Abstract
A-type lamins are intermediate filament proteins that provide a scaffold for protein complexes regulating nuclear structure and function. Mutations in the LMNA gene are linked to a variety of degenerative disorders termed laminopathies, whereas changes in the expression of lamins are associated with tumourigenesis. The molecular pathways affected by alterations of A-type lamins and how they contribute to disease are poorly understood. Here, we show that A-type lamins have a key role in the maintenance of telomere structure, length and function, and in the stabilization of 53BP1, a component of the DNA damage response (DDR) pathway. Loss of A-type lamins alters the nuclear distribution of telomeres and results in telomere shortening, defects in telomeric heterochromatin, and increased genomic instability. In addition, A-type lamins are necessary for the processing of dysfunctional telomeres by non-homologous end joining, putatively through stabilization of 53BP1. This study shows new functions for A-type lamins in the maintenance of genomic integrity, and suggests that alterations of telomere biology and defects in DDR contribute to the pathogenesis of lamin-related diseases.
Keywords: A-type lamins, DNA damage response, genomic instability, nuclear organization, telomeres
Introduction
Several studies have suggested that the 3D organization of the genome represents an additional level of regulation of genome function (Misteli, 2007; Dechat et al, 2008). The nuclear periphery, in particular, is viewed as a compartment actively participating in gene transcription and DNA repair (Therizols et al, 2006; Mekhail et al, 2008; Nagai et al, 2008). A-type lamins, intermediate filament proteins that form part of the nuclear lamina and a nucleoplasmic network, are key determinants of nuclear architecture, providing a scaffold for the organization of nuclear functions (Taddei et al, 2004; Dechat et al, 2008). Accordingly, roles for A-type lamins in DNA replication and repair, gene transcription and silencing, nuclear pore complex positioning, remodelling of chromatin, and nuclear envelope breakdown during mitosis have been suggested (Goldman et al, 2002; Gruenbaum et al, 2005). However, the causal relationship between alterations of A-type lamins and regulation of genome functions as well as the molecular mechanisms involved remain ill defined. Elucidating the functions of these proteins is a topical subject, as mutations in the LMNA gene are associated with various degenerative disorders termed laminopathies (Broers et al, 2006; Capell and Collins, 2006). In addition, changes in the expression of A-type lamins are associated with different types of human tumours (Agrelo et al, 2005; Prokocimer et al, 2006; Willis et al, 2008).
Most of the available data regarding A-type lamins functions come from studies on human cells or mouse models carrying disease-causing mutations in the LMNA gene, especially progerias (Mounkes and Stewart, 2004; Varela et al, 2005; Stewart et al, 2007). Common features include nuclear morphological abnormalities, disorganization of heterochromatin, and defects in the DNA damage response (DDR) pathway and in DNA repair (Liu et al, 2005; Capell and Collins, 2006). In particular, increased sensitivity to DNA damage has been molecularly linked to delayed recruitment of the DNA damage sensor 53BP1 to γ-H2AX-labelled DNA repair foci and defective formation of Rad 51 foci (Liu et al, 2005). Collectively, these data suggest that increased genomic instability contributes to the pathogenesis of disease-causing mutations in the LMNA gene.
Increased genomic instability is also a known contributor to tumourigenesis. Although expression of mutant forms of A-type lamins has not been associated with increased tumour susceptibility in humans or mice, an altered expression of A-type lamins has been observed in different types of human tumours (Agrelo et al, 2005; Prokocimer et al, 2006, 2009; Willis et al, 2008). The LMNA knockout mouse has provided insights into the cellular consequences of the loss of A-type lamins (Sullivan et al, 1999). Lmna−/− cells present alterations in the nuclear envelope, loss of heterochromatin from the nuclear periphery, and defects in the tethering and stabilization of members of two families of tumour suppressor proteins—the Retinoblastoma (Rb) and ING families—(Johnson et al, 2004; Han et al, 2008). Overall, these findings suggest common as well as different functional consequences of mutation versus silencing of the LMNA gene, with loss of A-type lamins being associated with uncontrolled proliferation. The impact that the loss of A-type lamins has on mechanisms responsible for maintaining genomic stability remains unknown.
Defects in DNA repair and the DDR pathway as well as alterations in telomere biology are among the leading causes of genomic instability in cancer and aging. Telomeres, heterochromatic structures sheltering the ends of linear chromosomes, are essential for the preservation of chromosome integrity and controlled cell proliferation (Blackburn, 2001; de Lange, 2002). A minimal length of telomeric DNA repeats and proper recruitment of telomere binding proteins are necessary to preserve telomere function (Liu et al, 2004; de Lange, 2005a). In addition, the acquisition of a heterochromatic structure at telomeres is crucial for the maintenance of telomere length homoeostasis (Blasco, 2007). The evidence linking A-type lamins with aging, cancer, and at the cellular level, with heterochromatin regulation, has lead investigators to suggest that alterations in telomere biology could contribute to the phenotypes of laminopathies.
The best evidence about a role for A-type lamins in telomere biology is that Hutchinson–Gilford progeria syndrome (HGPS) fibroblasts show faster telomere attrition than normal counterparts (Allsopp et al, 1992; Huang et al, 2008). HGPS fibroblasts also present defects in epigenetic marks characteristic of constitutive heterochromatin, such as decreased trimethylation of histone H3 at lysine 9 (H3K9me3) and increased trimethylation of histone H4 at lysine 20 (H4K20me3). However, the effect on specific domains, such as centromeres and telomeres is unknown (Scaffidi and Misteli, 2006; Shumaker et al, 2006). Further evidence of a crosstalk between A-type lamins and telomeres is the finding that proliferative defects of human fibroblasts expressing lamin A mutants are rescued by telomerase (Kudlow et al, 2008), and that alterations of the nuclear lamina contributing to senescence are associated with aggregation of telomeres at the nuclear periphery (Raz et al, 2008).
To understand the role that A-type lamins have in telomere biology, we determined the effect that loss of A-type lamins has on the nuclear distribution of telomeres, and on the maintenance of telomere structure, length, and function. Using Lmna knockout mouse fibroblasts as a model (Sullivan et al, 1999), we provide evidence for an essential role of A-type lamins in the 3D distribution of telomeres within the nucleus. Changes in the nuclear compartmentalization of telomeres upon loss of A-type lamins were accompanied by telomere shortening, defects in telomeric chromatin structure, and increased genomic instability. In addition, we show that A-type lamins are necessary for the processing of dysfunctional telomeres through the non-homologous end joining (NHEJ) repair pathway. Interestingly, loss of A-type lamins leads to a marked decrease in the cellular levels of 53BP1, a checkpoint protein that participates in the repair of double-strand breaks, and that has been recently implicated in the processing of dysfunctional telomeres by NHEJ (Dimitrova et al, 2008). Overall, this study shows new functions for A-type lamins in the maintenance of genomic stability through their participation in the maintenance of telomeres and 53BP1 stability. These findings suggest that alterations in telomere biology and defects in the DDR pathway are likely to contribute to the phenotypes of lamin-related diseases, especially those characterized by the loss of A-type lamins function.
Results
Loss of A-type lamins induces changes in the nuclear distribution of telomeres and telomere shortening
A 3D analysis of telomere positioning indicates that mammalian telomeres are distributed throughout the entire nuclear volume in G0/G1/S phases of the cell cycle, whereas they assemble into a telomeric disk at the centre of the nucleus during G2 (Chuang et al, 2004). The molecular mechanisms that ensure proper nuclear localization of mammalian telomeres and their relevance in telomere metabolism remain undefined. Interestingly, the 3D positioning of telomeres is altered in tumour cells (Chuang et al, 2004; Mai and Garini, 2006) and in senescent cells presenting defects in the nuclear lamina (Raz et al, 2008), suggesting a relationship between nuclear distribution of telomeres and alterations of telomere metabolism observed during senescence and immortality.
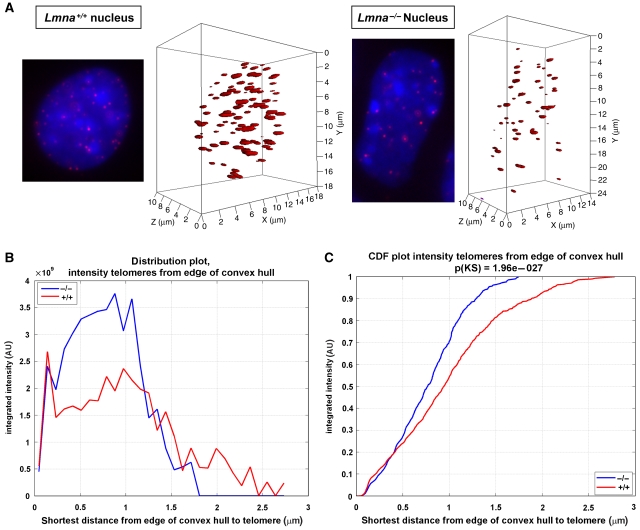
To determine whether A-type lamins have a role in the nuclear compartmentalization of telomeres, we compared the nuclear distribution of telomeres between wild-type (Lmna+/+) mouse embryonic fibroblasts (MEFs) and MEFs devoid of A-type lamins (Lmna−/−). MEFs were analysed after spontaneously bypassing senescence and becoming immortal. We carried out 3D telomere fluorescence in situ hybridization followed by quantitative analysis to monitor telomere distribution (See Supplementary data). The telomere distances to the nuclear edge were determined using the TeloView program (see Vermolen et al, 2005 and Figure 1A). Figure 1B shows a clear difference in telomere distribution between the two genotypes, with a shift in the localization of telomeres towards the nuclear periphery upon loss of A-type lamins. By calculating the cumulative distribution of telomere intensities (Figure 1C), we show that approximately 20% of telomere signals are found at the very edge of the nucleus (⩽0.4 μm) in both genotypes. However, although the remaining 80% of telomeres in Lmna−/− cells accumulate up to a distance of 1.75 μm from the edge, telomeres in Lmna+/+ MEFs expand throughout the nucleoplasm and up to the nuclear centre (2.75 μm). These results clearly show that A-type lamins participate in the correct distribution of telomeres throughout the entire nuclear volume, with a significant change in distribution towards the nuclear periphery and away from the nuclear centre on loss of A-type lamins. Fluorescence-activated cell sorting analysis carried out on proliferating Lmna+/+ and Lmna−/− immortalized fibroblasts indicates that changes in telomere distribution are not due to differences in cell-cycle profiles between genotypes (Supplementary Figure 1). In addition, chromatin immunoprecipitation (ChIP) assays performed using lamin A/C antibody show binding of lamins to telomeres (Supplementary Figure 2B), suggesting that tethering of telomeres to the lamins scaffold might regulate their nuclear distribution.
Figure 1.
Changes in nuclear distribution of telomeres in Lmna−/− cells. (A) Representative reconstruction images of nuclei from Lmna+/+ and Lmna−/− MEFs after carrying out 3D FISH using a fluorescent telomere probe. DAPI counterstain shown in blue. Telomere distribution in the 3D nuclear space is shown. (B) Distribution plot of the intensity of telomere signals with respect to distance of signals from the edge of nuclei. Integrated intensity represents sum intensity of all telomeres at a distance from edge ⩽x. Note the change in distribution of telomeres towards the periphery and away from the centre in Lmna−/− MEFs. (C) Cumulative distribution function (CDF) showing integrated intensity in relation to distance from nuclear edge. Integrated intensity is defined as: (sum intensity of telomeres at a distance from edge ⩽x)/(sum intensity of all telomeres). No changes in the pool of telomeres between the two genotypes are observed at the very edge of nuclei. In contrast, higher pools of telomeres are found at all peripheral distances in Lmna−/− MEFs, when compared with Lmna+/+ MEFs. 3D FISH was carried out three times. Telomeres in 85 nuclei of Lmna−/− MEFs and 81 nuclei of Lmna+/+ MEFs were analysed.
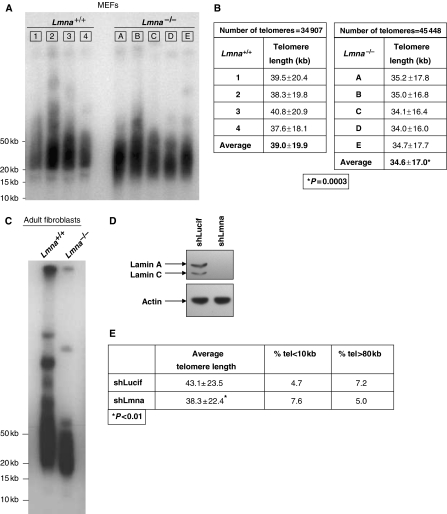
To determine whether changes in telomere distribution upon loss of A-type lamins are accompanied by alterations of telomere metabolism, we compared telomere length between multiple sets of Lmna−/− and Lmna+/+ MEFs of early passage (pre-senescent). On carrying out terminal restriction fragment (TRF) analysis (Garcia-Cao et al, 2002), we observed a faster migration of telomeres in all five Lmna−/− lines compared with Lmna+/+ lines, indicating a moderate but highly consistent telomere shortening on loss of A-type lamins (Figure 2A). These results were confirmed by quantitative fluorescence In situ hybridization (Q-FISH) of metaphase nuclei using a telomeric probe (Garcia-Cao et al, 2002). We found that mean telomere lengths of Lmna+/+ lines ranged between 37.6 and 40.8 kb, with an average telomere length of 39.0 kb. All five lines of Lmna−/− MEFs presented lower mean telomere length than any of the Lmna+/+ lines, ranging between 34.0 and 35.2 kb, with an average telomere length of 34.6 kb (Figure 2B). To determine whether these differences were statistically significant, we carried out a two-sided t-test considering the mean from each of the cell lines to be an independent sample of either Lmna−/− or Lmna+/+ genotype. We found that the mean telomere length is significantly different between these two genotypes (P=0.0003). In addition, TRF analysis of adult fibroblasts from Lmna−/− mice shows a more pronounced telomere shortening phenotype (Figure 2C). To test whether acute depletion of A-type lamins would also lead to telomere attrition, we lentivirally transduced wild-type MEFs with constructs carrying an shRNA specific for depletion of A-type lamins (shLmna) or an shRNA specific for luciferase (shLucif) as control. Transduction using shLmna led to undetectable levels of lamins A/C (Figure 2D). Q-FISH analysis carried out in these cells showed a marked decrease in telomere length after only five passages of the cells in culture (Figure 2E). All together, these data show that A-type lamins have a key role in the maintenance of telomere length homoeostasis.
Figure 2.
Defective maintenance of telomere length upon loss of A-type lamins. (A) Representative TRF analysis of several independent Lmna+/+ (1–4) and Lmna−/− (A–E) MEF lines of early passage (passages 3–5). Note the faster migration of telomeres in all Lmna−/− lines when compared with Lmna+/+ lines, indicating telomere shortening. (B) Telomere length distribution (expressed as average±s.d.) in Lmna+/+ (1–4) and Lmna−/− (A–E) MEFs as determined by Q-FISH. The individual values for each of the samples are shown. Q-FISH was carried out three times. For each experiment, at least 20 metaphases were analysed in every MEF line. Note how all Lmna−/− MEF lines presented shorter telomere length than the Lmna+/+ controls. (C) TRF analysis of adult fibroblasts from Lmna+/+ and Lmna−/− mice showing faster migration and thus, pronounced telomere shortening on loss of A-type lamins. (D) Western blot to detect the levels of lamins A/C on lentiviral transduction of wild-type MEFs with constructs carrying an shRNA specific for lamins A and C (shLmna) or an shLucif control. (E) Q-FISH analysis to measure telomere length in MEFs after depletion of A-type lamins (compare shLmna and shLucif). Note the decrease in telomere length on depletion of A-type lamins after only five passages of the cells in culture. *Represents P-value of statistical significance.
Telomere length homoeostasis is maintained primarily by telomerase (Blackburn et al, 2006). Furthermore, shelterin complex proteins, especially TRF1 and TRF2, regulate telomere length (Smogorzewska et al, 2000; Munoz et al, 2005). To further investigate the telomere shortening phenotype in Lmna−/− MEFs, we evaluated whether loss of A-type lamins affects these regulatory mechanisms. We found no significant changes in telomerase activity between Lmna+/+ and Lmna−/− MEFs (Supplementary Figure 2A). In addition, ChIP analysis showed no detectable alterations in the telomere-bound levels of TRF1 and TRF2 between genotypes (Supplementary Figure 2B). Our results indicate that the binding of these known regulators of telomere length homoeostasis is not affected by the loss of A-type lamins. We cannot rule out, however, that the accessibility of telomerase to telomeres, or the binding of other factors implicated in telomere metabolism, especially factors implicated in telomere replication, are affected by the loss of A-type lamins.
Loss of A-type lamins results in alterations of telomere chromatin structure
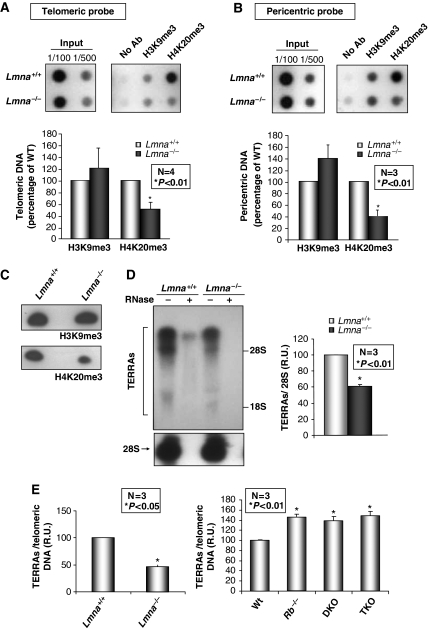
The maintenance of a heterochromatic structure at telomeres is important for telomere length homoeostasis (Gonzalo et al, 2005, 2006). A common feature in fibroblasts from HGPS patients and from old individuals expressing progerin is the alteration of histone marks characteristic of constitutive heterochromatin (Scaffidi and Misteli, 2006; Shumaker et al, 2006), although the effect on telomeres is unknown. To investigate whether loss of A-type lamins affects the assembly of telomeric heterochromatin, we carried out ChIP assays using antibodies recognizing well-established heterochromatic marks, H3K9me3 and H4K20me3. Although we found no changes in H3K9me3 levels, we observed a significant decrease in telomeric H4K20me3 levels in Lmna−/− MEFs (Figure 3A). These results show that A-type lamins participate in the maintenance of histone marks characteristic of telomeric heterochromatin. Interestingly, these defects were phenocopied by pericentric heterochromatin (Figure 3B), supporting the idea that alterations of A-type lamins function affect the epigenetic status of constitutive heterochromatin (Shumaker et al, 2006). Nonetheless, the changes reported here are different from those described in HGPS cells, indicating different functional implications of mutation or silencing of the LMNA gene. The epigenetic defects of Lmna−/− MEFs were confirmed by western blot analysis, showing a marked reduction in global H4K20me3 levels with no changes in H3K9me3 (Figure 3C).
Figure 3.
Alterations of telomeric heterochromatin structure in Lmna−/− MEFs. (A) Chromatin immunoprecipitation (ChIP) analysis with antibodies recognizing heterochromatic marks H3K9me3 and H4K20me3. DNA immunoprecipitated was hybridized to a telomeric probe. Note the marked decrease in H4K20me3 in Lmna−/− at this domain in one representative experiment. The bottom graph shows the quantification of immunoprecipitated telomeric repeats after normalization to input signals in four independent experiments. (B) Immunoprecipitated DNA was hybridized to a probe recognizing major satellite repeat sequences characteristic of pericentric heterochromatin. The graph shows the quantification of major satellite repeats signal after normalization to input signals in three independent experiments. Note that the decrease in H4K20me3 is phenocopied by pericentric heterochromatin. (C) Western blots to monitor global levels of H3K9me3 and H4K20me3 after acid extraction of histones from cell lysates of Lmna+/+ and Lmna−/− MEFs. The immunoblots show no changes in the levels of H3K9me3, but a significant decrease in global H4K20me3 levels on loss of A-type lamins. (D) Total RNA from Lmna+/+ and Lmna−/− cells was isolated, separated by gel electrophoresis and subjected to northern blotting with a telomeric probe to monitor changes in the levels of TERRAs on loss of A-type lamins. Hybridization with a probe specific for 28S ribosomal RNA shows equivalent levels of RNA isolated from both cell lines (bottom panels). Treatment with RNase A was done as a control for purity of RNAs. The graph shows the quantification of the levels of TERRAs after normalization to levels of 28S in three independent experiments. Note the decrease in TERRAs levels in Lmna−/− with respect to Lmna+/+ MEFs. (E) Quantification of TERRAs density in Lmna+/+, Lmna−/−, and MEFs devoid of one, two or three Rb family members. TERRAs levels are normalized to telomeric DNA content to correct for differences in telomere length (see Supplementary Figure 4). Note the increase in the levels of TERRAs per telomeric DNA content in Rb-deficient MEFs compared with wild type. In contrast, Lmna−/− MEFs show a significant decrease in TERRAs density. Bars represent standard error. *Represents P-value of statistical significance. R.U. stands for relative units.
Recently, new components of telomeric heterochromatin have been identified. These are non-coding telomeric RNAs (TERRAs or TelRNAs) that are transcribed by DNA-dependent RNA polymerase II (Azzalin et al, 2007; Schoeftner and Blasco, 2008). Although the function of TERRAs on telomere metabolism remains unclear, different lines of evidence suggest that TERRAs might inhibit telomerase activity (Luke et al, 2008; Ng et al, 2009). The fact that alterations of A-type lamins had been shown to impact RNA pol II-directed transcription (Kumaran et al, 2002; Spann et al, 2002), prompted us to evaluate whether maintenance of TERRA levels is affected by their loss. We observed a marked decrease in the levels of TERRAs in Lmna−/− MEFs (Figure 3D), indicating that loss of A-type lamins either inhibit RNA pol II-directed transcription of TERRAs or alters their stability. All together, our data show that A-type lamins have a role in the assembly of telomeric heterochromatin by stabilizing H4K20me3 and maintaining TERRA levels.
We had previously shown that stabilization of H4K20me3 at telomeres requires the action of histone methyltransferases, Suv4-20h1 and Suv4-20h2, as well as Rb family members (Gonzalo and Blasco, 2005; Benetti et al, 2007). In addition, Rb family members are transcriptional regulators, which could putatively modulate the transcription of TERRAs. Given that A-type lamins contribute to the stabilization of Rb family members (Johnson et al, 2004), we hypothesized that the defects in telomeric chromatin structure on loss of A-type lamins are due to Rb deficiency. We confirmed the decrease in the levels of Rb family members in our Lmna−/− MEF lines (Supplementary Figure 3), indicating that the lower levels of telomeric H4K20me3 are due, at least in part, to reduced Rb family function. We cannot rule out, however, that decreased Suv4-20h1/h2 HMTase activity also contributes to this phenotype upon loss of A-type lamins. Next, we tested whether the decrease in Rb family function could also be responsible for the reduction in TERRA levels on loss of A-type lamins. We monitored TERRA levels in Rb family-deficient MEFs (Rb−/−, Rb−/−p107−/− herein DKO, and Rb−/−p107−/−p130−/− herein TKO). Interestingly, we found increasing levels of TERRAs on loss of one, two, or three Rb family members (Supplementary Figure 4A). To control for differences in telomere length in Rb-deficient MEFs (Supplementary Figure 4B), we normalized TERRA levels to telomeric DNA sequences (Supplementary Figure 4C). Remarkably, although the loss of A-type lamins leads to decreased levels of TERRAs per telomeric DNA repeats, a similar increase in the density of TERRAs was observed in all Rb-deficient MEFs (Figure 3E), indicating that reduced Rb function cannot account for the phenotype observed in Lmna−/− MEFs. Our findings thereby indicate that A-type lamins play a role in the assembly of telomeric heterochromatin through Rb-dependent and Rb-independent mechanisms.
Intriguingly, Rb deficiency, decreased levels of H4K20me3, and decreased TERRA levels are consistent with a telomere elongation phenotype (Garcia-Cao et al, 2002; Gonzalo et al, 2005; Benetti et al, 2007; Schoeftner and Blasco, 2008 and Supplementary Figure 4B). The fact that loss of A-type lamins leads to telomere shortening despite Rb and H4K20me3 deficiency raises the possibility that A-type lamins might have an active function in telomere elongation in these contexts. In summary, the alterations of telomere chromatin structure described here cannot account for the telomere shortening phenotype observed on loss of A-type lamins. It is possible that a more global defect in chromatin structure and DNA metabolism because of alterations of nuclear architecture upon loss of A-type lamins is responsible for this phenotype.
Impact of loss of A-type lamins on telomere function and genomic stability
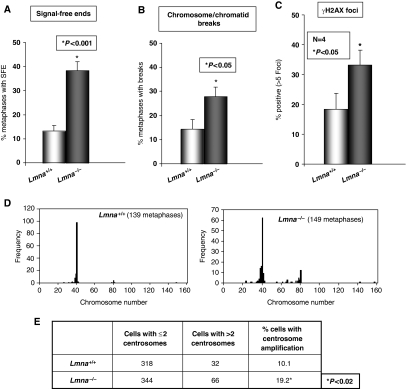
We evaluated whether the aforementioned changes in the nuclear distribution of telomeres, telomere length, and telomere chromatin structure translate into telomere dysfunction and genomic instability. We determined the loss of telomeric signals (signal-free ends), the frequency of chromosome/chromatid breaks and end-to-end fusions, and the presence of aneuploidy in pre-senescent Lmna−/− and Lmna+/+ MEFs (Figure 4 and Supplementary Figure 5). We found a threefold increase in the number of signal-free ends (Figure 4A and Supplementary Figure 5A) and a twofold increase in breaks (Figure 4B and Supplementary Figure 5B) in Lmna−/− MEFs, indicating increased genomic instability. This finding was supported by an increased number of nuclei presenting basal DNA damage, as indicated by a twofold increase in cells presenting γ-H2AX-labelled foci upon loss of A-type lamins (Figure 4C).
Figure 4.
Loss of A-type lamins impacts genomic stability. (A) Quantification of the percentage of metaphases with signal-free ends in the different lines of Lmna+/+ (1–4) and Lmna−/− (A–E) MEFs. Note the threefold increase in signal-free ends in Lmna−/− MEFs. (B) Quantification of the percentage of metaphases with chromosome and/or chromatid breaks. A twofold increase in breaks was observed in Lmna−/− lines. (C) Quantification of the percentage of cells presenting >5 γH2AX-labelled DNA repair foci. Note the increase in basal damage in Lmna−/− MEFs. Graphs show results from at least three independent experiments. Bars represent standard error. (D) Karyotype analysis of more than 100 metaphases shows a clear increase in the number of metaphases with an aberrant number of chromosomes in Lmna−/− MEFs. (E) Quantification of number and percentage of Lmna+/+ and Lmna−/− MEFs presenting more than two centrosomes by labelling with anti-γ tubulin antibody. *Represents P-value of statistical significance.
In normal cells, inappropriate recombination between telomeres of sister chromatids can serve to lengthen one telomere at the expense of another (Bailey et al, 2004; Laud et al, 2005). To test whether aberrant recombination involving telomeric repeats contributes to the loss of telomere signals in Lmna−/− MEFs, we carried out chromosome orientation fluorescence in situ hybridization (CO-FISH) (Bailey et al, 2004). This technique allows the differential labelling of leading and lagging strands of telomeres. In the absence of recombination events between telomeric sequences, only one telomere at each chromosome end is labelled with either the leading or the lagging strand probe. If recombination occurs, the labelling is split between both sister telomeres giving rise to telomeres labelled with both the leading and lagging strand probes. CO-FISH results indicate that loss of A-type lamins does not lead to an increase in the frequency of recombination events involving telomeres (Supplementary Figure 6). Thus, alternative mechanisms are responsible for the increased loss of telomere signals upon loss of A-type lamins.
In addition, karyotype analysis shows increased numbers of metaphases with abnormal chromosome dosage in Lmna−/− MEFs (Figure 4D). Earlier studies showed that reduced Rb family function leads to aneuploidy due, in part, to defects in centrosome duplication (Iovino et al, 2006). To test whether loss of A-type lamins affects centrosome duplication control, we quantified centrosome numbers in Lmna+/+ and Lmna−/− MEFs by carrying out immunofluorescence using γ-tubulin antibody. Our results indicate an increase in the number of cells presenting centrosome amplification (Figure 4E). Thus, loss of A-type lamins leads to aneuploidy, at least in part, by deregulation of centrosome duplication cycle. Overall, these results show increased genomic instability on loss of A-type lamins.
A-type lamins participate in the processing of dysfunctional telomeres by NHEJ
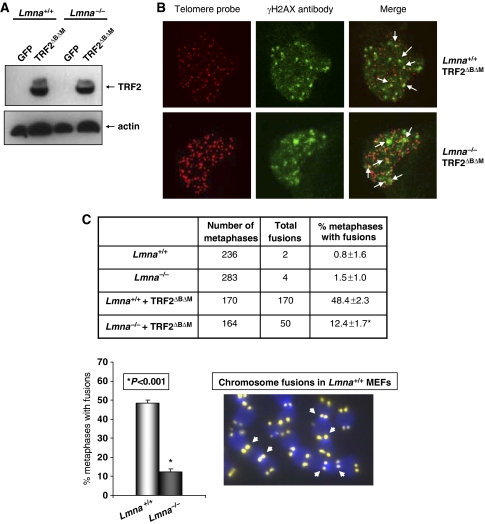
Surprisingly, despite telomere shortening and increased frequency of signal-free ends, we found no significant differences in the percentage of metaphases presenting chromosome end-to-end fusions in Lmna−/− MEFs (Supplementary Figure 5C). We hypothesized that loss of A-type lamins might hinder the recognition and/or processing of dysfunctional telomeres through the NHEJ DNA repair pathway. To test this hypothesis, we carried out retroviral transduction of Lmna+/+ and Lmna−/− MEFs with a dominant-negative mutant of the telomeric binding protein TRF2 (TRF2ΔBΔM) that induces telomere dysfunction and chromosome end-to-end fusions in the presence of an intact NHEJ repair pathway (Smogorzewska et al, 2002; Riha et al, 2006 and Figure 5A). Loss of telomere integrity on expression of TRF2ΔBΔM leads to the formation of DNA damage foci at telomeres, referred to as TIF (telomere dysfunction-induced foci) (Takai et al, 2003; De Lange, 2005b). The ability of Lmna−/− MEFs to form TIF upon expression of TRF2ΔBΔM was evaluated by carrying out Immuno-FISH using γ-H2AX antibody and telomeric probe. The expression of TRF2ΔBΔM leads to the formation of γ-H2AX-labelled TIF in Lmna+/+ MEFs and Lmna−/− MEFs (Figure 5B and Supplementary Figure 7), suggesting that the initial step in the sensing of dysfunctional telomeres is intact on loss of A-type lamins. In contrast, we found that the processing of dysfunctional telomeres by NHEJ is hindered by the loss of A-type lamins. In Lmna+/+ MEFs, expression of TRF2ΔBΔM resulted in chromosome fusions in approximately 50% of metaphases, consistent with previous reports (Smogorzewska et al, 2002 and Figure 5C). Conversely, we found a fourfold reduction in the number of fusions in Lmna−/− MEFs expressing equivalent levels of TRF2ΔBΔM (Figure 5A and C). These results show that NHEJ of dysfunctional telomeres depends on the expression of A-type lamins.
Figure 5.
Loss of A-type lamins hinders the processing of dysfunctional telomeres by non-homologous end joining. (A) Western blots with TRF2 and actin antibodies showing similar levels of expression of TRF2ΔBΔM protein in Lmna+/+ and Lmna−/− MEFs. Retroviral transduction with GFP was the negative control. (B) Immuno-FISH using a telomeric PNA probe (red) and an antibody specific for γ-H2AX (green). Images show the ability of Lmna+/+ and Lmna−/− MEFs to form TIFs on expression of TRF2ΔBΔM. (C) Quantification of the percentage of metaphases presenting chromosome end-to-end fusions in Lmna+/+ and Lmna−/− MEFs before and after retroviral transduction with TRF2ΔBΔM. The graph shows a fourfold decrease in end-to-end fusions in Lmna−/− MEFs on expression of TRF2ΔBΔM compared with Lmna+/+ MEFs. The image represents an example of TRF2ΔBΔM-induced chromosome end-to-end fusions in Lmna+/+ MEFs. Retroviral transduction was carried out three times, and FISH analysis four times. Bars represent standard error. *Represents P-value of statistical significance.
A-type lamins stabilize 53BP1 protein levels
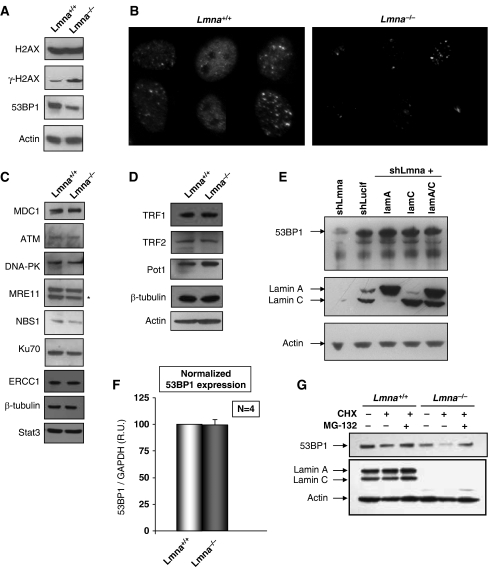
Similar to double-strand breaks, dysfunctional telomeres activate the DDR pathway (Takai et al, 2003). Loss of A-type lamins has been reported to affect the stability of nuclear factors, such as Rb family members and ING1 (Johnson et al, 2004; Nitta et al, 2006; Han et al, 2008). To gain insight into specific steps during DDR that could be affected by the loss of A-type lamins hindering the processing of dysfunctional telomeres, we tested whether the cellular levels of key components of the DDR pathway could be altered in Lmna−/− MEFs. First, we monitored the levels H2AX, γH2AX, and 53BP1, which have a function in the sensing of dysfunctional telomeres (Takai et al, 2003). Although we found no differences in the levels of H2AX upon loss of A-type lamins, we confirmed the increase in γH2AX levels in Lmna−/− MEFs, indicating increased basal DNA damage upon loss of A-type lamins. In addition, we observed a marked decrease in the levels of 53BP1 in Lmna−/− MEFs when compared with Lmna+/+ cells (Figure 6A). The reduced levels of 53BP1 were confirmed by immunofluorescence. As shown in Figure 6B, the intensity of label with 53BP1 antibody is clearly reduced in Lmna−/− MEFs with respect to Lmna+/+under the same exposure conditions. We then tested whether the stability of other factors involved in DDR and in DNA repair are also affected by the loss of A-type lamins. The cellular levels of MDC1, ATM, DNA-PK, Mre11, Nbs1, Ku70, and ERCC1 were compared between Lmna+/+ and Lmna−/− MEFs (Figure 6C). We found no detectable differences in the levels of all these factors between the two genotypes. In addition, no changes in the global levels of proteins—TRF1, TRF2, and POT1—with a key structural function at telomeres were found (Figure 6D). These results indicate that the loss of A-type lamins preferentially affects 53BP1. Interestingly, acute depletion of A-type lamins by shLmna also significantly reduced the 53BP1 levels (Figure 6E). Most importantly, reintroduction of either lamin A, lamin C, or both lamins A/C into A-type lamin-depleted cells rescued the levels of 53BP1, demonstrating that lamins A and C are able to stabilize 53BP1 cellular levels.
Figure 6.
A-type lamins stabilize 53BP1 protein. (A) Western blots showing cellular levels of factors involved in the initial sensing of DNA damage. No changes in H2AX levels, but an increase in γH2AX levels was observed in Lmna−/− MEFs. Note the marked decrease in 53BP1 levels on loss of A-type lamins in one representative blot. (B) Immunofluorescence with 53BP1 antibody confirms decreased 53BP1 levels in Lmna−/− MEFs. Images were acquired under equal exposure times. (C) Westerns blots showing that the levels of MDC1, ATM, DNA-PK, Mre11, Nbs1, Ku70, and ERCC1 remain unchanged on loss of A-type lamins. Immunoblotting with β-tubulin and Stat3 were performed as control for loading. (D) Westerns of members of the shelterin complex, such as TRF1, TRF2 and POT1 also remain unchanged. (E) Westerns blots after lentiviral transduction of wild-type MEFs with shLucif and shLmna to deplete A-type lamins (left two lanes), and subsequent retroviral transduction to reconstitute A-type lamins (right three lanes). Note how acute depletion of A-type lamins decreases 53BP1 levels (top). On reintroduction of lamins A and/or C (middle), 53BP1 levels are restored (top). (F) 53BP1 transcripts abundance normalized to GAPDH transcripts levels. The average of four different experiments is shown. (G) Cells incubated with vehicle (EtOH), CHX, or CHX and MG-132 were subjected to immunoblotting to monitor 53BP1 levels. Note the rescue of 53BP1 levels with proteasome inhibitor. *Represents non-specific bands in western blots.
To determine whether the decrease in the levels of this DNA damage sensor protein was the result of lower transcription, the levels of 53BP1 transcripts were monitored by quantitative PCR (Q-PCR). As shown in Figure 6F, no differences in the levels of 53BP1 transcripts were detected between the two genotypes, indicating that the loss of A-type lamins impacts the stability of the 53BP1 protein. Interestingly, incubation of Lmna+/+ and Lmna−/− MEFs with the proteasome inhibitor MG132 rescued the levels of 53BP1, implicating the proteasome pathway in the degradation of 53BP1 (Figure 6G). We hypothesize that similar to what has been reported for Rb family members (Johnson et al, 2004), A-type lamins might stabilize 53BP1 protein levels by preventing their degradation by the proteasome. Loss of 53BP1 function has been recently associated with defective processing of dysfunctional telomeres by the NHEJ repair pathway. On the basis of our results we speculate that the decrease in 53BP1 levels could be responsible, at least in part, for the hindered ability of Lmna−/− MEFs to process dysfunctional telomeres.
Discussion
Understanding the cellular functions of A-type lamins is a highly topical subject because of their implication in a number of disease states, including laminopathies, aging and cancer. In particular, reduced expression of A-type lamins is emerging as a factor contributing to tumourigenesis (Broers et al, 1993; Agrelo et al, 2005; Prokocimer et al, 2006). Our results indicate that A-type lamins play a fundamental role in the maintenance of telomeres and genomic stability. We show that loss of A-type lamins leads to a variety of alterations in telomere biology: (i) nuclear decompartmentalization of telomeres; (ii) impaired maintenance of telomere length homoeostasis; (iii) defects in telomere chromatin architecture; and (iv) a hindered ability to process dysfunctional telomeres by NHEJ. In addition, our results reveal a function for A-type lamins in the DDR pathway, the stabilization of 53BP1 protein levels. Given that alterations of telomere biology and the DDR pathway are hallmarks of cancer and aging, we envision that the observed alterations in these processes, upon loss of A-type lamins, are contributing to the pathogenesis of lamin-related diseases, especially premature aging syndromes, such as HGPS, and tumoural processes characterized by the silencing of the LMNA gene.
Various lines of evidence indicate that the nucleus is compartmentalized and that changes in the spatial organization of chromatin affect nuclear functions (Goldman et al, 2002; Gruenbaum et al, 2005; Misteli, 2007). The importance of telomere compartmentalization for telomere function has been clearly shown in yeast (Akhtar and Gasser, 2007). To date, the mechanisms arranging the nuclear distribution of mammalian telomeres remain to be identified. In addition, how the nuclear localization of mammalian telomeres influences telomere biology is unknown. Lamins, which are absent in yeast, can bind directly to DNA and core histones, which attributes them a major role in tethering chromatin to specific sub-nuclear compartments (Glass et al, 1993; Stierle et al, 2003). Our results show that A-type lamins associate with telomeres and contribute towards their proper nuclear localization. The profound impact that loss of A-type lamins has on different aspects of telomere biology suggests that the nuclear compartmentalization of telomeres could be fundamental for telomere metabolism. Our results do not indicate that changes in telomerase activity, the binding of TRF1 and TRF2, or aberrant recombination are the cause of the telomere shortening in Lmna−/− cells. It is possible that the accessibility of telomerase and/or activities participating in telomere metabolism is reduced on disruption of the scaffold for nuclear organization provided by A-type lamins. Studies aimed to elucidate the impact of loss of A-type lamins on telomere replication, and on the binding of other shelterin complex components or DNA repair factors to telomeres will be fundamental in understanding the mechanisms behind the alterations in telomere biology described here.
The change in localization of telomeres away from the nuclear centre and towards the periphery upon loss of A-type lamins is intriguing. A-type lamins are highly enriched at the nuclear periphery and are also found throughout the nucleoplasm (Schermelleh et al, 2008). We reasoned that loss of A-type lamins could lead to the detachment of telomeres from the nuclear periphery. In contrast, we found that the localization of telomeres shifts towards the nuclear periphery in the absence of A-type lamins, raising the possibility that the nuclear periphery might represent a default pathway for telomere localization. In this model, A-type lamins would have an active role in the localization of telomeres throughout the nucleoplasm in mouse cells. A recent study showing that alterations of the nuclear lamina during senescence are associated with increased aggregation of telomeres at the nuclear periphery supports this model (Raz et al, 2008). It remains to be investigated whether tumour cells with silenced LMNA gene also present alterations in telomere compartmentalization and telomere structure, length, and function. These types of studies will provide insights into the mechanisms altered upon loss of A-type lamins, which could contribute to tumourigenesis.
In addition to the effect on telomere biology, loss of A-type lamins impacts on other molecular mechanisms, such as stabilization of Rb and ING tumour suppressors (Johnson et al, 2004; Nitta et al, 2006; Han et al, 2008) and the DNA damage sensor 53BP1 (this study). These mechanisms could contribute to the telomere phenotypes in Lmna−/− MEFs and to the genomic instability that drives cancer and aging. Our results show that indeed, reduced Rb family function is likely to be responsible for histone modifications defects in Lmna−/− cells. We show, however, that the telomere-shortening phenotype and the decrease in TERRA density are Rb independent. Although the function of TERRAs in telomere biology is undefined, the observation that Rb family deficiency leads to increased levels of these new telomeric components is of great significance. Changes in TERRA density could contribute to the alterations in telomere metabolism previously described to occur upon loss of Rb family members. Future studies will need to address whether regulation of TERRAs and putatively, other non-coding RNAs are part of the tumour suppressor function of Rb family.
Previous studies had shown that expression of mutant lamin A isoforms leads to alterations in the DDR and defective repair, which translate into increased sensitivity to DNA-damaging agents (Liu et al, 2005; Scaffidi and Misteli, 2006; di Masi et al, 2008). To date, the effect that complete loss of A-type lamins has on DNA repair mechanisms remains unknown. Our results show that the fusions resulting from dysfunctional telomeres upon expression of TRF2ΔBΔM require A-type lamins, providing the first link between loss of A-type lamins and defective NHEJ repair. The fact that mutations in the LMNA gene were previously associated with increased NHEJ (Liu et al, 2005) indicates yet another functional difference between mutation and loss of A-type lamins. The increased genomic instability in Lmna−/− MEFs supports a role for A-type lamins in DNA repair. In light of a recent study showing that loss of 53BP1 inhibits processing of dysfunctional telomeres by NHEJ (Dimitrova et al, 2008), we hypothesize that destabilization of this protein occurring upon loss of A-type lamins is in part responsible for the observed phenotype. Similar to other nuclear factors, binding of 53BP1 to A-type lamins either directly or through lamin-associated proteins could contribute to its stability. The finding that loss of 53BP1 restricts the mobility of dysfunctional telomeres within the nucleus, lead to the proposal that 53BP1 has an active role in chromatin dynamics that facilitate the association and fusion of dysfunctional telomeres that might be far away within the nucleus (Dimitrova et al, 2008). We speculate that A-type lamins could provide a platform for the association of 53BP1 with dysfunctional telomeres, as well as contribute to the mobility of dysfunctional telomeres to reach specific subcompartments where they come in contact with each other and with NHEJ repair factors.
Materials and methods
Cell culture
Lmna+/+ and Lmna−/− MEFs and adult fibroblasts were generated in the laboratory of Colin L Stewart as described by Sullivan et al (1999). Rb family-deficient MEFs were generated in the laboratory of Julien Sage (Stanford University, CA). All lines were maintained in DMEM-Glutamax (Invitrogen) supplemented with 10% bovine growth serum, antibiotics, and antimycotics. For cycloheximide and proteasome inhibitor treatments, 0.5 × 106 cells were cultured for 6 h in media containing 10 μg/ml cycloheximide, 30 μM MG-132 (EMD Biochemicals) or EtOH as control.
ChIP assays
ChIP analyses were carried out exactly as described by Garcia-Cao et al (2004). Chromatin was immunoprecipitated using the following antibodies: anti-H3K9me3 (#07-442, Upstate); anti-H4K20me3 (#07-463, Upstate); anti-TRF1 (T1948, Sigma); anti-TRF2 (#05-521, Upstate); or anti-lamin A/C (SC-6215, Santa Cruz). Immunoprecipitated DNA was slot-blotted on a Hybond N+ membrane and hybridized to a telomeric probe (gift from T de Lange, Rockefeller University, NY, USA) or a major satellite probe. The quantification of the signal was done using the ImageQuant software (Molecular Dynamics). For input samples, serial dilutions of the unbound fraction of the no-antibody control were processed. We calculated the amount of telomeric or pericentric DNA immunoprecipitated relative to the signal of the corresponding inputs. In all cases, we represented the ChIP values as a percentage of the total input telomeric DNA, therefore correcting for differences in the number of telomere repeats.
Immunoprecipitation, immunoblotting and immunofluorescence
Rb family members. Cells were lysed in RIPA buffer (0.15 M NaCl, 0.05 M Tris–HCl pH 7.2, 1% Triton-X 100, 1% sodium deoxycholate, and 0.1% SDS), sonicated and 1 mg of total protein immunoprecipitated with antibodies bound to protein A-agarose beads: Rb (IF18), p107 (C-18), and p130 (C-20) from Santa Cruz. Protein detection was carried out using antibodies against Rb (BD Pharmingen), p107 and p130 (Santa Cruz).
Lamin A/C, actin, γ-tubulin, TRF2, and DDR factors. For immunoblotting, cells were lysed in RIPA buffer. Protein detection was carried out using Lamin A/C (SC-6215, Santa Cruz), actin (Clone C4, MPB), β-tubulin (Sigma), TRF1 (gift from Maria A Blasco), TRF2 (#05-521, Upstate), POT1 (gift from Qin Yang), 53BP1 (Bethyl Laboratories, A300-272A), MDC1 (gift from Junran Zhang), ATM (GTX7107, GeneTex), DNA-PKcs (MS-423-P, NeoMarkers), Mre11 (Novus, MB100-142), Nbs1 (Cell Signaling, 3002B), Ku70 (SC-1486, Santa Cruz), H2AX (Upstate, 07-164), γH2AX (Upstate, 07-164), and ERCC1 (SC-17809, Santa Cruz) antibodies. For immunofluorescence, cells were fixed using 4% paraformaldehyde and 0.1% Triton-X 100 for 10 min at RT. Incubations using lamin A/C, γ-tubulin (Sigma), and Alexa- or Cy3-labelled secondary antibodies were done for 1 h. Washes were carried out in PBS and 20 mM glycine. Slides were counterstained using DAPI in Vectashield (Vector). Fluorescent images were taken using a Nikon 90i upright microscope or with a confocal microscope Zeiss L510.
Histones. Histones were purified by acid extraction as previously described by Shechter et al (2007). Histone modifications were detected by immunoblotting using anti-H3K9me3 and anti-H4K20me3 (Upstate).
Isolation of TERRAs
Total RNA and genomic DNA were isolated from the same samples using the TRI Reagent (Sigma) and following manufacturer's instructions. For RNA, 5 μg of isolated sample per well was separated in 1.2% agarose–formaldehyde gels, blotted on a Hybond N+ membrane and hybridized to telomeric and 28S probes. When indicated, negative controls were generated treating samples with RNase A (ABgene). For DNA, 0.1–0.5 μg of DNA was dot blotted on a nylon membrane and hybridized to telomeric or pericentric probes.
Telomerase activity
Telomerase activity was determined using the TeloTAGGG Telomerase PCR ELISA* Kit (Roche) following manufacturer's instructions.
Telomere length measurements
TRF analysis. We prepared cells in agarose plugs and carried out TRF analysis as described by Blasco et al (1997).
Q-FISH on metaphases. We prepared metaphase stage chromosomal spreads for Q-FISH and hybridized them as described by Herrera et al (1999). Fluorescent images were taken using a Nikon 90i upright microscope and intensity of telomere fluorescence analysed using the TFL-Telo program (gift from P Lansdorp, Vancouver, Canada) (Zijlmans et al, 1997). Images and telomere fluorescence values were obtained from more than 50 metaphases in all cases.
Viral transduction with dominant-negative TRF2, lamin A, lamin C, and shRNAs (shLmna and shLucif)
A dominant-negative TRF2 mutant (TRF2ΔBΔM) cloned into the pLPC vector (gift from Titia de Lange, Rockefeller University) along with packaging and envelope plasmids pUMVC and pCMV-VSV-G (provided by Sheila Stewart, Washington University) were transfected in 293T packaging cells using Fugene. After 48 h post-transfection, virus-containing media was used to infect Lmna+/+ or Lmna−/− MEFs. Infection was repeated 24 h later. The same procedure was used to introduce lamins A/C into A-type lamins depleted cells. Retroviral vectors for expression of lamins A/C were a gift from Brian Kennedy (University of Washington, Seattle, WA). GFP was used to monitor the efficiency of 293T transfection and MEF infection. After selection in puromycin (1 μg/ml) for 36–48 h, cells were prepared for FISH and western blot to test the expression of TRF2ΔBΔM. Lentiviral transduction of shLmna and shLucif cloned into pLKO was carried out in the same manner, using the packaging plasmid pHR'8.2ΔR and the envelope plasmid pCMV-VSV-G. Infected cells were subjected to immunoblotting after selection. Both shLmna and shLucif were a gift from Didier Hodzic (Washington University, St Louis, MO).
Immuno-FISH
Cells growing in coverslips were first processed for immunofluorescence. Cells were fixed for 10 min in 3.7% paraformaldehyde/0.2% Tx-100/PBS at RT, followed by three washes in PBS. After blocking in 10% BGS/PBS for 1 h, cells were incubated with anti-γH2AX antibody 1 h at 37°C, washed three times and incubated with secondary antibodies. After washing extensively in PBS, cells were processed for FISH. Cells were fixed in 3.7% paraformaldehyde/PBS for 10 min at RT, washed in PBS, dehydrated in ethanol (70%, 90%, 100% ethanol, 5min each), and air dried. Hybridization solution containing the cy3-labeled telomeric probe was added to coverslips, which were heated to 80°C for 10 min, and incubated for 3 h in the dark at RT. Coverslips were washed twice for 15 min in washing solution, and three times in PBS. Cells were counterstained with DAPI and coverslips mounted on slides.
CO-FISH
CO-FISH analysis was carried out as described by Bailey et al (2001). Briefly, cells were cultured in 20 μM BrdU for 22 h and 0.1 μg/ml colcemid was added for the last 4 h to enrich for mitotic cells. Metaphase stage chromosomal spreads were prepared as for Q-FISH (Herrera et al, 1999). The slides were treated with RNase A (0.5 μg ml−1 for 10 min at 37°C, stained with Hoechst 33258 in 2 × SSC for 15 min at RT and exposed to 365 nm UV light for 30 min. Exonuclease III (3 U/μl) treatment for 20 min at RT was used to digest the BrdU-labelled strands. The slides were then processed as for Q-FISH but with two different probes added sequentially. Hybridizations were carried out first with the G-rich probe (labelling the leading strand) followed by the C-rich probe (labelling the lagging strand).
Quantitative real time PCR
Quantitative real-time PCR was carried out on a MyiQ Detection system (BIO-RAD, California, USA) using Taqman Universal PCR Master Mix (PE Applied Biosystems, California, USA). The cDNA was generated from 1 μg of total RNA using Geneamp RNA PCR kit (PE Applied Biosystems) following manufacturer's instructions. 53BP1 and GAPDH expression was determined using Taqman Gene expression assays (Mm00658689_m1 and Mm99999915_g1, respectively, PE Applied Biosystems). For the analysis, all reactions (in triplicate) were carried out by amplifying target gene and endogenous controls in the same plate. Relative quantitative evaluation of target gene was determined by comparing the threshold cycles.
Statistical analysis
A ‘two-tailed' Student's t-test, ‘two-samples of equal variance' was used to calculate statistical significance of the observed differences in telomere length. Microsoft Excel v.2001 and Graphpad Instat v3.05 were used for the calculations. A paired t-test to determine statistical significance was alternatively used when indicated. In all cases, differences were considered statistically significant when P<0.05.
Supplementary Material
Supplementary data
Review Process File
Acknowledgments
We thank S Stewart, J Zhang and J Roti-Roti of Washington University and J Davie of Manitoba Institute of Cell Biology for helpful discussions. We are grateful to T de Lange for providing TRF2ΔBΔM retroviral vector, D Hodzig for providing shLmna and shLucif vectors, and B Kennedy for the gift of lamins A/C expressing vectors. We thank A Searleman, L Revollo, K Chiappinelli, P Ramachandan, E Peterson, and C Franz for their contributions during their laboratory rotation. We are grateful to T Whitehead for his help with statistical analyses. CancerCare Manitoba Foundation supported the study carried out at the SM lab. Research in the laboratory of SG was supported by an institutional research grant from American Cancer Society, an Alzheimer's Disease Research Centre Pilot Grant, and Start-Up funds provided by the Radiation Oncology Department at Washington University.
Author contributions: IGS and AR carried out most of the experiments and contributed towards writing this paper; SMP observed the decrease in 53BP1 levels. BV, DL, and SM carried out the 3D analysis of nuclear distribution of telomere intensity, and wrote the corresponding Supplementary data. DG carried out western blots of DDR factors. LM carried out CO-FISH assays. EG and BPS carried out western blot analysis of ATM and DNA-PK. JS provided all Rb family-deficient MEFs. TS and CLS generated the Lmna−/− mouse model and provided the different lines of MEFs used in this study. SG supervised the research project and the preparation of the paper.
References
- Agrelo R, Setien F, Espada J, Artiga MJ, Rodriguez M, Perez-Rosado A, Sanchez-Aguilera A, Fraga MF, Piris MA, Esteller M (2005) Inactivation of the lamin A/C gene by CpG island promoter hypermethylation in hematologic malignancies, and its association with poor survival in nodal diffuse large B-cell lymphoma. J Clin Oncol 23: 3940–3947 [DOI] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM (2007) The nuclear envelope and transcriptional control. Nat Rev Genet 8: 507–517 [DOI] [PubMed] [Google Scholar]
- Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB (1992) Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci USA 89: 10114–10118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318: 798–801 [DOI] [PubMed] [Google Scholar]
- Bailey SM, Brenneman MA, Goodwin EH (2004) Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res 32: 3743–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH (2001) Strand-specific postreplicative processing of mammalian telomeres. Science 293: 2462–2465 [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA (2007) Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 178: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH (2001) Switching and signaling at the telomere. Cell 106: 661–673 [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Greider CW, Szostak JW (2006) Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 12: 1133–1138 [DOI] [PubMed] [Google Scholar]
- Blasco MA (2007) The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8: 299–309 [DOI] [PubMed] [Google Scholar]
- Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW (1997) Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91: 25–34 [DOI] [PubMed] [Google Scholar]
- Broers JL, Ramaekers FC, Bonne G, Yaou RB, Hutchison CJ (2006) Nuclear lamins: laminopathies and their role in premature ageing. Physiol Rev 86: 967–1008 [DOI] [PubMed] [Google Scholar]
- Broers JL, Raymond Y, Rot MK, Kuijpers H, Wagenaar SS, Ramaekers FC (1993) Nuclear A-type lamins are differentially expressed in human lung cancer subtypes. Am J Pathol 143: 211–220 [PMC free article] [PubMed] [Google Scholar]
- Capell BC, Collins FS (2006) Human laminopathies: nuclei gone genetically awry. Nat Rev Genet 7: 940–952 [DOI] [PubMed] [Google Scholar]
- Chuang TC, Moshir S, Garini Y, Chuang AY, Young IT, Vermolen B, van den Doel R, Mougey V, Perrin M, Braun M, Kerr PD, Fest T, Boukamp P, Mai S (2004) The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol 2: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T (2002) Protection of mammalian telomeres. Oncogene 21: 532–540 [DOI] [PubMed] [Google Scholar]
- de Lange T (2005a) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110 [DOI] [PubMed] [Google Scholar]
- De Lange T (2005b) Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol 70: 197–204 [DOI] [PubMed] [Google Scholar]
- Dechat T, Pfleghaar K, Sengupta K, Shimi T, Shumaker DK, Solimando L, Goldman RD (2008) Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev 22: 832–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Masi A, D'Apice MR, Ricordy R, Tanzarella C, Novelli G (2008) The R527H mutation in LMNA gene causes an increased sensitivity to ionizing radiation. Cell Cycle 7: 2030–2037 [DOI] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao M, Gonzalo S, Dean D, Blasco MA (2002) A role for the Rb family of proteins in controlling telomere length. Nat Genet 32: 415–419 [DOI] [PubMed] [Google Scholar]
- Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA (2004) Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36: 94–99 [DOI] [PubMed] [Google Scholar]
- Glass CA, Glass JR, Taniura H, Hasel KW, Blevitt JM, Gerace L (1993) The alpha-helical rod domain of human lamins A and C contains a chromatin binding site. EMBO J 12: 4413–4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP (2002) Nuclear lamins: building blocks of nuclear architecture. Genes Dev 16: 533–547 [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Blasco MA (2005) Role of Rb family in the epigenetic definition of chromatin. Cell Cycle 4: 752–755 [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, Blasco MA (2005) Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol 7: 420–428 [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA (2006) DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8: 416–424 [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL (2005) The nuclear lamina comes of age. Nat Rev Mol Cell Biol 6: 21–31 [DOI] [PubMed] [Google Scholar]
- Han X, Feng X, Rattner JB, Smith H, Bose P, Suzuki K, Soliman MA, Scott MS, Burke BE, Riabowol K (2008) Tethering by lamin A stabilizes and targets the ING1 tumour suppressor. Nat Cell Biol 10: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Herrera E, Samper E, Blasco MA (1999) Telomere shortening in mTR−/− embryos is associated with failure to close the neural tube. EMBO J 18: 1172–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Risques RA, Martin GM, Rabinovitch PS, Oshima J (2008) Accelerated telomere shortening and replicative senescence in human fibroblasts overexpressing mutant and wild-type lamin A. Exp Cell Res 314: 82–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovino F, Lentini L, Amato A, Di Leonardo A (2006) RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol Cancer 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BR, Nitta RT, Frock RL, Mounkes L, Barbie DA, Stewart CL, Harlow E, Kennedy BK (2004) A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc Natl Acad Sci USA 101: 9677–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlow BA, Stanfel MN, Burtner CR, Johnston ED, Kennedy BK (2008) Suppression of proliferative defects associated with processing-defective lamin A mutants by hTERT or inactivation of p53. Mol Biol Cell 19: 5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Muralikrishna B, Parnaik VK (2002) Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J Cell Biol 159: 783–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laud PR, Multani AS, Bailey SM, Wu L, Ma J, Kingsley C, Lebel M, Pathak S, DePinho RA, Chang S (2005) Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev 19: 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, Huang JD, Li KM, Chau PY, Chen DJ, Pei D, Pendas AM, Cadinanos J, Lopez-Otin C, Tse HF, Hutchison C, Chen J, Cao Y, Cheah KS, Tryggvason K et al. (2005) Genomic instability in laminopathy-based premature aging. Nat Med 11: 780–785 [DOI] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z (2004) Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem 279: 51338–51342 [DOI] [PubMed] [Google Scholar]
- Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J (2008) The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell 32: 465–477 [DOI] [PubMed] [Google Scholar]
- Mai S, Garini Y (2006) The significance of telomeric aggregates in the interphase nuclei of tumor cells. J Cell Biochem 97: 904–915 [DOI] [PubMed] [Google Scholar]
- Mekhail K, Seebacher J, Gygi SP, Moazed D (2008) Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456: 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T (2007) Beyond the sequence: cellular organization of genome function. Cell 128: 787–800 [DOI] [PubMed] [Google Scholar]
- Mounkes LC, Stewart CL (2004) Aging and nuclear organization: lamins and progeria. Curr Opin Cell Biol 16: 322–327 [DOI] [PubMed] [Google Scholar]
- Munoz P, Blanco R, Flores JM, Blasco MA (2005) XPF nuclease-dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet 37: 1063–1071 [DOI] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ (2008) Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM (2009) Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res 37: 1152–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta RT, Jameson SA, Kudlow BA, Conlan LA, Kennedy BK (2006) Stabilization of the retinoblastoma protein by A-type nuclear lamins is required for INK4A-mediated cell cycle arrest. Mol Cell Biol 26: 5360–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar D, Barkan R, Meshorer E, Gruenbaum Y (2009) Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med 13: 1059–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokocimer M, Margalit A, Gruenbaum Y (2006) The nuclear lamina and its proposed roles in tumorigenesis: projection on the hematologic malignancies and future targeted therapy. J Struct Biol 155: 351–360 [DOI] [PubMed] [Google Scholar]
- Raz V, Vermolen BJ, Garini Y, Onderwater JJ, Mommaas-Kienhuis MA, Koster AJ, Young IT, Tanke H, Dirks RW (2008) The nuclear lamina promotes telomere aggregation and centromere peripheral localization during senescence of human mesenchymal stem cells. J Cell Sci 121: 4018–4028 [DOI] [PubMed] [Google Scholar]
- Riha K, Heacock ML, Shippen DE (2006) The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet 40: 237–277 [DOI] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T (2006) Lamin A-dependent nuclear defects in human aging. Science 312: 1059–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, Leonhardt H, Sedat JW (2008) Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320: 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Blasco MA (2008) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB (2007) Extraction, purification and analysis of histones. Nat Protoc 2: 1445–1457 [DOI] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, Jenuwein T, Goldman RD (2006) Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA 103: 8703–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T (2002) DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol 12: 1635–1644 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T (2000) Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 20: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann TP, Goldman AE, Wang C, Huang S, Goldman RD (2002) Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J Cell Biol 156: 603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CL, Kozlov S, Fong LG, Young SG (2007) Mouse models of the laminopathies. Exp Cell Res 313: 2144–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierle V, Couprie J, Ostlund C, Krimm I, Zinn-Justin S, Hossenlopp P, Worman HJ, Courvalin JC, Duband-Goulet I (2003) The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry 42: 4819–4828 [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B (1999) Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 147: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Gasser SM (2004) The function of nuclear architecture: a genetic approach. Annu Rev Genet 38: 305–345 [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T (2003) DNA damage foci at dysfunctional telomeres. Curr Biol 13: 1549–1556 [DOI] [PubMed] [Google Scholar]
- Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, Fabre E (2006) Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol 172: 189–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, Tryggvason K, Freije JM, Lopez-Otin C (2005) Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature 437: 564–568 [DOI] [PubMed] [Google Scholar]
- Vermolen BJ, Garini Y, Mai S, Mougey V, Fest T, Chuang TC, Chuang AY, Wark L, Young IT (2005) Characterizing the three-dimensional organization of telomeres. Cytometry A 67: 144–150 [DOI] [PubMed] [Google Scholar]
- Willis ND, Cox TR, Rahman-Casans SF, Smits K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M, Wilson RG, de Bruine A, Hutchison CJ (2008) Lamin A/C is a risk biomarker in colorectal cancer. PLoS ONE 3: e2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlmans JM, Martens UM, Poon SS, Raap AK, Tanke HJ, Ward RK, Lansdorp PM (1997) Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci USA 94: 7423–7428 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Review Process File