telomere seeding plasmids containing 0 or 1 D4Z4 sequence, 94% of integrated T8X constructs form a de novo telomere (Supplementary Table S1) and the number of repeats remains stable after long-term culture (Supplementary Figure S5). As we hypothesized, T8X harbours the same subnuclear distribution than the D4Z4-less construct (T construct) and is more internal than telomeres carrying a single D4Z4 sequence (T1X construct) (Figure 4A and B). Interestingly, this loss of peripheral localization on multimerization correlates with a decrease in CTCF (Ottaviani et al, 2009) and A-type Lamins (Supplementary Figure S6) binding confirming the importance of CTCF for the peripheral positioning activity of D4Z4.
Figure 4.
Loss of CTCF binding upon D4Z4 multimerization abrogates peripheral tethering. (A) The position of the FISH signal was scored as described. The mean values±s.d. (y-axis) are presented for different populations compared with the Kruskal–Wallis statistic test (P<2.2 × 10−16, n=number of interphase nuclei). Multiple pairwise comparisons corrected for 5% FDR define four independent groups (identified by *; **; ***; **** above the brackets). (B) Mean positioning of the FISH signal for constructs with different number of D4Z4 from the centre (0%) to the outer (100%) edge of the nucleus.
The fact that the wild-type 4q telomere is not delocalized on CTCF knock-down in our cells together with the particular behaviour of the 4q35 locus, which remains at the nuclear periphery even in FSHD patients with only one copy of D4Z4 (Masny et al, 2004; Tam et al, 2004), suggests the existence of at least two Lamins-dependent pathways for the perinuclear targeting of the 4q35 region. The first one is CTCF-independent and likely acts through a sequence located upstream of the D4Z4 array on the 4q35 region, as suggested earlier (Masny et al, 2004) (Figure 5A). According to our results, the second pathway might be linked to a gain of function of CTCF in patients carrying a single or a weakly oligomerized D4Z4 array (Ottaviani et al, 2009). In this configuration, a region located upstream of D4Z4 (Masny et al, 2004; Guelen et al, 2008; Petrov et al, 2008) might cooperate with the remaining D4Z4 repeats bound to CTCF to tighten the bonds between the corresponding region and the nucleus rim (Figure 5B). Consistent with the existence of two positioning elements at the 4q35 end, the proximal 4q35 FISH probe (D4S139) that hybridizes 215 kb upstream of the region containing the repeats is more peripheral than the D4Z4 array in normal cells (Masny et al, 2004) where CTCF is absent (Ottaviani et al, 2009). The putative existence of another region upstream of D4Z4 mediating perinuclear positioning and the inability of long D4Z4 arrays to bind CTCF and A-type Lamins in normal cells might also explain why the D4Z4 repeats alone do not tether the 10q26 telomere to the rim of the nucleus (Masny et al, 2004) and why LMNA knock-down does not affect the 4q35 positioning.
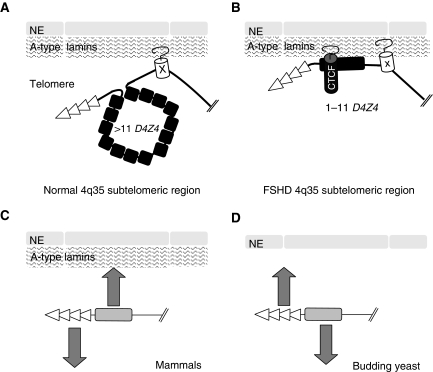
Figure 5.
Model for the tethering of the 4q35 locus at the nuclear periphery. (A) In normal cells, D4Z4 repeats are methylated (van Overveld et al, 2005) and not bound by CTCF (Ottaviani et al, 2009). On the basis of our data and those from Masny et al (2004); Tam et al (2004); Guelen et al (2008), we propose that this subtelomeric region is attached to the periphery through a Lamin-A-dependent tethering site localized outside the D4Z4 array, likely on the centromere side. (B) A model for the tethering of 4q35 at the nuclear periphery in FSHD cells. The contraction of D4Z4 allows the binding of CTCF (Ottaviani et al, 2009) and changes the functional organization of the 4q35 region. The D4Z4 repeats become specifically attached to the periphery (our work). We propose that this higher-order switch modifies the expression of FSHD-associated genes probably by preventing their repression (Gabellini et al, 2002). The involvement of CTCF opens new strategies for the identification of these genes. (C) In human cells, most of the telomeres are localized within the nucleoplasm, probably anchored at defined subnuclear sites. These sites might serve to nucleate heterochromatin at telomeres. The peripheral localization might be caused by specific subtelomeric elements. (D) In baker's yeast, the nuclear periphery acts as a subnuclear compartment where telomeres are clustered to recruit and bind key heterochromatic factors. The telomere-NE association is antagonized by subtelomeric insulator elements called STAR, linking subnuclear localization and transcriptional insulation (Magdinier et al, 2008). Similarities with yeast suggest that D4Z4 could represent a human equivalent of S. cerevisiae STAR elements.
Both loci are 98% homologous for the regions extending 40 kb proximal to D4Z4 and at least 10 kb distal to the repeat at 4q35 but does not contain the element located upstream the region of homology between the two chromosomes speculated earlier (Tam et al, 2004; Guelen et al, 2008). Consistent with this hypothesis, we observed a stronger degree of internal location (25.29%) than the de novo formed telomeres (Figure 4A) for the 10q terminus that does not possess the proximal positioning element postulated for 4q35 (see above). A second explanation might be linked to the D4Z4 sequence itself. Indeed, D4Z4 from 4q show a 98% homology to the one found on 10q but the major differences between the two types of repeat are found in the vicinity of the CTCF site that we identified earlier (Ottaviani et al, 2009) and consists in the insertion of 7–8 bases in the D4Z4 from 10q (Supplementary Figure S7). CTCF and A-type Lamins, together with yet unidentified factors might thus be present only on the type 4 D4Z4. However, one cannot exclude at this step that the level of LMNA KD might not be sufficient as we and others were only able to see the repositioning of the 4q35 locus can only be observed in cells with homozygous invalidation of the LMNA gene (Masny et al, 2004) (Ottaviani et al, unpublished observations).
Consistent with a key role for the 4q-specific repeat, it is interesting to note that in the Caucasian population at least 20% of individuals carry 10-type 3.3 kb repeats on chromosome 4 and vice versa (van Deutekom et al, 1996; van Overveld et al, 2000). However, the disease is always associated with a low number of four-type repeats (<11) on the 4q locus only, regardless of the size of the 10q–4q-hybrid array.
In summary, our data together with Masny et al (2004) and Guelen et al (2008), suggest that a normal 4q35 locus is attached to the nuclear rim through a D4Z4-independent anchor, whereas a pathogenic 4q35 locus is also tethered through a second anchor depending on D4Z4. A corollary of this hypothesis is that the peripheral environment of the pathogenic 4q35 allele might be different for the wild-type allele, leading to a differential expression of genes causing the disease. In agreement with this model, the FSHD displays some features of muscular laminopathies (Bakay et al, 2006). Moreover, we showed that A-type Lamins contribute to the regulation of D4Z4 activity (Ottaviani et al, 2009) but also to D4Z4-associated telomere positioning, suggesting that the Lamins-dependence of D4Z4 might contribute to the epigenetic regulation of the FSHD-associated gene(s) and put A-type Lamins as key regulators of D4Z4 function (Figure 5B).
D4Z4 as a prototype of CTCF/Lamins-dependant perinuclear anchoring elements
As the perinuclear positioning cis-acting element of D4Z4 also behaves as a transcriptional insulator (Ottaviani et al, 2009), we propose that this element defines a particular class of CTCF-dependent insulator that acts in cooperation with A-type Lamins to position certain chromosome regions at the nuclear periphery. The in vivo binding of Lamins A/C to D4Z4 suggests that the nuclear lamina directly recruits the repeat (Ottaviani et al, 2009). In this case, CTCF and A-type Lamins could be part of the same complex positioning short D4Z4 arrays to the nuclear lamina. Consistent with the hypothesis of a functional Lamins/CTCF interaction, these proteins were previously copurified in soluble extracts from HeLa cells overexpressing CTCF (Yusufzai et al, 2004) and A-type Lamins depletion reduces D4Z4 anti-silencing activity (Ottaviani et al, 2009).
Alternatively, CTCF might counteract the internal anchoring pathway acting on telomeres, for instance through its insulator activity, thereby allowing its associated telomere to be displaced towards the periphery. Interestingly, a recent report revealed that A- and B-type Lamins form separate meshworks with a preferential association of B-type Lamins with silenced regions and A-type Lamins with euchromatin (Shimi et al, 2008), suggesting that CTCF-Lamins A/C interactions might control a subset of chromosomal domains regulated at the nuclear rim.
Our findings, together with the colocalization between some CTCF insulators and the nucleolus (Yusufzai et al, 2004) and recent data showing that Cohesins (Parelho et al, 2008; Wendt et al, 2008) or Emerin and B-type Lamins (Guelen et al, 2008) can colocalize with CTCF throughout the genome, suggests that CTCF acts as a global regulator of nuclear architecture by interacting with different key components of human chromosomes. In agreement with the possible existence of different classes of CTCF-dependent insulators, we have been unable to detect a significant association of D4Z4-tagged telomeres and the nucleolus (data not shown). Strikingly, we found that a 30 bp GA-rich motif, that is included into the 80 bp perinuclear positioning sequence and binds CTCF in vitro (Ottaviani et al, 2009) is present in different unrelated regions of the human genome (Supplementary Figure S8; Supplementary Table S2). Therefore, this motif might be the signature of a subset of CTCF-binding sites able to function together with A-type Lamins to position various chromosomal segment to the nuclear envelope.
Modulation of telomere perinuclear anchoring: a conserved feature of subtelomeric insulators
This work identifies, for the first time, a human subtelomeric element that positions a telomere at the nuclear periphery. Hence, it emerges that a basic principle governing telomere subnuclear organization in human cells is the ability of specific subtelomeric sequences to counteract the tendency of human telomeres to remain in the innermost nuclear space (Figure 5C). These findings corroborate the notion that the localization of a telomere within the nucleoplasm is determined, at least in part, by the nature of its subtelomeric region. Moreover, our work suggests that A-type lamins play a role in the function of telomeres by contributing to their subnuclear distribution. In agreement with this view, Gonzalez-Suarez et al (2009) recently showed that A-type lamins play a general role in telomere function and localization in the mouse. However, if the main pool of nuclear lamins is found at the nuclear periphery, soluble lamins and lamin speckles have also been observed in the inner nuclear space (Kumaran et al, 2002) and the integration of A-type lamins at the nuclear periphery depends on the phase of the cell cycle (Muralikrishna et al, 2001). Therefore, together with the components of the lamina, internal lamins might also contribute to the subnuclear positioning of a subset of telomeres (this work and Gonzalez-Suarez et al, 2009).
It is worth noting that the opposite happens in budding yeast, that is, specific subtelomeric sequences antagonize the tendency of telomeres to be at the periphery (Berthiau et al, 2006; Hediger et al, 2006) (Figure 5D). Strikingly, in both organisms, the subtelomeric sequences modulating telomere positioning are coupled to transcriptional insulation (Fourel et al, 1999; Hediger et al, 2006; Ottaviani et al, 2009). Therefore, subnuclear positioning and insulation might have been conserved in subtelomeric regions during evolution to fulfill critical functions for telomere maintenance, transcriptional programming and chromosome organization.
Materials and methods
Cell culture
The human epithelial cervix carcinoma C33-A cell line was maintained in Dulbecco's Modified Eagle's Medium with L-alanyl-L-glutamine (GlutaMAXTMI), D-glucose and sodium pyruvate (Invitrogen). Media were supplemented with 10% FBS (Invitrogen) and 1% Penicillin-Streptomycin (10 000 units/ml; Invitrogen) and grown at 37°C, 5% CO2, in a humidified atmosphere.
Constructs, transfections and telomeric fragmentation procedure
The constructs containing or not a telomeric seed are described in Ottaviani et al (2009). Linearization downstream of the 1.2 kb (TTAGGG)n seed at the BstXI site (B) allows the non-targeted introduction of cloned telomeres into mammalian cells. Details are available on request. The conditions of transfection of the linearized vectors with a modified calcium phosphate method (Koering et al, 2002) were optimized for each construct to obtain a single integration per cell. Three days after transfection, Hygromycin B is added to the medium. The percentage of eGFP-positive cells and the average level of eGFP is monitored by flow cytometry (FACS) (data not shown). Successful de novo formation of eGFP-tagged telomeres and single integration were confirmed in the polyclonal population of transfected cells and in a set of clones by FISH on metaphase spreads (Figure 1A, photographs 1, 2; Supplementary Table S1) and by detection of a diffuse hybridization signal in Southern blot (data not shown).
RNA interference
We used pre-annealed small interfering RNAs (siRNAs): siGENOME SMARTpool reagent, human CTCF (M-020165-01); human LMNA (NM_170707), Silencer Negative Control #1 siRNA (Ambion). Transfections were performed with DharmaFECT 1 (Dharmacon) with 200 pmoles siRNA for 2 × 105cells.
3D immuno-FISH
To detect our constructs, the pCMV vector was used as a probe and labelled with the DIG-Nick Translation Kit (Roche Diagnostics). TelVysion 4q and 10q probes (Vysis) and Star FISH X-painting (Cambio) were used for the detection of natural chromosomes. All probes were denatured at 80±1°C for 5 min before hybridization. Conditions for slides preparation, hybridization and immunodetection are available on request. For detection, we used mouse anti-DIG antibodies (Roche Diagnostics), rabbit anti-Lamin A/C antibodies (H110, Santa-Cruz) and goat anti-Lamin B antibodies (M-20, Santa-Cruz), all diluted 1/50, followed by incubation with secondary donkey antibodies coupled with different ALEXA fluorochromes, diluted 1/300 (Molecular Probes). Nuclei were counterstained with DAPI (Sigma) diluted to 1 μg/ml in PBS and mounted in Vectashield (Vector Laboratories).
Images were acquired with the confocal scanning laser system, LSM510, from Zeiss (Germany). A 63 × Plan-APOCHROMAT, oil immersion, NA 1.40 objective (Zeiss) was used to record optical sections at intervals of 0.48 μm. The pinhole was set the closest to 1 Airy with optical slices in all wavelengths with identical thickness (0.8 μm). Images were averaged four times to improve the signal to noise ratio. Generated. lsm files had a voxel size of 0.1 μm × 0.1 μm × 0.48 μm and were processed through the Imaris software (Bitplane AG). After 3D analysis, data sets were presented as the distribution of FISH signals between three concentric zones of equal volume or as the mean ratio between two volumes. P-values were obtained by performing non-parametrical statistic tests (Statistics were done with R. http://www.R-project.org). After Kruskal–Wallis tests, subsequent pairwise comparisons were performed using the Mann–Whitney test and the significance threshold (α=0.05) was corrected for multiple comparisons using the Simes method for false discovery rate (FDR) determination. Regular Mann–Whitney test was used to compare knock-down with control conditions.
Supplementary Material
Supplementary data
Supplementary data
Acknowledgments
We thank Dr Gary Felsenfeld for the kind gift of the pJC5-4 (5′ HS4) vector and helpful discussion and Dr Susana Gonzalo for sharing unpublished results on telomere and A-type lamins. We acknowledge the facilities of the IFR 128 for microscopy (PLATIM) and flow cytometry. We are grateful to Susan Gasser, Edith Heard, the members of the FSHD consortium coordinated by the Association Française contre les Myopathies (AFM), the members of the laboratory for their helpful discussion and Dr Gaël Yvert for assistance with the statistical analysis of the 3D-FISH experiments. The work in EG's laboratory was supported by grants from AFM, Association pour la Recherche contre le Cancer (ARC) and Programme Emergence de la Région Rhône-Alpes (to FM).
Footnotes
The authors declare that they have no conflict of interest.
References
- Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R (1998) Perinuclear localization of chromatin facilitates transcriptional silencing. Nature 394: 592–595 [DOI] [PubMed] [Google Scholar]
- Bakay M, Wang Z, Melcon G, Schiltz L, Xuan J, Zhao P, Sartorelli V, Seo J, Pegoraro E, Angelini C, Shneiderman B, Escolar D, Chen YW, Winokur ST, Pachman LM, Fan C, Mandler R, Nevo Y, Gordon E, Zhu Y et al. (2006) Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain 129 (Pt 4): 996–1013 [DOI] [PubMed] [Google Scholar]
- Berthiau AS, Yankulov K, Bah A, Revardel E, Luciano P, Wellinger RJ, Geli V, Gilson E (2006) Subtelomeric proteins negatively regulate telomere elongation in budding yeast. EMBO J 25: 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG (1997) Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91: 845–854 [DOI] [PubMed] [Google Scholar]
- Chung JH, Whiteley M, Felsenfeld G (1993) A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell 74: 505–514 [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S (1996) Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature 381: 529–531 [DOI] [PubMed] [Google Scholar]
- Farr CJ, Stevanovic M, Thomson EJ, Goodfellow PN, Cooke HJ (1992) Telomere-associated chromosome fragmentation: applications in genome manipulation and analysis. Nat Genet 2: 275–282 [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA (2008) Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4: e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel G, Revardel E, Koering CE, Gilson E (1999) Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J 18: 2522–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz P, De Jong JH, Lysak M, Castiglione MR, Schubert I (2002) Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc Natl Acad Sci USA 99: 14584–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A (2000) Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature 407: 1018–1022 [DOI] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R (2002) Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110: 339–348 [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G (2006) Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet 7: 703–713 [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Corces VG (2001) Chromatin insulators and boundaries: effects on transcription and nuclear organization. Annu Rev Genet 35: 193–208 [DOI] [PubMed] [Google Scholar]
- Gilson E, Laroche T, Gasser SM (1993) Telomeres and the functional architecture of the nucleus. Trends Cell Biol 3: 128–134 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez I, Redwood AB, Perkins SM, Vermolen B, Lichtensztejin D, Grotsky DA, Morgado-Palacin L, Gapud EJ, Sleckman BP, Sullivan T, Sage J, Stewart CL, Mai S, Gonzalo S (2009) Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J advance online publication 23 July 2009; doi:10.1038/emboj.2009.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM (1996) The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol 134: 1349–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Hediger F, Berthiau AS, van Houwe G, Gilson E, Gasser SM (2006) Subtelomeric factors antagonize telomere anchoring and Tel1-independent telomere length regulation. EMBO J 25: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, van Deutekom JC, Francis F, Sharpe PT, Hofker M, Frants RR, Williamson R (1994) Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet 3: 1287–1295 [DOI] [PubMed] [Google Scholar]
- Koering CE, Pollice A, Zibella MP, Bauwens S, Puisieux A, Brunori M, Brun C, Martins L, Sabatier L, Pulitzer JF, Gilson E (2002) Human telomeric position effect is determined by chromosomal context and telomeric chromatin integrity. EMBO Rep 3: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Muralikrishna B, Parnaik VK (2002) Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J Cell Biol 159: 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL (2008) A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180: 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderus ME, van Steensel B, Chong L, Sibon OC, Cremers FF, de Lange T (1996) Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol 135: 867–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle R, Wright TJ, Clark LN, Hewitt JE (1995) The FSHD-associated repeat, D4Z4, is a member of a dispersed family of homeobox-containing repeats, subsets of which are clustered on the short arms of the acrocentric chromosomes. Genomics 28: 389–397 [DOI] [PubMed] [Google Scholar]
- Magdinier F, Ottaviani A, Gilson E (2008) Telomere position effect and the evolution of the genome. In Origin and Evolution of Telomeres, Tomaska L, Nosek J (eds), Vol. 9, pp 128–142. New York, NY: Eureka.com and Kluwer Academic/Plenum Publishers [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM (1996) Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev 10: 1796–1811 [DOI] [PubMed] [Google Scholar]
- Malhas A, Lee CF, Sanders R, Saunders NJ, Vaux DJ (2007) Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J Cell Biol 176: 593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masny PS, Bengtsson U, Chung SA, Martin JH, van Engelen B, van der Maarel SM, Winokur ST (2004) Localization of 4q35.2 to the nuclear periphery: is FSHD a nuclear envelope disease? Hum Mol Genet 13: 1857–1871 [DOI] [PubMed] [Google Scholar]
- Muralikrishna B, Dhawan J, Rangaraj N, Parnaik VK (2001) Distinct changes in intranuclear lamin A/C organization during myoblast differentiation. J Cell Sci 114 (Pt 22): 4001–4011 [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ (2008) Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottaviani A, Rival-Gervier S, Boussouar A, Foerster AM, Rondier D, Sacconi S, Desnuelle C, Gilson E, Magdinier F (2009) The D4Z4 macrosatellite repeat acts as a CTCF and A-type lamins-dependent insulator in facio-scapulo-humeral dystrophy. PLoS Genet 5: e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschlager M (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132: 422–433 [DOI] [PubMed] [Google Scholar]
- Petrov A, Allinne J, Pirozhkova I, Laoudj D, Lipinski M, Vassetzky YS (2008) A nuclear matrix attachment site in the 4q35 locus has an enhancer-blocking activity in vivo: implications for the facio-scapulo-humeral dystrophy. Genome Res 18: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B (2006) Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38: 1005–1014 [DOI] [PubMed] [Google Scholar]
- Rawlins DJ, Shaw PJ (1990) Localization of ribosomal and telomeric DNA sequences in intact plant nuclei by in-situ hybridization and three-dimensional optical microscopy. J Microsc 157(Pt 1): 83–89 [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H (2008) Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247 [DOI] [PubMed] [Google Scholar]
- Scherthan H (2007) Telomere attachment and clustering during meiosis. Cell Mol Life Sci 64: 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Pfleghaar K, Kojima S, Pack CG, Solovei I, Goldman AE, Adam SA, Shumaker DK, Kinjo M, Cremer T, Goldman RD (2008) The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev 22: 3409–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Gartenberg MR, Neumann FR, Hediger F, Gasser SM (2005) Multiple pathways tether telomeres and silent chromatin at the nuclear periphery: functional implications for sir-mediated repression. Novartis Found Symp 264: 140–156; discussion 156–165, 227–130 [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM (2006) Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778 [DOI] [PubMed] [Google Scholar]
- Tam R, Smith KP, Lawrence JB (2004) The 4q subtelomere harboring the FSHD locus is specifically anchored with peripheral heterochromatin unlike most human telomeres. J Cell Biol 167: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham WH, Wyithe JS, Ko Ferrigno P, Silver PA, Zakian VA (2001) Localization of yeast telomeres to the nuclear periphery is separable from transcriptional repression and telomere stability functions. Mol Cell 8: 189–199 [DOI] [PubMed] [Google Scholar]
- Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, Fabre E (2006) Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol 172: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela L, Kamakaka RT (2006) Chromatin insulators. Annu Rev Genet 40: 107–138 [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Bakker E, Lemmers RJ, van der Wielen MJ, Bik E, Hofker MH, Padberg GW, Frants RR (1996) Evidence for subtelomeric exchange of 3.3 kb tandemly repeated units between chromosomes 4q35 and 10q26: implications for genetic counselling and etiology of FSHD1. Hum Mol Genet 5: 1997–2003 [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Wijmenga C, van Tienhoven EA, Gruter AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR (1993) FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 2: 2037–2042 [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Enthoven L, Ricci E, Rossi M, Felicetti L, Jeanpierre M, Winokur ST, Frants RR, Padberg GW, van der Maarel SM (2005) Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann Neurol 58: 569–576 [DOI] [PubMed] [Google Scholar]
- van Overveld PG, Lemmers RJ, Deidda G, Sandkuijl L, Padberg GW, Frants RR, van der Maarel SM (2000) Interchromosomal repeat array interactions between chromosomes 4 and 10: a model for subtelomeric plasticity. Hum Mol Genet 9: 2879–2884 [DOI] [PubMed] [Google Scholar]
- Weierich C, Brero A, Stein S, von Hase J, Cremer C, Cremer T, Solovei I (2003) Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res 11: 485–502 [DOI] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451: 796–801 [DOI] [PubMed] [Google Scholar]
- Winokur ST, Bengtsson U, Feddersen J, Mathews KD, Weiffenbach B, Bailey H, Markovich RP, Murray JC, Wasmuth JJ, Altherr MR, Schutte BC (1994) The DNA rearrangement associated with facioscapulohumeral muscular dystrophy involves a heterochromatin-associated repetitive element: implications for a role of chromatin structure in the pathogenesis of the disease. Chromosome Res 2: 225–234 [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G (2004) CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell 13: 291–298 [DOI] [PubMed] [Google Scholar]
- Zalenskaya IA, Bradbury EM, Zalensky AO (2000) Chromatin structure of telomere domain in human sperm. Biochem Biophys Res Commun 279: 213–218 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data