Abstract
DNA vaccines have tremendous potential for rapid deployment in pandemic applications, wherein a new antigen is ‘plugged’ into a validated vector, and rapidly produced in a validated, fermentation - purification process. For this application, it is essential that the vector and fermentation process function with a variety of different antigen genes. However, many antigen genes are unpredictably ‘toxic’ or otherwise low yielding in standard fermentation processes. We report cell bank and fermentation process unit operation innovations that reduce plasmid-mediated metabolic burden, enabling successful production of previously known toxic influenza hemagglutinin antigen genes. These processes, combined with vector backbone modifications, doubled fermentation productivity compared to existing high copy vectors, such as pVAX1 and gWIZ, resulting in high plasmid yields (up to 2220 mg/L, 5% of total dry cell weight) even with previously identified toxic or poor producing inserts.
Keywords: DNA vaccine, plasmid, vector, fermentation, Escherichia coli, gene therapy
Introduction
Plasmid DNA is increasingly finding its way into new experimental non-viral gene therapeutics, including DNA vaccines, gene replacement vectors, and seed constructs for viral vector production. Numerous human plasmid products are in the pipeline. Kutzler and Weiner (2008) identified 72 Phase I, 20 Phase II and 2 Phase III clinical trials of human DNA vaccine products. Four DNA plasmid products are currently licenced for veterinary applications. This includes two infectious disease vaccines for West Nile virus in horses (Ft Dodge Animal Health) and infectious haematopoietic necrosis virus in salmon (Novartis). As well, a melanoma cancer vaccine for dogs (Merial) and a growth hormone releasing factor therapy for pigs (VGX Animal Health) are approved. In order to commercialize DNA medicines, it is essential to devise industrial processes whereby plasmid DNA can be manufactured to meet the quality, economy, and scale requirements projected for future products.
Gene therapy or DNA vaccine plasmids usually contain the pUC temperature inducible replication origin. A number of pUC plasmid vector design criteria that impact plasmid yield and quality have been identified. The orientation of elements within a vector can affect plasmid quality and yield [Williams et al., 2006; Levy, J.: US20046709844 (2004)]. Z DNA forming regions are unstable while intra-molecular triplex regions reduce supercoiling in batch fermentation (Cooke et al., 2004). Direct repeats are mutational hot spots, especially if cells are grown to stationary phase during propagation (Ribeiro et al., 2008). Critically, even with optimized vector backbones, gene inserts may bestow low yield or toxicity onto a plasmid for largely unknown reasons (reviewed in Carnes and Williams, 2007).
Plasmid DNA production is typically performed in E. coli K12 strains such as DH5α (Carnes et al., 2006), DH5 (Listner et al., 2006), DH1 (Cooke et al., 2004), JM108 [Huber, H., Pacher, C., Necina, R., Kollmann, F., Reinisch, C.:WO2005098002 (2005)] or DH10B (Lahijani et al., 1996). The reasons for this are historical, and based on plasmid productivity in shake flasks which is generally not predictive of fermentation productivity. For example, JM108 was selected as the best strain cell line evaluation of a panel of 7 strains (DH1, DH5, DH5α, JM83, JM101, HB101, and JM108) for production of three different plasmids, using LB shake flask media. One of the plasmids in these strains was then evaluated in 37°C semi-defined batch fermentation. DH5α was one of the poorer fermentation producers in this study (1.3 mg/OD550/L; 65% supercoiled), with DH1 intermediate (3.0 mg/OD550/L; 84% supercoiled) while JM108 was the highest (8.2 mg/OD550/L; 91% supercoiled) [Huber, H., Pacher, C., Necina, R., Kollmann, F., Reinisch, C. WO2005098002 (2005)]. By contrast, in an inducible 30°C to 42°C semi-defined batch fermentation process, DH1 and DH5α were similarly high producing strains (Carnes et al., 2006) with DH5α producing 96% supercoiled plasmid (Plasmid DNA for vaccine manufacture: high-yield production and rapid analysis. Downstream 41: 8–11. 2007 General Electric Company). An extensive shake flask evaluation of plasmid specific yield and percent supercoiling in 17 host strains with three different pUC origin based plasmids (5.8 kb gWIZ-GFP, 6.9 kb pSVβ, 20 kb pQR150) has been reported (Yau et al., 2008). HB101 and DH5α had near 100% supercoiling with all 3 plasmids and high specific yields with 2 of 3 plasmids. By contrast, the other 15 strains were found to have poor supercoiling or production yields with one or more plasmids. For example, while BL21(DE3) plasmid yields were high, % supercoiling was inconsistent (gWIZ-GFP = 98%, pSVβ = 82% and pQR150 = 45%). Recently, BL21 recA endA has been demonstrated to produce high yields (1904 mg/L) of predominantly supercoiled plasmid with a gWIZ derived plasmid in a 30°C to 42°C inducible process. In contrast to the Yau et al., 2008 shake flask results, unmodified BL21 was a poor producer (Phue et al., 2008). These studies demonstrate plasmid productivity in shake flask is poorly predictive of fermentation productivity, and strain performance varies between different fermentation processes.
Increasing yield and productivity, while decreasing manufacturing cost, are essential goals for plasmid production. As well, the fermentation process dramatically impacts the quality of plasmid DNA at harvest, as well as the quality of plasmid DNA after downstream lysis. In general, plasmid quality is higher from fed-batch rather than batch fermentation cultures (reviewed in O’Kennedy et al., 2003). As well, differences in growth media carbon:nitrogen ratio can dramatically affect cell harvest and lysis characteristics (O’Kennedy et al., 2000). Thus, fermentation media and process needs to be carefully optimized for plasmid yield, plasmid quality (% supercoiling), and compatibility of the resultant cells for harvest and lysis.
To date, most plasmid fermentation media and processes result in low yields (<200 mg/L), imposing a cost and purity burden on commercialization of plasmid DNA production (reviewed in Carnes and Williams, 2007; Hoare et. al., 2006; Carnes, 2005; Prather et al., 2003). A few high yield processes (500–1500 mg/L) have been described [Carnes et al., 2006; Listner et al., 2006; Singer et al., 2009; Huber, H., Weigl, G., Buchinger, W.: WO2005097990 (2005)] that enable cost effective plasmid manufacture. These processes all couple reduced growth rate (which generally increases copy number) with high copy replication origins (reviewed in Carnes and Williams, 2007).
These fermentation processes were developed and evaluated only with ‘optimal plasmids’ with relatively benign effects on the host strain. However, many antigen producing vector backbones contain detrimental antigen insert sequences that limit plasmid copy number, resulting in low fermentation productivity. Regrettably, this is often discovered late in development when plasmid production is scaled from shake flasks into fermentors. Generally, this problem cannot be fixed without vector reengineering. This is an insurmountable problem for rapid deployment applications to curb an arising pandemic, since a validated vector backbone and fermentation process must work with the new selected antigen. Plasmids, strains, and fermentation processes are therefore needed that allow useable production of both optimal and suboptimal antigens. Protein over-expression, and high copy plasmid production, both place a metabolic load on the host cell that may limit productivity (reviewed in Ricci and Hernandez, 2000; Glick, 1995). Fermentation of a kanamycin resistant (kanR) plasmid at 37°C in a batch defined media fermentation process resulted in reduced growth rate and biomass, and overflow metabolism; overexpression of the kanR gene product to 18% of total protein was speculated as the cause of these effects (Rozkov et al., 2004). Engineering a strain to overproduce intermediaries of the pentose-phosphate pathway, required for plasmid replication and protein production, by overexpression of glucose-6-phosphate dehydrogenase (G6PDH, encoded by zwf) partially alleviated plasmid-mediated growth rate reduction (Flores et al., 2004). Thus, reducing the metabolic burden associated with high copy number plasmids may increase tolerable plasmid copy numbers, and reduce plasmid toxicity.
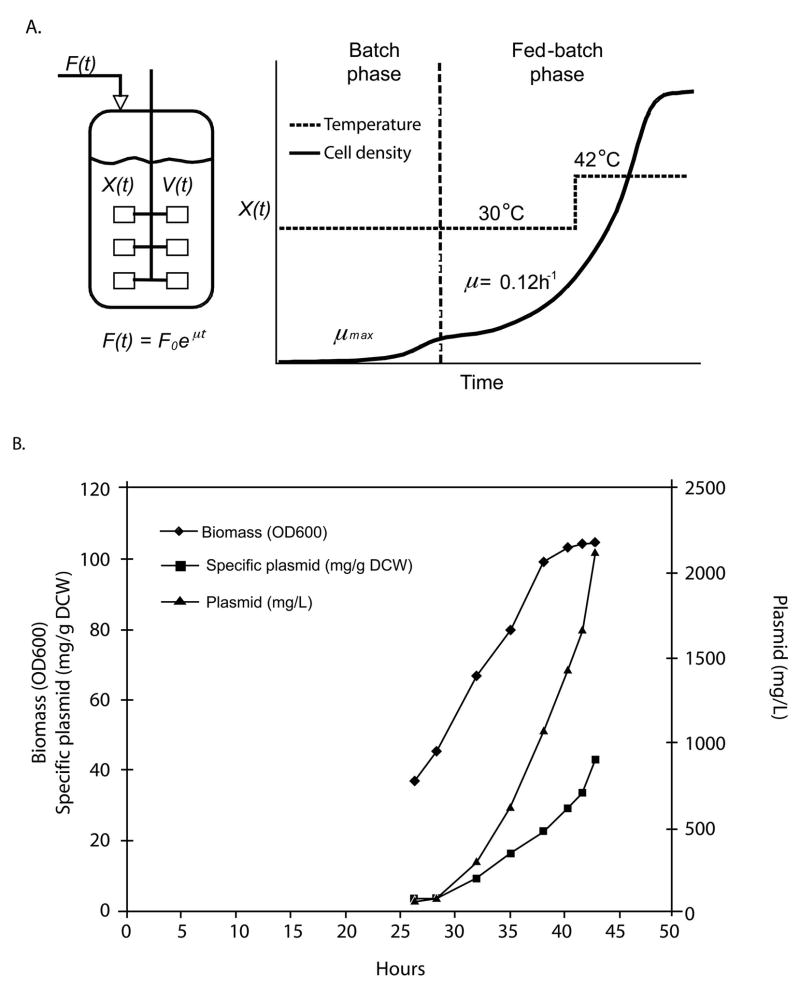
Recently, we developed pUC plasmid fermentation control parameters that reduce metabolic burden by utilizing conditions that both restrict growth rate and maintain reduced plasmid copy number during the biomass accumulation phase prior to a plasmid accumulation phase immediately before harvest (Figure 1A; Carnes et al., 2006). Plasmid-mediated protein production during the biomass accumulation phase is also reduced, since the kanR gene product is not significantly overproduced (Carnes et al., 2006).
Figure 1.
A. Inducible fed-batch plasmid fermentation process. Plasmids containing the pUC origin are induced by the 30°C to 42°C shift. B. E. coli strain DH5α containing pNTCUltra1 plasmid inducible fed-batch fermentation growth and plasmid yield profiles. Induction (30°C to 42°C shift) was at 29 h, plasmid yield reached 2130 mg/L.
We now report optimized vectors and further pUC plasmid fermentation process innovations that increase plasmid yields up to 2.2 g/L, with plasmid comprising 5% of the total dry cell weight at harvest. We demonstrate that plasmid copy number (specific yield) within a strain is largely set by process and vector-intrinsic factors. We also describe improved toxic plasmid production in fermentation culture, using a cell bank process that reduces plasmid-mediated cell stress and metabolic burden. Strain engineering of DH5α to overexpress key replication proteins was utilized to study kinetics of plasmid productivity. These data highlight differences between plasmid replication in shake flask culture compared to high yield fermentation culture.
Materials and Methods
Strains and Plasmids
Fermentations were performed with E. coli strain DH5α [F- Φ 80dlacZΔM15 Δ(lacZYA -argF) U169 recA1 endA1 hsdR17(rk−, mk+) phoA supE44 λ-thi-1 gyrA96 relA1] and derivatives. DH5α includes the recA mutation, which minimizes recombination of cloned DNA, the endA1 mutation, eliminating non-specific digestion of plasmid, and relA, which allows slow growth without induction of the stringent response.
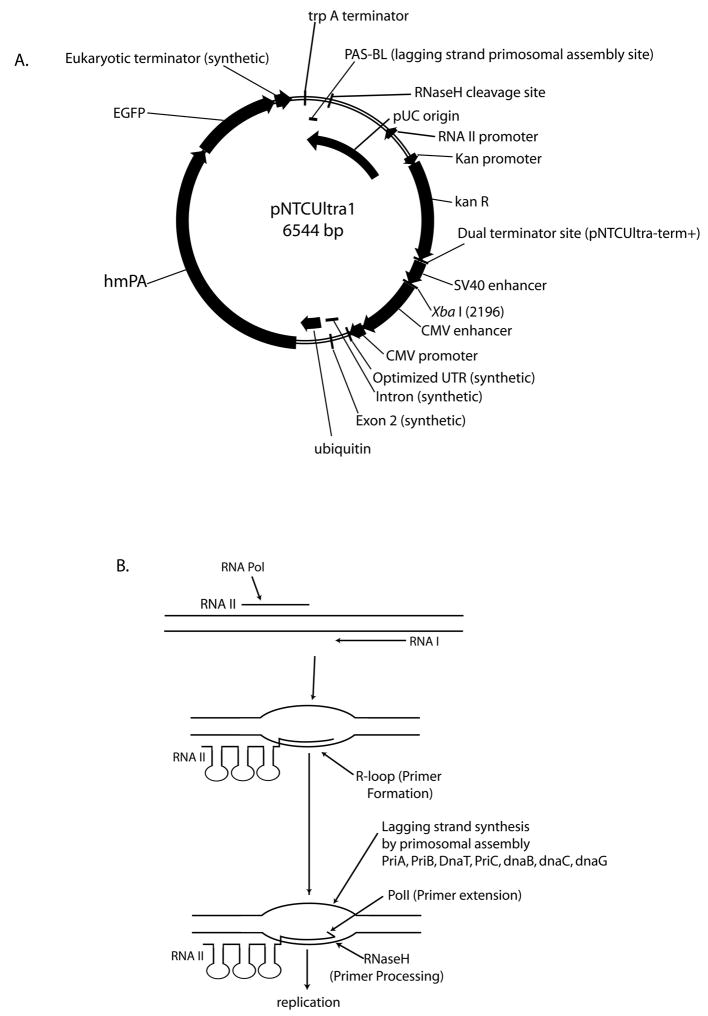
Plasmid gWiz-GFP is a 5757 bp kanR pUC origin DNA vaccine plasmid vector (Gene Therapy Systems, San Diego, CA). gWiz-D is a 6338 bp gWiz-derived plasmid (modifications to the CMV promoter and antigen). pNTCUltra1-3 plasmids are kanR pUC type pDNAVACCultra DNA vaccine plasmids (Williams et al., 2006) with different promoters and antigen genes with or without immunostimulatory RNA elements [eRNA; Williams, JA. World Patent Application WO2008153733 (2008)]: NTCUltra1 = 6544 bp NTC7164-hmPA-EGFP (SV40-CMV promoter, ubiquitin targeting vector, anthrax protective antigen; Figure 3A); NTCUltra2 = 5895 bp pDNAVACCUltra5 SV40 CMVR HA eRNA41J (SV40-CMV-HLTV-1 R-U5 chimeric promoter, immunostimulatory RNA producing, secreted influenza H5 hemagglutinin antigen); NTCUltra3 = 6490 bp NTC7162-hmPA-EGFP (SV40-CMV promoter, TPA secretion targeting vector, anthrax protective antigen). The presence of the pUC high copy origin mutation was sequence verified in all plasmids.
Figure 3.
A. Annotated pNTCUltra-1 restriction map. Critical features such as the trpA terminator, dual terminator site from pNTCUltra1-term+ and functional domains in the origin (PAS-BL, RNAII promoter, RNase H cleavage site) are indicated. B. Plasmid DNA replication steps are shown, with critical gene products indicated. For the leading strand, a RNA polymerase transcript (RNAII) forms an R-loop (RNA DNA hybrid) that is cleaved by RNase H, the resulting 3′ OH serving as a primer for DNA polymerase I (Pol I) DNA synthesis. DNA synthesis then converts to DNA Pol III about 400 bp downstream of the origin. In pBR322, Pol III leading strand synthesis may be initiated by primosomal assembly at either phiX174 type (at PAS-BH, a leading strand primosomal assembly site; Minden and Marians, 1985) or dnaA type (at dnaA binding sites within the origin; Parada and Marians, 1991) assembly sites. PAS-BH is deleted in pUC origin vectors. Replication of the leading strand exposes a phiX174 type primosome assembly site on the lagging strand (PAS-BL), which binds priA, which, in turn, recruits the remaining proteins to form the preprimosome [priB, dnaT, recruits dnaB (delivered by dnaC)], which then also recruits primase (dnaG), which then makes a short RNA substrate for DNA polymerase III. PriC may also be involved in this pathway. Lagging strand replication initiated at PAS-BL terminates at a termination site (ter) within the origin. Leading strand synthesis, with dnaB/dnaC bound to polIII generating lagging strand Okazaki fragments every 1–2 kb, proceeds around the plasmid, also stopping at the ter site. DNA Pol I and DNA ligase are required after Pol III replication to remove primers and seal nicks, respectively. DNA gyrase then negatively supercoils the resultant covalently closed circular (CCC) plasmid. Plasmid replication is different than chromosomal replication, wherein initial replication is set by OriC priming initiated with dnaA (reviewed in Masai and Arai, 1996).
The pNTCUltra1-term+ plasmid contains a 100 bp ApaI-KpnI dual terminator containing the fd gene VIII terminator-tonB bidirectional terminator of the following sequence:
GGGCCCACCGATACAATTAAAGGCTCCTTTTGGAGCCTTTTTTTTTGGACTGCAAACAAATAGTCAAAAGCCTCCGGTCGGAGGCTTTTGACTTTGGTACC.
The underlined dual terminator sequence is not present in pNTCUltra1. pNTCUltra1-PAS-BH contains the pUC19 replication origin extended to include an additional 282 bp of the pBR322 origin (starting at the PvuII site immediately downstream of the ROP gene) that includes the PAS-BH site. pNTCUltra1 (-SV40) has the SV40 enhancer deleted by removal of the 324 bp ApaI-XbaI term-SV40 enhancer region from pNTCUltra1-term+.
Seed Stock Creation
Transformations were performed using either Z competent cells (Zymo Research, Orange CA) or electrocompetent cells using standard methodologies. Briefly, electrocompetent cells (in 10% glycerol) were electroporated with ≤ 1 ng pDNA using the BioRad (Hercules, CA) Micropulser electroporator per manufacturer’s instructions (2.5 kV, 25 μF, 100Ω, 0.1 cm cuvette, 25 μL cells) and cells were transferred to 0.5 mL SOC media in a 15ml Falcon tube. Transformed cells were outgrown for 1 h in a shaker incubator, and then plated onto Luria-Bertani (LB) + kanamycin plates. Z competent cells were made and transformed according to the manufacturer’s instructions (Zymo Research, Orange, CA). SOC media was added to the transformed competent cells after ½ h incubation on ice and the cells were outgrowth for 1 h in a shaker incubator prior to plating. LB media cultures were inoculated with well isolated single colonies and grown to saturation. Glycerol was added to 10% final volume, and seed stocks were created. Incubations (post transformation outgrowth, plating and culturing) were performed at either 30°C or 37°C.
Molecular Biology
Vectors were made by standard restriction digestion-mediated transfer of fragments between vectors. Bacterial or bacteriophage genes were amplified from genomic DNA using the polymerase chain reaction (PCR) with primers that incorporated restriction sites compatible with cloning sites in target vectors. PCR products were digested with restriction enzymes and cloned into compatibly cleaved vectors. Alternatively, when these restriction sites were present in the gene, the PCR primers were modified to incorporate AarI (Fermentas, Vilnius Lithuania) type IIS enzyme sites. This enzyme cleaves after +4 and +8 bp downstream of the AarI recognition sequence to create any designed 4 bp 3′-recessed sticky end. Methods for use of AarI in cloning are described [Williams, JA. WO2006078979 (2006)]. As needed, the primers also incorporated ribosome binding sites to create polycistronic gene fusions.
Chromosome Engineering
Chromosome engineering to delete the relA, spoT, iciA and topA genes was performed using lambda Red recombination as described (Datsenko and Wanner, 2000). Briefly, a chloramphenicol resistance (chlorR) gene was PCR-amplified from the R6K origin conditional replication plasmid pKD3 using gene targeting primers that add 50 bp of homology to each end of the PCR product. The homology regions correspond to the 5′ and 3′ flanking regions of the target gene such that recombination between the PCR product and the genome precisely replaces the target gene with the chlorR selectable marker. The pKD3plasmid requires an engineered pir+ host cell strain such as BW23474 for propagation and cannot propagate in DH5α. The DH5α strain was transformed with the ampicillin resistant (ampR) lambda Red recombination function-containing plasmid pKD46-RecA+ (pKD46 modified to include a constitutively produced recA gene) and Red recombinase protein production was induced with arabinose. Electrocompetent cells were prepared and electroporated with the PCR fragment. Homologous recombinants were selected with chloramphenicol and cured of the pKD46 helper plasmid by shifting to the non-permissive temperature (pKD46 has a temperature sensitive R-plasmid replication origin). Transformation of the spoT, iciA and topA cell lines with the pCP20 FLP recombinase expression plasmid resulted in FRT-mediated excision of the chlorR gene. The pKD3, pCP20 and pKD46 plasmids and pir+ replication host BW23474 were obtained from the E. coli Genetic Stock Center (New Haven, CT).
Heat Inducible Integration Vectors
The pAH144 and pAH63 vectors (obtained from the E. coli Genetic Stock Center, New Haven, CT) are R6K origin conditional replication plasmids that are site specifically integrated into the genome at bacteriophage attachment sites utilizing bacteriophage recombinase-expressing plasmids (Haldimann and Wanner 2001). The pAH144 vector contains a multiple cloning site, a streptomycinR and spectinomycinR resistance marker (specR/strepR) and the HK022 attachment site. pCAH63 is a chlorR modified version of the pAH63 plasmid, and contains the lambda attachment site.
Chromosome engineering to over-express genes was performed using pAH144 or pAH63 integration vectors modified to allow expression of cloned genes from the bacteriophage lambda pL pR promoters. These vectors overproduce target proteins specifically during the 42°C induction phase of the inducible plasmid fermentation process and were constructed as described below.
pAH144-Lambda C1857tsrepressor-PR PL (pAH144-Lambda Repressor)
The pND213 (Love et al., 1996) native stuffer protein expression vector was digested with BamHI/PstI and the 1.35 kb fragment containing the phage lambda C1857ts lambda repressor, PR and PL promoters upstream of a multiple cloning site was purified. pAH144 was digested with BamHI/PstI and the linear vector (2.45 kb) was purified. The two fragments were ligated and transformed into the BW23474 cell line. Recombinants were selected on spec/strep (35 μg/mL each) and confirmed by restriction digestion and sequencing with the RgnB-f and tL3-r primers described in Haldimann and Wanner (2001). The pCAH63-lambda repressor construct (chlorR vector for integration at phage lambda attachment site) was constructed using a similar strategy. DH5α cell lines with phage attachment site integrated pAH144-lambda repressor or pCAH63-lambda repressor derived vectors have been verified to drive heat-inducible protein production during the 42°C plasmid production stage of the inducible fed-batch plasmid fermentation process, without affecting plasmid DNA productivity (Carnes et al., manuscript in preparation).
pAH144-Lambda repressor-zwf, and pAH144-Lambda repressor-zwf-rpiA
To make the pAH144 lambda repressor zwf construct, the E. coli zwf gene was PCR amplified and digested to generate an NcoI/EcoRI fragment and cloned into NcoI/EcoRI compatible pAH144-Lambda Repressor. The ligation was transformed into the BW23474 cell line, and was selected on strep/spec and sequence-verified (pAH144-Lambda Repressor-zwf).
For multiple cistron vectors, a polycistronic linker was included between the genes. For example:
Zwf-rpiA
TAActcgAGGAGATATACATATG
TAA is the zwf stop codon, AGGA is second cistron ribosome binding site (RBS), ATG is the rpiA start codon. ctcgAG is the XhoI site, CATATG is the NdeI site.
The pAH144-zwf-rpiA heat-inducible, dual cistron construct was made by PCR amplification of the E. coli rpiA gene with XhoI-EcoRI compatible ends (including the polycistronic linker shown above), and was cloned downstream of zwf in pAH144-lambda-repressor zwf. DH5α cell lines with a pAH144-lambda repressor gene1-polycistronic linker-gene2 vector integrated at the HK022 attachment site has been verified to express gene 2 protein during the 42°C plasmid production stage of the inducible fed-batch plasmid fermentation process (Carnes et al., manuscript in preparation).
pAH144-Lambda Repressor-priA-priB-dnaT-priC
The E. coli priA gene was PCR amplified from DH5α genomic DNA. It was digested to generate an NcoI/EcoRI fragment and cloned into NcoI/EcoRI compatible pAH144-Lambda repressor to create pAH144-Lambda repressor-priA. The priB gene was PCR amplified from DH5α genomic DNA, and was digested to generated NdeI and EcoRI compatible ends. It was cloned downstream of priA in pAH144-Lambda repressor-priA (NdeI/EcoRI digested) to make pAH144-Lambda repressor-priA-priB. The priB reverse PCR primer was designed to allow cloning a third gene downstream of priB. It contained a Kozak consensus start site, the 5′ cloning KpnI site (GGTACC) upstream of third gene ATG, and an XbaI (TCTAGA) site next to an EcoRI site. The third gene (dnaT) was PCR-amplified, digested with XbaI and KpnI, and cloned into KpnI/XbaI-cleaved pAH144-Lambda repressor-priA-priB, to generate pAH144-lambda repressor-priA-priB-dnaT. The pAH144-lambda Repressor-priA-priB-dnaT-priC vector was made by cloning a PCR product of the priC gene from DH5α genomic DNA (replaced GTG start codon with ATG). The PCR product was digested with SacI/XbaI and cloned into SacI/XbaI digested pAH144-lambda repressor-priA-priB-dnaT plasmid downstream of the dnaT gene.
The zwf-trxA (thioredoxin), gyrA-gyrB (gyrAB), polA (DNA Pol I), priA-rpiA and priA-dnaG heat-inducible vectors were constructed in pAH144-lambda repressor using similar strategies. The ligA (DNA ligase) and rnhA (RNase H), genes were constructed using similar methodologies in the heat-inducible pCAH63-lambda repressor vector.
Integrated Cell Lines
Sequence-verified pAH144-lambda repressor overexpression clones were integrated into the phage HK022 attachment site using the pAH69 heat-inducible phage HK022 integrase helper plasmid. pAH69 is an ampR temperature sensitive R plasmid replicon that, like pKD46, is lost at 42°C. Transformation of the DH5α + pAH69 cell line with the pAH144 derivative plasmid followed by heat shock results in integration of the pAH144 plasmid into the genome at the phage HK022 attachment site and loss of the pAH69 plasmid. Recombinants were selected with spec/strep, and integration was verified using PCR as described (Haldimann and Wanner 2001). The pINT-ts integrase plasmid was used to integrate the pCAH63-lambda repressor plasmids into the phage lambda attachment site, and recombinants were selected with chloramphenicol. The ligA-polA cell line contain single integrated copies of both pCAH63-ligA and pAH144-polA plasmids.
Fermentation
Fermentations were performed using proprietary fed-batch media (NTC3019) in New Brunswick BioFlo 110 bioreactors as described (Carnes et al., 2006). The seed cultures were started from glycerol stocks and streaked onto LB agar plates containing 50 μg/mL kanamycin. The plates were grown at 30°C, cells were resuspended in media and used to provide approximately 0.1% inoculums for the fermentations.
Analytical Methods
Culture samples were taken at key points and at regular intervals during all fermentations. Samples were analyzed immediately for biomass (OD600) and for plasmid yield. Plasmid yield was determined by quantification of plasmid obtained from Qiagen Spin Miniprep Kit preparations, total DNA analysis was performed using detergent lysis, and agarose gel electrophoresis analysis (AGE) was performed on 0.8–1% Tris/acetate/EDTA (TAE) gels as described previously (Carnes et al., 2006). Two stage AGE was performed as follows. In stage one, unstained plasmid DNA was resolved on a TAE agarose gel and poststained with a 1:10,000 dilution of SYBR Green I (Sigma, St. Louis, MO) in TAE. The gel was destained, and further electrophoresed (stage 2). The different plasmid isoforms (e.g. nicked versus supercoiled or linear) migrate differently in the presence or absence of SYBR Green I; resolution on a single gel using both conditions enables exquisite resolution of the different isoforms. As well, open circular plasmid fluorescence after SYBR Green I post-stain is approximately 10 fold stronger than supercoiled plasmid (data not shown) dramatically increasing the sensitivity of nicked plasmid detection. Dry cell weight (DCW) was determined by drying cell pellets from 1 mL of culture in a speed vacuum at high drying rate to constant weight.
EGFP Analysis Post Transformation
Vector backbones with EGFP genes were transformed into DH5α. Cultures containing the plasmids were grown at 37°C until saturated, the cells were pelleted and washed twice with PBS, and clarified cell lysates were made by sonication of the cells in TE buffer, which is compatible with EGFP. EGFP was quantified in cell lysates, compared to control cell lines without the plasmid, using an FLX800 microplate fluorescence reader with samples in black 96 well assay plates. The assay was repeated using lysates from electro-transformed cells that were outgrown by shaking either 1 h at 37°C or 2 h at 30°C, and were stored at 4°C to allow EGFP protein maturation prior to lysate preparation.
Results
Process-Intrinsic Factors
The fermentation process improvements reported herein are modifications of the fed-batch fermentation process described in Carnes et al., 2006 (Figure 1A). This process takes advantage of the temperature sensitivity of high copy number pUC plasmids. Briefly, plasmid-containing E. coli cells are grown at a reduced temperature during the batch phase and part of the fed-batch phase. Typically, the temperature setpoint is 30°C, at which the plasmid is maintained stably at low levels while biomass can accumulate efficiently. During the fed-batch portion of this period, the specific growth rate is controlled at approximately μ = 0.12 h−1 by an exponential feeding strategy. Induction of plasmid accumulation is performed by shifting the temperature to 42°C when the cell density is in the range of 25–60 OD600, and by continued exponential nutrient feeding for up to 15 hours. The temperature shift at restricted growth rate improves plasmid yield and purity such that the plasmid DNA from the process is essentially 100% supercoiled.
Post Plasmid Induction Hold Step
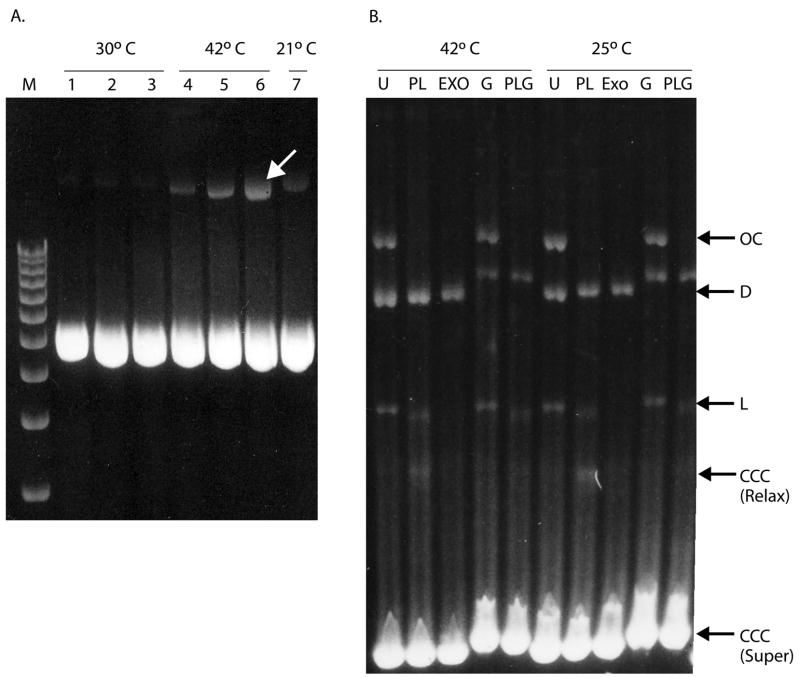
After induction of plasmid production at 42°C, the fermentor is cooled to 15°C prior to harvest. With certain high yield vector backbones, it was unexpectedly found that a 0.5 h to >3 h hold at 25°C following plasmid production, and prior to cooling to 15°C for cell harvest improved plasmid yield (Table I). The observed percentage of nicked and dimer plasmid was also reduced after the hold (Figure 2A). Agarose gel analysis of total cellular DNA (plasmid and genomic) from before and after the hold did not reveal loss of specific plasmid isoforms during the hold (data not shown), but a general reduction in nicked and dimerized plasmid was observed (Figure 2A). These results demonstrated that holding cells prior to harvest at a temperature which is non-permissive for extensive plasmid replication (i.e. 25–30°C) improved yields and quality. This hold may allow plasmid repair since in vitro addition of DNA Pol I and DNA ligase mimics this effect by repairing residual post production nicked plasmid (Figure 2B). DNA ligase addition alone did not produce these effects (data not shown), demonstrating that nicked plasmid was either gapped or otherwise blocked from ligation, perhaps through retention of replication primers that are normally removed by DNA Pol I.
Table I.
Process hold step post 42°C plasmid induction stage increases plasmid yield
| Run ID # |
Host | Plasmid* | Induction T=0 OD600 |
Induction Time |
Induction end T=E OD600 |
Post induction hold |
Specific Yield T=E (mg/L/ OD600) |
Specific Yield T=E+hold (mg/L/ OD600) |
Specific yield Δ (hold) (mg/L/ OD600) |
|---|---|---|---|---|---|---|---|---|---|
| RF41 | DH5α | pNTCUltra1 | 32 | 15 | 80 † | 2 h 25°C | 15.7 | 16.3 | 0.6 |
| RF36 | DH5α | pNTCUltra1 | 40 | 11 | 93 | 1 h 25°C | 13.9 | 16.2 | 2.3 |
| RF48 | DH5α | pNTCUltra1 | 40 | 14 | 98 | 1 h 25°C | 14.7 § | 16.9 § | 2.2 |
| RF73 | DH5α | pNTCUltra1 | 49 | 12.5 | 104 | 1 h 25°C | 16.0 | 20.4 | 4.4 |
| RF96 | DH5α | pNTCUltra2 | 44 | 12 | 102 | 1 h 25°C | 15.5 | 21.2 | 4.7 |
| RF47 | DH5α | gWiz-D | 52 | 8 | 86 | 1 h 25°C | 8.4 | 9.6 | 1.2 |
| RF6A | DH5α | gWiz-D | 39 | 8.5 | 85 | 1 h 10°C | 8.7 | 8.4 | −0.3 |
| LS-01016 | DH5α | gWiz-D | 54 | 7 | 97 | 0.5h 10°C | 8.6 | 8.5 | −0.1 |
| RF84 ‡ | DH5α spoT- | pNTCUltra1 | 38 | 12 | 95 | 1 h 25°C | 12.8 | 16.7 | 3.9 |
| RF86 ‡ | DH5α topA- | pNTCUltra1 | 38 | 12 | 78 | 1 h 25°C | 15.6 | 18.8 | 3.2 |
| RF67 ‡ | DH5α+ zwf | pNTCUltra1 | 31 | 14 | 76 | 1 h 30°C | 14.9 | 16.3 | 1.4 |
| RF33 | DH5α | pNTCUltra3 | 41 | 11.5 | 87 | 3 h 25°C | 14.0 | 17.5 | 3.5 |
All plasmids contain pUC origin.
Partial lysis resulted in loss of ≥ 5 OD600 from previous timepoint.
DH5α derivative with a spoT mutation (relA/spoT eliminates stringent response), topA mutation (removes DNA topoisomerase) or overexpressing zwf gene products, respectively.
See Figure 2B for two stage AGE analysis of these samples
Figure 2.
Agarose gel analysis of plasmid DNA from sample timepoints from inducible fed-batch fermentation process. A. Agarose gel analysis of gWiz-D plasmid DNA from timepoints during 30°C growth restriction phase (Samples 1–3) 42°C plasmid amplification phase (Samples 4–6) and after holding at 21°C (Sample 7). Arrow indicates dimer and nicked plasmid band that is repaired during hold. B. Two stage agarose gel analysis of pNTCUltra1 RF48 fermentation plasmid DNA samples (see Table I), from end of 42°C induction (1432 mg/L, 14.7 mg/L/OD600 specific yield) and after 1 h at 25°C (1560 mg/L, 16.9 mg/L/OD600 specific yield). U = untreated plasmid in 1 × DNA gyrase buffer; PL = plasmid was incubated in 1× DNA gyrase buffer with dNTP, E. coli DNA Pol I (P) and T4 DNA ligase (L), for 1 h at 37°C prior to analysis; Exo = plasmid was incubated with T5 exonuclease (digests linear, nicked and denatured DNA) in 1× DNA gyrase buffer for 1 h at 37°C prior to analysis; G = plasmid was incubated 1× DNA gyrase buffer with E. coli DNA gyrase (G), for 1 h at 37°C prior to analysis; PLG = plasmid was incubated in 1× DNA gyrase buffer with dNTP, E. coli DNA Pol I (P), T4 DNA ligase (L) and E. coli DNA gyrase (G)(Topogen, Port Orange, FL) for 1 h at 37°C prior to analysis. OC = open circular nicked plasmid, D = supercoiled dimer plasmid, L = linear plasmid, CCC = covalently closed circular monomer plasmid, relaxed = not supercoiled, super = supercoiled. Note that addition of DNA gyrase (G, PLG lanes) increases negative supercoiling of CCC plasmids, slowing mobility of the monomer and dimer.
Reduced Cell Stress Process for Toxic Plasmid Production
Creation of seed stocks from transformations performed at 30°C, rather than at 37°C, unexpectedly increased plasmid yield, productivity and seed stock viability with low yield plasmids. Three kanamycin resistant DNA Vaccine plasmids containing proprietary influenza hemagglutinin (HA) genes from serotypes that obtain low (H1, H3) or high (H5) plasmid yields were evaluated. Seed banks of the influenza DNA vaccine plasmids were manufactured at either 30°C or at 37°C (see Materials and Methods). Shake flask and fermentation plasmid yields are summarized in Table II. The minipreps at 30°C and 37°C were grown from the seed stock immediately after manufacture. Glycerol stock viability was determined after >1 week at −80°C. The high yielding H5 construct was insensitive to temperature, while plasmids with the H1 and H3 inserts had much higher yields and seed stock viability after creation of cryo-preserved seed stocks at 30°C, rather than at 37°C. No yield differences were observed between seed stocks made with Z competent or electrocompetent cells (data not shown).
Table II.
Improved performance of reduced temperature manufactured seed stocks
| Plasmid antigen* | Seed stock preparation | Specific yield in 30°C miniprep (mg/L/OD600) | Specific yield in 37°C miniprep (mg/L/OD600) | Glycerol stock viable | Fermentation yield |
|---|---|---|---|---|---|
| H1 influenza | 30°C | 1.4 | 1.3 | Yes | 252 mg/L |
| 37°C | 2.0 | 2.5 | No | Inviable cell line | |
| H3 influenza | 30°C | 1.3 | 3.7 | Yes | 570 mg/L † |
| 37°C | 1.6 | 0.7 | No | Inviable cell line | |
| H5 influenza | 30°C | 1.5 | 9.8 | Yes | 1,290 mg/L |
| 37°C | 1.7 | 10.4 | Yes | 1,260 mg/L |
H1 and H5 are in pNTCUltra backbone. H3 is in gWiz-D backbone.
Fermentation yield from 30°C glycerol stock with H3 antigen in pNTCUltra backbone was 1005 mg/L
As indicated above, the DNA vaccine plasmids tested in Table II are eukaryotic expression vectors, and are not designed to express encoded genes in E. coli. To confirm this, versions of the vector backbones with EGFP rather than influenza HA were created, and transformed into DH5α cells. No detectable EGFP was present in cell lysates, compared to control cell lines without the plasmid (see Materials and Methods).
Plasmid- Intrinsic Factors
Sequences and vector configurations that affect plasmid quality (% supercoiled, replication intermediates) have been described (reviewed in Carnes and Williams, 2007) and are not discussed herein. However, in the process of developing high yielding vector backbones, sequences that dramatically affect yield have been observed, and are described below.
Positioning of Prokaryotic Terminator Sequences
Certain vector backbone modifications are dramatically lower-producing than the parent backbone. For example, a pNTCUltra1 plasmid, with a dual terminator sequence inserted upstream of the SV40 enhancer (Figure 3A; see Materials and Methods), was inexplicably a poor producer (data not shown). The slow growth, poor induction and low overall yield of 119 mg/L at harvest (2.5 mg/L/OD600) was in contrast to the control pNTCUltra1 plasmid without the terminator region (Figure 1B; Table I: 1500–2100 mg/L, 16–20 mg/L/OD600). This is not a general feature of transcriptional terminators: deletion of the trpA terminator, present in pNTCUltra1 downstream of the replication origin (Figure 3A), had no effect on plasmid yield (data not shown).
Chimeric SV40-CMV promoter
Dramatic differences in yield were obtained using currently defined ‘optimal’ plasmid backbones. Fermentation yields exceed 2000 mg/L with pNTCUltra vectors (Figure 1B). In this 2100 mg/L fermentation, plasmid DNA represented 75% of the total DNA at harvest. Surprisingly, the pNTCUltra vectors yielded approximately twice the production yield of gWiz-derived plasmids (Genlantis, San Diego CA) (Table I; gWIZ RF47, RF6A, and LS-01016 versus pNTCUltra1 RF41, RF36, RF48 and RF73 and pNTCUltra2 RF96) or a pVAX1-derived plasmid (Invitrogen Corp., Carlsbad, CA) (data not shown). The pNTCUltra improvement compared to gWiz was not insert-specific, and yields in these ranges were obtained with a variety of insert sequences within these vector backbones (data not shown). These vectors were all eukaryotic expression vectors containing the same elements in different orientations: pUC origin, a kanR selectable marker, and a eukaryotic expression cassette comprising the CMV promoter-gene of interest-eukaryotic terminator. Deletion analysis demonstrated that the SV40 enhancer in pNTCUltra was critical for improved productivity (Table III; RF157 versus RF158).
Table III.
Plasmids containing the SV40 enhancer and PAS-BH have superior final yields
| Run ID # | Vector/host* | Modification | Fermentation yield (mg/L) | Fermentation specific yield (mg/L/OD600) |
|---|---|---|---|---|
| RF157 | pNTCUltra1 | SV40-CMV | 1930 | 17.3 |
| RF158 | pNTCUltra1 (-SV40) | CMV | 1050 | 9.9 |
| RF160 RF167 ‡ |
pNTCUltra1-PAS-BH | SV40-CMV +PAS-BH origin | 2210 2220 ‡ |
22.9 25.3 ‡ |
| RF166 | pNTCUltra1-PAS-BH DH5α + priA-rpiA |
SV40-CMV +PAS-BH origin | 1820 | 17.7 |
| RF154 | pNTCUltra4 † | PAS-BH-CMV | 1230 | 12.3 |
| RF161 | pNTCUltra4 (UP deletion) † | CMV | 800 | 10.2 |
| RF164 | gWIZ-D | PAS-BH-CMV | 1210 | 14.4 |
| RF162 | gWIZ-D DH5α + priA-rpiA |
PAS-BH-CMV | 1140 | 12.6 |
All fermentations were in DH5α, except RF162 and RF166 (DH5α + priA-rpiA) and were induced at 50 OD600 for 10.5 h at 42°C, 1 h 25°C hold
pNTCUltra4 = 5895 bp NTC7382 HA eRNA41H UP (CMV-HLTV-1 R-U5 chimeric promoter, immunostimulatory RNA producing, secreted influenza H5 hemagglutinin antigen; UP = PAS-BH site from pUC19 replaces SV40 upstream of CMV promoter)
12 h induction, 88 OD600. Projected specific yield of 51 mg plasmid/g DCW (estimate 50 g/L DCW with 100 OD600 culture)
PAS-BH sites
Therapeutic plasmids such as VR1012 (Vical) and derivative gWIZ (Gene Therapy Systems), pPJV7563 (PowderMed/Pfizer), V1JNS (Merck), pVAX1 (Invitrogen), pLL14 (Merial) pNTCUltra1 (Nature Technology Corporation) were all constructed using origin regions derived from high copy pUC plasmids. These vectors therefore deleted the leading strand primosomal assembly site (PAS-BH) present in pBR322 (Marians et al., 1982). The function of this site is unknown, since its deletion does not dramatically affect plasmid copy number in vivo (Masai and Arai, 1989). Addition of PAS-BH to pNTCUltra1 through extension of the pUC origin to include the deleted 300 bp region of pBR322 including PAS-BH further improved productivity (Table III; RF160 versus RF157) up to 2220 mg/L and 25.3 mg/L/OD600 (51 mg/gDCW) with a 12 hr induction (Table III; RF167). Improved yields may be due to enhanced primosomal assembly on the leading strand when the assembly site is within the PolI replication region. Interestingly, yield improvement was also observed in plasmids containing the PAS-BH site 3 kb downstream of the origin (Table III RF154, pNTCUltra4 PAS-BH-CMV versus RF161, CMV). This long-range effect is vector specific since deletion of PAS-BH from the gWiz vector (a pUC vector fragment containing PAS-BH is naturally present immediately upstream of the CMV enhancer in VR1012 and gWIZ) had no effect on plasmid productivity (data not shown).
Strain-Intrinsic Factors
Strain Matrix Evaluation
The inducible plasmid production process of Carnes et al., 2006 was performed in various relA-, relaxed response cell lines, including DH1, DH5α, JM109, and TG1 (relA). The relA mutation is necessary because the fed-batch growth rate was below the threshold necessary to invoke the stringent response. relA+ strains can be converted to relA− by deletion of the relA+ gene as was performed to create TG1 relA (see Materials and Methods). Other mutations that eliminate the stringent response, such as spoT, could also be used in place of relA. Indeed, similar plasmid yields were obtained with DH5α (relA) and the spoT/relA double mutant DH5α-spoT which completely eliminates the stringent response (Table I; RF84 versus RF36).
Fermentation yields in DH5α and DH1 (Carnes et al., 2006) were superior to JM109 or TG1 (relA) (data not shown). By contrast, in a shake culture evaluation, TG1 was found to have very high copy numbers of gWiz and pSVβ consistent with runaway replication of these plasmids (Yau et al., 2008). Recently, plasmid production yields of 1904 mg/L (189 OD600, 10.1 mg/L/OD600) have been reported from strain BL21 recA- endA-, using an ampR derivative of plasmid VRC5737 in a 30°C to 42°C inducible fed-batch fermentation process. Yields of 991 R mg/L, 141 OD600, 7.0 mg/L/OD600 were obtained in DH5α (Phue et al., 2008). The amp VRC5737 derivative has a backbone similar to the gWiz-D plasmid reported here, which obtained higher specific and volumetric yields of 8.4, 8.7, 8.6 mg/L/OD600 inpre-hold DH5α (Table I; RF47, RF6A, LS-01016), and 14.4 mg/L/OD600 (1210 mg/L, 84 OD600) in post-hold DH5α (Table III; RF164).
Replication Engineered DH5α Cell Lines
After temperature induction, the rate of accumulation of plasmid follows a linear amplification curve (Figure 1B), prior to plateau (gWiz-D backbones). The amplification slope is the same for pNTCUltra1 and gWiz-D backbones, and it appears to be runaway replication, as the plasmid replicates to copy number equilibrium (plateau). The plateau is probably determined by: 1) the efficiency of the replication origin (e.g. gWiz-D compared to pNTCUltra1); and 2) the percent of initiated replication cycles that are completed.
The amplification slope may be set by a limiting factor, perhaps a protein or nucleotide necessary for plasmid DNA replication (Figure 3B). To test the effects of deletion or overexpression of key replication genes, DH5α strains with gene knockouts or heat inducible gene over-expression were constructed using chromosome engineering (see Materials and Methods). The integrated cell lines (overexpression, or knockout, of genes that may improve plasmid productivity) were transformed with a kanamycin resistant pNTCUltra plasmid and tested for improved productivity in shake flask and fermentation culture. The results are summarized in Tables IV and V.
Table IV.
Engineered cell line shake culture yields*
| Name(+ = heat induced gene) | Description | Final OD600 Uninduced (30°C) | Final OD600 Induced (30–42°C) | Specific Yield Uninduced (mg/L/OD600) | Specific Yield induced (mg/L/OD600) |
|---|---|---|---|---|---|
| DH5α | CONTROL | 2.09 | 1.075 | 0.6 | 2.88 |
| DH5α + gyrAB | ↑ negative supercoiling | 1.62 | 1.3 | 0.771 | 1.85 |
| DH5α + ligA-polA | ↑ nick repair | 1.89 | 1.195 | 0.66 | 2.09 |
| DH5α + polA | ↑ primer extension | 2.025 | 1.755 | 0.99 | 4.36 |
| DH5α + ligA | ↑ nick repair | 1.935 | 1.675 | 0.904 | 4.03 |
| DH5α + ligA polA P4α | P4α is phage dnaG dnaB fusion | 1.65 | 0.785 | 0.67 | 3.31 |
| DH5α + priA | ↑ Primosomal assembly | 1.76 | 1.07 | 0.97 | 3.97 |
| DH5α + priA-dnaG | ↑ Primosomal assembly | 1.855 | 0.995 | 0.97 | 4.21 |
| DH5α + priA-priB | ↑ Primosomal assembly | 1.96 | 1.345 | 0.893 | 3.79 |
| DH5α + priA-priB- dnaT | ↑ Primosomal assembly | 1.945 | 1.545 | 1.03 | 3.62 |
| DH5α + priA-priB- dnaT-priC | ↑ Primosomal assembly | 1.71 | 1.705 | 1.02 | 4.34 |
Yields are from 5 mL cultures grown in 15 mL Falcon tubes and are either uninduced (grown to saturation in LB + kanamycin at 30°C) or induced (grown to midlog in LB + kanamycin at 30°C, and plasmid production induced by shifting to 42°C and continued growth until saturated)
Table V.
Engineered cell line inducible fermentation yields
| Run ID # | Name (+ = heat induced, − = gene knockout) | Description | Induction (T=0) OD600 | T=9 OD600 | T=9 Volume tric yield | Specific Yield T=0 (mg/L/OD600) | Specific Yield T=9 (mg/L/OD600) | Total Specific yield increase |
|---|---|---|---|---|---|---|---|---|
| RF41 | DH5α | Control | 32 | 1.64 | 9.53 | 7.89 | ||
| RF48 | 40 | 1.34 | 10.5 | 9.16 | ||||
| RF73 | 49 | 99 | 1075 | 1.76 | 10.84 | 9.08 | ||
| RF62 | DH5α +gyrAB | ↑ negative supercoiling | 36 | 71 | 295 | 1.80 | 4.15 | 2.35 |
| RF86 | DH5α–topA | ↓ positive supercoiling | 38 | 79 | 872 | 2.24 | 11.09 | 8.85 |
| RF61 | DH5α +polA | ↑ primer extension | 30 | 69 | 565 | 1.61 | 8.16 | 6.55 |
| RF59 | DH5α +ligA polA | ↑ nick repair | 38 | 71 | 390 | 0.84 | 5.53 | 4.69 |
| RF60 | DH5α +ligA polA P4α | P4α is phage dnaG dnaB fusion | 40 | 57 | 92 | 0.76 | 1.62 | 1.54 |
| RF78 | DH5α +rnhA | ↑ RNaseH primer cleavage | 31 | 68 | 570 | 1.16 | 8.38 | 7.22 |
| RF77 | DH5α +priA-priB- dnaT-priC | ↑ Primosomal assembly | 32 | 65 | 805 | 1.01 | 12.35 | 11.34 |
| RF67 | DH5α +zwf | ↑ nucleotide precursors | 31 | 62 | 572 | 1.41 | 9.29 | 7.88 |
| RF68 | DH5α +zwf-trxA | ↑ nucleotide pool and redox | 40 | 76 | 676 | 1.33 | 8.92 | 7.59 |
| RF74 | DH5α +zwf-rpiA | ↑ nucleotide precursors | 49 | 96 | 1135 | 1.45 | 11.88 | 10.43 |
| RF84 | DH5α–spoT | Eliminate stringent response | 38 | 81 | 833 | 1.68 | 10.25 | 8.57 |
| RF81 | DH5α–iciA | Eliminate chromatin binding protein | 27 | 78 | 700 | 1.03 | 9.02 | 7.99 |
Replicative Enzyme Overexpression Does Not Improve Fermentation Plasmid Production
One of the first steps in plasmid replication is formation of the RNAII R-loop. In shake culture, increased R-loop formation using rho or nusG mutations increases copy number (Harinarayanan and Gowrishankar, 2003). DNA topoisomerase I (topA) mutations also increase R-loop formation (Masse and Drolet, 1999). No effect of topA knockout on fermentation productivity was observed (Table V, RF86 compared to RF48) suggesting R-loop formation is not a limiting factor in plasmid replication.
RNase H (rnhA) is required to cleave the RNAII R-loop into a primer template for DNA Pol I (polA). RNase H appears not limiting since overexpression of rnhA was mildly detrimental to fermentation productivity (Table V, RF78 compared to RF41).
Overexpression of components that extend the RNA primer template (DNA Pol I; polA), remove RNA primers post replication (polA), seal nicks (DNA ligase; ligA) or negatively supercoil the resultant covalently closed circular plasmid (DNA gyrase; gyrAB) may increase yield and quality if limiting. In shake culture overexpression of DNA polymerase I (polA) and DNA ligase (ligA) improved yields, while overproduction of DNA gyrase (gyrAB) reduced yield (Table IV). In the fermentor, a similar yield decrease with gyrAB overexpression was observed (Table V, RF62 compared to RF41 and RF48). In contrast to shake cultures, overexpression of polA or ligA/polA in the fermentor was detrimental. Overexpression of polA reduced fermentation productivity (Table V, RF61 compared to RF41) while overexpression of ligA and polA further reduced fermentation productivity (Table V, RF59 compared to RF48). Deletion of a key chromatin binding protein (iciA) did not alter fermentation yields (Table V, RF81 compared to RF41 and RF48).
Increasing Primosomal Assembly by priA Overexpression Improves Plasmid Amplification Rate
The requirement for primosome assembly on the plasmid (one lagging strand primosome per plasmid per replication) is very high compared to the requirement for the chromosome. PriA primosomal assembly sites are utilized only for replication restart (damage) and are non essential for chromosomal replication except in restart conditions (reviewed in Sandler, 2005). The basic primosome assembly components (dnaB, dnaC) are probably not limiting, since they are used to create Okasaki fragments every 1–2 kb on the genomic DNA. Potentially limiting components include priA, priB, dnaT-priC. Decreased priB has been shown to reduce plasmid copy number in shake culture (Berges et al., 1997). Overexpression of DNA primase (dnaG) or P4α, a phage dnaG-dnaB fusion protein improved ColE1-type plasmid yield in shake culture (Diaz-Orejas et al., 1997). Overexpression of P4α slowed growth in both shake flasks and fermentation but strongly reduced plasmid yield only in the fermentor (Table V, ligA polA P4α RF60, compared to ligA polA RF59).
However, overexpression of primosomal assembly proteins PriA priB, dnaT-priC improved shake flask and fermentation culture plasmid amplification rate during plasmid induction. In shake culture overexpression of priA and other primosomal assembly genes improved yields (Table IV). Overexpression of priA, which recruits other primosomal assembly proteins (priA, priB, dnaT, priC) to PAS-BL was probably the critical primosomal component, since the priA overexpression cell line had equivalently improved yield as priA-dnaG, priA-priB, priA-priB-dnaT, and priA-priB-dnaT-priC.
In the fermentor, similar plasmid amplification rate improvement with priA primosomal assembly gene over-expression (priA-priB-dnaT-priC) was observed (Table V RF77, compared to RF41).
Increasing Nucleotide Precursor Pools by rpiA Overexpression Improves Plasmid Amplification Rate
Nucleotide pool levels are highly controlled and deoxyribonucleotides are made on demand during aerobic growth by allosterically regulated ribonucleotide reductase (NrdA/B). NrdA/B is associated with the replication apparatus, and is stoichiometric with replication (reviewed in Guzman et al., 2003). Cofactors necessary for ribonucleotide reductase activity may be limiting under certain conditions. Thioredoxin (trxA) and glutaredoxin (grx1) (dithiol forms) are hydrogen donors for ribonucleotide reductase (reviewed in Miranda-Vizuete et al., 1996). Overexpression of thioredoxin has been reported to dramatically increase ColE1 plasmid copy number (to 1700/cell) in shake flask culture stationary phase cells (Lunn et al., 1984). As well, key enzymes necessary for creation of nucleotide precursor pools may be limiting. For example, overexpression of glucose-6-phosphate dehydrogenase (G6PDH, encoded by zwf) decreased plasmid-mediated metabolic burden (Flores et al., 2004). Overexpression of ribose-5-phosphate isomerase A (RpiA) has been demonstrated to improve plasmid copy number (Wang et al., 2006).
Overexpression of zwf and zwf-trxA had no effect on fermentation productivity (Table V RF67 and RF68, compared to RF41 and RF48; Table VI, RF67 versus RF41). In contrast, overexpression of zwf-rpiA improved fermentation plasmid amplification rate during plasmid induction (Tables V RF74, compared to RF73).
Table VI.
Engineered cell line productivity*
| Run ID # | Name (+ = heat induced, − = gene knockout) | Induction (T=0) OD600 | Specific Yield T=0 (mg/L/OD600) | Specific Yield T=6 (mg/L/OD600) | Specific Yield T=9 (mg/L/OD600) | Specific Yield T=12 (mg/L/OD600) | Total Specific yield increase(0–9 h) | Total Specific yield increase(6–12 h) |
|---|---|---|---|---|---|---|---|---|
| RF41 | DH5α | 32 | 1.64 | 7.4 | 9.53 | 12.3 | 7.89 | 4.9 |
| RF77 | DH5α + priA-priB-dnaT-priC | 32 | 1.01 | 8.21 | 12.35 | 15.82 | 11.34 | 7.61 |
| RF67 | DH5α + zwf | 31 | 1.41 | 7.35 | 9.29 | 12.41 | 7.88 | 5.06 |
| RF48 | DH5α | 40 | 1.34 | 8.04 | 10.5 | 12.55 | 9.16 | 4.51 |
| RF84 | DH5α spoT | 38 | 1.68 | ND | 10.25 | 12.76 | 8.57 | ND † |
| RF73 | DH5α | 49 | 1.76 | 7.74 | 10.84 | 16.03 | 9.08 | 8.29 |
| RF74 | DH5α + zwf-rpiA | 49 | 1.45 | 8.87 | 11.88 | 12.02(lysis) | 10.43 | 3.15 |
| RF97 | DH5α + 10% xylose feed | 49 | 1.19 | 8.52 | 12.99 | 12.91(lysis) | 11.8 | 4.39 |
It is critical to compare to control fermentations induced at a similar optical density (separated by gray spacer). In general, inductions at 50–55 OD600 result in higher specific yields
ND = Not determined
priA and rpiA Overexpression Does Not Increase Fermentation Copy Number (Plateau)
Further evaluation of primosomal assembly gene over-expression (priA-priB-dnaT-priC) and nucleotide precursor genes zwf-rpiA revealed plasmid amplification rate improvement occurred during the first 9 hours of induction (Table VI RF77, compared to RF41; RF74, compared to RF73) and did not increase the final copy number (plateau) compared to DH5α. This was in contrast to shake cultures, where the final copy number was increased (Table IV).
A strain overexpressing priA-rpiA during the plasmid amplification stage was evaluated for improvement in final plateau. The harvest yield was reduced in this strain with both the gWIZ-D and pNTCUltra1-PAS-BH vector backbones (Table III; RF166 versus RF160; RF162 versus RF164). This demonstrated that overexpression of two factors that increased the amplification slope (plasmid amplification rate) did not increase the final copy number (plateau) in the fermentor.
Discussion
The inducible 30°C to 42°C fed-batch fermentation process described herein results in a lower metabolic burden than alternative high yield plasmid production processes that constitutively maintain elevated plasmid copy number by growth at 37°C [Listner et al.., 2006; Huber, H., Weigl, G., Buchinger, W.: WO2005097990 (2005)]. Critically, the specific yields of plasmid using the inducible process were very high (Tables I, III); High specific yields are desirable, since increased plasmid yield per gram of bacteria leads directly to higher final product purities (Carnes et al., 2006). As well, the resultant cells are compatible with a variety of downstream lysis processes, including alkaline lysis (Carnes et al.., 2006), heat lysis, and autolysis (Carnes et al.., manuscript in preparation).
The reduced metabolic burden inducible fermentation process may also be utilized to produce high quality plasmid DNA with traditionally unstable vectors containing direct repeats, long terminal repeats or palindromic hairpin inverted repeats, eliminating the need for specialized stabilizing cell lines for production of traditionally unstable vectors (Williams et al., manuscript in preparation).
For application to rapid deployment vaccine production, a fermentation process must consistently produce acceptable amounts of plasmid DNA with potentially toxic inserts designed de novo from sequences of novel emerging pathogen serotypes. We have determined that plasmid production processes which utilize 37°C growth conditions during seed stock generation do not perform well with toxic plasmids perhaps due to increased copy number/metabolic burden/instability during early growth. Application of reduced temperature seed stock manufacture, coupled with the inducible fermentation process, solved the problem of obtaining manufacturing cell lines for toxic plasmids, and resulted in successful production of all three influenza DNA vaccine plasmids compared to only one of three using standard transformation at 37°C (Table II). Collectively, these data demonstrated that the reduced metabolic burden inducible fermentation process, coupled with reduced metabolic burden seed stock creation, has application for production of rapid deployment vaccines.
The improvement in yield from seed stocks manufactured at 30°C compared to 37°C was not due to a transient or constitutive burst of vector-driven antigen expression after transformation or growth at 37°C. However, cryptic promoters in HA sequences could drive transient or constitutive burst expression of toxic peptides (from the sense or antisense strand) in E. coli. Likewise, RNA sequences or structures could have contributed. As well, growth at 30°C lowers metabolic burden by reducing copy number and preventing cells from entering late stationary phase, perhaps eliminating stationary phase SOS induction (Lee et al., 2002) that is detrimental with toxic plasmids. Regardless, the steps of reducing metabolic burden gave rise to improved yield, quality and stability. Application of low temperature seed stock growth to seed stock creation for defined media fermentation may eliminate the costly and time consuming step of extensive prescreening of colonies to identify rare high producing cell lines (Prather et al., 2006).
We also report that plasmid yield and quality is improved by holding fermentation cells at reduced temperature post the 42°C induction phase. One hypothesis is that improved yield results from conversion of replication intermediates into covalently closed circular (CCC) plasmid, and improved quality from DNA Pol I, DNA ligase and DNA gyrase enzyme-mediated creation of supercoiled covalently closed circular (CCC) plasmid, by: 1) removal of replication primers (DNA Pol I); 2) ligation of the resultant nicked plasmid (DNA ligase); and 3) plasmid supercoiling (DNA gyrase). Interestingly, overproduction of DNA Pol I and DNA ligase during the 42°C induction phase actually reduced yield and final plasmid % supercoiling (unpublished observations). This indicated that while these gene products may become limiting during runaway plasmid replication at 42°C, they are not limiting factors during the process hold step.
A vector backbone, pNTCUltra1 (Figure 3A), was identified that is twice the productivity of previously characterized high-yielding vectors such as pVAX1 and gWiz. Productivity enhancement was determined to be due to an SV40 enhancer present in the pNTCUltra1 vector. Productivity was further improved by extension of the pUC origin to include a leading-strand primosomal assembly site that was deleted during creation of the pUC vector family from pBR322.
These new modifications result in dramatically higher fermentation yield increases than previously defined modifications, such as optimizing the orientation of the kanR gene relative to the replication origin (Williams et al., 2006) or reduction of kanR gene expression [pLL14; Audonnet, J.: WO2008136790 (2008)].
While the basis for yield improvement is unknown, optimal vectors are expected to have a higher plateau (copy number) due to reduction of replication inhibiting factors and incorporation of replication promoting factors. Completion of replication can be confounded by many factors, including: sequences that cause inhibition of DNA Pol III replication [such as protein-DNA complexes assembled on replication termini (Ter) or repressor binding sequences (McGlynn and Guy, 2008)]; stable RNA-DNA hybrids; unusual DNA structures [e.g. internal-ribosome entry sites; Levy, J. US20046709844 (2004)]; or head on transcription-replication collisions (reviewed in Mirkin and Mirkin, 2007). As well, yield may be improved by replication promoting factors that increase RNA primer synthesis rate and/or replication primer utilization. For example, optimization of the orientation of the selectable marker relative to the replication origin (Williams et al., 2006) or reducing the strength of the selectable marker promoter [Audonnet, J.: WO2008136790 (2008)] may reduce marker-mediated transcriptional interference of replication primer synthesis and/or reduce head on transcription-replication collisions. Inclusion of a primosomal assembly site such as PAS-BH may be beneficial at a later step, through increasing the percent of replication primers that successfully initiate DNA Pol III-mediated replication cycles.
DH5α fermentation cells were found to be distinct from shake flask cells in their replicative capacity such that shake flask yields were not predictive of fermentation productivity. Perhaps surprisingly, fermentation plasmid copy number (specific yield at plateau) within DH5α appears largely set by process and vector-intrinsic factors. While the rate of plasmid replication during runaway replication (42°C stage) can be influenced by increasing levels of key replication proteins, the final copy number plateau is insensitive to such alterations. The observed limitations of certain factors during runaway replication can be compensated by process-modification (i.e. post production reduced temperature hold step). The impact of this observation is that in a well designed fermentation process, replication protein levels within a cell are sufficient to fully replicate a vector to its inherent copy number.
These conclusions are supported by the results from fermentation evaluation of overexpression or knockout of a number of gene products involved in plasmid replication (Tables V and VI). The effect of altering replication gene product levels in various plasmid replication steps (Figure 3B) was generally neutral or negative. Collectively, only overexpression of primosomal assembly gene priA and nucleotide precursor genes zwf-rpiA improved plasmid productivity in fermentation culture. The improved plasmid amplification rate of zwf-rpiA was mimicked by inclusion of xylose in the feed (Table VI, RF97 compared to RF74 and RF73). Both of these mechanisms would be expected to increase levels of Ribulose 5 phosphate (Ru5P), a nucleotide precursor. Glycerol is metabolized into G3P (Glyceraldehyde 3-phosphate). Glucose-6-phosphate dehydrogenase (G6PDH, encoded by zwf) and ribose-5-phosphate isomerase A (RpiA) will increase metabolic flux of G3P towards Ru5P. Zylose is directly metabolized into R5P which is converted to Ru5P in a single step by RpiA. (reviewed in Wang et al., 2006). Of these options, strain engineering is the more cost effective mechanism to increase nucleotide precursors.
Overexpression of two factors (priA and rpiA) that increased the amplification slope (plasmid amplification rate) did not increase the final copy number (plateau) in the fermentor (Table III; RF166 versus RF160; RF162 versus RF164). This is in contrast to the dramatic improvements obtained by optimization of vector-intrinsic (i.e. pNTCUltra versus gWiz-D) or process-intrinsic (i.e. post production 25°C hold) factors. Copy number (plateau) thus appears to be set by vector and process-intrinsic factors, independent from primosomal assembly or nucleotide pool flux, or perhaps even strain-dependent variations in various replication protein levels.
DNA polymerase III, the main replicative polymerase in E. coli, is a complex assembly of 10 different protein subunits and multiple accessory proteins (reviewed in Schaeffer et al., 2005). Improving DNA polymerase III capacity would be difficult to model, and this activity may be strain-specific. While we did not attempt herein to engineer strains with increased DNA polymerase III activity, we did employ process controls to reduce growth rate and slow chromosome replication, allowing a higher percent of the replicative capacity to be diverted to plasmid replication. This effect may be enhanced by prolonged growth restriction, since induction at a higher OD600 of 50–55 resulted in higher specific yields, especially in the later stage of induction (6–12 hrs) (Table VI, compare RF41, RF48, and RF73, induced at 32, 40, and 49 OD600, respectively).
DH5α maintains plasmid quality while tolerating plasmid yields up to 5% total DCW using the low metabolic burden, inducible fermentation process. We define high plasmid capacity and quality as the critical attributes to evaluate in alternative plasmid production hosts, such as BL21 recA- endA- (Phue et al., 2008), that produce higher biomass in fermentation culture than DH5α.
Acknowledgments
This paper described work supported by NIH grant R44GM072141.
Footnotes
Conflict of Interest Statement
JAW, JL, SA, AEC, and CPH have an equity interest in Nature Technology Corporation
References
- Berges H, Oreglia J, Joseph-Lianuzun E, Fayet O. Isolation and characterization of a priB mutant of Escherichia coli influencing plasmid copy number of Δrop ColE1-type plasmids . J Bacteriol. 1997;179:956–958. doi: 10.1128/jb.179.3.956-958.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes AE. Fermentation design for the manufacture of therapeutic plasmid DNA. BioProcess Intl. 2005;3:36–44. [Google Scholar]
- Carnes AE, Hodgson CP, Williams JA. Inducible Escherichia coli fermentation for increased plasmid DNA production. Biotechnol Appl Biochem. 2006;45:155–66. doi: 10.1042/BA20050223. [DOI] [PubMed] [Google Scholar]
- Carnes AE, Williams JA. Plasmid DNA manufacturing Technology. Recent Patents on Biotechnology. 2007;1:151–166. doi: 10.2174/187220807780809436. [DOI] [PubMed] [Google Scholar]
- Cooke JR, McKie EA, Ward JM, Keshavarz-Moore E. Impact of intrinsic DNA structure on processing of plasmids for gene therapy and DNA vaccines. J Biotechnol. 2004;114:239–254. doi: 10.1016/j.jbiotec.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Orejas R, Lurz R, Ruckert B. Enhancement of ColE1-type replication promoted by the GPα initiator protein of bacteriophage P4. FEMS Microbiology Letter. 1997;155:11–15. doi: 10.1111/j.1574-6968.1997.tb12679.x. [DOI] [PubMed] [Google Scholar]
- Flores S, Anda-Herrera R, Gosset G, Bolivar FG. Growth-rate recovery of Escherichia coli cultures carrying a multicopy plasmid, by engineering of the pentose-phosphate pathway Biotechnol. Bioeng. 2004;87:485–494. doi: 10.1002/bit.20137. [DOI] [PubMed] [Google Scholar]
- Glick BR. Metabolic load and heterologous gene expression. Biotechnol Adv. 1995;13:247–61. doi: 10.1016/0734-9750(95)00004-a. [DOI] [PubMed] [Google Scholar]
- Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nature Reviews Genetics. 2008;9:776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman EC, Guarino E, Riola J, Caballero JL, Jimenez-Sanchez A. Ribonucleotide diphosphate reductase is a functional and structural component of the replication hyperstructure in Escherichia coli Recent Res. Devel Mol Biol. 2003;1:29–43. doi: 10.1046/j.1365-2958.2002.02761.x. [DOI] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harinarayanan R, Gowrishankar J. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J Mol Biol. 2003;332:31–46. doi: 10.1016/s0022-2836(03)00753-8. [DOI] [PubMed] [Google Scholar]
- Hoare M, Levy S, Bracewell DG, Doig SD, Kong S, Titchener-Hooker N, Ward JM, Dunnill P. Bioprocess engineering issues that would be faced in producing a DNA vaccine at up to 100 m3 fermentation scale for an influenza pandemic. Biotechnol Prog. 2005;21:1577–1592. doi: 10.1021/bp050190n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahijani R, Hulley G, Soriano G, Horn NA, Marquet M. High-yield production of pBR322-derived plasmids intended for human gene therapy by employing a temperature-controllable point mutation. Hum Gene Ther. 1996;7:1971–1980. doi: 10.1089/hum.1996.7.16-1971. [DOI] [PubMed] [Google Scholar]
- Lee J, Him HC, Kim SW, Hong SI, Park YH. Interplay of SOS induction, recombinant gene expression, and multimerization of plasmid vectors in Escherichia coli. Biotechnol Bioeng. 2002;80:84–92. doi: 10.1002/bit.10354. [DOI] [PubMed] [Google Scholar]
- Listner K, Bentley L, Okonkowski J, Kistler C, Wnek R, Caparoni A, Junker B, Robinson D, Salmon P, Chartrain M. Development of a highly productive and scalable plasmid DNA production platform. Biotechnol Prog. 2006;22:1335–45. doi: 10.1021/bp060046h. [DOI] [PubMed] [Google Scholar]
- Love CA, Lilley PE, Dixon NE. Stable high-copy-number bacteriophage lambda promoter vectors for overproduction of proteins in Escherichia coli. Gene. 1996;176:49–53. doi: 10.1016/0378-1119(96)00208-9. [DOI] [PubMed] [Google Scholar]
- Lunn CA, Kathju S, Wallace BJ, Kushner SR, Pigiet V. Amplification and purification of plasmid-encoded thioredoxin from Escherichia coli K12. J Biol Chem. 1984;259:10469–10474. [PubMed] [Google Scholar]
- Marians KJ, Soeller W, Zipursky SL. Maximal limits of the Escherichia coli replication factor Y effector site sequences in pBR322 DNA. J Biol Chem. 1982;257:5656–5662. [PubMed] [Google Scholar]
- Masai H, Arai KI. Escherichia coli dnaT gene function is required for pBR322 plasmid replication but not for R1 plasmid replication. J Bacteriol. 1989;171:2975–2980. doi: 10.1128/jb.171.6.2975-2980.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Arai K. Mechanisms of primer RNA synthesis and D-loop/R-loop-dependent DNA replication in Escherichia coli. Biochimie. 1996;78:1109–1117. doi: 10.1016/s0300-9084(97)86737-5. [DOI] [PubMed] [Google Scholar]
- Masse E, Drolet M. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J Biol Chem. 1999;274:16659–16664. doi: 10.1074/jbc.274.23.16659. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Guy CP. Replication forks blocked by protein-DNA complexes have limited stability in vitro. J Mol Biol. 2008;381:249–255. doi: 10.1016/j.jmb.2008.05.053. [DOI] [PubMed] [Google Scholar]
- Merkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Molec Biol Reviews. 2007;71:13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minden JS, Marians KJ. Replication of pBR322 DNA in vitro with purified proteins: Requirement for topoisomerase I in the maintenance of template specificity J Biol. Chem. 1985;260:9316–9325. [PubMed] [Google Scholar]
- Miranda-Vizuete A, Rodriguez-Ariza A, Toribio F, Holmgren A, Lopez-Barea J, Pueyo C. The levels of ribonucleotide reductase, Thioredoxin, glutaredoxin 1, and GSH are balanced in Escherichia coli K12 J Biol. Chem. 1996;271:19099–19103. doi: 10.1074/jbc.271.32.19099. [DOI] [PubMed] [Google Scholar]
- O’Kennedy R, DBaldwin C, Keshavarz-Moore E. Effects of growth medium selection on plasmid DNA production and initial processing steps. J Biotechnol. 2000;76:175–183. doi: 10.1016/s0168-1656(99)00187-x. [DOI] [PubMed] [Google Scholar]
- O’Kennedy RD, Ward JM, Keshavarz-Moore E. Effects of fermentation strategy on the characteristics of plasmid DNA production. Biotechnol Appl Biochem. 2003;37:83–90. doi: 10.1042/ba20020099. [DOI] [PubMed] [Google Scholar]
- Parada CA, Marians KJ. Mechanism of DNA A protein-dependent pBR322 DNA replication. J Biol Chem. 1991;266:18895–18906. [PubMed] [Google Scholar]
- Phue JN, Lee SJ, Trinh L, Shiloach J. Modified Escherichia coli B (BL21), a superior producer of plasmid DNA compared with Escherichia coli K (DH5α) Biotechnol Bioeng. 2008;101:831–836. doi: 10.1002/bit.21973. [DOI] [PubMed] [Google Scholar]
- Prather KJ, Edmonds MC, Herod JW. Identification and characterization of IS1 transposition in plasmid amplification mutants of E. coli clones producing DNA vaccines. Appl Microbiol Biotechnol. 2006;73:815–826. doi: 10.1007/s00253-006-0532-1. [DOI] [PubMed] [Google Scholar]
- Prather KJ, Sagar S, Murphy J, Chartrain M. Industrial scale production of plasmid DNA for vaccine and gene therapy: plasmid design, production and purification. Enzyme Microbial Technol. 2003;33:865–883. [Google Scholar]
- Ribeiro SC, Oliveira PH, Prazeres DM, Monteiro GA. High frequency plasmid recombination mediated by 28 bp direct repeats. Mol Biotechnol. 2008;40:252–260. doi: 10.1007/s12033-008-9082-3. [DOI] [PubMed] [Google Scholar]
- Ricci JCD, Hernandez ME. Plasmid effects on Escherichia coli metabolism. Crit Rev Biotechnol. 2000;20:79–108. doi: 10.1080/07388550008984167. [DOI] [PubMed] [Google Scholar]
- Rozkov A, Avignone-Rossa CA, Ertl PF, Jones P, O’Kennedy RD, Smith J, Dale JW, Bushell ME. Characterization of the metabolic burden on Escherichia coli DH1 cells imposed by the presence of a plasmid containing a gene therapy sequence Biotechnol. Bioeng. 2004;88:909–915. doi: 10.1002/bit.20327. [DOI] [PubMed] [Google Scholar]
- Sandler SJ. Requirements for replication restart proteins during constitutive stable DNA replication in Escherichia coli. K-12 Genetics. 2005;169:1799–806. doi: 10.1534/genetics.104.036962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer PM, Headlam MJ, Dixon NE. Protein-protein interactions in the eubacterial replisome. IUBMB Life. 2005;57:5–12. doi: 10.1080/15216540500058956. [DOI] [PubMed] [Google Scholar]
- Singer A, Eiteman MA, Altman E. DNA plasmid production in different host strains of Escherichia coli. J Ind Microbiol Biotechnol. 2009 doi: 10.1007/s10295-008-0522-7. epub. [DOI] [PubMed] [Google Scholar]
- Wang Z, Xiang L, Shao J, Wegrzyn A, Wegrzyn G. Effects of the presence of ColE1 plasmid DNA in Escherichia coli on the host cell metabolism. Microbial Cell Factories. 2006;5:34. doi: 10.1186/1475-2859-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Luke J, Johnson L, Hodgson C. pDNAVACCultra vector family: high throughput intracellular targeting DNA vaccine plasmids. Vaccine. 2006;24:4671–4676. doi: 10.1016/j.vaccine.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Yau SY, Keshavarz-Moore E, Ward J. Host strain influences on supercoiled plasmid DNA production in Escherichia coli: Implications for efficient design of large-scale processes. Biotechnol Bioeng. 2008;101:529–544. doi: 10.1002/bit.21915. [DOI] [PubMed] [Google Scholar]