Abstract
Cyp26A1 is a major enzyme that controls retinoic acid (RA) homeostasis by metabolizing RA into bioinactive metabolites. Previous research revealed that the mouse Cyp26A1 promoter has two canonical RA response elements (RAREs) that underlie the regulation of the gene by RA. Analyzing the 2,533-base pairs (2.5 k) genomic sequence upstream of zebrafish cyp26a1 start codon, we report that the two RAREs are conserved in zebrafish cyp26a1 promoter. Mutagenesis demonstrated that the two RAREs work synergistically in RA inducibility of cyp26a1. Fusing the 2.5 k (kilobase pairs) fragment to the enhanced yellow fluorescent protein (eYFP) reporter gene, we have generated two transgenic lines of zebrafish [Tg(cyp26a1:eYFP)]. The transgenic zebrafish display expression patterns similar to that of cyp26a1 gene in vivo. Consistent with the in vitro results, the reporter activity is RA inducible in embryos. Taken together, our results demonstrate that the 2.5 k fragment underlies the regulation of the zebrafish cyp26a1 gene by RA.
Keywords: zebrafish, cyp26a1, retinoic acid, transgenic zebrafish, retinoic acid response element, promoter
INTRODUCTION
Cyp26, also known as cytochrome P450 retinoic acid (RA), plays essential roles in vertebrate embryogenesis by metabolizing RA into inactive polar forms to prevent the specific tissues and cells from RA exposure (Glover et al., 2006; Niederreither and Dollé, 2008). Three Cyp26s (Cyp26A1, Cyp26B1, and Cyp26C1) have been characterized in vertebrates (Gu et al., 2005, 2006; Glover et al., 2006; Hernandez et al., 2007). Cyp26A1 is a major enzyme that is responsible for RA degradation. It was the first identified Cyp26 member and was isolated from zebrafish (White et al., 1996). Its orthologs have been characterized in human (White et al., 1997), mouse (Abu-Abed et al., 1998), rat (Wang et al., 2002), chick (Swindell et al., 1999), and African clawed frog (Hollemann et al., 1998). In mouse, embryos with a knock out of Cyp26A1 die from caudal regression associated with exencephaly, spina bifida, agenesis of the caudal portions of the digestive and urogenital tracts, malformed lumbosacral skeletal elements, and lack of caudal tail vertebrae (Abu-Abed et al., 2001; Sakai et al., 2001). The Cyp26A1 null phenotype closely resembles that of wild-type mouse embryos treated with teratogenic doses of RA at embryonic day (E) 8.5–E9.5 (Alles and Sulik, 1990; Kessel, 1992; Padmanabhan, 1998). When Cyp26A1 −/− homozygous mice were bred into a Raldh2 −/+ background (Raldh2, the major enzyme that synthesizes RA during early development), the Cyp26A1 null phenotypes were shown to be suppressed (Niederreither et al., 2002). The results suggest that a function of Cyp26A1 could be to titrate the local level of RA to prevent inappropriate RA signaling. In zebrafish, the mutant embryo with cyp26a1 null mutation (giraffe) displays developmental defects in the common cardinal vein, pectoral fin, tail, hindbrain, and spinal cord (Emoto et al., 2005). Its rostral spinal cord territory was expanded at the expense of the hindbrain territory. The phenotype is the opposite of that of the mutant for raldh2 (Begemann et al., 2001; Grandel et al., 2002).
Consistent with its essential roles in embryogenesis, Cyp26A1 is tightly regulated and exhibits a complex spatiotemporal expression pattern during early development (Fujii et al., 1997; Hollemann et al., 1998; De Roos et al., 1999; MacLean et al., 2001; Kudoh et al., 2002; Wang et al., 2002; Dobbs-McAuliffe et al., 2004). Although certain tissues express Cyp26A1 in an apparent RA-independent manner (Dobbs-McAuliffe et al., 2004), recent work demonstrates that endogenous RA involves controlling the expression of cyp26a1 in cells within or adjacent to the presumptive hindbrain during gastrulation to establish a robust RA gradient that assigns positional identities of zebrafish hindbrain rhombomeres (White et al., 2007). In the gir mutant, cyp26a1 expression in the rostral spinal cord was strongly upregulated, suggesting a strong feedback control of its expression by RA signaling (Dobbs-McAuliffe et al., 2004; Emoto et al., 2005). Cyp26A1 is also highly inducible by exogenous RA treatment in early embryos (Hollemann et al., 1998; Iulianella et al., 1999; Dobbs-McAuliffe et al., 2004), certain adult tissues (White et al., 1996; Winterfield et al., 2003) as well as in cell lines (Abu-Abed et al., 1998; Ozpolat et al., 2002) in a time and concentration-dependent manner. Taken together, these results suggest that Cyp26A1 is RA inducible by both endogenous and exogenous RA.
RA signaling plays crucial roles in vertebrate embryogenesis. In the absence of RA, the heterodimers of nuclear receptors RAR (RA receptor) and RXR (retinoid X receptor) bind to DNA sequences known as RA response elements (RAREs), and unliganded RARs can recruit co-repressors to actively repress transcription. Binding of RA to the heterodimers leads to a conformational change of the RAR ligand-binding domain, releasing the co-repressors and recruiting co-activators to activate transcription (Niederreither and Dollé, 2008). Typically, a RARE is composed of two direct repeats of the motif, 5′-PuG(G/T)TCA-3′, separated by a 1-base pair (bp), 2-bp, 5-bp spacer (DR1, DR2, and DR5; Chambon, 1996). Previously, research on the mouse Cyp26A1 promoter revealed that the promoter contains 2 DR5 RAREs, one lying in the proximal region (R1) and the other (R2) occurring approximately 2 k (kilobase pairs) upstream of the transcription start site. The two RAREs were shown to function synergistically to provide maximal induction of Cyp26A1 expression in response to RA (Loudig et al., 2000, 2005). In this study, we report that like the mouse Cyp26A1 promoter, the zebrafish cyp26a1 promoter contains two RAREs. Mutagenesis reveals that the two RAREs are crucial to the RA inducibility of the promoter. By microinjecting the construct of the promoter driving eYFP (enhanced yellow florescent protein) reporter gene, we established a stable transgenic line [Tg(cyp26a1:eYFP)] in which the reporter displays an expression pattern similar to that of endogenous cyp26a1 and is RA-inducible during early development. Our results provide a useful tool for further studying the regulation of cyp26a1 gene in zebrafish development.
RESULTS
Two Canonical DR5 RAREs’ Sequences Are Highly Conserved in the cyp26a1 Promoter Among Zebrafish, Mouse, and Human
Using polymerase chain reaction (PCR) amplification, we cloned a 2,533-bp (2.5 k) genomic DNA fragment upstream of cyp26a1 start codon site using as template a genomic DNA PAC clone containing zebrafish cyp26a1 gene (Fig. 1A). The sequence was deposited into GenBank under the accession number EU636200. Sequence analysis by comparing the zebrafish sequence to the mouse Cyp26A1 promoter sequence revealed that this fragment contains a promoter and the presumptive TATA box is −102 bp upstream of start codon site (Loudig et al., 2000; Fig. 1B). Analyzed by MatInspector (http://www.genomatix.de/products/MatInspector/MatInspector2.html), the predicted promoter is found to contain two canonical RAREs (5′-AGTTCA-(N)5-AGTTCA-3′). One of the RAREs is the proximal site of the promoter (R1), locating in −166 to −150 and the other is the distal site of the promoter (R2), locating in −1963 to −1947 (Fig. 1A). The genomic location of the two RAREs is similar to that of human and mouse Cyp26A1 promoters. In the mouse Cyp26A1 promoter, the two RAREs are present in −134 to −118 (R1) and −2001 to −1985 (R2) and the core sequences of the two RAREs are 5′-GGGTCA-(N)5-GGGTCA-3′ and 5′-AGTTCA-(N)5-AGTTCA-3′, respectively. In the human Cyp26A1 promoter, two RAREs are present in −132 to −116 and −2018 to −2002, respectively, and the core sequences of the RAREs are the same as that of the mouse Cyp26A1 promoter, respectively (Fig. 1B). The results indicate that the two RAREs are conserved in Cyp26A1 promoters among the different animals except the core sequences of the distal RARE are different (Loudig et al., 2000, 2005).
Fig. 1.
Genomic organization of zebrafish cyp26a1 promoter. A: Genomic DNA sequence that is upstream of zebrafish cyp26a1 gene start codon site. The start codon of zebrafish cyp26a1 gene is bolded and underlined. The 1st nucleotide upstream of zebrafish cyp26a1 gene start codon site is numbered as −1. The core sequences of the two retinoic acid response elements (RAREs) are bolded and underlined. B: Schematic diagram showing the genomic organizations of 5′-flanking sequence upstream of zebrafish, mouse and human Cyp26A1 gene start codon sites. Gray rectangle represents the presumptive TATA box and black rectangle represents the RAREs. The genomic location of the cis-elements is numbered. The sequences of mouse and human Cyp26A1 promoters were retrieved from GenBank (http://www.ncbi.nlm.nih.gov).
Activity of the 2.5 k DNA Fragment Containing cyp26a1 Promoter Is RA Inducible In Vitro
The fact that the 2.5 k genomic fragment of cyp26a1 gene has two canonical RAREs suggests that its activity should be RA inducible. To test this hypothesis, we examined the activity of the 2.5 k fragment in 293T cells by fusing the fragment with the luciferase reporter gene. As shown in Figure 2, the luciferase activities at 10 nM, 100 nM, and 1,000 nM RA treatment are significantly higher than control experiment (P < 0.05, P < 0.01, and P < 0.01, respectively). Statistical analysis also reveals that the luciferase activity at 100 nM treatment is significantly higher than that at 10 nM treatment (P < 0.05) and that the luciferase activity at 1,000 nM treatment is significantly higher than that at 100 nM treatment (P < 0.01). The results demonstrate that the activity of the 2.5 k DNA fragment is RA inducible and the promoter response to RA is dose-dependant.
Fig. 2.
Luciferase activity assay showing that the activity of the 2.5 k sequence upstream of the zebrafish cyp26a1 start codon site is retinoic acid (RA) inducible. X-axis: the different concentrations of RA that are administrated. Y-axis: the relative luciferase activity. Units represent ratios of luciferase activity to control renilla luciferase activity for each sample. With all different concentrations of RA added, the relative luciferase activities driven by the 2.5 k fragment were significantly increased as compared with controls (*P < 0.05; **P < 0.01).
Both of the Two Predicted RAREs Are Required for RA Inducibility of cyp26a1 Gene In Vitro and In Vivo
To confirm the roles of the predicted RAREs in the RA inducibility, we mutated the core sequences of the two RAREs or truncated the promoter, respectively (Fig. 3A), and then examined the promoters’ activities after RA treatment in 293T cells (Fig. 3B). As shown in Figure 3C, the promoter activity of the 2.5 k fragment (WT) is very low when there is no RA treatment. The WT fragment shows a significant higher activity when it is treated with 100 nM RA. When the core sequences of either R1 or R2 are mutated, the responses of the mutated promoters (R1mut or R2mut) to 100 nM RA treatment are significantly reduced (P < 0.01) than that of WT promoter. Of interest, the activity of R1mut promoter with the mutated R1 and normal R2 at 100 nM RA treatment is similar to that of WT promoter at no RA treatment. The result indicates that R1 is essential to the activity of the 2.5 k fragment. However, the truncated fragment with only R1 present (DF379) has nearly no response to RA treatment and the R2mut promoter with the mutated R2 and normal R1 is still RA responsive, although its RA inducing activity is reduced. The results indicate R1 itself is not enough for the activity of the 2.5 k fragment and R1 and R2 function synergistically to provide maximal induction of zebrafish cyp26a1 expression in response to RA.
Fig. 3.
In vitro and in vivo assays showing that the two retinoic acid response elements (RAREs) are required for retinoic acid (RA) inducibility of cyp26a1 gene. A: Sequences of wild-type and mutated RAREs of zebrafish cyp26a1 promoter. The core sequences of wild-type RARE and mutated RAREs are underlined. R1 and R2 represent the proximal and the distal site RARE of cyp26a1 promoter, respectively. B: Schematic diagram showing the different promoter fragments of cyp26a1 gene driving luciferase reporter gene. WT: the 2.5 k fragment with wild-type R1 and R2; R1mut: the 2.5 k fragment with mutated R1 and normal R2; R2mut: the 2.5 k fragment with mutated R2 and normal R1; DF379: truncated fragment with only 379 bp of 2,533 bp. C: Luciferase activity assay showing that R1 and R2 are required for the RA-inducible activity of zebrafish cyp26a1 promoter. X-axis: the different promoter fragments shown in (B) are examined for their RA inducibilities. The activities of WT, R1mut, R2mut, and DF379 fragments were tested at 100 nM RA. No RA CTL represents that the activity of WT is measured when there is no RA treatment. Y-axis: the relative luciferase activity. Units represent ratios of luciferase activity to control renilla luciferase activity for each sample. Mutating either of two RARE sites or truncating promoter fragment with only R1 present results in that the RA-inducible activities of the promoter fragments are significantly lower than that of wild-type 2.5k promoter fragment (**P < 0.01). D–G,D′–G′: In vivo transient expression assay showing the activities of the fragments with different lengths or mutations of cyp26a1 regulatory sequence. The linearized constructs of p2.5kcyp26a1pr_eYFP (WT) (D, D′), pR1mut_eYFP (E,E′), pR2mut_eYFP (F,F′), and pDF379_eYFP (G,G′) were microinjected into 1- to 4-cell stage embryos, respectively. The injected eggs were grown to 6 hours postfertilization (hpf) and then treated with 100 nM RA for 6 hr for examining the transient eYFP expression at 12 hpf (D′: WT; E′: R1mut; F′: R2mut; G′: DF379). The injected embryos incubated at 0.1% dimethyl sulfoxide (DMSO) were used as control (D, WT; E, R1mut; F, R2mut; G, DF379). Embryos are laterally viewed and positioned anterior top. Fluorescent images are photographed by the DP70 digital camera (Olympus, Japan) with 10-sec exposure time. Scale bar = 100 µM.
To explore whether the two RAREs play the essential roles in the RA inducibility in vivo, we microinjected the mutated constructs that drive the eYFP gene into zebrafish embryos at one- to four-cell stage. The RA induction of the eYFP transient expression was then examined on the embryos at 12 hours postfertilization (hpf) that had been incubated with 100 nM RA for 6 hr. As shown in Figure 3D,D′, the eYFP expression of the 2.5k_eYFP construct is significantly higher in the RA-treated embryos than in the control embryos. The expressions of R1mut_eYFP and DF379_eYFP in the RA-treated embryos (Fig. 3E′,G′) are similar to those in the control embryos (Fig. 3E,G). The expression of R2mut-eYFP is slightly higher in the RA-treated embryos than in the control embryos (Fig. 3F′,F). The in vivo expression results are consistent with the above in vitro results that both R1 and R2 are required for the RA induced expression. Unexpectedly, we found that the expression of R2mut_eYFP construct in the control embryos (Fig. 3F) is higher than the expression of WT_eYFP construct in the control embryos (Fig. 3D). The result suggests that R2 may involve repression of the gene somehow.
Generation of Transgenic Zebrafish Lines [Tg(cyp26a1: eYFP)] Reveals That Expression Pattern of the Reporter Driven by the cyp26a1 Promoter Closely Mimics That of Endogenous cyp26a1 Gene
In vitro and in vivo results show that the activity of the 2.5 k genomic fragment upstream of cyp26a1 start codon site is RA inducible. To investigate whether the 2.5 k sequence controls the dynamic expression pattern of cyp26a1 in vivo, we made stable transgenic fish lines by microinjection of the 2.5 k fragment driving the eYFP reporter gene into zebrafish one-cell embryos. In total, we have obtained two transgenic zebrafish lines (Line A and Line B) from two different founders. We have passed the transgenic zebrafish to generation 3 (F3) and have fish homozygous for the reporter gene. Generally, the two lines of zebrafish display similar expression patterns. Here, we describe the reporter expression pattern in Line A.
When a transgenic female zebrafish (heterozygous for the transgene) is mated with a male wild-type zebrafish, the nontransgenic embryos are all fluorescent in cytoplasm at one-cell and two-cell stages (Fig. 4A,B). The results indicate that the reporter gene has been transcribed and possibly translated during oogenesis. The fluorescent signal is distributed evenly in all embryonic cells of embryos at gastrulation stage (Fig. 4C,D) and even at somite stage, although the signal becomes weak (Fig. 4E).
Fig. 4.
Expression of enhanced yellow fluorescent protein (eYFP) in embryos from a female transgenic zebrafish. Nontransgenic embryos are produced by a female transgenic zebrafish (heterozygous for the transgene) mated with a male wild-type zebrafish. Images are photographed by the DP70 digital camera (Olympus, Japan) with 5-sec exposure time. Embryos are laterally viewed and positioned animal pole top (A–D) or anterior top (E). A: Fertilized egg at one-cell stage. B: Two-cell. C: 50% epiboly. D: 75% epiboly. E: Three-somite stage (11 hours postfertilization [hpf]). eYFP expressions appearing in these stages are due to maternal loading of eYFP protein (A–C) or both eYFP protein and mRNA (D,E). Scale bar = 100 µM.
When a male transgenic fish mates with a wild-type female, the positive embryos display a transgene expression pattern similar to cyp26a1 expression pattern (Kudoh et al., 2002; Dobbs-McAuliffe et al., 2004; Fig. 5). Initially, the reporter signals are detected from 75% epiboly (8 hpf) embryos at the presumptive neural plate and around the blastoderm margin (Fig. 5A). The presence of the reporter signal at 90% epiboly is the same as that at 75% epiboly (Fig. 5B). Like the transgene expression pattern, whole-mount in situ hybridization reveals that cyp26a1 is endogenously expressed in the presumptive anterior neural ectoderm and around the blastoderm margin at 75% and 90 epiboly stages (Fig. 5A′,B′). At 11 hpf (three-somite stage), the transgene signals are mainly at the presumptive brain regions and the tail bud that correspond to the endogenous expression domains of cyp26a1 in presumptive forebrain, midbrain and tail bud (Fig. 5C,C). At 16 hpf (14-somite), the transgene is expressed in tail bud, developing eyes and pharyngeal arches (Fig. 5D) like cyp26a1 expression (Fig. 5D′). However, nonspecific expression of the transgene is present in brains and somites (Fig. 5D,D′). At 24 hpf, the expression of cyp26a1 is detected in retina, cells in olfactory vesicle, epidermis, pharyngeal arches, anterior dorsal spinal cord, proctodeum, caudal notochord, and tail bud (Fig. 5E′). Consistent with the cyp26a1 expression, the fluorescent signals are mainly present in retina, cells in olfactory vesicle, anterior dorsal spinal cord, proctodeum and caudal notochord except that nonspecific signals are present in midbrain region (Fig. 5E). At 30 hpf, the expression pattern of cyp26a1 is very similar to embryos at 24 hpf except lower expression levels (Fig. 5F′,K′) and the expression pattern of the transgene is similar to that of cyp26a1 expression, mainly in retina (Fig. 5G), cells in olfactory vesicle (Fig. 5G), anterior dorsal spinal cord (Fig. 5H), proctodeum (Fig. 5I), caudal notochord (Fig. 5J), and pharyngeal arches (Fig. 5K). Taken together, our results suggest that the transgene is expressed in a very similar pattern to that of cyp26a1 during zebrafish early development.
Fig. 5.
Expression pattern of transgenic reporter in zebrafish early development. Transgenic embryos are produced by a female wild-type zebrafish mated with a male transgenic zebrafish. A–K: Fluorescent images are photographed by the DP70 digital camera with 10-sec exposure time. A′–F′,K′: Whole-mount in situ hybridization is performed to reveal the expression pattern of cyp26a1. Embryos are laterally viewed (except that K′ is dorsal viewed) and positioned animal pole top (A,B; A′,B′), anterior top (C,D; C′,D′), or anterior left (E–K; E′–K′). A,B: Enhanced yellow fluorescent protein (eYFP) is initially detected at presumptive neural plate and around the blastoderm margin of the embryo at 75% epiboly (A) and then 95% epiboly (B). The fluorescent expression patterns mimic the expression of cyp26a1 at 75% epiboly (A′) and 95% epiboly (B′), respectively. C,C′: At 11 hours postfertilization (hpf), eYFP is strong at the presumptive brain region and the tail bud (C), corresponding to the cyp26a1 expression at presumptive forebrain, midbrain and the tail bud (C′). D,D′: At 16 hpf, the transgene signals are present in developing eyes, pharyngeal arches and tail bud (D) where the cyp26a1 is also expressed (D′). E,E′: At 24 hpf, the fluorescent signals are mainly present in retina, cells in olfactory vesicle, anterior dorsal spinal cord, proctodeum, and caudal notochord (E) where the cyp26a1 is also expressed (E′). F,F′,K′: At 30 hpf, the expression pattern of the transgene (F) is similar to that of cyp26a1 (F′, K′). G–K: High magnification reveals that the fluorescent signals are mainly present in retina (G), cells in olfactory vesicle (G), anterior dorsal spinal cord (H), proctodeum (I), caudal notochord (J), and pharyngeal arches (K). de, developing eye; dp, developing pharyngeal arch; ep, epidermis; fr, presumptive forebrain; mr, presumptive midbrain; nc, caudal notochord; of, cells in olfactory vesicle; ph, pharyngeal arch; pr, proctodeum; rt, retina; sc, anterior dorsal spinal cord; tb, tail bud. Scale bar = 100 µM.
Reporter Expression of the Transgenic Zebrafish Is RA Inducible
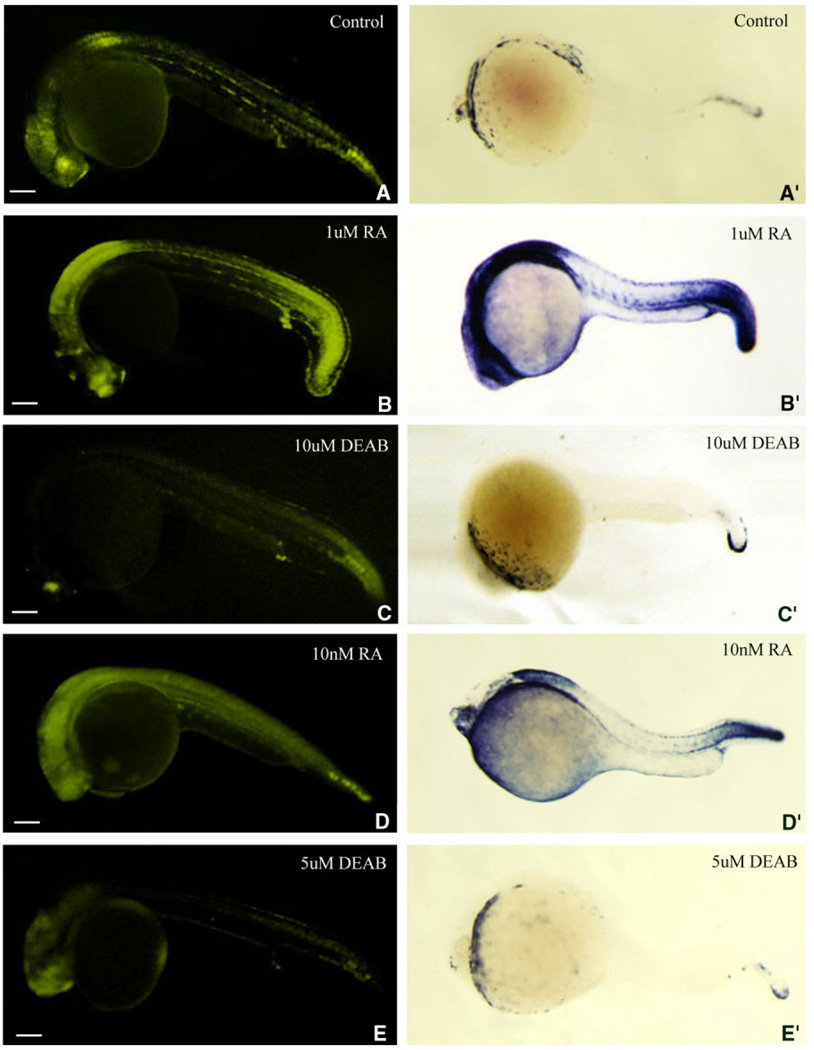
As described above, our results show that the dynamic expression pattern of eYFP in the transgenic fish closely matches the in vivo expression pattern of the cyp26a1. Because other studies have shown that zebrafish cyp26a1 gene is RA inducible (Dobbs-McAuliffe et al., 2004), we, therefore, asked whether our transgenic reporter gene could mimic this feature in vivo. To answer this question, we first increased RA signaling by treating the transgenic embryos with 1 µM RA for 6 hr starting from 18 hpf or 10 nM RA for 24 hr starting from 0 hpf. When viewing the treated embryos at 24 hpf, we found that the eYFP signal is globally up-regulated (Fig. 6A,B,D). For the embryos treated with 1 µM RA for 6 hr, the up-regulated expression is mainly seen in forebrain, anterior spinal cord, proctodeum, and whole tail (Fig. 6A,B). The expression patterns are consistent with that of cyp26a1 expression in the embryos after RA treatment (Fig. 6A′,B′). As to the embryos treated with 10 nM RA for 24hr, the up-regulated expression is seen in almost whole body but mainly in forebrain, anterior spinal cord, and caudal notochord (Fig. 6A,D). The main expression patterns of the transgene are roughly consistent with that of cyp26a1 expression in the embryos after RA treatment (Fig. 6A′,D′). The results demonstrate that the transgene is RA inducible in vivo. We then reduced RA signaling by treating the transgenic embryos with 10 µM diethylaminobenzaldehyde (DEAB) for 6 hr starting from 18 hpf or 5 µM DEAB for 24 hr starting from 0 hpf. When viewing the treated embryos at 24 hpf, we found that the eYFP signals are generally down-regulated in certain tissues including retina, anterior spinal cord and caudal notochord (Fig. 6A,C,E). The expression pattern is similar to endogenous down-regulation of cyp26a1 in the embryos after DEAB treatment (Fig. 6A′,C′,E′). The results demonstrate that like the down-regulation of cyp26a1 after reducing endogenous signaling, reducing RA signaling could decrease the expression of the transgene in specific tissues during zebrafish early development.
Fig. 6.
Retinoic acid (RA) inducibility of enhanced yellow fluorescent protein (eYFP) expression in transgenic zebrafish embryos is similar to that of cyp26a1. B–E′: The transgenic embryos produced by a female wild-type zebrafish mated with a male transgenic zebrafish were treated with 1 µM RA (B,B′) or 10 µM DEAB (C,C′) for 6 hr starting from 18 hours postfertilization (hpf), or 10 nM RA (D,D′), or 5 µM DEAB (E,E′) for 24 hr starting from 0 hpf to examine whether the RA-induced expression of the transgene (B–E) is similar to that of cyp26a1 (B′–E′; revealed by whole-mount in situ hybridization) during zebrafish early development. A,A′: Untreated transgenic embryos were used as controls. Fluorescent images were photographed by a DP70 digital camera with 10-sec exposure time. Embryos are laterally viewed and positioned anterior left. In the embryos that were treated with 1 µM RA for 6 hr (B,B′), the greatly up-regulated expression was mainly seen in forebrain, anterior spinal cord, proctodeum, and whole tail (A,B,A′,B′). In the embryos treated with 10 nM RA for 24 hr treatment (D,D′), the up-regulated expression was almost in whole body but mainly seen in retina, anterior spinal cord and caudal notochord (A,D,A′,D′). When the embryos are treated with 10 µM DEAB for 6 hr (C, C′) or 5 µM DEAB for 24 hr (E,E′), the expressions of the transgene and cyp26a1 both are down-regulated mainly in retina, anterior spinal cord, and caudal notochord compared with wild-type embryos (A,A′). Scale bar = 100 µM.
In summary, we have constructed two stable transgenic lines of zebrafish being driven by the zebrafish cyp26a1 promoter. The transgenic expression patterns correspond well to the expression patterns of in vivo dynamic expression pattern of cyp26a1 gene and are RA inducible. The transgenic fish is named as Tg(cyp26a1: eYFP), in accord with the rules set by Zebrafish Nomenclature Committee (http://zfin.org).
DISCUSSION
Cyp26A1 is a major enzyme that is responsible for limiting RA activity and plays essential roles in RA homeostasis in vivo (Dobbs-McAuliffe et al., 2004). Like many of the RA-regulated genes owning RAREs (Niederreither and Dollé, 2008), previously research revealed that the mouse Cyp26A1 promoter has two canonic RAREs and the two RAREs work synergistically to direct Cyp26A1 gene response to RA signal to form a RA-free zone (Loudig et al., 2000, 2005). In this study, we report that the zebrafish cyp26a1 promoter owns two RAREs with a similar genomic location to the mouse Cyp26A1 promoter. Mutation analyses on the zebrafish cyp26a1 promoter reveal that the two RAREs are both essential to the promoter’s activity for RA regulation. In vivo and in vitro assays both confirm that the promoter is RA inducible. These results suggest that the zebrafish cyp26a1 could be a direct target gene of RA signal and the two RAREs underlie an aspect of the regulation of gene by RA. However, our results also show that there are some different features between zebrafish and mouse cyp26a1 promoters. In mouse, mutated Cyp26A1 promoter with the mutated R1 and normal R2 is still RA-inducible while our results show that the mutated zebrafish cyp26a1 promoter with the mutated R1 and normal R2 completely lost its response to RA (Fig. 3C,E,E′; Loudig et al., 2005). Additionally, the mouse Cyp26A1 promoter with a mutated R2 and a normal R1 has a very weak response to RA induction while the mutated zebrafish cyp26a1 promoter with a mutated R2 and a normal R1 remains responsive to RA, although its activity is reduced compared with the wild-type promoter (Figs. 3C,F,F′; Loudig et al., 2005). The results suggest there might be other regulatory cis-element(s) in the zebrafish promoter between R1 and R2 that are responsible for the remaining response to RA. Taken together, our results suggest that, although the canonic RAREs are conserved among Cyp26a1 promoters of different animals, species-specific features of Cyp26a1 promoters can be present to ensure its specific roles in the regulation of this gene during early development.
In addition to RA induction, recent researches have suggested that the expression of Cyp26A1 gene is tightly regulated during development by many other factors or genes. Cyp26A1 is transcriptionally activated by progesterone in mice and by estrogen in humans (Fritzsche et al., 2007). It is up-regulated by a Wnt-dependent mechanism in tissues deficient in the colon tumor suppressor gene, adenomatous polyposis coli (APC), in zebrafish, mouse, and human (Shelton et al., 2006). In zebrafish, tgif is found to be necessary for normal initiation of cyp26a1 (Gongal and Waskiewicz, 2008) and the expression of cyp26a1 in the tail bud is controlled by Su(H) (Suppressor of Hairless, downstream of Notch Signaling) activity (Echeverri and Oates, 2007). In mouse, Cyp26A1 is identified as a target of Tbx1 gene (Roberts et al., 2006). In acute promyelocytic leukemia (APL) cells, highly RA-sensitive APL blasts expressed higher levels of Cyp26A1 than low RA-sensitive APL blasts and the enhanced expression of Cyp26A1 is mediated by Hoxa10v2, a homeobox transcription factor, after RA treatment (Quere et al., 2007). Collectively, these data show that the transcriptional activation of Cyp26A1 gene cannot be simply ascribed to the simple substrate-driven feedback mechanism that generally regulates the intracellular concentrations of RA. The mechanism underlying its regulation appears to be more complex and not totally dependent upon RA (Dobbs-McAuliffe et al., 2004). Therefore, investigating the underlying mechanism of cyp26a1 regulation is very important for understanding the role of cyp26a1 in zebrafish early development. Because the fluorescent reporter’s expression can be assessed by confocal microscopy, a variety of transgenic zebrafish lines have been developed with different reporter genes expressed in tissue-restricted patterns or under regulation of inducible promoters to allow one to observe the dynamic changes of gene expression and the detailed morphologic movements in developing, living zebrafish embryos with a real time course (Udvadia and Linney, 2003). In this study, we report a stable transgenic zebrafish line with the cyp26a1 promoter driving an eYFP as a reporter. Like results found in other transgenic fish (Linney et al., 1999; Perz-Edwards et al., 2001), we noticed that the 1-cell embryos produced from the female transgenic zebrafish are fully fluorescent. This is obviously due to maternal storage of transgenic reporter mRNA and/or protein that is stored in oocytes. If the embryos are produced by male transgenic zebrafish and female wild-type zebrafish, the eYFP signal should reflect the expression of zygotic transgenic reporter. In our results, we show that the dynamic expression pattern of the reporter in the transgenic fish closely match that of endogenous cyp26a1 expression during early development. If the transgenic embryos are treated with RA, the reporter expression is greatly enhanced (Fig. 6B,D) whereas the reporter expression is reduced in certain tissues (Fig. 6C,E) if the embryos are treated with DEAB. The expression changes of transgene were similar to that of endogenous cyp26a1 in the embryos that RA signaling is altered. Taken together, our results indicate that the 2.5 k sequence upstream of start codon site underlies the regulation of cyp26a1 gene. Therefore, our transgenic fish [Tg(cyp26a1:eYFP)] provide a useful model for further addressing the regulation of cyp26a1 gene during development, tracing the developmental fates of cyp26a1-expressing cells, and establishing biological indices of environmental retinoid pollutants.
EXPERIMENTAL PROCEDURES
Molecular Cloning of the Zebrafish cyp26a1 Promoter
A zebrafish genomic PAC clone library (IncyteGenomics, Wilmington, DE) was screened using a probe complementary to the cDNA of zebrafish cyp26a1 (GenBank accession no. U68234). Three positive clones were obtained. By directly sequencing upstream of cyp26a1 gene in one PAC clone, a 2,533-bp sequence upstream of the start codon (ATG) of the gene was identified. The 2,533-bp fragment (2.5 k) was then amplified with high fidelity DNA polymerase (BD Advantage HF2, Takara, Shiga, Japan) and subcloned into pGEM-T vector (Promega, Madison, WI).
Construction of DNA Plasmids for In Vitro Transfection and In Vitro Luciferase Assay
The reporter constructs with different lengths or mutations of cyp26a1 regulatory sequence were all cloned into the pGL3 reporter vector for luciferase assays (Promega). The cyp26a1-2533 (WT) and cyp26a1-379 (DF379) fragments were generated by PCR amplifications with the primers containing SacI and NcoI sites, respectively. The sequences of the forward primers are 5′-atccgagctcGAGCTCCAGATTTGGCTTCA-3′ (for WT) and 5′-atccgagctcAGAGTAATGAGTGTAAACG-3′ (for DF379), and the sequence of the reverse primer is 5′-CCCATGGGTTGAAGCGCGCAACTGATC-3′. The PCR reactions were conducted with BD Advantage HF2 (Takara). The PCR condition was 94°C 1 min, 30 cycles of (94°C 30 sec, 58°C 45 sec, 68°C 4 min), followed by a 68°C 6-min extension. The fragments containing mutations of the predicted RAREs were generated by PCR amplification using QuikChange II-E Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer’s instruction. The primers used to generate R1mut fragment (mutations in the proximal RARE) are 5′-TTAAAGAgctACTTTGTGactACTAATTTATC-3′ (forward) and 5′-GATAAATTAGTagtCACAAAGTagcTCTTTAA-3′ (reverse). The primers used to generate R2mut fragment (mutations in the distal RARE) are 5′-TCCTCTGgctACTATCCTactACTTCAGAGTT-3′ (forward) and 5′-AACTCTGAAGTagtAGGATAGTagcCAGAGGAA-3′ (reverse). The PCR condition was 95°C 30 sec, 18 cycles of (95°C 30 sec, 60°C 1 min, 68°C 8 min), followed by a 68°C 8-min extension. All constructs were sequenced to confirm their sequence identities.
In vitro luciferase assays were performed as previously described (Lassiter et al., 2002; Zhao et al., 2005) using Lipofectamine 2000 for transfection (Invitrogen, Carlsbad, CA) and the Dual Luciferase Reporter Assay System for measuring luciferase activity (Promega) according to the manufacturers’ instructions. Briefly, 293T cells (ATCC, Mannassas, VA, USA) were transfected when reaching 70–80% confluence. The 375 ng of tested DNA plasmid, 25 ng of SV40-renilla luciferase expression plasmid (Promega), and 100 ng of Rarα2.B expression plasmid (Perz-Edwards et al., 2001) were co-transfected into one well of the 24-well plate. Transfection was done in Dulbecco’s MEM (DMEM, Invitrogen) for 4 hr. After transfection, each well was changed with 1 ml of DMEM containing 15% stripped serum (final concentration; Invitrogen) and the various concentrations of all-trans RA (Sigma, St. Louis, MO) or vehicle dimethyl sulfoxide (DMSO; Sigma; the final vehicle concentration was always 0.1% for all wells). After 24 hr treatment, the wells were washed with cold PBS twice and the cells were then lysed for dual luciferase activity assay using the Passive Lysis Buffer (Promega). Luciferase activity was measured using a Berthold Luminometer (Promega). Firefly luciferase activity was normalized by dividing by the activity of renilla luciferase. Each treatment was repeated three times with two independent transfections. Results were subjected to the Student t-test.
Construction of the DNA Fragments for Microinjection and Generation of the Transgenic Zebrafish Lines
The fragments with different lengths or mutations of cyp26a1 regulatory sequence were cloned into the RGYn expression vector (Perz-Edwards et al., 2001) for performing in vivo transient expression assay on the activities of the cyp26a1 promoters and making transgenic zebrafish. The RGYn plasmid is an eYFP expression vector derived from pGEM-9Zf(−) (Promega). It contains RARE enhancer, GT2 basal promoter, eYFP gene, cDNA for coding nuclear translocation domain (ntd), the SV40 intron, and poly(A) addition signal in the multiple cloning sites of pGEM-9Zf(−). The eYFP gene driven by the RARE enhancer and GT2 basal promoter in the RGYn vector had been modified by means of PCR to remove the translational stop codon and insert an in-frame coding sequence of the SV40 ntd, for nuclear localization of the eYFP protein after expression (Linney et al., 1999). To perform the cloning, we inserted the different cyp26a1 regulatory fragments including WT, R1mut, R2mut and DF379 into the RGYn plasmid that had been cut with SacI and NcoI to remove the RARE enhancer and GT2 basal promoter. The resulting expression plasmids were named as p2.5kcyp26a1pr_eYFP, pR1mut_eYFP, pR2mut_eYFP and pDF379_eYFP.
Before microinjection, the expression plasmids were linearized with SacI and NotI to remove the vector sequence, respectively. Microinjection was performed on one-cell stage (for making transgenic zebrafish) or one-to four-cell stage embryos (for transient assay) and approximately 2 nl of linearized DNA at a concentration of 50 ng/µl was microinjected into the cytoplasm of the fertilized egg as previously described (Linney et al., 1999). For transient assay, the injected eggs were grown to 6 hpf at 28.5°C and then treated with 100 nM RA (Sigma) for 6 hr to examine the transient eYFP expression. The injected embryos incubated at 0.1% DMSO were used as control. For making transgenic zebrafish, the injected eggs were incubated at 28.5°C and examined for transient eYFP expression at 24 hpf under a fluorescent dissecting microscope (Leica, Wetzlar, Germany). The embryos expressing eYFP ubiquitously (except in yolk) were raised to adults for the screening of transgenic fish founders.
The screen for transgenic fish was first carried out on the potential founder (G0) by PCR as described before (Linney et al., 1999). The positive candidates were then confirmed by visual screening using a fluorescent dissecting microscope. The transgenic offspring (F1) of the founder was raised to sexual maturity. The transgenic offspring (F2) of F1 were sibling mated to produce stable transgenic fish line (F3).
Observation of eYFP Expression under Fluorescent Dissecting Microscopes
The eYFP signal was monitored under a fluorescent dissecting microscope (Leica) equipped with a DP70 digital imaging system (Olympus, Tokyo, Japan) with a YFP filter set. Images were collected using IPP software (Olympus). Color micrographs were pseudo-colored to yellow.
Embryos younger than 18 hpf were placed on a Petri dish to observe directly. Embryos older than 18 hpf were anaesthetized by 0.1% tricain (Sigma) before observation. To investigate whether the eYFP signal of the transgenic fish is RA inducible, the fish embryos were treated with 10 nM RA or 5 (µM DEAB (Sigma) from one-cell to 24 hpf, or with 1 µM RA or 10 [µM DEAB from 18 hpf to 24 hpf.
Whole-Mount In Situ Hybridization
Whole-mount in situ hybridization analyses on the embryos to examine the expression pattern of cyp26a1 was performed as described previously (Dobbs-McAuliffe et al, 2004).
ACKNOWLEDGMENTS
Q.Z. and X.Ga. were funded by China MOST, Q.Z. and X.Gu. were funded by China NSFC, and Q.Z. was funded by Fund for Distinguished Scholars in Six Scientific Fields of Jiangsu Province, China, and E.L. was funded by the National Institutes of Health.
Grant sponsor: China MOST; Grant number: 2005CB522501; Grant number: 2006CB943500; Grant number: 2007CB947101; Grant sponsor: China NSFC; Grant number: 30771071; Grant number: 30671046; Grant sponsor: Fund for Distinguished Scholars in Six Scientific Fields of Jiangsu Province, China; Grant sponsor: NIH; Grant number: U19 ES11375.
REFERENCES
- Abu-Abed SS, Beckett BR, Chiba H, Chithalen JV, Jones G, Metzger D, Chambon P, Petkovich M. Mouse P450RAI (CYP26) expression and retinoic acid-inducible retinoic acid metabolism in F9 cells are regulated by retinoic acid receptor g and retinoid X receptor a. J Biol Chem. 1998;273:2409–2415. doi: 10.1074/jbc.273.4.2409. [DOI] [PubMed] [Google Scholar]
- Abu-Abed SS, Dolle P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev. 2001;15:226–240. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alles AJ, Sulik KK. Retinoic acid-induced spina bifida: evidence for a pathogenetic mechanism. Development. 1990;108:73–81. doi: 10.1242/dev.108.Supplement.73. [DOI] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- De Roos K, Sonneveld E, Compaan B, Ten Berge D, Durston AJ, Van der Saag PT. Expression of retinoic acid 4-hydroxylase (CYP26) during mouse and Xenopus laevis embryogenesis. Mech Dev. 1999;82:205–211. doi: 10.1016/s0925-4773(99)00016-7. [DOI] [PubMed] [Google Scholar]
- Dobbs-McAuliffe B, Zhao Q, Linney E. Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev. 2004;121:339–350. doi: 10.1016/j.mod.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Oates AC. Coordination of symmetric cyclic gene expression during somitogenesis by Suppressor of Hairless involves regulation of retinoic acid catabolism. Dev Biol. 2007;301:388–403. doi: 10.1016/j.ydbio.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Emoto Y, Wadab H, Okamotob H, Kudoa A, Imaia Y. Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish. Dev Biol. 2005;278:415–427. doi: 10.1016/j.ydbio.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Fritzsche B, Vermot J, Neumann U, Schmidt A, Schweigert FJ, Dollé P, Rühl R. Regulation of expression of the retinoic acid metabolizing enzyme CYP26A1 in uteri of ovariectomized mice after treatment with ovarian steroid hormones. Mol Reprod Dev. 2007;74:258–264. doi: 10.1002/mrd.20526. [DOI] [PubMed] [Google Scholar]
- Fujii H, Sato T, Kaneko S, Gotoh O, Fujii-Kuriyama Y, Osawa K, Kato S, Hamada H. Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. EMBO J. 1997;16:4163–4173. doi: 10.1093/emboj/16.14.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JC, Renaud JS, Rijli FM. Retinoic acid and hindbrain patterning. J Neurobiol. 2006;66:705–725. doi: 10.1002/neu.20272. [DOI] [PubMed] [Google Scholar]
- Gongal PA, Waskiewicz AJ. Zebrafish model of holoprosencephaly demonstrates a key role for TGIF in regulating retinoic acid metabolism. Hum Mol Genet. 2008;17:525–538. doi: 10.1093/hmg/ddm328. [DOI] [PubMed] [Google Scholar]
- Grandel H, Lun K, Rauch GJ, Rhinn M, Piotrowski T, Houart C, Sordino P, Kuchler AM, Schulte-Merker S, Geisler R, Holder N, Wilson SW, Brand M. Retinoic acid signalling in the zebrafish embryo is necessary during pre-segmentation stages to pattern the anterior-posterior axis of the CNS and to induce a pectoral fin bud. Development. 2002;129:2851–2865. doi: 10.1242/dev.129.12.2851. [DOI] [PubMed] [Google Scholar]
- Gu X, Xu F, Wang X, Gao X, Zhao Q. Molecular cloning and expression of a novel CYP26 gene (cyp26d1) during zebrafish early development. Gene Expr Patterns. 2005;5:733–739. doi: 10.1016/j.modgep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Gu X, Xu F, Song W, Wang X, Hu P, Yang Y, Gao X, Zhao Q. A novel cytochrome P450, zebrafish Cyp26D1, is involved in metabolism of all-trans retinoic acid. Mol Endocrinol. 2006;20:1661–1672. doi: 10.1210/me.2005-0362. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollemann T, Chen Y, Grunz H, Pieler T. Regionalized metabolic activity established boundaries of retinoic acid signalling. EMBO J. 1998;17:7361–7372. doi: 10.1093/emboj/17.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iulianella A, Beckett B, Petkovich M, Lohnes D. A molecular basis for retinoic acid-induced axial truncation. Dev Biol. 1999;205:33–48. doi: 10.1006/dbio.1998.9110. [DOI] [PubMed] [Google Scholar]
- Kessel M. Respecification of vertebral identities by retinoic acid. Development. 1992;115:487–501. doi: 10.1242/dev.115.2.487. [DOI] [PubMed] [Google Scholar]
- Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- Lassiter CS, Kelley B, Linney E. Genomic structure and embryonic expression of estrogen receptor beta a (ERbetaa) in zebrafish (Danio rerio) Gene. 2002;299:141–151. doi: 10.1016/s0378-1119(02)01050-8. [DOI] [PubMed] [Google Scholar]
- Linney E, Hardison NL, Lonze BE, Lyons S, DiNapoli L. Transgene expression in zebrafish: a comparison of retroviral-vector and DNA-injection approaches. Dev Biol. 1999;213:207–216. doi: 10.1006/dbio.1999.9376. [DOI] [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI (CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–1497. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Loudig O, Maclean GA, Dore NL, Luu L, Petkovich M. Transcriptional cooperativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility. Biochem J. 2005;392:241–248. doi: 10.1042/BJ20050874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean G, Abu-Abed S, Dolle P, Tahayato A, Chambon P, Petkovich M. Cloning of a novel retinoic-acid metabolizing cytochrome P450, Cyp26B1, and comparative expression analysis with Cyp26A1 during early murine development. Mech Dev. 2001;107:195–201. doi: 10.1016/s0925-4773(01)00463-4. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dolle P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat Genet. 2002;31:84–88. doi: 10.1038/ng876. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Dollé P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Ozpolat B, Mehta K, Tari AM, Lopez-Berestein G. All-trans-Retinoic acid-induced expression and regulation of retinoic acid 4-hydroxylase (CYP26) in human promyelocytic leukemia. Am J Hematol. 2002;70:39–47. doi: 10.1002/ajh.10099. [DOI] [PubMed] [Google Scholar]
- Padmanabhan R. Retinoic acid-induced caudal regression syndrome in the mouse fetus. Reprod Toxicol. 1998;12:139–151. doi: 10.1016/s0890-6238(97)00153-6. [DOI] [PubMed] [Google Scholar]
- Perz-Edwards A, Hardison NL, Linney E. Retinoic acid-mediated gene expression in transgenic reporter zebrafish. Dev Biol. 2001;229:89–101. doi: 10.1006/dbio.2000.9979. [DOI] [PubMed] [Google Scholar]
- Quere R, Baudet A, Cassinat B, Bertrand G, Marti J, Manchon L, Piquemal D, Chomienne C, Commes T. Pharmacogenomic analysis of acute promyelocytic leukemia cells highlights CYP26 cytochrome metabolism in differential all-trans retinoic acid sensitivity. Blood. 2007;109:4450–4460. doi: 10.1182/blood-2006-10-051086. [DOI] [PubMed] [Google Scholar]
- Roberts C, Ivins S, Cook AC, Baldini A, Scambler PJ. Cyp26 genes a1, b1 and c1 are down-regulated in Tbx1 null mice and inhibition of Cyp26 enzyme function produces a phenocopy of Di-George Syndrome in the chick. Hum Mol Genet. 2006;15:3394–3410. doi: 10.1093/hmg/ddl416. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev. 2001;15:213–225. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton DN, Sandoval IT, Eisinger A, Chidester S, Ratnayake A, Ireland CM, Jones DA. Up-regulation of CYP26A1 in adenomatous polyposis coli-deficient vertebrates via a WNT-dependent mechanism: implications for intestinal cell differentiation and colon tumor development. Cancer Res. 2006;66:7571–7577. doi: 10.1158/0008-5472.CAN-06-1067. [DOI] [PubMed] [Google Scholar]
- Swindell EC, Thaller C, Sockanathan S, Petkovich M, Jessell TM, Eichele G. Complementary domains of retinoic acid production and degradation in the early chick embryo. Dev Biol. 1999;216:282–296. doi: 10.1006/dbio.1999.9487. [DOI] [PubMed] [Google Scholar]
- Udvadia AJ, Linney E. Windows into development: historic, current, and future perspectives on transgenic zebrafish. Dev Biol. 2003;256:1–17. doi: 10.1016/s0012-1606(02)00083-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zolfaghari R, Ross AC. Cloning of rat cytochrome P450RAI (CYP26) cDNA and regulation of its gene expression by all-trans-retinoic acid in vivo. Arch Biochem Biophys. 2002;401:235–243. doi: 10.1016/S0003-9861(02)00043-7. [DOI] [PubMed] [Google Scholar]
- White JA, Guo Y, Baetz K, Beckett-Jones B, Bonasoro J, Hsu K, Dilworth J, Jones G, Petkovich M. Identification of the retinoic acid-inducible all-trans-retinoic acid 4-hydroxylase. J Biol Chem. 1996;271:29922–29927. doi: 10.1074/jbc.271.47.29922. [DOI] [PubMed] [Google Scholar]
- White JA, Beckett-Jones B, Guo YD, Dilworth FJ, Bonasoro J, Jones G, Petkovich M. cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J Biol Chem. 1997;272:18538–18541. doi: 10.1074/jbc.272.30.18538. [DOI] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterfield L, Cather J, Menter A. Changing paradigms in dermatology: nuclear hormone receptors. Clin Dermatol. 2003;21:447–454. doi: 10.1016/j.clindermatol.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Dobbs-McAuliffe B, Linney E. Expression of cyp26b1 during zebrafish early development. Gene Expr. Patterns. 2005;5:363–369. doi: 10.1016/j.modgep.2004.09.011. [DOI] [PubMed] [Google Scholar]