Glycan arrays are increasingly popular tools for probing the ligand specificities of glycan-binding receptors,[1] antibodies,[2] and enzymes.[1c,d,f,3] In a typical architecture, the glycan components are attached to the array surface using a linker appended to the glycan's reducing end. The multivalency of surface display can mirror, to a limited extent, the environment of the cell surface. Therein, glycans are organized into multivalent collections that are often critical for high-avidity binding to cognate receptors.[4] However, the spatial arrangement of glycans on a cell surface is surely different from that on a synthetic glycoarray. On cells, many glycans are displayed on glycoprotein scaffolds with three-dimensional geometries that cannot be emulated on a two-dimensional substrate. It is widely appreciated that the relative positioning of glycans can profoundly influence recognition by oligomerized receptors.[5] Additionally, changes in linker length,[1f] glycan density,[1d,6] and mode of glycan immobilization can[7] alter the avidity and specificity of glycan–protein interactions. New modes of glycan display within array platforms should be explored to better approximate native glycoconjugate structures.
Natural glycoproteins can be more closely mimicked by attachment of glycans to synthetic scaffolds with defined structures. Glycopolymers have been skillfully employed as soluble multivalent ligands that bind cell-surface receptors and activate biological processes.[8] Previous studies established that, as in native glycoconjugates, the shape and size uniformity of these synthetic architectures[9] as well as the density and relative positioning of their glycan appendages[10] can significantly alter the outcome of their interactions with protein receptors. Recently, surface-bound multivalent glycan ligands were shown to exhibit higher avidity to protein receptors compared to immobilized monomeric glycans,[11] and glycodendrimers were integrated into a microarray platform to study how multivalency affects lectin binding.[12] Inspired by these examples, we have sought to develop glycoconjugate mimetics that spatially position the pendant glycans similarly to natural glycoproteins. Integration of such constructs into arrays may create a more physiologically authentic platform for probing glycan-binding proteins.
We were interested in generating glycopolymers that mimic mucins, glycoproteins participating in numerous cell-surface interactions.[13] Mucins possess dense clusters of serine and threonine residues to which complex glycans are attached using a core N-acetylgalactosamine (GalNAc) moiety (Figure 1a). We previously designed mucin mimetic glycopolymers comprising a poly(methylvinylketone) backbone to which aminooxy glycans were conjugated using oxime linkages (shown schematically in Figure 1b).[14] Like natural mucins, the synthetic glycopolymers possessed rigid extended structures. These polymers were displayed on synthetic materials[14a–d] and on live cells,[14e] where their pendant glycans bound multivalent lectins. However, unlike natural mucins, the synthetic polymers were highly polydisperse with limited scope of end-functionalization, reflecting the classic limitations of the free radical polymerization process that was used to generate them.[15]
Figure 1.
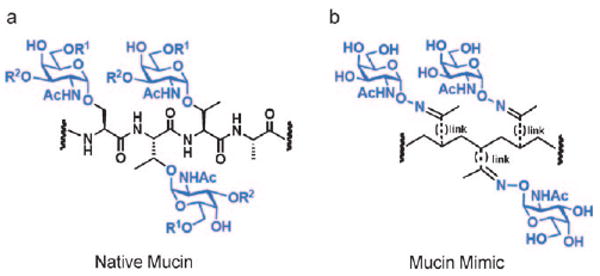
Design of a synthetic mucin mimic. a) Representative fragment of a native mucin glycoprotein. R1, R2 = glycan extensions from the core GalNAc residue. b) Schematic of a mucin mimic glycopolymer containing oxime-linked α-GalNAc residues.
Herein, we report on a new class of mucin mimetic glycopolymers that are suitable for integration with current microarray technology platforms. As shown in Figure 2a, the polymers comprise three functional domains: a central mucin mimetic domain on which glycans are displayed similarly to native mucins, one terminal domain bearing a surface attachment element, and a second terminal domain outfitted for conjugation of a reporter group. We exploited the reversible addition-fragmentation chain transfer (RAFT) polymerization technique[16] to generate polymers of low polydispersity, while simultaneously introducing chemically orthogonal capping groups onto each terminus. The RAFT technique is robust, tolerates the presence of myriad functional groups, and accordingly, has been employed previously in the synthesis of glycopolymers.[17] We envisioned using the RAFT process to generate key intermediate P1 (Figure 2b) containing pendant ketones for the attachment of aminooxy glycans, as well as terminal pentafluorophenyl (PFP) ester and trithiocarbonate groups. The PFP ester offers a reactive site for amine-functionalized surface anchors, while the trithiocarbonate can be cleaved to release a free sulfhydryl group for conjugation of the probe.
Figure 2.
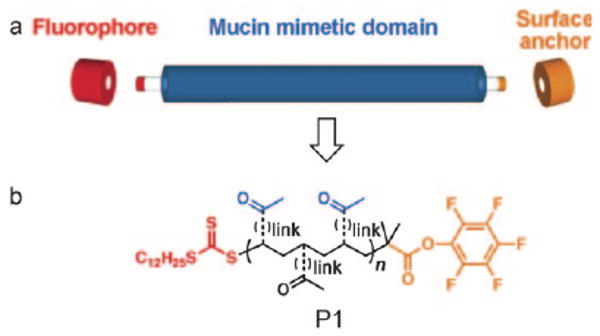
Design of a synthetic mucin mimic. a) Modular approach for the assembly of dual end-functionalized mucin mimetics. b) Schematic of a key synthetic intermediate (P1) possessing orthogonal chemical functionality in each domain.
The synthesis of intermediate P1 is shown in Scheme 1. We found that (2-oxopropyl)acrylate (1) efficiently polymerized in the presence of a PFP-ester and trithiocarbonate-functionalized chain transfer agent (2) and a radical initiator, 4,4′-azobis(4-cyanovaleric acid) (ACVA), to give polymer scaffold P1 of low polydispersity (PDI<1.15). Reaction of P1 with propargylamine afforded alkyne-functionalized polymer P2, which was primed for surface attachment by using a copper-catalyzed alkyne-azide cycloaddition reaction (CuAAC).[18] This highly selective reaction has been previously employed for surface attachment of individual glycans.[19] The end-functionalization reaction was determined to be quantitative from 19F and 1H NMR spectroscopy analysis of isolated P2 (see the Supporting Information). Glycopolymers α- and β-P3 were then assembled by condensation of the pendant ketones of P2 with α- and β-aminooxy-GalNAc, respectively.[20] The ligation reaction proceeded efficiently (>70%) to give glycopolymers that adopted mucin-like extended structures, as evidenced by TEM analysis (see the Supporting Information). Trapping of the newly released free sulfhydryl group in P3 with a maleimide-conjugated Texas Red fluorophore then afforded the target dual end-functionalized glycopolymer P4 (Scheme 1).
Scheme 1.
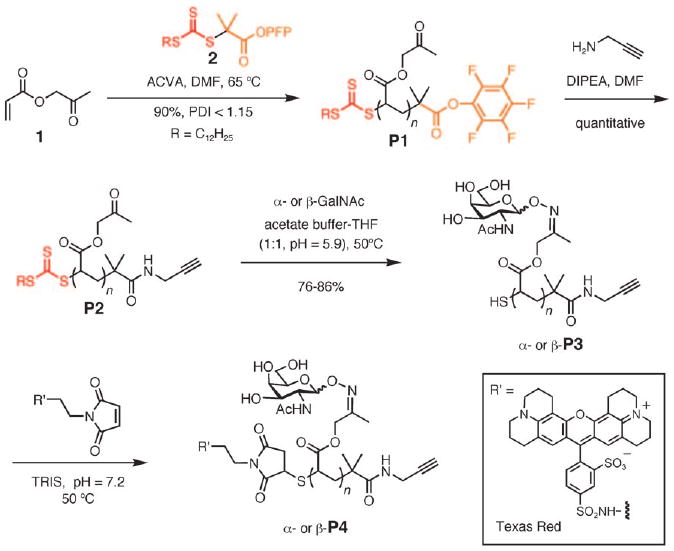
Synthesis of dual-end-functionalized mucin mimics using polymer scaffold P1.
The covalent printing of alkyne-terminated glycopolymers by CuACC required the preparation of chips containing surface azido groups. For this purpose we chose silicon wafers with a thin layer of oxide (500 nm) treated with (3-azidopropyl)trimethoxy silane (Figure 3a). Successful surface modification was apparent from a significant change in contact angle. We observed an increase in the contact angle of 80° after functionalization, indicative of a significantly more hydrophobic surface. X-ray photoelectron spectroscopy (XPS) analysis of the functionalized wafers showed a new emission at 401 eV corresponding to the 1 s transition of the newly introduced nitrogen atoms. The presence of surface azide groups was further corroborated by FTIR analysis. A characteristic azide N—N stretch absorption was clearly evident at 2100 cm−1 (see the Supporting Information).
Figure 3.
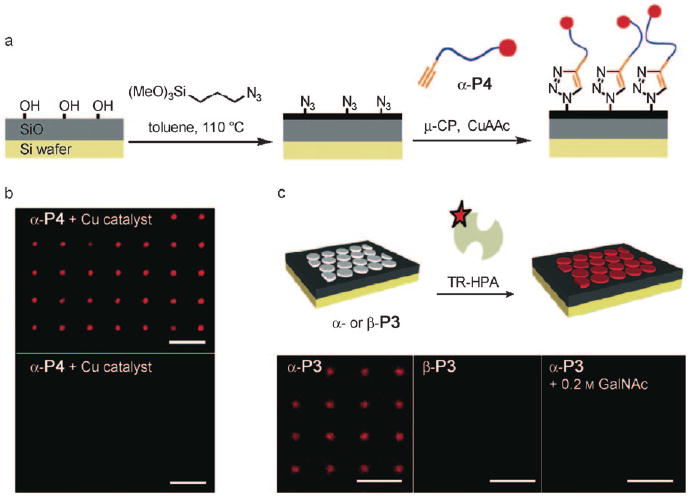
Glycopolymer microarrays for probing specific glycan–protein interactions. a) Schematic of the preparation of azide-functionalized silicon oxide wafers and covalent microcontact printing (μ-CP) of glycopolymer α-P4. b) Fluorescence micrograph of wafers printed with α-P4 in the presence (top) and in the absence (bottom) of a copper catalyst. c) Binding of Texas Red-labeled HPA lectin (TR-HPA) to microarrayed patterns of nonfluorescent glycopolymers α- and β-P3 (all scale bars are 10 μm).
We explored the microcontact printing (μCP) technique[21] as a means to array the glycopolymers on azide-functionalized chips in well-defined patterns. In a typical experiment, a poly(dimethylsiloxane) stamp with approximately 2 μm circular features was inked with a solution of a glycopolymer (0.2mgmL−1), copper sulfate (50 μm), tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA; 50 μm), and sodium ascorbate (250 mm) in a mixture of water and DMSO (1:1). The stamp was dried in a stream of nitrogen, brought to contact with the chip surface, and pressure was applied (see the Supporting Information). In the presence of the copper catalyst, the terminal alkyne of the glycopolymers reacted with the surface azides to form stable triazol linkages and, thus, the resulting patterns became covalently attached to the surface. After 30 minutes, the stamps were removed; the chips were extensively washed with water and incubated for one hour in a solution of bovine serum albumin (BSA) in phosphate-buffered saline (PBS; 50 μgmL−1). The resulting patterns of fluorescent glycopolymer α-P4 could be directly observed using fluorescent microscopy (Figure 3b, top). Notably, fluorescence was not detected when the printing was performed in the absence of copper catalyst (Figure 3b, bottom).
For microarray applications, it is important that the printed glycopolymers interact in a ligand-specific manner with glycan-binding proteins. To ascertain whether our platform fulfills this requirement, we tested the binding of Helix pomatia agglutinin (HPA), a lectin that recognizes α-GalNAc, to the printed glycopolymers (Figure 3c, top). We incubated separate silicon wafers displaying nonfluorescent α- and β-P3 polymers with Texas Red-HPA (0.25 μgmL−1, TRIS buffer, pH 7.2) for 20 minutes. Fluorescence microscopy analysis of the surfaces revealed specific binding of HPA only to the α-GalNAc-modified polymer α-P3 (Figure 3c, bottom). HPA binding to printed α-P3 was inhibited in the presence of free GalNAc (200 mm), further confirming that this process is indeed glycan specific. These observations establish that microcontact-printed glycopolymers on silicon wafers can distinguish glycan-binding proteins based on their ligand specificity.
In summary, we have developed a modular approach for the assembly of a new class of orthogonally end-functionalized mucin-mimetic glycopolymers. Micropatterns of these polymers can be generated by microcontact printing followed by CuACC, and the surface-displayed glycopolymers bind lectins specific for their pendant glycans. A key element of this approach is that the densities and orientations of the glycan ligands are determined by the polymer structure rather than by poorly understood features of the underlying surface. These parameters should therefore be more controllable than is the case with conventional glycan arrays. The utility of glycopolymer microarrays as a platform for profiling glycan-binding proteins is a matter of considerable future interest.
Supplementary Material
Footnotes
This work was supported by the Director, Office of Energy Research, Office of Basic Energy Sciences, Division of Materials Sciences, of the U.S. Department of Energy under Contract No. DE-AC03-76SF00098, within the Interfacing Nanostructures Initiative and NIH (K99M080585-01). Portions of this work were performed at the Molecular Foundry, Lawrence Berkeley National Laboratory, which is supported by the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We thank Dr. Ramesh Jasti for stimulating discussions, Dr. Marian Snauko for technical support, and Prof. Pil J. Yoo of SKKU Advanced Institute of Nanotechnology for graciously providing PDMS stamps.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200805756.
Contributor Information
Kamil Godula, Department of Chemistry, University of California and The Molecular Foundry, Lawrence Berkeley National Laboratory Berkeley, CA 94720 (USA).
David Rabuka, Department of Chemistry, University of California and The Molecular Foundry, Lawrence Berkeley National Laboratory Berkeley, CA 94720 (USA).
Ki Tae Nam, Department of Chemistry, University of California and The Molecular Foundry, Lawrence Berkeley National Laboratory Berkeley, CA 94720 (USA).
Carolyn R. Bertozzi, Email: crb@berkeley.edu, Departments of Chemistry and Molecular and Cell Biology and Howard Hughes Medical Institute, University of California and The Molecular Foundry, Lawrence Berkeley National Laboratory Berkeley, CA 94720 (USA), Fax: (+1)510-643-2628
References
- 1.a) Wang D, Liu SY, Trummer BJ, Deng C, Wang A. Nat Biotechnol. 2002;20:275–281. doi: 10.1038/nbt0302-275. [DOI] [PubMed] [Google Scholar]; b) Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. Nat Biotechnol. 2002;20:1011–1017. doi: 10.1038/nbt735. [DOI] [PubMed] [Google Scholar]; c) Fazio F, Bryan MC, Blixt O, Paulson JC, Wong CH. J Am Chem Soc. 2002;124:14397–14402. doi: 10.1021/ja020887u. [DOI] [PubMed] [Google Scholar]; d) Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Park S, Lee MR, Pyo SJ, Shin I. J Am Chem Soc. 2004;126:4812–4819. doi: 10.1021/ja0391661. [DOI] [PubMed] [Google Scholar]; f) Adams EW, Ratner DM, Bokesch HR, McMahon JB, O'Keefe BR, Seeberger PH. Chem Biol. 2004;11:875–881. doi: 10.1016/j.chembiol.2004.04.010. [DOI] [PubMed] [Google Scholar]; g) Bochner BS, Alvarez RA, Mehta P, Bovin NV, Blixt O, White JR, Schnaar RL. J Biol Chem. 2005;280:4307–4312. doi: 10.1074/jbc.M412378200. [DOI] [PubMed] [Google Scholar]; h) Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. Nat Biotechnol. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 2.a) Liang PH, Wang SK, Wong CH. J Am Chem Soc. 2007;129:11177–11184. doi: 10.1021/ja072931h. [DOI] [PubMed] [Google Scholar]; b) Wang CC, Huang YL, Ren CT, Lin CW, Hung JT, Yu JC, Yu AL, Wu CY, Wong CH. Proc Natl Acad Sci USA. 2008;105:11661–11666. doi: 10.1073/pnas.0804923105. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) de Boer AR, Hokke CH, Deelder AM, Wuhrer M. Glycoconj J. 2008;25:75–84. doi: 10.1007/s10719-007-9100-x. [DOI] [PubMed] [Google Scholar]
- 3.a) Houseman BT, Mrksich M. Chem Biol. 2002;9:443–454. doi: 10.1016/s1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]; b) Park S, Shin I. Org Lett. 2007;9:1675–1678. doi: 10.1021/ol070250l. [DOI] [PubMed] [Google Scholar]
- 4.a) Mammen M, Choi SK, Whitesides GM. Angew Chem. 1998;110:2908–2953. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]; b) Lundquist JJ, Toone EJ. Chem Rev. 2002;102:555–578. doi: 10.1021/cr000418f. [DOI] [PubMed] [Google Scholar]
- 5.a) Kitov PI, Sadowska JM, Mulvey G, Armstrong GD, Ling H, Pannu NS, Read RJ, Bundle DR. Nature. 2000;403:669–672. doi: 10.1038/35001095. [DOI] [PubMed] [Google Scholar]; b) Cairo CW, Gestwicki JE, Kanai M, Kiessling LL. J Am Chem Soc. 2002;124:1615–1619. doi: 10.1021/ja016727k. [DOI] [PubMed] [Google Scholar]; c) Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J Am Chem Soc. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]; d) Houseman BT, Mrksich M. Host-Guest Chemistry (Top Curr Chem) Vol. 218. Springer; Heidelberg: 2002. pp. 1–44. [Google Scholar]; e) Dam TK, Brewer CF. Biochemistry. 2008;47:8470–8476. doi: 10.1021/bi801208b. [DOI] [PubMed] [Google Scholar]
- 6.Horan N, Yan L, Isobe H, Whitesides GM, Kahne D. Proc Natl Acad Sci USA. 1999;96:11782–11786. doi: 10.1073/pnas.96.21.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Galanina OE, Mecklenburg M, Nifantiev NE, Pazynina GV, Bovin NV. Lab Chip. 2003;3:260–265. doi: 10.1039/b305963d. [DOI] [PubMed] [Google Scholar]; b) Xia B, Kawar ZS, Ju T, Alvarez RA, Sachdev GP, Cummings RD. Nat Methods. 2005;2:845–850. doi: 10.1038/nmeth808. [DOI] [PubMed] [Google Scholar]
- 8.a) Manning DD, Hu X, Beck P, Kiessling LL. J Am Chem Soc. 1997;119:3161–3162. [Google Scholar]; b) Bovin NE. Glycoconj J. 1998;15:431–446. doi: 10.1023/a:1006963717646. [DOI] [PubMed] [Google Scholar]; c) Gordon EJ, Gestwicki JE, Strong LE, Kiessling LL. Chem Biol. 2000;7:9–16. doi: 10.1016/s1074-5521(00)00060-0. [DOI] [PubMed] [Google Scholar]; d) Mowery P, Yang ZQ, Gordon EJ, Dwir O, Spencer AG, Alon R, Kiessling LL. Chem Biol. 2004;11:725–732. doi: 10.1016/j.chembiol.2004.03.027. [DOI] [PubMed] [Google Scholar]; e) Fleming C, Maldjian A, Da Costa D, Rullay AK, Haddleton DM, St John J, Penny P, Noble RC, Cameron NR, Davis BG. Nat Chem Biol. 2005;1:270–274. doi: 10.1038/nchembio730. [DOI] [PubMed] [Google Scholar]; f) Guan R, Sun XL, Hou S, Wu P, Chaikof EL. Bioconjugate Chem. 2004;15:145–151. doi: 10.1021/bc034138t. [DOI] [PubMed] [Google Scholar]; g) Rawat M, Gama CI, Matson JB, Hsieh-Wilson LC. J Am Chem Soc. 2008;130:2959–2961. doi: 10.1021/ja709993p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J Am Chem Soc. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 10.Sigal GB, Mammen M, Dahmann G, Whitesides GM. J Am Chem Soc. 1996;118:3789–3800. [Google Scholar]
- 11.Gestwicki JE, Cairo CW, Mann DA, Owen RM, Kiessling LL. Anal Biochem. 2002;305:149–155. doi: 10.1006/abio.2002.5652. [DOI] [PubMed] [Google Scholar]
- 12.Branderhorst HM, Ruijtenbeek R, Liskamp RMJ, Pieters RJ. ChemBioChem. 2008;9:1836–1844. doi: 10.1002/cbic.200800195. [DOI] [PubMed] [Google Scholar]
- 13.a) Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hang HC, Bertozzi CR. Bioorg Med Chem. 2005;13:5021–5034. doi: 10.1016/j.bmc.2005.04.085. [DOI] [PubMed] [Google Scholar]
- 14.a) Chen X, Lee GS, Zettl A, Bertozzi CR. Angew Chem. 2004;116:6237–6242. doi: 10.1002/anie.200460620. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2004;43:6111–6116. doi: 10.1002/anie.200460620. [DOI] [PubMed] [Google Scholar]; b) Chen X, Tam UC, Czlapinski JL, Lee GS, Rabuka D, Zettl A, Bertozzi CR. J Am Chem Soc. 2006;128:6292–6293. doi: 10.1021/ja060276s. [DOI] [PubMed] [Google Scholar]; c) Rabuka D, Parthasarathy R, Lee GS, Chen X, Groves JT, Bertozzi CR. J Am Chem Soc. 2007;129:5462–5471. doi: 10.1021/ja067819i. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Parthasarathy R, Rabuka D, Bertozzi CR, Groves JT. J Phys Chem B. 2007;111:12133–12135. doi: 10.1021/jp072136q. [DOI] [PubMed] [Google Scholar]; e) Rabuka D, Forstner MB, Groves JT, Bertozzi CR. J Am Chem Soc. 2008;130:5947–5953. doi: 10.1021/ja710644g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Odian G. Principles of Polymerization. John Willey; Hoboken, NJ: 2004. pp. 198–331. [Google Scholar]
- 16.a) Chiefari J, Chong YK, Ercole F, Krstina J, Jeffery J, Le TPT, Mayadunne RTA, Meijs GF, Moad CL, Moad G, Rizzardo E, Thang SH. Macromolecules. 1998;31:5559–5562. [Google Scholar]; b) Moad G, Chong YK, Postma A, Rizzardo E, Thang SH. Polymer. 2005;46:8458–8468. [Google Scholar]
- 17.a) Albertin L, Stenzel MH, Barner-Kowollik C, Foster LJR, Davis TP. Macromolecules. 2005;38:9075–9084. [Google Scholar]; b) Ladmiral V, Mantovani G, Clarkson GJ, Cauet S, Irwin JL, Haddleton DM. J Am Chem Soc. 2006;128:4823–4830. doi: 10.1021/ja058364k. [DOI] [PubMed] [Google Scholar]
- 18.Kolb HC, Finn MG, Sharpless KB. Angew Chem. 2001;113:2056–2075. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 19.a) Bryan MC, Fazio F, Lee HK, Huang CY, Chang A, Best MD, Calarese DA, Blixt O, Paulson JC, Burton D, Wilson IA, Wong CY. J Am Chem Soc. 2004;126:8640–8641. doi: 10.1021/ja048433f. [DOI] [PubMed] [Google Scholar]; b) Zhang Y, Luo S, Tang Y, Yu L, Hou KY, Cheng JP, Zheng X, Wang PG. Anal Chem. 2006;78:2001–2008. doi: 10.1021/ac051919+. [DOI] [PubMed] [Google Scholar]; c) Sun XL, Stabler CL, Cazalis CS, Chaikof EL. Bioconjugate Chem. 2006;17:52–57. doi: 10.1021/bc0502311. [DOI] [PubMed] [Google Scholar]; d) Rozkiewicz DI, Janczewski D, Verboom W, Ravoo BJ, Reinhoudt DN. Angew Chem. 2006;118:5418–5422. doi: 10.1002/anie.200601090. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2006;45:5292–5296. doi: 10.1002/anie.200601090. [DOI] [PubMed] [Google Scholar]; e) Michel O, Ravoo BJ. Langmuir. 2008;24:12116–12118. doi: 10.1021/la802304w. [DOI] [PubMed] [Google Scholar]
- 20.Marcaurelle LA, Shin Y, Goon S, Bertozzi CR. Org Lett. 2001;3:3691–3694. doi: 10.1021/ol0166247. [DOI] [PubMed] [Google Scholar]
- 21.Xia Y, Whitesides GM. Angew Chem. 1998;110:568–594. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 1998;37:550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


