Abstract
Immune protection by the complement system critically depends on assembly of C3 convertases on the surface of pathogens and altered host cells. These short-lived protease complexes are formed through pro-convertases, which for the alternative pathway consist of the complement component C3b and the pro-enzyme factor B (FB). Here, we present the crystal structure at 2.2-Å resolution, small-angle X-ray scattering and electron microscopy (EM) data of the pro-convertase formed by human FB and cobra venom factor (CVF), a potent homologue of C3b that generates more stable convertases. FB is loaded onto CVF through its pro-peptide Ba segment by specific contacts, which explain the specificity for the homologous C3b over the native C3 and inactive products iC3b and C3c. The protease segment Bb binds the carboxy terminus of CVF through the metal-ion dependent adhesion site of the Von Willebrand factor A-type domain. A possible dynamic equilibrium between a ‘loading' and ‘activation' state of the pro-convertase may explain the observed difference between the crystal structure of CVFB and the EM structure of C3bB. These insights into formation of convertases provide a basis for further development of complement therapeutics.
Keywords: complement system, convertase, immunology, protein complex, structure
Introduction
The complement system is a key part of the innate and adaptive immune system and is critical for the resistance to infection and clearance of altered host cells. This intricate host defence system consists of over 30 plasma and cell-surface proteins that enables the host to recognize pathogens or immunogenic particles and eliminates them from the host's system (Muller-Eberhard, 1988; Walport, 2001). In the central step of the proteolytic cascade of the complement system, cells are covalently labelled, or opsonized, for B-cell stimulation, clearance by phagocytosis and cell lysis. On activation of the recognition pathways, protease complexes called C3 convertases form on the target cell surface that cleave and activate C3 into the large fragment C3b and a small fragment C3a that mediates inflammation (Walport, 2001). C3b molecules react indiscriminately with hydroxyls and hence bind covalently to the targeted surface, in which they act as labels for recognition by macrophages and B-cells (Muller-Eberhard, 1988). Two homologous surface-bound C3 convertases are formed. One through the antibody-mediated classical and lectin-binding pathways; and, one through the alternative pathway formed by C3b and pro-enzyme factor B (FB) that is used in the central amplification step of the complement response (Muller-Eberhard, 1988) (see Figure 1A).
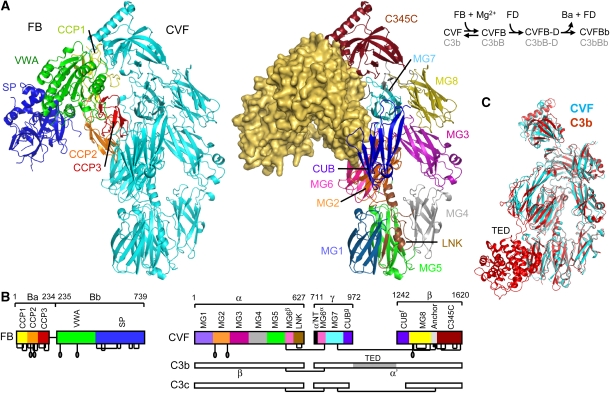
Figure 1.
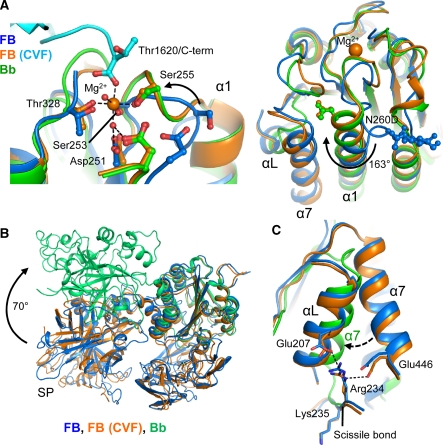
Structure of the CVFB complex at 2.2-Å resolution. (A) Ribbon representation of CVFB with FB coloured by domain and CVF coloured cyan (left) and of CVF coloured by domain with FB in wheat surface representation (right). The proteolytic assembly process of the C3 convertase is shown schematically. (B) Domain compositions, including disulphide bridges and glycan positions, of FB and CVF are indicated, together with the topology of C3b and C3c for clarity. (C) Comparison of CVF (cyan) with C3b (Janssen et al, 2006) (red) (see also Supplementary Table IIA).
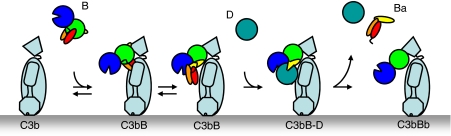
Control over the activity of the complement system is of critical importance to the homeostasis of the organism and depends on formation and dissociation of the central convertases. Uncontrolled complement activity may lead to host tissue damage and is associated with several pathological conditions such as age-related macular degeneration, atypical haemolytic uraemic syndrome (aHUS) and rejection of transplants (Ricklin and Lambris, 2007). Recently, mutations in both C3 and FB have been associated with aHUS (Goicoechea de Jorge et al, 2007; Fremeaux-Bacchi et al, 2008). On the other hand, lack of function, due to deficiencies or mutations in complement proteins, may predispose individuals to infectious diseases. Formation of the convertase complexes depends on a proteolytic assembly process, which starts with proteolytic activation of C3 into C3b. Next, FB binds surface-bound C3b forming the pro-convertase C3bB. When bound to C3b, FB becomes susceptible to proteolysis by factor D (FD). Cleavage by FD removes the pro-peptide fragment Ba (residues 1–234) and yields the active and labile convertase C3bBb (consisting of C3b and the protease fragment Bb (residues 235–739)), which amplifies the complement response by cleaving C3 into C3b (Muller-Eberhard, 1988). Similarly, the venom of the Indian cobra contains a C3 homologue called cobra venom factor (CVF) (49% identical in sequence to C3) (Fritzinger et al, 1994), which is processed by proteases in the venom gland into a three-chain molecule, which has C3b-like activity and forms soluble convertases (Vogt et al, 1974) (see Figure 1B). The CVF-containing convertases are far more stable (with a half lifetime of ∼7 h) (Vogel and Muller-Eberhard, 1982) than C3bBb convertases (half lifetime of ∼90 s) (Fishelson et al, 1984) and cleave C3 and in some instances also C5 to consume complement components of the prey (von Zabern et al, 1980). This prolonged convertase activity underpins the putative therapeutic use of humanized CVF in pathological conditions in which tissue damage may be prevented by complement depletion (Vogel and Fritzinger, 2007).
Like C3bBb, the CVFBb convertase assembles in two steps, which are (i) Mg2+-dependent binding of FB to CVF (Kd of 1 μM) (Hensley et al, 1986) and (ii) subsequent cleavage of FB by FD (Figure 1A). In recent years, crystal structures have been reported of C3b (Janssen et al, 2006; Wiesmann et al, 2006), pro-enzyme FB (Milder et al, 2007) and of the isolated fragment Bb (Ponnuraj et al, 2004). C3b consists of 12 domains (see Figure 1B). The structure of FB is formed by five domains, three N-terminal complement-control protein domains (CCP1–3; also called short consensus repeat or SCR), a Von Willebrand factor A-type (VWA) domain and a C-terminal serine protease (SP) domain. Mutagenesis and binding studies located putative binding sites for FB on the C345C domain and the α′ chain N-terminal tail (α′NT) of C3b (Taniguchi-Sidle and Isenman, 1994; Kolln et al, 2005; Fritzinger et al, 2009) and for C3b or CVF at or near the metal-ion dependent adhesion site (MIDAS) of the VWA domain and on the CCP domains of FB (Hourcade et al, 1995, 1999; Tuckwell et al, 1997; Hinshelwood et al, 1999; Thurman et al, 2005). An allosteric model for the activation of the pro-enzyme FB was proposed based on 1H NMR spectroscopy studies (Hinshelwood and Perkins, 2000a, 2000b). The crystal structure of the pro-enzyme FB (Milder et al, 2007) allowed a more detailed hypothesis for FB binding to C3b or CVF and exposure of the scissile loop in FB for cleavage by FD. Putatively, binding of C3b or CVF to the Mg2+ ion of the MIDAS in the VWA domain of FB relocates the CCP1–3 domains and the linker helix αL (which together form the Ba pro-peptide segment). Dislocation of helix αL putatively allows docking of the activation helix α7 of the VWA domain into its canonical groove as observed in the structure of Bb and related integrin inserted domains. In the pro-enzyme FB, the scissile bond (Arg234–Lys235) is partially occluded with the Arg234 (the P1 residue of the scissile bond) interacting with both helices αL and α7. Alterations in the αL–α7 arrangement may disrupt this interaction leading to exposure of the scissile loop for FD cleavage. A very recent electron microscopy (EM) model of C3bB at ∼27-Å resolution is consistent with the predicted C3b–B binding sites and supports the rearrangement of the CCP1–3 domains (Torreira et al, 2009). However, details of the C3b–FB or CVF–FB interactions and possible induced structural changes are unknown. Here, we study the CVFB complex at 2.2-Å resolution to determine the CVF–FB interaction sites, the conformational state of the MIDAS and the associated allosteric changes, which addresses the composite roles of the multiple domains of CVF and FB that underlie convertase formation and activation.
Results
Structure determination of the CVFB complex
We determined the crystal structure of the pro-convertase complex using CVF (1266 residues) purified from cobra venom and recombinant human FB (739 residues). To enhance complex formation and increase the chances of successful crystallization, we used a double gain-of-function mutation (D254G/N260D) that increases stability of the pro-convertase (Hourcade et al, 1999) and eliminates the glycan moiety on N260 (see Materials and methods and Supplementary Figure 1; all amino acids are numbered according to mature, secreted protein thus excluding the 22 and 25 residue long signal peptides of CVF and FB, respectively). First, we solved the structure successfully at 8.5-Å resolution using glycosylated CVF; and, later at 3.0 and 2.2-Å resolution using deglycosylated CVF (Figure 1; Supplementary Figure 2; Supplementary Table I). Crystal structures were validated at low resolution by small-angle X-ray scattering (SAXS) and negative stain EM. SAXS data were collected on the CVFB complex consisting of glycosylated CVF and wild-type FB in solution (see Materials and methods and Figure 2). The experimental molecular mass of the solute (220±20 kDa) agreed with the expected molecular mass of the complex (230 kDa), proving homogeneous complex formation in the samples. The radius of gyration of the crystal structure of CVFB accounting for the hydration shell (45.4 Å) matched the experimental SAXS value (45.8±0.5 Å). Moreover, the computed SAXS curve from the crystal structure neatly fitted the measured scattering of CVFB indicating that the quaternary crystal structure is preserved in solution up to a resolution of ca 25 Å (Figure 2B). Furthermore, single-particle reconstruction using EM negative stained images of CVFB also correlated well with the crystal structures of CVFB (Figure 2A). Overall, the structure of the pro-convertase CVFB is characterized by an extensive interface that involves four out of five domains of pro-enzyme FB and five out of 11 domains of CVF.
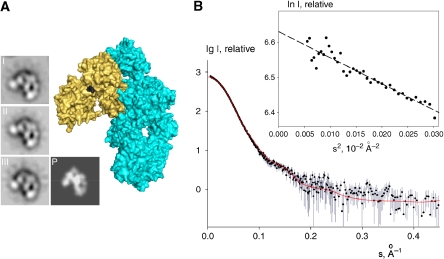
Figure 2.
The CVFB crystal structures correlate well with EM and SAXS of CVFB in solution. (A) EM class averages of the pro-convertase CVFB (I–III) correlate well with the crystal structure of CVFB, shown in surface representation (cyan and wheat, respectively, with the FB scissile bond black) and as a low-resolution projection (P). (B) The computed scattering curve of the 2.2-Å crystal structure of CVFB (red line) fitted with CRYSOL (Svergun et al, 1995) to the measured scattering data of 2 mg/ml CVFB (black dots with experimental errors) with good correlation (χ2 of 1.2). The computed scattering curves of the 3.0-Å crystal structures of CVFB gave good correlation (both χ2 of 1.2) with the measured scattering data as well (not shown). Inset; Guinier plot for CVFB from X-ray scattering.
CVF structurally resembles C3b
The domain composition of mature, three-chain CVF falls in between that of the biologically active C3b (12 domains) and inactive C3c (10 domains) molecules. Mature CVF contains macroglobulin (MG) domains 1–8, a linker domain, a ‘complement C1r/C1s, Uegf, Bmp1' (CUB) domain and a C-terminal C345C domain, but lacks a thioester-containing domain (TED). Overall, the 11 domains of CVF are arranged very similar to the corresponding domains in C3b (Janssen et al, 2006) and C3c (Janssen et al, 2005) (see Figure 1C). After submission of this study, the crystal structure of free CVF was published (Krishnan et al, 2009). The structures of free CVF and CVF bound to FB are very similar, with domain arrangements and conformations more similar between the two CVF structures than between CVF and C3b (Supplementary Table IIA). An exception is formed by the C345C domain, which has adopted an orientation closer to the CUB domain in free CVF (Krishnan et al, 2009). In the structure of FB bound CVF, this domain has an orientation more similar to that observed in structures of C3b. Reorientations of the CUB and C345C domains (Supplementary Table IIA) likely reflect inherent flexibility of the molecules (Janssen et al, 2007; Krishnan et al, 2009).
Five domains of CVF contact FB
FB binds to the ‘top' part of CVF, away from the cell-surface attachment site in C3b. Domains MG2, MG6, MG7, CUB, C345C and the α′NT region (using C3 chain names) of CVF form a concave ‘clasp' that grabs around domains CCP1–3 and VWA of FB (Figure 1). This observation is supported by C3/CVF chimera studies in which more stable C3bBb complexes were generated by replacing the C345C domain of C3b with that of CVF (Kolln et al, 2005; Fritzinger et al, 2009). In addition, mutagenesis studies in the α′NT of C3b that affect the ability of FB to bind to C3b underscore the role for the central α′NT in this interaction (Taniguchi-Sidle and Isenman, 1994). Proteolytic activation of C3 into C3b induces large rearrangements of α′NT, MG7 and CUB (Janssen et al, 2005, 2006) that are required to form the observed FB-binding site, which explains that FB binds to C3b and not to native C3. Inactivation of C3b is caused by cleavages in the CUB domain by factor I (FI) yielding iC3b and finally C3c, which do not bind FB and cannot form convertases (Ross et al, 1983). The FB-binding site is virtually present in C3c except for the CUB domain, which is missing in C3c. Furthermore, FB contacts Arg1262–Glu1263 of the CUB domain in CVF, which correspond to Arg1281–Ser1282 in C3b that is the first scissile bond cleaved by FI when forming iC3b (Figure 3A). The structural data, therefore, indicate that pro-convertase formation depends on an arrangement of five domains in CVF or C3b with a critical role for an intact CUB domain, which is used in the regulation of complement activity.
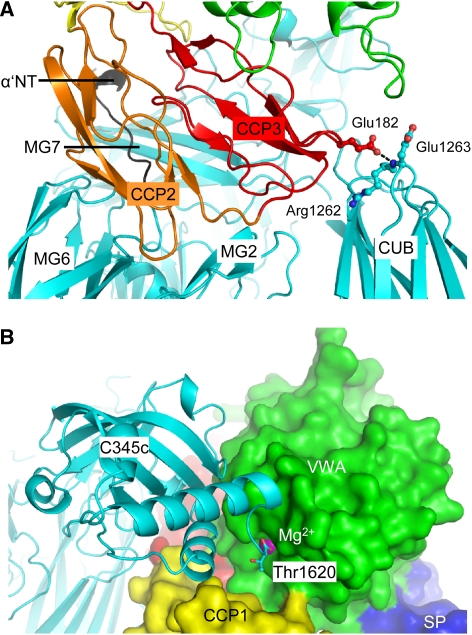
Figure 3.
The CVFB interface consists of two patches. (A) Ribbon representation of CCP2–3 (coloured orange and red, respectively) of the Ba segment interacting with MG2, MG6, MG7, CUB (all coloured cyan) and α′NT (coloured black) of CVF. Glu182 of FB interacts with Arg1262 and Glu1263 of CVF (ball-and-stick representation), which correspond to the first FI-cleavage site in C3b. (B) VWA of the Bb segment, shown in green surface representation, interacts with C345C of CVF shown in cyan ribbon representation. The C-terminus (Thr1620) of CVF binds to the Mg2+ ion (purple sphere) in FB.
The FB interface consists of two distinct functional patches
The FB interface is divided into a large contact site formed by the pro-peptide segment Ba and a small contact site formed by the protease segment Bb (∼3600 and ∼1300 Å2 buried surface areas, respectively). The anti-parallel arranged CCP2–3 domains of the Ba segment contact α′NT and MG2, MG6, MG7 and CUB domains of CVF (see Figures 3 and 4). This binding site includes epitopes of antibodies that block pro-convertase formation (Hourcade et al, 1995; Thurman et al, 2005), and explains the effects of FB/C2 chimeras in which replacement of several short parts in Ba of FB with those of C2 increased the binding of FB to C3b (Hourcade et al, 1995) (Figure 4E). The orientation of CCP1 is variable in the structures determined at 3.0- and 2.2-Å; CCP1 contributes only 30–600 Å2 buried surface area, respectively, to the CVF–B interface (Supplementary Figures 2 and 3). The Bb segment contacts the C345C domain of CVF through its VWA domain (Figure 3B). This is supported by previous biochemical and mutagenesis studies in the VWA domain of FB that identified the VWA domain to be involved in pro-convertase formation (Tuckwell et al, 1997; Hinshelwood et al, 1999; Hourcade et al, 1999). In contrast, no contacts are made to CVF by the SP domain of FB, as predicted earlier (Smith et al, 1982; Pryzdial and Isenman, 1987). A positive charged patch on FB, centred on VWA, complements a negative charged patch on C345C of CVF, in an otherwise largely neutral interface (Figure 4D). In conclusion, the CCP domains of the Ba segment and the VWA domain of the Bb segment form two distinct functional interfaces with CVF in which Ba makes specific contacts that discriminate C3b from native C3 and inactive cleavage products iC3b and C3c and in which the VWA-C345C interface is likely important for the activity of the active convertase.
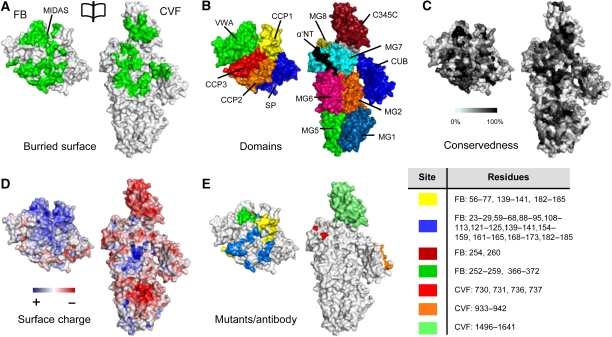
Figure 4.
Surface representation of the CVFB interface coloured functionally. (A) An opened view of the 4900 Å2 footprint of the FB–CVF interface is highlighted in green. (B) Domains of FB and CVF coloured according to Figure 1. (C) FB and CVF colour-coded to residue conservation; from non-conserved (white) to conserved (black). Figure is produced using CONSURF (Glaser et al, 2005). (D) FB and CVF coloured by electrostatic potential from red (−10 kbT/ec) to blue (−10 kbT/ec). The VWA:C345C interface consists of conserved complementary electrostatic patches. (E) Previously proposed sites involved in complex formation. The yellow coloured patches are epitopes to which antibody binding decreases complex formation (Hourcade et al, 1995; Thurman et al, 2005). The other patches are based on FB to C2 chimeras that increase binding of FB to C3b >150% (Hourcade et al, 1995) (blue) or decrease binding <10% (Tuckwell et al, 1997) (green); on C3 to CVF chimeras that increase C3bBb complex stability (Kolln et al, 2005; Fritzinger et al, 2009) (lime); on an alternative proteolytic product of C3, that supports activation of FB (O'Keefe et al, 1988) (orange) or on single site mutants (single numbers) that increase complex formation (Hourcade et al, 1999) (dark red) or decrease complex formation (Taniguchi-Sidle and Isenman, 1994) (red). Legend for colour-coding and residue numbers are presented in the table. CVF residue numbering is according to human C3.
The FB MIDAS adopts a high-affinity state
On binding to CVF, the distorted MIDAS in free FB has rearranged into a canonical high-affinity ligand-bound state, as in fragment Bb (Ponnuraj et al, 2004) and the isolated VWA domain (Bhattacharya et al, 2004) (Figure 5). MIDAS residues Ser255 and Asp364 move up to 7.4 Å and together with residues Asp251, Ser253, Thr328 and two water molecules coordinate the Mg2+ ion. The COO− terminus (Thr1620) of CVF is the sixth chelating ligand of the Mg2+ ion (Figure 5A). Thus, the Bb segment binds CVF through its MIDAS, in which the carboxy terminus of CVF completes the coordination sphere of the Mg2+ ion. These details confirm the prominent role for Mg2+-dependent MIDAS-mediated complex formation, which has been shown earlier by mutagenesis studies in which replacement of two MIDAS loops (252–259 and 366–372) of FB with those of C2 decreased the binding of FB to CVF (Tuckwell et al, 1997), by a combined affinity and mass spectrometry approach that identified two segments that contain the MIDAS (229–265 and 355–381) to be involved in pro-convertase formation (Hinshelwood et al, 1999), and by gain-of-function mutations (D254G and N260D) near the MIDAS of FB that increased stability of the pro-convertase (Hourcade et al, 1999) (Figure 4E). Furthermore, C3/CVF chimera studies underscore the role for the C345C domain in this interaction (Kolln et al, 2005; Fritzinger et al, 2009) (Figure 4E). Reduction of steric hindrance explains the D254G gain-of-function mutation in FB. Deletion of the glycan in the N260D gain-of-function mutant possibly facilitates rotation by 163° and elongation of VWA helix α1 that is coupled to the MIDAS loop rearrangements (Figure 5A). Similarly, mutation F261L, which is located in the refolding region of helix α1, may favour this rearrangement and hence enhance pro-convertase formation causing atypical haemolytic uremic syndrome (Goicoechea de Jorge et al, 2007). In conclusion, FB binding to CVF induces a local but functionally important rearrangement in the MIDAS and surrounding loops from a distorted to a high-affinity ligand-binding state.
Figure 5.
Conformational rearrangements in the VWA domain of FB. (A) The MIDAS site in VWA rearranges from distorted in free FB (Milder et al, 2007) (blue) to a high-affinity Mg2+-bound conformation in CVFB (cyan and orange) similar to free Bb (Ponnuraj et al, 2004) (green) (left panel). Helix α1 elongates and glycan-linked Asn260 (mutated to Asp in CVFB) rotates 163° (right panel). (B) Comparison of free FB (blue), FB bound to CVF (orange) and free Bb (lime). (C) Helixes αL and α7 and the Arg234–Lys235 scissile bond do not rearrange on FB binding to CVF. Arg234 remains hydrogen bonded to Glu207 and Glu446. In Bb α7 has swapped with αL that is removed. Colour scheme as in (A).
CVF does not induce domain rearrangements in FB
We observe no further large conformational changes in FB on binding to CVF neither in the crystal structures (Figure 5B; Supplementary Table IIB), SAXS data or EM classes (Figure 2). Apparently, FB provides sufficient space for the C-terminal tail of CVF to access the MIDAS without dislocating the CCP1 domain. Correspondingly, helix αL and α7 do not relocate and the scissile bond 234–235 remains partially occluded, when FB binds CVF (Figure 5C). Detailed comparison of the VWA domain in CVFB and in free Bb (Ponnuraj et al, 2004) shows that activation of FB into Bb repositions helix α7 into the groove previously occupied by helix αL, away from the elongated helix α1. This additional rearrangement may further stabilize the high-affinity MIDAS configuration that is observed in Bb (and the isolated VWA domain (Bhattacharya et al, 2004)). Possibly the VWA domain of FB adopts this conformation in the active convertase CVFBb and C3bBb. This rearrangement may prevent release of Mg2+ from C3bBb (in which the MIDAS is locked into a stable configuration) and allows Mg2+ release and dissociation of C3bB or CVFB, when treated with EDTA (Harris et al, 2005). We observe no structural changes in the active site of the SP domain between free FB, the pro-convertase and free Bb (Supplementary Figure 4), indicating that enzyme activity is possibly controlled by quaternary changes in the enzyme complex; that is by substrate (C3) binding to the convertase. Thus, except for the MIDAS and surrounding loops, FB binding to CVF does not induce major conformational changes in either FB or CVF.
Discussion
The central amplification step of the complement system is crucial for the defence against invading pathogens and the homeostasis of the host. Lack of activity may result in recurrent bacterial infections, whereas unregulated activation may lead to tissue damage. Convertase formation and activity is, therefore, a tightly regulated process and reduced control, for example caused by mutations or deficiencies in proteins that form the convertase (C3b and FB) or in complement regulators that dissociate convertases, have been associated with several immune-related diseases (Ricklin and Lambris, 2007). Detailed understanding of the formation, activity and regulation of the convertase will be instrumental in the development of therapeutics to control complement related diseases. Here, we show for the first time in atomic detail the interactions that underlie pro-convertase formation.
Restricting the assembly of the convertase in place and time is an important regulatory mechanism of the complement system. FB only binds to the activated form C3b and not to C3, iC3b or C3c. The data presented here reveal that the Ba segment of FB determines this specificity. Segment Ba has specific interactions with α′NT, MG7 and CUB, which undergo conformational rearrangements in the activation of C3 to C3b (Janssen et al, 2005, 2006; Wiesmann et al, 2006). Complex formation depends on an intact CUB domain, which is degraded in the conversion of C3b to iC3b and C3c as shown structurally by EM (Nishida et al, 2006). The α′NT has also been implied in binding complement regulators factor H (FH) and CR1 (CD35) to C3b (Weiler et al, 1976; Pryzdial and Isenman, 1987). This overlapping binding site for FB, FH and CR1 results in steric hindrance, which explains the observed competitive binding (Weiler et al, 1976; Pryzdial and Isenman, 1987). The proposed binding sited for decay accelerating factor (DAF/CD55), identified by mutagenesis studies on helix 4 and 5 (Hourcade et al, 2002) and at aHUS-related residue K298 (Goicoechea de Jorge et al, 2007) located on the VWA domain of FB, is exposed in the complex. This is in line with the over 10-fold higher affinity of DAF for the C3bB complex compared with the individual components C3b and B (Pangburn, 1986; Harris et al, 2005). Both segments Ba and Bb contribute to the binding interface, but segment Ba provides 73% of the total ∼4900 Å2 buried surface area and is apparently essential to load Bb onto CVF or C3b, as the Bb fragment itself cannot bind to either. Thus, although segment Ba itself is not part of the active convertase, it has a crucial function in its assembly and regulation.
Several studies have indicated that the CVFB and C3bB complexes are functionally similar (Vogt et al, 1974; Smith et al, 1982; Vogel and Muller-Eberhard, 1982; Vogel et al, 1984; Hensley et al, 1986). And although similarity was inferred from sequence homology (Fritzinger et al, 1994) we and others (Krishnan et al, 2009) also show in structural detail that CVF is very similar to C3b. It, therefore, seems likely that the structure of CVFB resembles that of C3bB. After completion of this study, a low-resolution (∼27 Å) structure of negative stained C3bB was published (Torreira et al, 2009). In agreement with our high-resolution data, these EM data indicated that FB binds to the top part of C3b involving CUB, C345C and α′NT of C3b and that C3b does not change conformation on binding of FB; however, the details of the C3b–FB interaction could not be resolved due to the low resolution of the EM images. In contrast to our observations on FB binding to CVF, Torreira et al show that FB undergoes a large conformational change on binding to C3b involving a relocation of the Ba segment towards CUB, although it was not possible to reveal the details of the conformational change nor could it be resolved whether the Bb part of FB undergoes a conformational change on binding to C3b (Torreira et al, 2009). Therefore, the X-ray and EM data of CVFB and C3bB indicate that there is a difference in conformation of FB when bound to CVF and bound to C3b. This difference in conformation is unlikely to arise from crystal packing events as the CVFB crystal structure is very similar to the structure determined by solution SAXS and EM studies. Possibly, this difference illustrates two functional different states of the pro-convertase structure, which may exist in a dynamic equilibrium.
In this equilibrium, the CVFB structure may represent the ‘loading' state of FB to the cofactor, whereas the C3bB structure indicates the form that is activated for cleavage by FD. Activation of FB by FD occurs solely when FB forms a pro-convertase. In the CVFB structure, the scissile bond 234–235 has remained partially occluded similar to its position in free FB. In the C3bB structure, relocation of the Ba segment may have exposed the scissile bond and helix α7 has possibly relocated to the position previously occupied by helix αL. Unfortunately, these details could not be resolved due to the low resolution of the EM data (Torreira et al, 2009). Our data suggest that FB bound to CVF remains predominantly in the initial ‘loading' state, whereas FB bound to C3b adopts more frequently an ‘activation' state with an exposed scissile loop that can be cleaved by FD. In support of this hypothesis, we observe a much slower activation rate of FB by FD in CVFB than in C3bB (Supplementary Figure 5) and Harris et al demonstrate a conformation change in the C3bB complex in surface plasmon resonance experiments (Harris et al, 2005). Although a difference in the initial ‘loading' of FB to CVF and C3b cannot be excluded at present, the data presented here and the observation that FB in the CVFB complex is very similar in domain arrangements to free FB indicate that FB is likely to bind initially in a similar manner to C3b as it binds to CVF. Thus, there may be two distinct, ‘loading' and ‘activation', states of the pro-convertase structure represented by the CVFB and C3bB structures. However, the details of the differences between the CVFB and C3bB structures can only be resolved with high-resolution data of the C3bB complex.
In conclusion, the CVFB complex indicates that the CCP domains of the Ba segment of FB provide a scaffold for the protease segment Bb to be loaded onto the C-terminal C345C of CVF or C3b in an Mg2+-dependent manner (Figure 6). By binding to MG7, CUB and α′NT, the Ba segment determines the specificity of FB for CVF and C3b. Complex formation may be followed by a conformational change in FB in which the scissile bond is exposed and which enables FD to activate FB. Conversion of the pro-convertase into the active convertase releases the Ba fragment and affects the orientation of the SP domain (Torreira et al, 2009). Therefore, control over convertase activity is determined by pro-convertase complex formation and activation in which release of the Ba fragment provides conformational freedom for C3 activation. These detailed structural insights into pro-convertase formation are instrumental to the developmental of novel therapeutic approaches that modulate this central step in the complement system.
Figure 6.
Convertase formation and activation model. Schematic representation of the assembly of the pro-convertase complex and its subsequent activation by FD into the active C3 convertase (colour scheme as in Figure 1A, left panel).
Materials and methods
Protein expression and purification
CVF was purified from lyophilized Indian cobra (Naja kaouthia) venom as described (Vogel and Muller-Eberhard, 1984) including an additional final size-exclusion chromatography step. Purified CVF (7 mg/ml) was deglycosylated by incubation for 4 days at 37°C, in phosphate-buffered saline (PBS), 0.1% w/v azide, 10 mg/ml soybean trypsin inhibitor, 5 mM ethylenediaminetetraacetic acid, 5 mM benzamidine, 1 mM phenylmethylsulphonyl fluoride and 0.3 Units/ml endo-beta-N-acetylglucosaminidase F (Endo-F3). Deglycosylation of CVF does not have an effect on its activity (Gowda et al, 1994) but in our hands improves the quality of crystals considerably. As a final step, deglycosylated CVF was purified by size-exclusion chromatography. To promote a more stable pro-convertase and to enhance the rate of successful crystallization, we used a double gain-of-function mutant (D254G/N260D) of FB in which the glycan moiety on N260 is removed (Hourcade et al, 1999). The large and flexible glycan moieties present on glycoproteins are often detrimental to crystallization. In fact, deglycosylation of CVF was necessary to obtain crystals that diffracted to high resolution. The double gain-of-function mutant (D254G/N260D) (Hourcade et al, 1999) was fused to a N-terminal His-tag containing a TEV cleavage site, entirely sequenced to confirm a correct DNA sequence and expressed in N-acetylglucosaminyltransferase I (GnTI) deficient human embryonic kidney 293 cells that stably express EBNA1 (HEK293ES). The deficiency of GnTI results in production of homogeneous N-linked glycosylation (Reeves et al, 2002). FB was purified by metal-affinity chromatography as described (Milder et al, 2007). The His-tag was removed by TEV protease cleavage, followed by a second column passage on Ni-NTA superflow beads and a size-exclusion chromatography step. To verify the crystal structure of the CVFB complex and to validate that the (D254G/N260D) mutation in FB and the deglycosylation of CVF do not have an effect on the structure of the CVFB complex, we used wt FB and glycosylated CVF in solution SAXS and negative stain EM studies. Wild-type human FB was expressed in HEK293 cells stably expressing EBNA1 (HEK293E) (Durocher et al, 2002) and purified as described (Milder et al, 2007).
Crystallization and data collection
Glycosylated CVF (14 mg/ml) and FB mutant D254G/N260D (11 mg/ml) were mixed at a molar ratio of 1:1 to a final concentration of 12 mg/ml (52 μM) and 10 mM Tris pH 7.4, 5 mM MgCl2 and 13 mM NaCl. Crystals were grown in sitting drops from mother liquor containing 12% w/v PEG-monometylether 2000, 50 mM malic acid 2-(N-morpholino)ethanesulfonic acid tris (hydroxymethyl)aminomethane buffer (MMT) pH 6.8, at 30°C, to typical dimensions of 80 × 30 × 20 μm within 2 weeks. For cryo-protection, 20% v/v glycerol was added to the mother liquor, and crystals were flash-cooled in liquid nitrogen. Crystals displayed space group P212121 (a=129.8, b=134.0, c=291.8 Å), contained two complexes per asymmetric unit and diffracted to 8.5-Å resolution at European synchrotron radiation facility (ESRF) beamline ID23-2. Purified deglycosylated CVF (12 mg/ml) and FB mutant D254G/N260D (15 mg/ml) were mixed to a molar ratio of 1:1 and dialyzed against 10 mM Tris pH 7.4, 5 mM MgCl2 and 10 mM NaCl resulting in a final concentration of 12 mg/ml (50 μM). This sample yielded two different crystal forms. Crystals with space group C2221 (a=128.9, b=283.5, c=134.4 Å) were grown in sitting drops from mother liquor containing 6.6% w/v PEG 3350, 16 mM Bis-Tris propane pH 6.5, 66 mM Na/K Phosphate, at 18°C, to typical dimensions of 400 × 100 × 80 μm within 1 week. For cryo-protection, 30% v/v PEG 400 was added to the mother liquor, and crystals were flash-cooled in liquid nitrogen. These crystals contained one complex per asymmetric unit and diffracted to 2.2-Å resolution at ESRF beamline ID23-1. Crystals with space group P212121 (a=134.0, b=137.0, c=283.7 Å) were grown in sitting drops from mother liquor containing 8.25% w/v PEG 1500, 33 mM MMT pH 9.0, at 18°C, to typical dimensions of 200 × 60 × 40 μm within 1 week. For cryo-protection, 12% v/v 2,3 butanediol was added to the mother liquor, and crystals were flash-cooled in liquid nitrogen. These crystals contained two complexes per asymmetric unit and diffracted to 3.0-Å resolution at ESRF beamline ID23-1. All diffraction data was integrated and scaled with MOSFLM (Leslie, 1992) and SCALA (Evans, 2006) in CCP4 (CCP4, 1994).
Structure determination
Initially the 8.5-Å resolution structure was solved by molecular replacement with PHASER (McCoy et al, 2007) using the isolated structures of C3b (Janssen et al, 2006) (pdb code: 2I07) with the TED domain omitted and FB (Milder et al, 2007) (pdb code: 2OK5) as the search models. Owing to limited resolution, only rigid body refinement (using 18 groups) was performed in REFMAC (Murshudov et al, 1997) resulting in Rwork/Rfree values of 34.4/34.9%. At a later stage, the 2.2-Å resolution structure was solved by molecular replacement in PHASER (McCoy et al, 2007) starting with the β-chain and MG6 part of the α-chain from C3b followed step-by-step by SP, VWA and CCP2–3 from FB and α′NT-MG7-anchor, MG8, CUB and C345C from C3b and CCP1 from FB, respectively, using molecular graphics in COOT (Emsley and Cowtan, 2004) with rigid-body refinement in PHASER. This partial model was re-built automatically by ARP/wARP (Perrakis et al, 1999) and completed by several cycles of manual rebuilding in COOT and refinement in PHENIX (Adams et al, 2002) to a final Rwork/Rfree value of 18.0/22.6%. The 3.0-Å structure was solved by molecular replacement in PHASER using the refined 2.2-Å structure and was completed by several cycles of manual rebuilding in COOT and refinement in PHENIX to a final Rwork/Rfree value of 18.9/24.3%. All molecular graphics figures were generated with pymol (W Delano; http://www.pymol.org/).
EM and image classification
FB (from HEK293E cells) and CVF were mixed to a molar ratio of 1.2:1 and a final concentration of 1 mg/ml in PBS, 5 mM MgCl2 and 1 mM NiCl2 and incubated for 15 min at room temperature. The sample was diluted to 5 μg/ml in water and immediately applied to a freshly glow-discharged carbon layer supported by an EM grid and negatively stained with 0.75% uranyl formate as described (Ohi et al, 2004). Micrographs were recorded under low-dose conditions on a 4k × 4k CCD camera with a FEI Tecnai 12 transmission electron microscope operating at 120 kV and a magnification of 39 000 × resulting in a pixel size of 3.8 Å. A total of 10 943 particles were selected from the micrographs, respectively, using the program ‘Boxer' from the EMAN software package (Ludtke et al, 1999). The contrast transfer function of the microscope for each micrograph was estimated and corrected using XMIPP (Sorzano et al, 2004). The extracted particles were classified into 20 classes with the maximum-likelihood multireference refinement protocol in XMIPP as described (Scheres et al, 2008). The three most populous or representative classes are shown in Figure 2. The projection was generated from the crystal structure of CVFB after low-pass filtering to 25-Å resolution in EMAN (Ludtke et al, 1999).
Small angle X-ray scattering
FB (from HEK293E cells) and CVF were mixed to a molar ratio of 1:1 in Tris-buffered saline and 5 mM MgCl2 and incubated for at least 15 min at room temperature. Synchrotron X-ray scattering data from a 1 mg/ml and a 2 mg/ml solution of the CVFB complex were collected following standard procedures at the European Molecular Biology Laboratory (EMBL) beamline X33 (Roessle et al, 2007) (storage ring DORIS-III, Deutsches Elektronen-Synchrotron (DESY), Hamburg) using a Pilatus 500K detector (DECTRIS, Switzerland). To monitor for radiation damage, four successive 30 s exposures on the same sample were compared, and no changes were detected. The data were processed using PRIMUS (Konarev et al, 2003). The molecular mass of the solute was calculated by normalization against the scattering from a reference solution of bovine serum albumin. The SAXS curves were computed from the 2.2- and 3.0-Å crystal structures of CVFB by CRYSOL (Svergun et al, 1995).
Accession codes
Coordinates and structure factors of the CVFB complex have been deposited in the Protein Data Bank with succession numbers 3HRZ (2.2-Å resolution) and 3HS0 (3.0-Å resolution).
Supplementary Material
Supplementary data
Review Process File
Acknowledgments
We thank Catharina Verheij for help with initial crystallization experiments, Hans Meeldijk (Cellular Architecture and Dynamics, Utrecht) for help with EM grid preparation and testing, Sjors Scheres (National Center for Biotechnology, Madrid) for suggestions on EM image processing in XMIPP and Roland Romijn (U-Protein Express, Utrecht) for preparation of FB for SAXS analysis. We acknowledge the ESRF and the EMBL/DESY for the provision of synchrotron radiation facilities and thank David Flot (ESRF, Grenoble) and Adam Round (EMBL, Hamburg) for beamline assistance. This work was supported by the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (NWO-CW) and the US National Institutes of Health (to PG).
Author contributions: DCF purified CVF. LG cloned, expressed and purified FB and deglycosylated CVF. BJCJ crystallized the complex, determined and analysed the structures. PG, LG and DIS collected, processed and analysed the SAXS data. RIK and BJCJ prepared and tested the EM grids. RIK collected the EM data under supervision of AJK. BJCJ processed the EM data. RIK and BJCJ analysed the EM data. BJCJ, C-WV and PG conceived the project. BJCJ and PG wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Bhattacharya AA, Lupher ML Jr, Staunton DE, Liddington RC (2004) Crystal structure of the A domain from complement factor B reveals an integrin-like open conformation. Structure (Camb) 12: 371–378 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Durocher Y, Perret S, Kamen A (2002) High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res 30: E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evans P (2006) Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Fishelson Z, Pangburn MK, Muller-Eberhard HJ (1984) Characterization of the initial C3 convertase of the alternative pathway of human complement. J Immunol 132: 1430–1434 [PubMed] [Google Scholar]
- Fremeaux-Bacchi V, Miller EC, Liszewski MK, Strain L, Blouin J, Brown AL, Moghal N, Kaplan BS, Weiss RA, Lhotta K, Kapur G, Mattoo T, Nivet H, Wong W, Gie S, Hurault de Ligny B, Fischbach M, Gupta R, Hauhart R, Meunier V et al. (2008) Mutations in complement C3 predispose to development of atypical hemolytic uremic syndrome. Blood 112: 4948–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzinger DC, Bredehorst R, Vogel CW (1994) Molecular cloning and derived primary structure of cobra venom factor. Proc Natl Acad Sci USA 91: 12775–12779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzinger DC, Hew BE, Thorne M, Pangburn MK, Janssen BJ, Gros P, Vogel CW (2009) Functional characterization of human C3/cobra venom factor hybrid proteins for therapeutic complement depletion. Dev Comp Immunol 33: 105–116 [DOI] [PubMed] [Google Scholar]
- Glaser F, Rosenberg Y, Kessel A, Pupko T, Ben-Tal N (2005) The ConSurf-HSSP database: the mapping of evolutionary conservation among homologs onto PDB structures. Proteins 58: 610–617 [DOI] [PubMed] [Google Scholar]
- Goicoechea de Jorge E, Harris CL, Esparza-Gordillo J, Carreras L, Arranz EA, Garrido CA, Lopez-Trascasa M, Sanchez-Corral P, Morgan BP, Rodriguez de Cordoba S (2007) Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc Natl Acad Sci USA 104: 240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda DC, Petrella EC, Raj TT, Bredehorst R, Vogel CW (1994) Immunoreactivity and function of oligosaccharides in cobra venom factor. J Immunol 152: 2977–2986 [PubMed] [Google Scholar]
- Harris CL, Abbott RJ, Smith RA, Morgan BP, Lea SM (2005) Molecular dissection of interactions between components of the alternative pathway of complement and decay accelerating factor (CD55). J Biol Chem 280: 2569–2578 [DOI] [PubMed] [Google Scholar]
- Hensley P, O'Keefe MC, Spangler CJ, Osborne JC Jr, Vogel CW (1986) The effects of metal ions and temperature on the interaction of cobra venom factor and human complement factor B. J Biol Chem 261: 11038–11044 [PubMed] [Google Scholar]
- Hinshelwood J, Perkins SJ (2000a) Conformational changes during the assembly of factor B from its domains by (1)H NMR spectroscopy and molecular modelling: their relevance to the regulation of factor B activity. J Mol Biol 301: 1267–1285 [DOI] [PubMed] [Google Scholar]
- Hinshelwood J, Perkins SJ (2000b) Metal-dependent conformational changes in a recombinant vWF-A domain from human factor B: a solution study by circular dichroism, fourier transform infrared and (1)H NMR spectroscopy. J Mol Biol 298: 135–147 [DOI] [PubMed] [Google Scholar]
- Hinshelwood J, Spencer DI, Edwards YJ, Perkins SJ (1999) Identification of the C3b binding site in a recombinant vWF-A domain of complement factor B by surface-enhanced laser desorption-ionisation affinity mass spectrometry and homology modelling: implications for the activity of factor B. J Mol Biol 294: 587–599 [DOI] [PubMed] [Google Scholar]
- Hourcade DE, Mitchell L, Kuttner-Kondo LA, Atkinson JP, Medof ME (2002) Decay-accelerating factor (DAF), complement receptor 1 (CR1), and factor H dissociate the complement AP C3 convertase (C3bBb) via sites on the type A domain of Bb. J Biol Chem 277: 1107–1112 [DOI] [PubMed] [Google Scholar]
- Hourcade DE, Mitchell LM, Oglesby TJ (1999) Mutations of the type A domain of complement factor B that promote high-affinity C3b-binding. J Immunol 162: 2906–2911 [PubMed] [Google Scholar]
- Hourcade DE, Wagner LM, Oglesby TJ (1995) Analysis of the short consensus repeats of human complement factor B by site-directed mutagenesis. J Biol Chem 270: 19716–19722 [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P (2006) Structure of C3b reveals conformational changes that underlie complement activity. Nature 444: 213–216 [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Halff EF, Lambris JD, Gros P (2007) Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J Biol Chem 282: 29241–29247 [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Huizinga EG, Raaijmakers HC, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P (2005) Structures of complement component C3 provide insights into the function and evolution of immunity. Nature 437: 505–511 [DOI] [PubMed] [Google Scholar]
- Kolln J, Bredehorst R, Spillner E (2005) Engineering of human complement component C3 for catalytic inhibition of complement. Immunol Lett 98: 49–56 [DOI] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MH, Svergun DI (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Cryst 36: 1277–1282 [Google Scholar]
- Krishnan V, Ponnuraj K, Xu Y, Macon K, Volanakis JE, Narayana SV (2009) The crystal structure of cobra venom factor, a cofactor for C3- and C5-convertase CVFBb. Structure 17: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AG (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography 26 [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milder FJ, Gomes L, Schouten A, Janssen BJ, Huizinga EG, Romijn RA, Hemrika W, Roos A, Daha MR, Gros P (2007) Factor B structure provides insights into activation of the central protease of the complement system. Nat Struct Mol Biol 14: 224–228 [DOI] [PubMed] [Google Scholar]
- Muller-Eberhard HJ (1988) Molecular organization and function of the complement system. Annu Rev Biochem 57: 321–347 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nishida N, Walz T, Springer TA (2006) Structural transitions of complement component C3 and its activation products. Proc Natl Acad Sci USA 103: 19737–19742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe MC, Caporale LH, Vogel CW (1988) A novel cleavage product of human complement component C3 with structural and functional properties of cobra venom factor. J Biol Chem 263: 12690–12697 [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T (2004) Negative staining and image classification—powerful tools in modern electron microscopy. Biol Proced Online 6: 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangburn MK (1986) Differences between the binding sites of the complement regulatory proteins DAF, CR1, and factor H on C3 convertases. J Immunol 136: 2216–2221 [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Ponnuraj K, Xu Y, Macon K, Moore D, Volanakis JE, Narayana SV (2004) Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol Cell 14: 17–28 [DOI] [PubMed] [Google Scholar]
- Pryzdial EL, Isenman DE (1987) Alternative complement pathway activation fragment Ba binds to C3b. Evidence that formation of the factor B-C3b complex involves two discrete points of contact. J Biol Chem 262: 1519–1525 [PubMed] [Google Scholar]
- Reeves PJ, Callewaert N, Contreras R, Khorana HG (2002) Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc Natl Acad Sci USA 99: 13419–13424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Lambris JD (2007) Complement-targeted therapeutics. Nat Biotechnol 25: 1265–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev PV, Round A, Friedler S, Hermes S, Svergun DI (2007) Upgrade of the small angle X-ray scattering Beamline X33 at the European Molecular Biology Laboratory, Hamburg. J Appl Cryst 40: s190–s194 [Google Scholar]
- Ross GD, Newman SL, Lambris JD, Devery-Pocius JE, Cain JA, Lachmann PJ (1983) Generation of three different fragments of bound C3 with purified factor I or serum. II. Location of binding sites in the C3 fragments for factors B and H, complement receptors, and bovine conglutinin. J Exp Med 158: 334–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R (2008) Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc 3: 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Vogel CW, Muller-Eberhard HJ (1982) Ultrastructure of cobra venom factor-dependent C3/C5 convertase and its zymogen, factor B of human complement. J Biol Chem 257: 9879–9882 [PubMed] [Google Scholar]
- Sorzano CO, Marabini R, Velazquez-Muriel J, Bilbao-Castro JR, Scheres SH, Carazo JM, Pascual-Montano A (2004) XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol 148: 194–204 [DOI] [PubMed] [Google Scholar]
- Svergun DI, Barberato C, Koch MH (1995) CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst 28: 768–773 [Google Scholar]
- Taniguchi-Sidle A, Isenman DE (1994) Interactions of human complement component C3 with factor B and with complement receptors type 1 (CR1, CD35) and type 3 (CR3, CD11b/CD18) involve an acidic sequence at the N-terminus of C3 alpha'-chain. J Immunol 153: 5285–5302 [PubMed] [Google Scholar]
- Thurman JM, Kraus DM, Girardi G, Hourcade D, Kang HJ, Royer PA, Mitchell LM, Giclas PC, Salmon J, Gilkeson G, Holers VM (2005) A novel inhibitor of the alternative complement pathway prevents antiphospholipid antibody-induced pregnancy loss in mice. Mol Immunol 42: 87–97 [DOI] [PubMed] [Google Scholar]
- Torreira E, Tortajada A, Montes T, Rodriguez de Cordoba S, Llorca O (2009) 3D structure of the C3bB complex provides insights into the activation and regulation of the complement alternative pathway convertase. Proc Natl Acad Sci USA 106: 882–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckwell DS, Xu Y, Newham P, Humphries MJ, Volanakis JE (1997) Surface loops adjacent to the cation-binding site of the complement factor B von Willebrand factor type A module determine C3b binding specificity. Biochemistry 36: 6605–6613 [DOI] [PubMed] [Google Scholar]
- Vogel CW, Fritzinger DC (2007) Humanized cobra venom factor: experimental therapeutics for targeted complement activation and complement depletion. Curr Pharm Des 13: 2916–2926 [DOI] [PubMed] [Google Scholar]
- Vogel CW, Muller-Eberhard HJ (1982) The cobra venom factor-dependent C3 convertase of human complement. A kinetic and thermodynamic analysis of a protease acting on its natural high molecular weight substrate. J Biol Chem 257: 8292–8299 [PubMed] [Google Scholar]
- Vogel CW, Muller-Eberhard HJ (1984) Cobra venom factor: improved method for purification and biochemical characterization. J Immunol Methods 73: 203–220 [DOI] [PubMed] [Google Scholar]
- Vogel CW, Smith CA, Muller-Eberhard HJ (1984) Cobra venom factor: structural homology with the third component of human complement. J Immunol 133: 3235–3241 [PubMed] [Google Scholar]
- Vogt W, Dieminger L, Lynen R, Schmidt G (1974) Alternative pathway for the activation of complement in human serum. Formation and composition of the complex with cobra venom factor that cleaves the third component of complement. Hoppe Seylers Z Physiol Chem 355: 171–183 [DOI] [PubMed] [Google Scholar]
- von Zabern I, Hinsch B, Przyklenk H, Schmidt G, Vogt W (1980) Comparison of Naja n. naja and Naja h. haje cobra-venom factors: correlation between binding affinity for the fifth component of complement and mediation of its cleavage. Immunobiology 157: 499–514 [DOI] [PubMed] [Google Scholar]
- Walport MJ (2001) Complement. First of two parts. N Engl J Med 344: 1058–1066 [DOI] [PubMed] [Google Scholar]
- Weiler JM, Daha MR, Austen KF, Fearon DT (1976) Control of the amplification convertase of complement by the plasma protein beta1H. Proc Natl Acad Sci USA 73: 3268–3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C, Katschke KJ, Yin J, Helmy KY, Steffek M, Fairbrother WJ, McCallum SA, Embuscado L, DeForge L, Hass PE, van Lookeren Campagne M (2006) Structure of C3b in complex with CRIg gives insights into regulation of complement activation. Nature 444: 217–220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Review Process File