Abstract
Some neuroscientists argue that detailed maps of synaptic connectivity - wiring diagrams - will be needed if we are to understand how the brain underlies behavior and how brain malfunctions underlie behavioral disorders. Such large-scale circuit reconstruction, which has been called connectomics, may soon be possible, owing to numerous advances in technologies for image acquisition and processing. Yet, the community is divided on the feasibility and value of the enterprise. Remarkably similar objections were voiced when the Human Genome Project, now widely viewed as a success, was first proposed. We revisit that controversy to ask if it holds any lessons for proposals to map the connectome.
Introduction
con.nec.to.mics
Pronunciation: kə-něk-'tO-miks, kə-něk-'tahm-iks
Function: n pl but sing in constr
: a branch of biotechnology concerned with applying the techniques of computer-assisted image acquisition and analysis to the structural mapping of sets of neural circuits or to the complete nervous system of selected organisms using high-speed methods, with organizing the results in databases, and with applications of the data (as in neurology or fundamental neuroscience)— compare GENOMICS
see also con nec tome (From Merriam-Webster Unabridged Dictionary, 2012)
It is possible that some version of this definition will appear in a dictionary at some point. Before that happens, however, the idea of applying an `omics-scale' approach to neural circuit tracing will require considerable vetting. Here, in contrast to common practice in this journal, we take the charge of providing our current opinion seriously. We compare and contrast connectomics with genomics to ask whether mapping neural connections could ultimately have the same kind of value as sequencing genes.
A (very) short history of connectomics
As with almost everything in neuroscience, the idea of mapping circuits can be traced back to Cajal. He enunciated both the neuron doctrine and the law of dynamic polarization [1]. These two ideas, along with Sherrington's electrophysiological analysis of reflexes, provided the rationale for thinking about brain mechanisms in terms of circuits: Neurons were nodes that were electrically connected to each other via two types of wires, axons and dendrites. The dendrites sent information toward the cell body and the axon sent information toward other cells. Many of Cajal's drawings contained small arrows that let the viewer know exactly how he imagined the electricity would course in the circuit (Figure 1a). His drawings, based on the Golgi stain, could not reveal that some of the connections were inhibitory, nor did he imagine backpropagated action potentials or reciprocal synapses. Remarkably, however, many of his detailed conjectures of how information flowed remain unsurpassed today.
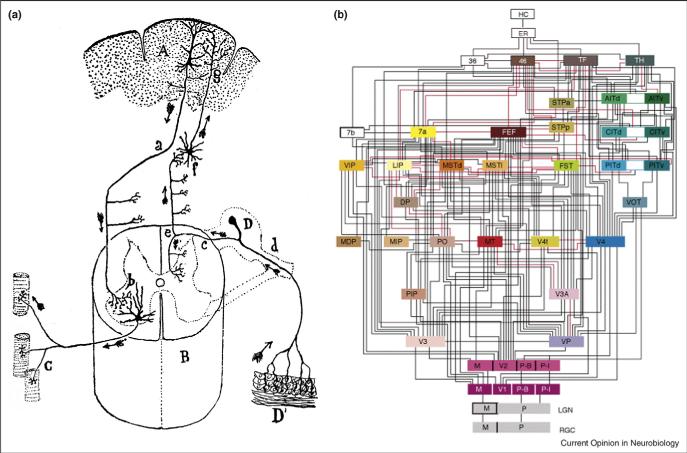
Figure 1.
Scaffolds on which a connectome can be built. (a) A schematic wiring diagram. The best known are Cajal's. An example is his diagram showing the flow of information from peripheral sensory receptors to the spinal cord and brain, then back to motoneurons and muscles. He used arrows to `indicate the direction of descending motor impulses and ascending sensory impressions'. This wiring diagram was pieced together from observations on multiple samples and the direction of current flow was inferred from the structure [35]. (b) A projectome. A well-known example is Van Essen and Felleman's summary of connections among cortical areas associated with vision and some other modalities. Their diagram was compiled from results of tracing and physiological studies by several groups [25].
Is this because they were unsurpassable? Despite Cajal's genius, his views were neither complete nor entirely correct. This is understandable, in that he had to contend with the fact that the Golgi stain labels only a very small fraction of the circuit elements. Cajal therefore had to reconstruct complex patterns of connectivity by mentally combining many connected pairs, each observed in a different piece of tissue. The essential stumbling block, then, was the lack of technologies for observing many or all elements and their connections in a single sample. That limitation has yet to be overcome.
Over much of the twentieth century, extension of Cajal's approach augmented his brain-wide cellular menagerie, and added greatly to the census of connections. The first technology that made a direct attack on the brain's circuit diagram possible was electron microscopy, applied to the nervous system in the 1950s. By the 1960s, attempts began to map microcircuits both in very small invertebrate nervous systems and in tractable regions of the mammalian nervous system. Among the earliest of these was the serial electron microscopic reconstruction of the same identified neuron in multiple isogenic animals in the water flea, Daphnia [2]. The first nominally complete connectome was the reconstruction of all ~300 neurons in the nervous system of the roundworm, C. elegans by White, Brenner and colleagues [3••]. There were several shortcomings in this attempt including an inability to identify synapses as excitatory or inhibitory and the thick sections (75 nm), which caused some processes running along the plane of the sections to be lost. Nevertheless this full reconstruction, reported in 1980, is a benchmark that has yielded the first glimpses of what connectomics might be (Figure 2). This effort has not been matched since.
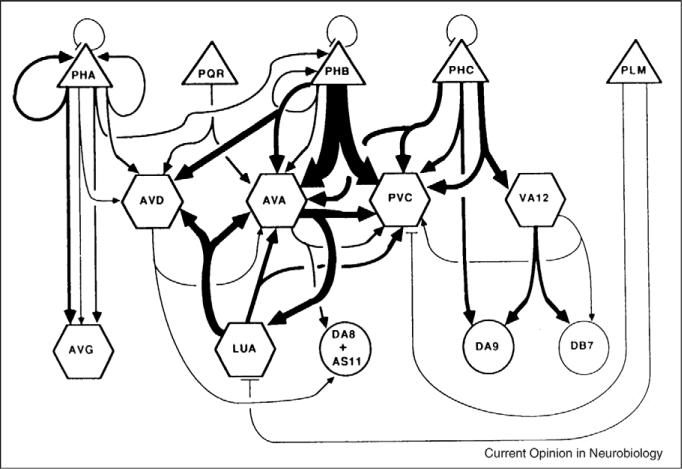
Figure 2.
A connectome. Wiring diagram for the tail of C. Elegans, reconstructed from serial electron micrographs. The width of individual lines is proportional to the relative frequency of synaptic contacts. Lines that end in arrowheads show chemical synapses; lines that end in bars show electrical synapses. Connections to other parts of the nervous system are not included in this diagram, but were reconstructed [36, 3••].
Our opinion is that the next decade will see another period of rapid advance in circuit tracing methods, possibly culminating in elucidation of a mammalian connectome. These advances include innovations in light and electron microscopy, genetically encoded probes, and computational methodology [4•,5•,6•,7•,8•]. The prospect of connectomic data is not being met, however, with unalloyed glee. We hear from some colleagues that it may be of limited utility or at least insufficient utility to justify the enormous expense and effort that will be required. To those of us of a certain age, these objections have a familiar ring. We heard them when the Human Genome Project was proposed.
The Human Genome Project
The DNA cloning revolution of the 1970s led to a flood of technical innovations for DNA manipulation. These included methods for sequencing DNA (Sanger, Maxam, and Gilbert), automating the sequencing (Hunkapillar and Hood), cloning (Olson) and fractionating (Olson and Cantor) large DNA segments, and PCR (Mullis) [9,10]. Once the first full sequence of a genome (phage phiX174) had been determined, many scientists began wondering whether these methods could be combined to sequence larger genomes. The idea of tackling the human genome was proposed in a series of highly publicized meetings beginning in 1985. Scientific, political, and bureaucratic considerations were debated, often acrimoniously, over the next five years, and the Human Genome Project (in caps) was formally inaugurated in 1990 [9-13]. By this time, the program had been expanded to include support for improving sequencing and computational technology, a progression from linkage maps to physical maps to sequence (described below) and sequencing of key model organisms' genomes. The pace of sequencing over the next decade exceeded the project's initial optimistic goals: genome sequences were completed for the bacterium E. coli (1997), the yeast, Saccharomyces cerevisiae (1996), the nematode, C. elegans (1998), the insect, Drosophila melanogaster (2000), and finally the human (2001) [10,14,15]. Since then, the pace has increased still more, with sequences now available for numerous species, ranging from sea urchin to mouse to dog to chimpanzee to eleven species of Drosophila [16]. Sequences have also been obtained for individual humans [17], and the era of the $1000 personal genome is not far off [18].
Objections
The proposal to sequence the human genome was met with skepticism and hostility in many quarters. Contentious issues involved bureaucracy (would the `wrong' agency be chosen to run the program?) and ethics (would the program threaten privacy or access to health care?) as well as scientific merit [9,11,13]. Here we focus on just the latter concerns and ask whether similar objections apply to connectomics.
It will provide limited insight
Of course no one thought genome sequence would be useless, and there was general agreement that it would be helpful for identifying mutations underlying hereditary diseases. On the other hand, there was considerable diversity of opinion on how much it would contribute to a more general understanding of biology. For example, some thought there could be no insight into developmental biology unless one knew when and where genes were expressed. Likewise, understanding cell biology might require knowing the inventory of proteins in a cell, their subcellular localization, and their binding partners.
Although these arguments were entirely true, the blind spot was in failing to understand how much genome sequence could contribute to obtaining this knowledge. Thus, one prominent early critic of the Genome Project argued that “sequencing a disease-causing gene will provide medical insight only in proportion to how much basic biochemistry or physiology is already known.” [19]. Now, even he might agree that that sequence can lead to biochemical, physiological, cell biological, and developmental insights. To give just two examples, sequences of genes' regulatory regions provide the substrate for models of developmental regulation [20], and protein-protein interactions have now been mapped globally by `inter-actome' methods based on genome-wide sets of protein-coding sequences [21].
Similar objections to the connectome are now heard. How can a static wiring diagram help us understand information processing if we do not know the spatiotemporal dynamics of electrical signals in the circuit? How can we understand learning or plasticity without knowing how synaptic efficacy changes with age and experience? We argue by analogy to genetic regulatory networks that circuit diagrams may enable predictions of circuit behavior. Indeed, the increasing success of such inferential reasoning in genomics is encouraging. Moreover it now seems likely that at least some alterations in synaptic efficacy will have a clear physical basis in synaptic size or shape. Such structural flags may serve to reveal parts of circuits that are especially active or silent.
It is merely descriptive
For over a century, descriptive and interventional approaches to knowledge acquisition in the sciences have vied for ascendancy. Those whose manuscripts or grant applications have been termed `merely descriptive' or `not hypothesis-driven' know fully well that the experimental or deductive approach is often deemed superior. In that the genome project was entirely descriptive and based on no specific hypothesis, it is unsurprising that some experimentalists thought it deficient. Rechsteiner [19], for example, viewed the genome project as `mediocre science' because `the finest science is characterized by relevant hypotheses tested with elegantly designed and competently performed experiments.'
In fact, the genome, together with many other `omes' that have succeeded it, have been instrumental in making the case for the value of hypothesis-free data gathering. The simplest argument is that comprehensive, high-quality data sets are essential for developing well thought-out hypotheses. Indeed, molecular biology today relies heavily on genomic data for hypothesis building. A sign of this altered view is the replacement of the derogatory term `descriptive science' by the much more attractive `discovery science', as in Leroy Hood's statement that “discovery science has absolutely revolutionized biology...giving us new tools for doing hypothesis-driven research.” [22].
For neuroscience, it should not be necessary to argue for the value of structural description. Cajal proved that pure description (at least when practiced by a genius) can lead to amazing revelations about the nervous system that equal or surpass any experimental result. Thus, although connectomes are undoubtedly `descriptive', they are no more `merely' descriptive than the observations used by Cajal to generate the neuron doctrine or law of dynamic polarization.
Excessive and unsubstantiated claims
Early critics of the genome project accused its proponents of overselling their project, claiming it was a `holy grail' that would lead to enormous medical breakthroughs along with vast fundamental knowledge [9]. In one sense the accusation was justified: like any other scientists convinced that their vision was worth funding, and faced with the need to convince skeptical funders, they needed to present an optimistic view. Here, we can only say that in our opinion, their claims have turned out to be, if anything, understated. For connectomics, some now claim circuit diagrams will provide insights into a new class of nervous system disorders (connectopathies), information storage and organization, and the seat of consciousness. This may be a bit too sanguine; time will tell. Our sense is that many fundamental questions about the development, aging, and variability of the brain cannot be answered without such data.
A poor use of scarce resources
While the genome project was being debated, the NIH was going through one of its periodic contractions in funding levels. Understandably, many scientists argued that diversion of funds from individual `R01' type grants to large-scale consortia would threaten curiosity-driven basic research as well as graduate education. In fact, funding for genome research never exceeded a few percent of the NIH budget. More important, however, is whether sequencing genomes in a concerted fashion ended up saving money in the long run. We cannot answer this question, but imagine that the cost of sequencing numerous genomic fragments piecemeal, as required by specific experimental agendas, would have ended up exceeding that of the genome project. In addition, no one would doubt that the availability of genome sequence has allowed individual researchers to tackle issues more complex and more interesting than they could have otherwise.
Perhaps a similar situation exists for neural circuits. In the first six months of 2008, more than 200 papers pop up when PubMed is queried with the term `neural circuits'. Many of those studies would presumably benefit from connectome data. At the moment, however, each lab must go it alone.
Full sequence is wasteful
The genome contains several types of sequence, including protein-coding exons, intervening introns, associated regulatory regions, and vast intergenic spans. A frequent criticism of the genome project was that it would be wasteful to spend time and money sequencing the entirety, when 99% of the value would be in the ~1% that encodes proteins. To us, this argument is faulty for two reasons. First, the inefficiency was less than the `1%' figure implies: it would have cost much more than 1% of the total to sequence the protein-coding 1%. Second, non-coding regions have turned out to be more interesting than anyone would have imagined: they contain mysterious conserved regions, microRNAs, distal enhancers, and much more. Perhaps most important, some of these features would surely remain unappreciated today if sequence had not been obtained and made publicly available.
Similarly for the connectome, some argue that analyzing the whole brain would be wasteful. Analysis should be targeted to specific areas, such as the retina or a single cortical column. Given that neuroscientists have found interesting nuggets in virtually every part of the brain of every animal studied, it seems shortsighted to assume that some brain regions are intrinsically more important than others. A second criticism comes from the many neuroscientists who are of the opinion that neural connectivity is largely statistical or stochastic in its fine details. If this were the case, they argue, there is little to be gained in mapping out every last connection. Accordingly, what we should focus on are not microcircuits but long tracts or intra-areal connections [23-25] (Figure 1b). This is a serious objection, but one that can only be evaluated after connectomic information is available.
It cannot be done
We remember naysayers worrying that proponents of the human genome project had bitten off more than they could chew—data sets would ultimately be thousands of megabytes (i.e., gigabytes)! Intelligent robots would be needed to do the nearly infinite amount of pipetting, among others. Obviously these concerns seem ridiculous in retrospect. Technological advances were rapid, spurred partly by the genome project.
Similarly, looking at how difficult the C. elegans reconstruction was, some now say that mapping much larger circuits is infeasible. But at the time, limitations in automation of specimen handling, data collection, and data analysis required that most of the effort be done by hand. The digital revolution had not yet occurred. Given the pace of progress in technology, most of us realize that nothing is really out of bounds, even amassing data sets that might exceed thousands (or even, gulp...millions?) of terabytes.
Lessons
Even those who objected to the Genome Project initially are likely to concede its success now. But how did it succeed? Here we consider lessons learned from the Project, including some that could not have been, or at least were not, fully anticipated at the time.
Things get better
It took around 15 years to sequence the human genome, from the first organized efforts in the late 1980s to the release of a complete sequence in 2003. (The `declaration' in 2000 that the genome had been sequenced actually corresponded to an incomplete `draft'.) Although the first genome took 15 years, now the whole sequence can be obtained in a matter of days, and it may soon be a matter of hours. For one of us, it took a month to obtain 500 base pairs of finished sequence in 1983, with gels being poured and loaded manually and nucleotide calls made by a student sitting with a ruler at a light box. Now a massively parallel machine generates 20 megabases in 5 h and delivers it to the spreadsheet of a student sitting in an office perhaps a continent away [17,26]. Even making allowances for post-processing, this is a speed-up of over 106-fold.
Likewise the C. elegans connectome would probably take an order of magnitude less time now than it did in 1980, given currently available automatic microscopes, digital cameras, and image acquisition, montaging and registration programs. Ongoing developments in these and other aspects of automated high throughput imaging and analysis promise to accelerate connectomics by at least another order of magnitude. Thus, within a few years, a new C. Elegans connectome will probably be reconstructed within a month. And given the extent to which progress in genome technology exceeded predictions, it seems not unlikely that more orders of magnitude will follow.
The value of multiple methods
In the end, two groups announced simultaneously that they had sequenced the human genome. One was a large public consortium, the other a private venture [14,15]. They used very different methods—placing sequences of large clones on an excellent physical map in one case, using computational methods to assemble long sequences from oversampled, overlapping short stretches (`whole-genome shotgun sequencing') in the other. The competition between these two groups was intense, but in retrospect, beneficial in some respects. First, given human nature, scientific competitions accelerate the rate of progress (for example, the double helix, or the `space race' in the 1950s). Second, different approaches may have complementary strengths that are not apparent in advance. For example, whole-genome shotgun sequencing did work well for the Drosophila genome, but many claimed that its apparent success for the human genome depended on the availability of maps and contigs provided by the public effort [27].
The lesson for connectomics is that there is no reason to decide now on the `best' way to trace circuits. The avalanche of new imaging approaches is not likely to slow anytime soon. For example the advent of super-resolution light microscopy may permit some tracing to be accomplished with fluorescence for which electron microscopy was previously required [5•,28,29]. Indeed, technologies that initially seem like alternative solutions such as optical and electron microscopic imaging may be combined.
Start small(er)
The human genome was built on technical and conceptual insights obtained by sequencing, the smaller genomes of viruses, bacteria, fungi, and C. elegans [10]. For the connectome, one might likewise imagine projects that would not only be immensely valuable themselves but also provide ways to develop, test, and optimize new methods.One idea would be to revisit C. elegans, whose wiring diagram has been (mostly) determined but only in the hermaphrodite [3••]. It would be fascinating and important to know how much variation exists among multiple isogenic individuals, and we would also learn how much faster we can now accomplish the same feat. Alternatively, one could focus initially on tractable or well-studied regions of a mammalian nervous system. For example, the circuit diagram of the retina could be put to immediate use in testing hypotheses about information processing derived from physiological measurements that have already been made. At the same time, such a project could help us see how new technologies can be combined into a seamless pipeline for high-throughput reconstruction.
Proceed in steps
The human genome was not sequenced by beginning with a telomere of Chromosome 1 and continuing through to the end of the X chromosome. Instead, it was assembled in a series of well thought-out steps that proceeded from coarse- to fine-grained [9]. First, it made use of a decades-long effort to generate a genetic or linkage map. Next, libraries of long DNA clones were arranged in order by sequencing small regions at their overlapping ends, and this physical map was aligned with the genetic map. Next, clones were subdivided and sequenced, and the pieces were reassembled. The clone-by-clone sequences were then arranged using the physical map as a guide, to generate `draft' sequence. Finally, gaps and areas of uncertainty were targeted to generate `finished' sequence.
For connectomes of any but the smallest organisms,such a step-wise approach will also be required. Distinct imaging modalities may be useful. Light microscopic tracttracing and MRI methods can be used to map long tracts and inter-areal connections [23-25]. In some regards, these so-called `projectomes' provide useful skeletons, much like physical maps of genomes, upon which to build a more detailed map. Higher resolution light microscopy may be useful for tracing axonal and dendritic arbors, and inventorying cell types. Super-resolution optical imaging or electron microscopy will surely be needed to identify synapses, and only ultrastructural methods are currently sufficient to document details within synapses. This multi-scale imaging approach allows fine details to be built on a coarser but more global map.
It takes a village
Much of the science we admire the most has been done by individuals or small groups. For molecular biology, one thinks of Watson and Crick, Brenner, and Benzer. By contrast, the papers reporting the human genomes each had >200 co-authors [14,15]. Moreover, the publicly funded effort, as well as consortia responsible for sequencing genomes of numerous other species involved groups at dozens of sites or institutions. The multi-institutional consortia increased the speed by parallel processing to generate huge data sets. In addition, different expertise in different institutions allowed a pipeline to be developed.
Even more than molecular biology, neurobiology has been dominated by small or personalized science: think of Hodgkin and Huxley, Katz, Neher, Sakmann, Hubel, and Wiesel.Yet the requisite transgenic lines, optical and electron microscopy imaging regimens, sectioning tools, immunohistochemistry, data storage, and data analysis almost certainly will not all be found in the same institution. Multi lab consortia will probably be a central feature of this effort from its inception. Although we are beginning to see a few `big' neurobiology projects [30,31], a connectome project would require a culture shift in the field.
Unanticipated benefits
The genome project and its sequelae have been of incalculable benefit to the biotechnology and pharmaceutical industries. On a smaller yet significant scale they provided a market for machines and computers, and an impetus to develop better ones. In short, the genome project has had economic benefits that were unanticipated as well as biomedical benefits that extended far beyond those its early proponents hoped for. For connectomics, too, even the anticipated benefits are still being debated, so it would be absurd (as well as oxymoronic) to discuss unanticipated ones. Nonetheless, it is a safe bet that results will be useful in ways we cannot yet imagine.
Success succeeds
In 1986, David Baltimore `shivered at the thought' that the genome project was gaining momentum [13]. In 1988, he judged that the increased momentum was `a ploy to raise money...justified for its public relations value, not its scientific value [12].' In an editorial accompanying the release of the draft human genome in 2001, he felt differently. He noted that "chills ran down my spine [32]" when he read the paper, but this time they were good chills, not bad ones. Now, he realized, "Biology today enters a new era" allowing new answers to fundamental questions such as, "Mommy, why am I different from Sally". We would like to imagine that 10 or 20 years from now, Professor Baltimore might see that the fruits of connectomics will also help edify Sally's sibling.
Differences
So far, we have outlined ways in which the history of genomics may help predict the future of connectomics. Given the success of the genome project, these analogies lead to a generally optimistic view. One needs to be cautious in drawing parallels, however, because there are fundamental differences between the two, as well as uncertainties about the latter, which will make mapping the connectome an even more daunting task than mapping the genome.
Dimensionality
One of the reasons the genome was tractable was that DNA is one-dimensional, a linear sequence of nucleotides. A connectome, by contrast, comprises information in three dimensions. Moreover the molecular identity, the fine structural details (like number of vesicles at synapses, spine shape etc.) add additional dimensions to the data set. These problems are especially daunting if one realizes that, unlike the genome, there is probably no level of resolution that actually completes the connectome. While one scientist might like a diagram that shows connections between neurons, another might want to know the number of vesicles per synapse and a third the molecular subtypes of neurons involved. Ultimately we imagine that a full connectome would contain not only a connectional wiring diagram but also expose the molecular heterogeneity and functional variations of the interconnections.
Variability
Classical genetics made clear that heritable differences among individuals resulted from differences among their genomes. As methods for DNA analysis improved, increased amounts of inter-individual variability were found, so that by the mid-1980s, polymorphisms at an array of genomic sites were being used as `DNA finger-prints' to identify or exonerate criminal suspects. Nonetheless, at no point was there a serious challenge to the idea that the genome of any one human would provide a useful guide to the genome of all other humans. This faith in constancy was well placed: the current estimate is that sequence similarity between the genomes of any two people is about 99.9% [17]. Thus, in practical terms, any single genome is a useful `reference genome'.
For connectomes, the situation is much less clear. In only a few cases, nearly all from invertebrates, have high-resolution maps been obtained for a defined region of multiple individuals. In the few comprehensive reconstructions of invertebrates (Daphnia and C. Elegans) the main processes of neurons varied little from animal to animal, but even in isogenic animals, fine details of shape varied among individuals [2,3••,36]. More recent analysis in insects corroborates the idea that single neurons have recognizable arbors but in fine details can be different [33,34]. Few of these studies assayed synaptic connections, but it seems likely that there will be variability there as well. For vertebrates, we do not even have, with rare exceptions (e.g. the Mauthner cell) the concept of identified neurons upon which to build, and we know almost nothing about the degree to which neuronal shape or connectivity differs between individuals. In our view, this is the most serious challenge that connectomics will face; hopefully early partial connectomes will reveal what is in store.
Stability
We know of no challenges to the view that having sequenced James Watson's genome past year [17], there is no reason whatsoever to sequence it again next year. By contrast, the connectome surely changes with maturation and aging and is likely to change even during adulthood, in response to experience. All of these changes would be important to understand but they initially are confounds in obtaining connectomes.
Conclusions
We envision a time when the idea of studying the brain's function without knowing how its cells are interconnected will seem as absurd as the idea of studying genetics without knowing genome sequence. Circuit information will be required if we are to understand how differences in brains underlie differences in behaviour—among healthy individuals, in disease, and within single individuals as they mature, write reviews and age. It is premature to predict that a `connectome' of the brain will ever exist in the sense that a genome exists. Issues of scale, variability and stability may be insurmountable. Nonetheless, at some point, extensive, publicly available information on neural circuits may do for neuroscience, neurology and psychiatry what genome sequence has done for many other areas of biology and medicine.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
••of outstanding interest
- 1.Ramón y, Cajal S. Histology of the nervous system. Oxford University Press; 1995. Originally published in 1909, translated into French in 1928 and translated from French to English by N Swanson and LW Swanson. [Google Scholar]
- 2.Macagno ER, Levinthal C, Sobel I. Three-dimensional computer reconstruction of neurons and neuronal assemblies. Annu Rev Biophys Bioeng. 1979;8:323–351. doi: 10.1146/annurev.bb.08.060179.001543. [DOI] [PubMed] [Google Scholar]
- 3••.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode C. elegans. Philosophical Transactions of the Royal Society of London - Series B: Biological Sciences. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]; Published over 20 years ago, this remains the only connectome to be reported to date. It was compiled from partial reconstructions of five individual worms
- 4•.Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review, along with references [5•, 6•, 7•, 8•] provides summaries of recent methodological advances that may soon allow completion of more complex connectomes that of C. Elegans, as well as connectomes from multiple individuals of a species
- 5•.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]; See reference [4•]. This review, from a pioneer in the new field of nanoscopy, shows how optical microscopes can attain resolution previously limited to electron microscopy
- 6•.Briggman KL, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr Opin Neurobiol. 2006;16:562–570. doi: 10.1016/j.conb.2006.08.010. [DOI] [PubMed] [Google Scholar]; See reference [4•]. This manuscript reviews Denk's imaginative new approach to high-throughput serial section electron microscopy, along with other related developments
- 7•.Luo L, Callaway EM, Svoboda K. Genetic dissection of neural circuits. Neuron. 2008;57:634–660. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; See reference [4•]. Three experts in developing and applying new genetic tools for visualizing neurons and their connections provide an exhaustive review of this fast-moving field
- 8•.Smith SJ. Circuit reconstruction tools today. Curr Opin Neurobiol. 2007;17:601–608. doi: 10.1016/j.conb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; See reference [4•]. In this review, Stephen Smith describes new techniques that will make possible molecular characterization of synaptic subtypes in large, high-resolution data sets
- 9.Cook-Deegan R. The Gene Wars. WW Norton; New York: 1994. [Google Scholar]
- 10. http://www.ornl.gov/sci/techresources/Human_Genome/project/hgp.shtml.
- 11.Roberts L. The human genome. Controversial from the start. Science. 2001;291:1182–1188. doi: 10.1126/science.291.5507.1182a. [DOI] [PubMed] [Google Scholar]
- 12.Burris J, Cook-Deegan R, Alberts B. The Human Genome Project after a decade: policy issues. Nat Genet. 1998;20:333–335. doi: 10.1038/3803. [DOI] [PubMed] [Google Scholar]
- 13.Lewin R. Proposal to sequence the human genome stirs debate. Science. 1986;232:1598–1600. doi: 10.1126/science.3715466. [DOI] [PubMed] [Google Scholar]
- 14.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 16.Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 17.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 18.Bonetta L. Getting up close and personal with your genome. Cell. 2008;133:753–756. doi: 10.1016/j.cell.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Rechsteiner MC. The Human Genome Project: misguided science policy. Trends Biochem Sci. 1991;16:455–457. 459. doi: 10.1016/0968-0004(91)90178-x. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Tabou de-Leon S, Davidson EH. Gene regulation: gene control network in development. Annu Rev Biophys Biomol Struct. 2007;36:191. doi: 10.1146/annurev.biophys.35.040405.102002. [DOI] [PubMed] [Google Scholar]
- 21.Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum Mol Genet. 2005;14(Spec No 2):R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- 22.Service RF. The human genome. Objection #1: big biology is bad biology. Science. 2001;291:1182. doi: 10.1126/science.291.5507.1182b. [DOI] [PubMed] [Google Scholar]
- 23.Kasthuri N, Lichtman JW. The rise of the `projectome'. Nat Methods. 2007;4:307–308. doi: 10.1038/nmeth0407-307. [DOI] [PubMed] [Google Scholar]
- 24.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- 26.Hutchison CA., 3rd DNA sequencing: bench to bedside and beyond. Nucleic Acids Res. 2007;35:6227–6237. doi: 10.1093/nar/gkm688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterston RH, Lander ES, Sulston JE. On the sequencing of the human genome. Proc Natl Acad Sci U S A. 2002;99:3712–3716. doi: 10.1073/pnas.042692499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 30.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 31.Stone JL, O'Donovan MC, Gurling H, Kirov GK, Blackwood DH, Corvin A, Craddock NJ, Gill M, Hultman CM, Lichtenstein P, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baltimore D. Our genome unveiled. Nature. 2001;409:814–816. doi: 10.1038/35057267. [DOI] [PubMed] [Google Scholar]
- 33.Chen BE, Kondo M, Garnier A, Watson FL, Püettmann-Holgado R, Lamar DR, Schmucker D. The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila. Cell. 2006;125:607–620. doi: 10.1016/j.cell.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 34.Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr, Luo L. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 2007;128:1187–1203. doi: 10.1016/j.cell.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.y Cajal SRamon. New Ideas on the Structure of the Nervous System in Man and Vertebrates translated from French to English by N Swanson and LW Swanson. MIT Press; Cambridge Massachusetts: 1990. [Google Scholar]
- 36.Hall DH, Russell RL. The posterior nervous system of the nematode Caenorhabditis elegans: serial reconstruction of identified neurons and complete pattern of synaptic interactions. J Neurosci. 1991;11:1–22. doi: 10.1523/JNEUROSCI.11-01-00001.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]