Abstract
The GAAA tetraloop-receptor is a commonly occurring tertiary interaction motif in RNA. This motif usually occurs in combination with other tertiary interactions in complex RNA structures. Thus, it is difficult to measure directly the contribution that a single GAAA tetraloop-receptor interaction makes to the folding properties of an RNA. To investigate the kinetics and thermodynamics for the isolated interaction, a GAAA tetraloop domain and receptor domain were connected by a single-stranded A7 linker. Fluorescence resonance energy transfer (FRET) experiments were used to probe intramolecular docking of the GAAA tetraloop and receptor. Docking was induced using a variety of metal ions, where the charge of the ion was the most important factor in determining the concentration of the ion required to promote docking ([Co(NH3)63+] ≪ [Ca2+], [Mg2+], [Mn2+] ≪ [Na+], [K+]). Analysis of metal ion cooperativity yielded Hill coefficients of ≈ 2 for Na+- or K+-dependent docking versus ≈ 1 for the divalent ions and Co(NH3)63+. Ensemble stopped-flow FRET kinetic measurements yielded an apparent activation energy of 12.7 kcal/mol for GAAA tetraloop-receptor docking. RNA constructs with U7 and A14 single-stranded linkers were investigated by single-molecule and ensemble FRET techniques to determine how linker length and composition affect docking. These studies showed that the single-stranded region functions primarily as a flexible tether. Inhibition of docking by oligonucleotides complementary to the linker was also investigated. The influence of flexible versus rigid linkers on GAAA tetraloop-receptor docking is discussed.
RNA is an essential biological molecule that functions in numerous cellular processes, including catalyzing such critical reactions as protein synthesis and RNA splicing (1–3). To achieve its various functions, RNAs must adopt complex, well-defined three-dimensional structures, and determining how these RNA structures are formed and stabilized is critical to understanding their biological function. The process by which RNAs fold to these three-dimensional structures is complex. Unlike most protein folding, RNA folding often proceeds by a hierarchical pathway, where the secondary structure forms prior to and independently of the tertiary structure, and the tertiary structure is stabilized by interactions between the secondary structural elements (4).
A variety of RNA tertiary interaction motifs have been identified, including loop-loop interactions between hairpin or internal loops, A-minor interactions, and pseudoknots (5–10). One of the most frequently occurring tertiary motifs in RNAs is the interaction of a GNRA hairpin tetraloop domain with an asymmetric internal loop receptor domain (11). The best-characterized example of this motif is the interaction between a GAAA tetraloop and 11-nucleotide tetraloop receptor, which has been shown to stabilize the folded structures of group I and group II introns and the specificity domains of RNase P (11–14). Structures for the free GAAA tetraloop and free receptor have been solved by NMR1 spectroscopy (15–17). The structure of the GAAA tetraloop-receptor complex has been solved by X-ray crystallography as part of the P4-P6 domain of the Tetrahymena group I ribozyme (18) and the specificity domain of RNase P (19) and has been further characterized by NMR spectroscopy (20). These structures show the tetraloop contacting the minor groove of the receptor domain, with numerous stabilizing hydrogen bonding and base stacking interactions.
The folded structures of natural RNAs are typically stabilized by multiple tertiary contacts. For example, the tertiary fold of the P4-P6 domain is stabilized by a GAAA tetraloop-receptor interaction and an interaction between the A-rich bulge of the P5a domain with the P4 helix (18), and additional tertiary contacts are formed between this domain and other regions in the full Tetrahymena ribozyme (21). These multiple contacts make it difficult to determine the contribution of a single tertiary interaction to the stability of the folded structure. Previous studies have indirectly probed the GAAA tetraloop-receptor interaction by measuring the folding of P4-P6 domains (12, 22) and group II introns (23) that contain changes in the sequence of the tetraloop and/or receptor.
Several studies have investigated the GAAA tetraloop-receptor interaction in the absence of other tertiary interactions. For example, a simple bimolecular GAAA tetraloop-receptor system has been shown to associate with a KD of 0.4 mM at 125 mM Mg2+ (24). Molecules termed “tecto RNAs” have been designed to form homodimers stabilized by two intermolecular GAAA tetraloop-receptor interactions, and these homodimers form with KDs as tight as 4 nM at 15 mM Mg2+(25, 26). We previously measured the Mg2+ dependence for the equilibrium and kinetics for intramolecular docking of a GAAA tetraloop with its receptor using a construct where these two domains are connected by an A7 single-stranded linker, and the study yielded a docking equilibrium constant of ≈ 20 at 10 mM Mg2+ (27).
Mg2+ is critical for folding of most RNAs because these ions help neutralize charge-charge repulsions in compact RNA structures. In vitro, other divalent cations can often substitute for Mg2+. For example, the Tetrahymena ribozyme shows high activity in the presence of Mn2+ ions, whereas this RNA appears to have a native fold but is not catalytically active with Ca2+ ions (28). Not all RNAs absolutely require Mg2+ for folding, and RNAs such as the hairpin and hepatitis delta virus ribozymes are catalytically active in the absence of Mg2+ (but only at high monovalent ion concentrations) (29). Thus, RNA folding has a complex dependence on metal cations. The metal dependence can be particularly complex in larger RNAs, which have multiple tertiary interactions that can interact with metal ions in different ways.
In this work we investigate the metal ion dependence of a simple tertiary interaction, the GAAA tetraloop-receptor. As previously discussed, connecting the GAAA tetraloop and receptor with a flexible single-stranded linker allows intramolecular docking to be studied independently of other tertiary interactions (27). The effect of various multivalent and monovalent cations on GAAA tetraloop-receptor docking was studied here by ensemble FRET techniques. The results show that docking is stabilized by a variety of cations, but with different apparent affinities and cooperativities. The kinetics of GAAA tetraloop-receptor docking were investigated using ensemble stopped-flow fluorescence experiments, demonstrating a significant apparent activation energy for docking. The sequence and length of the single-stranded region were varied in this RNA, and the effects on the docking equilibrium and kinetics were measured using both single-molecule and ensemble fluorescence methods. These results show that the single-stranded linker simply tethers the two domains together but does not orient them for docking. These data are compared with previous studies where the linker optimally orients the tetraloop and receptor domains. Ensemble and single-molecule FRET studies were also performed where oligonucleotides complementary to the single-stranded linker were used to reversibly inhibit docking, and potential applications of these inhibitors are discussed. The results here lead to a better understanding of the factors that affect the stability and folding of the frequently occurring GAAA tetraloop-receptor tertiary interaction.
EXPERIMENTAL PROCEDURES
RNA Constructs
Chemically synthesized 5′-amino modified RNA oligonucleotides were purchased from Dharmacon (Lafayette, CO) and the 5′-biotin modified DNA oligonucleotide from Integrated DNA Technologies (Coralville, IA)2. The sequence and secondary structure of the A7 construct used here is shown in Figure 1. Fluorescence labeling of the RNAs was achieved by coupling Cy3 (FRET donor) or Cy5 (FRET acceptor) N-succinimidyl esters (Amersham Biosciences) to amino groups on the 5′ end of the oligos. All oligos were purified by microfiltration to remove unreacted Cy3 or Cy5, followed by C-18 reverse-phase HPLC. Stock solutions of the GAAA tetraloop-receptor complexes were made by mixing 1 μM of the appropriate Cy3-RNA strand, 1.5 μM of the complementary Cy5-RNA strand, and 2.0 μM of the biotin-DNA strand, heating to 70 °C for 3 min and slow cooling to room temperature. Unless otherwise noted, all experiments were performed at 25 °C using standard buffer conditions of 50 mM HEPES pH 7.5, 100 mM NaCl, 0.1 mM EDTA.
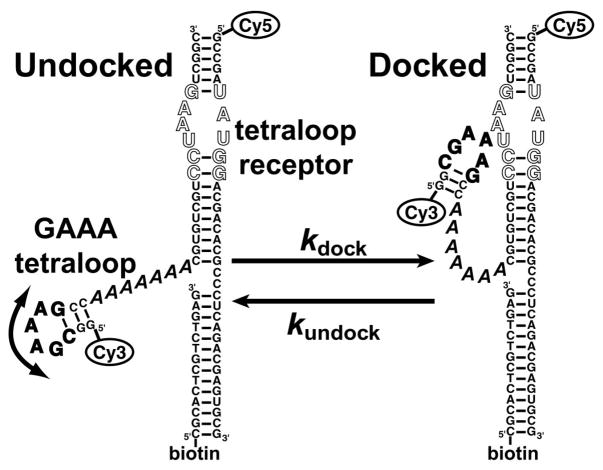
Figure 1.
RNA construct for probing intramolecular GAAA tetraloop-receptor docking. The GAAA tetraloop (bold) and the tetraloop-receptor (outlined) are connected by an A7 single-stranded linker (italics). The curved arrow illustrates the freedom of motion between the tetraloop and receptor domains allowed by the single-stranded linker. The docking and undocking reactions were monitored by FRET between the Cy3 and Cy5 fluorophores, where the Cy3–Cy5 distance is ≈ 35 Å in the docked state (18) and up to ≈ 70 Å in the undocked state. The 5′-biotin strand facilitates immobilization on a surface via interaction with streptavidin.
Docking of the GAAA Tetraloop-Receptor Monitored by Ensemble FRET
Fluorescence spectra were recorded for 20 nM RNA solutions using a Photon Technology International spectrofluorometer. Excitation was set at 500 nm (4 nm bandwidth) and emission measured in 1 nm steps from 525 to 700 nm (8 nm bandwidth). The FRET efficiency, EFRET, for each fluorescence spectrum was approximated as EFRET = IA/(ID + IA), where IA is the total acceptor intensity and ID is the total donor intensity. ID and IA were determined by summing the intensity at each wavelength from 549–620 and 655–700 nm, respectively.
Metal cation titrations were performed by manually pipeting small volumes of a concentrated solution of the appropriate metal chloride directly into the cuvettes, mixing for ≈ 1 min with a magnetic stir bar, and recording the fluorescence spectra. The resulting EFRET values determined at various concentrations for each metal ion were fit to the equation
| (1) |
where [M] is the metal ion concentration, ei is the FRET efficiency with no metal ion added, Δe is the maximum change in FRET efficiency induced by addition of metal ion, K1/2 is the apparent dissociation constant for the cation, and n is the Hill coefficient. This fit assumes the simple metal binding model:
such that the observed EFRET results from the population-weighted EFRET of the undocked and docked states, with no significant contribution from intermediate states.
The procedure for titration of the oligonucleotides complementary to the single-stranded linker was essentially the same as for the metal cation titrations. The resulting FRET data was fit assuming the following reaction model
such that
| (2) |
where ei is the EFRET prior to addition of any oligonucleotide inhibitor, Δe is the maximum change in EFRET upon addition of the inhibitor (in this case a negative number), and KD is the apparent dissociation constant for binding of the oligo to the RNA construct.
Ensemble Stopped-flow Fluorescence Experiments of GAAA Tetraloop-Receptor Docking Kinetics
Ensemble measurements of docking kinetics were performed on an Applied Photophysics model SX.17MV stopped-flow fluorometer. All experiments were performed under standard buffer conditions. Docking was initiated by rapid mixing with various concentrations of Mg2+. For each experiment, 4000 data points were collected over 100 ms, with a 2 ms dead time between mixing and collection. At least four runs were averaged for each data point. Fluorescence was excited at 532 nm, which primarily excites the Cy3 donor. Measurement of emission corresponding to the Cy5 acceptor was achieved using a 645 nm long pass filter. The increase in FRET efficiency from docking resulted in a time-dependent increase in Cy5 emission. These data were used to determine the apparent rate constant for docking, kobs, by fitting the to the equation:
| (3) |
Where FA is the observed acceptor fluorescence as a function of time, FAo is acceptor fluorescence at t = 0, and ΔFA is the change in fluorescence between t = 0 and t = ∞ The temperature dependence of kobs was measured at 10 mM final Mg2+ concentration and fit to the Arrhenius equation to determine the apparent activation energy:
| (4) |
Where Ea is the apparent activation energy, R is the gas constant, T is the absolute temperature, and A is the Arrhenius pre-exponential factor. The Mg2+ dependence of kobs was also measured at a constant temperature of 23 °C.
Single-molecule FRET Experiments on the GAAA Tetraloop-Receptor
Single-molecule fluorescence was measured on optically resolved single molecules using a water-immersion confocal fluorescence microscope system with excitation at 532 nm, described elsewhere (27). To enable long observation times for each molecule (≈ 2–30 s, limited by dye photobleaching), the RNA samples (initially at ≈ 25 pM) were immobilized on a glass microscope cover slip via biotin-streptavidin association. A nano-piezo scanning stage was used to locate molecules on the glass, then donor and acceptor emission were collected simultaneously to determine EFRET in real time, where EFRET = IA/(ID + IA). Single-step photobleaching of the dyes confirmed the observation of single molecules. For each RNA construct, time-resolved EFRET traces were recorded for 30–50 molecules. The rate constants kdock and kundock were calculated from the time-resolved FRET data as previously described (27). Raster-scanned images of surface immobilized single molecules were collected as previously described (27). All single-molecule experiments were performed at ambient temperature in standard buffer with an enzymatic oxygen scavenger to reduce photobleaching (30).
RESULTS
Effect of Various Metal Cations on GAAA Tetraloop-Receptor Docking
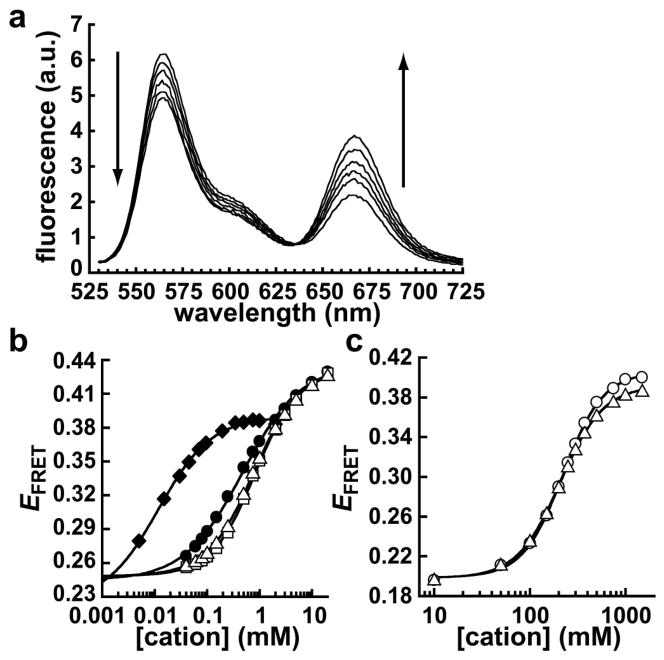
The docked state for this GAAA tetraloop-receptor tertiary interaction is stabilized by Mg2+ ions (24, 25, 27). Docking of the GAAA tetraloop-receptor construct with an A7 linker (Figure 1) was monitored by FRET, where there is an increase in acceptor (Cy5) fluorescence and decrease in donor (Cy3) fluorescence upon Mg2+-induced docking (Figure 2a). These spectra have a clear isosbestic point, which is consistent with a two-state system. The FRET efficiency, EFRET, as a function of Mg2+ concentration is shown in Figure 2b. The EFRET data were fit to Equation 1 to yield the apparent dissociation constant for binding of Mg2+ and the Hill coefficient (Table 1). This fluorescence assay was used to monitor the metal ion dependence for docking with two other divalent cations, Ca2+ and Mn2+ (Figure 2b). All three divalent cations promoted GAAA tetraloop-receptor docking with similar sub-millimolar K1/2 values and Hill coefficients close to 1 (Table 1).
Figure 2.
Metal ion-induced docking of the GAAA tetraloop-receptor. (a) Ensemble fluorescence spectra at Mg2+ concentrations between 0 and 10 mM, with fluorescence excited at 500 nm. The fluorescence emission from 530–600 nm is primarily from the Cy3 (donor), whereas emission greater than ≈ 640 nm is from the Cy5 (acceptor). Arrows indicate the direction of change in the donor and acceptor regions of the spectra as Mg2+ concentration increases. (b) Ensemble FRET measurements of GAAA tetraloop-receptor docking induced by Mg2+ (open triangles), Ca2+ (open squares), Mn2+ (filled circles), or Co(NH3)63+ (filled diamonds). (c) Ensemble FRET measurements of GAAA tetraloop-receptor docking induced by Na+ (circles) or K+ (triangles). Errors bars are approximately within the size of the symbols. The solid lines in (b) and (c) represent fits to Equation 1 (see Table 1).
Table 1.
Metal ion dependence of GAAA tetraloop-receptor dockinga
| Cation | K1/2 (mM) b | nb |
|---|---|---|
| Mg2+ | 0.87 ± 0.11 | 0.95 ± 0.05 |
| Ca2+ | 0.83 ± 0.02 | 1.05 ± 0.05 |
| Mn2+ | 0.44 ± 0.07 | 0.77 ± 0.05 |
| Co(NH3)63+ | 0.017 ± 0.03 | 0.99 ± 0.1 |
| Na+ | 220 ± 9 | 2.04 ± 0.1 |
| K+ | 200 ± 19 | 1.89 ± 0.1 |
Measured for the A7 construct.
Values for each cation represent the mean of at least three ensemble fluorometer-based experiments and the errors are one standard deviation.
To test the requirement for direct coordination between metal cations and the RNA, docking was also measured with Co(NH3)63+, an analogue for hydrated Mg2+ (31, 32). The data show that Co(NH3)63+ effectively promotes tetraloop-receptor docking (Figure 2b), with a K1/2 of 0.017 mM that is ≈ 50-fold tighter than the divalent cations (Table 1).
The monovalent cations K+ and Na+ also promote docking, with K1/2 values of ≈ 200 mM (Figure 2c, Table 1). Thus, there is a significant population of docked RNAs at physiological monovalent concentrations, which is consistent with our previous single-molecule observations (27). In contrast to the other cations, the RNA binds Na+ and K+ cooperatively, with Hill coefficients ≈ 2 (Table 1).
Activation Energy and Docking Kinetics for the GAAA Tetraloop-Receptor
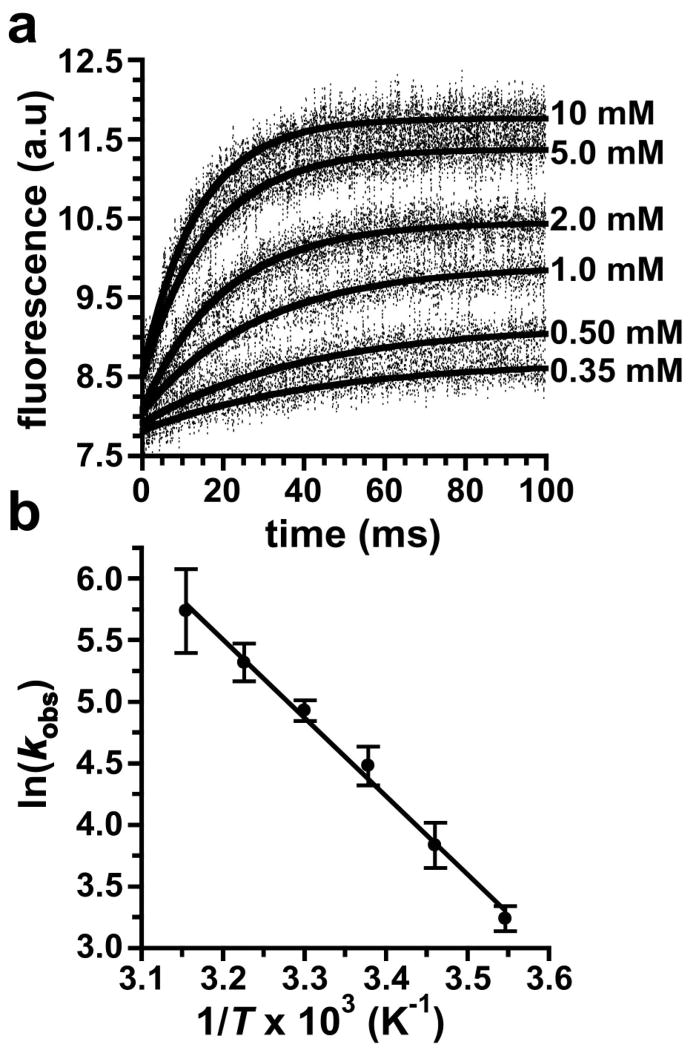
The kinetics for tetraloop-receptor docking were measured using ensemble stopped-flow fluorescence experiments, with docking initiated by rapid mixing with Mg2+. The time-resolved fluorescence data fit well to a single exponential (Equation 3), yielding an apparent first-order rate constant for docking, kobs. To determine the effect on kobs, experiments were performed with various Mg2+ concentrations at 23 °C (Figure 3a, Table 2). These stopped-flow data do not resolve docking and undocking events, so the measured kobs corresponds to the approach to equilibrium, kdock + kundock. As seen in Table 2, the kobs values measured here are all within 1.5-fold of the kdock + kundock values previously measured by single-molecule techniques (27).
Figure 3.
Kinetics of GAAA tetraloop-receptor docking from stopped-flow fluorescence measurements. (a) Time dependence of the acceptor fluorescence after rapid mixing with Mg2+, with the final Mg2+ concentrations given next to each curve. The solid lines represent fits to Equation 3 (see Table 2). (b) Arrhenius plot (Equation 4) of the temperature dependence of kobs at 10 mM Mg2+. An Ea for docking of 12.7 ± 2.6 kcal/mol was determined from a linear fit of the data (solid line).
Table 2.
Mg2+ dependence of GAAA tetraloop-receptor docking kineticsa
| [Mg2+] (mM) | kobsb (s−1) | kdock + kundockc (s−1) |
|---|---|---|
| 0.35 | 26 ± 4 | 18.2 ± 0.3 |
| 0.50 | 29 ± 5 | 24.5 ± 0.5 |
| 1.0 | 46 ± 12 | 37.3 ± 1.3 |
| 2.0 | 62 ± 13 | 44.1 ± 1.3 |
| 5.0 | 70 ± 12 | 55.4 ± 1.1 |
| 10 | 90 ± 14 | 66.4 ± 1.9 |
Measured for the A7 construct.
Determined from ensemble stopped-flow FRET experiments.
Previously determined from single-molecule FRET experiments (27).
To determine the apparent Ea for docking, kobs was measured between 9 and 44 °C at 10 mM Mg2+. The resulting Arrhenius plot yields an Ea of 12.7 ± 2.6 kcal/mol (Figure 3b). Because kobs is the approach to equilibrium, this value does not represent a true Ea. However, at room temperature kdock ≫ kundock (27), so the kobs from the stopped-flow data is ≈ kdock. The docking equilibrium will be shifted towards the undocked state at higher temperatures, making kobs greater than kdock. As a result, the Ea determined from kobs (Figure 3) may be slightly larger than the true value, and 12.7 kcal/mol represents an upper limit for the Ea for docking.
Effect of the Length and Composition of the Single-Stranded Linker on GAAA Tetraloop-Receptor Docking
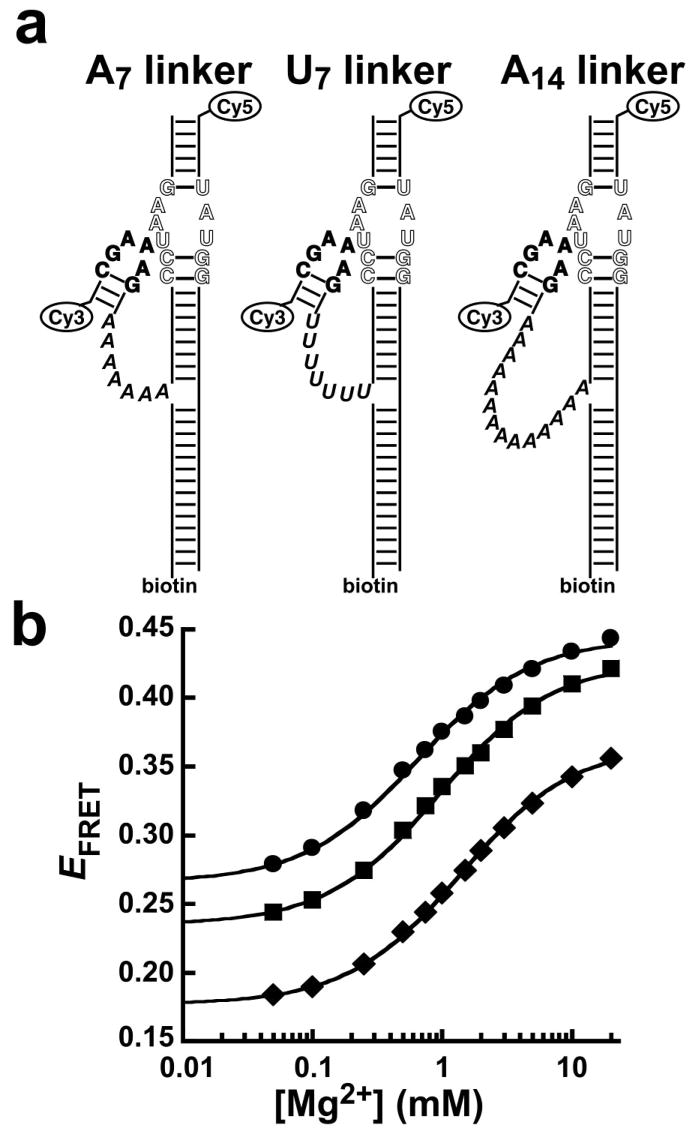
Two other GAAA tetraloop-receptor constructs were investigated by ensemble FRET experiments. In these constructs, the single-stranded linker was changed from A7 to U7 or A14 (Figure 4a), and the effect on tetraloop-receptor docking was tested. The Mg2+ dependence of EFRET was measured for each construct to determine the K1/2 (Figure 4b, Table 3). The Hill coefficient was close to 1 for all three constructs, indicating noncooperative binding of Mg2+ ions associated with docking. Therefore, for more direct comparison, the K1/2 values in Table 3 represent data fits with n defined as 1. The K1/2 values for the three constructs are similar, ranging from 0.63 to 1.4 mM (Table 3). Thus, the linkers have a very small effect on the free energy for the docking reaction (ΔΔGoMg < 0.3 kcal/mol, Table 3).
Figure 4.
The effect of the length and composition of the single-stranded linker on Mg2+-dependent GAAA tetraloop-receptor docking. (a) The original A7 linker (left) is modified in composition to U7(center) and in length to A14 (right). The RNA sequences for the three constructs are otherwise identical. (b) Ensemble FRET measurements of Mg2+-induced GAAA tetraloop-receptor docking for the U7 (circles), A7 (squares), and A14 (diamonds) RNAs. Error bars are approximately within the size of the symbols. The data are fit to Equation 1 with n = 1 (solid lines, see Table 3).
Table 3.
Thermodynamic and kinetic parameters GAAA tetraloop-receptor constructs with different single-stranded linkers
| RNA | K1/2 for Mg2+ (mM) a | ΔΔ Go Mga,b (kcal/mol) | Kdockc (tdock/tundock) | ΔΔGodockb (kcal/mol) | kdockd (s−1) | kundockd (s−1) | Kdockd,e (kdock/kundock) |
|---|---|---|---|---|---|---|---|
| A7 | 0.85 ± 0.09 | --- | 1.4 ± 0.1 | --- | 10.5 ± 0.2 | 7.7 ± 0.2 | 1.4 ± 0.1 |
| U7 | 0.63 ± 0.07 | −0.17 ± 0.06 | 2.7 ± 0.1 | −0.38 ± 0.05 | 19.5 ± 0.6 | 6.9 ± 0.2 | 2.8 ± 0.1 |
| A14 | 1.36 ± 0.07 | 0.28 ± 0.07 | 0.95 ± 0.1 | 0.22 ± 0.08 | 8.2 ± 0.2 | 8.0 ± 0.2 | 1.0 ± 0.1 |
K1/2 was determined from ensemble FRET experiments with data fit to Equation 1 with n = 1.0.
ΔΔGo values are calculated relative to the A7 construct.
Kdock determined from the ratio of the total docked and undocked times using single-molecule FRET data measured at 0.35 mM Mg2+.
Rate constants determined from single-molecule FRET data measured at 0.35 mM Mg2+.
Kdock determined from the ratio of the rate constants.
Similar docked states are expected for all three of these RNA constructs because they differ only in the single-stranded linker. However, the RNAs did not show the same minimum and maximum EFRET values in the ensemble FRET experiments (Figure 4b). Native gel electrophoresis experiments revealed multiple bands for each RNA construct, indicating heterogeneity in the secondary and/or tertiary structures of the molecules (See Figure S1, Supplementary Information). Significant populations of nondocking molecules (presumably molecules with inactive alternate conformations), as well as molecules with inactive Cy5, were previously observed for this RNA by single-molecule experiments (27). These effects lead to a background fluorescence that contributes to the measured EFRET but does not affect the shape of the curves in Figure 4b or the corresponding K1/2 and n values.
Single-molecule FRET Measurements of GAAA Tetraloop-Receptor Docking for RNAs with Different Single-Stranded Linkers
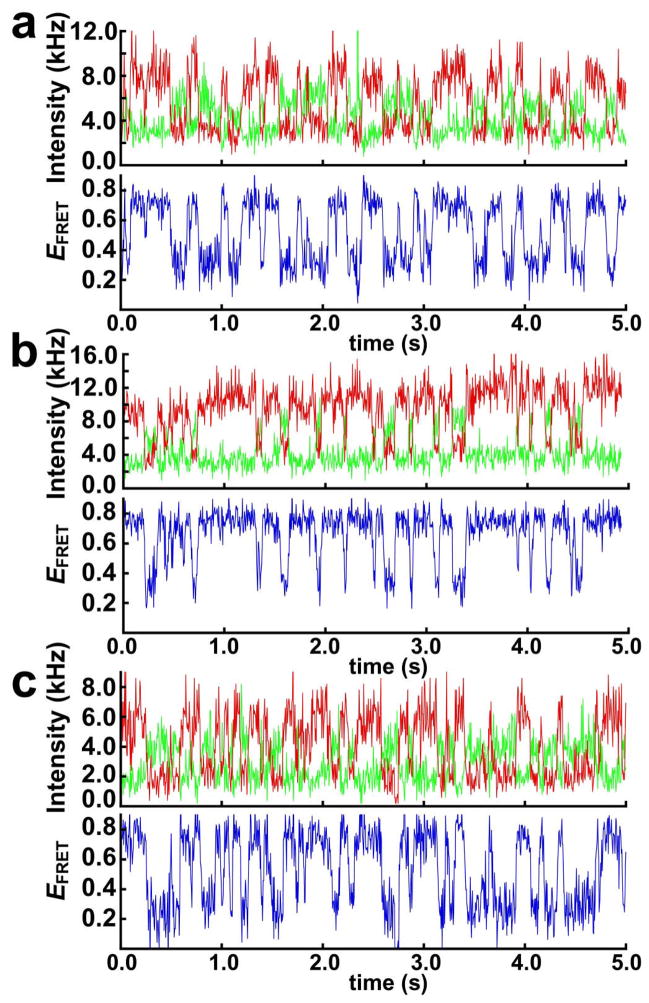
Single-molecule FRET experiments can directly resolve the docked and undocked states and thus determine the equilibrium constant for GAAA tetraloop-receptor docking, Kdock. Figure 5 shows plots of time-resolved single-molecule donor and acceptor fluorescence intensity and the corresponding EFRET for the A7, U7, and A14 constructs (at 0.35 mM Mg2+). All three constructs have a significant population (≈ 30%) of molecules that remain permanently undocked during the 2–30 s observation time (data not shown), similar to what was previously observed for the A7 construct (27). These nondocking species likely have inactive conformations of the tetraloop or receptor and are omitted from further analysis. The actively docking molecules exhibit two-state behavior, with each of the three constructs fluctuating between docked (EFRET ≈ 0.75) and undocked (EFRET ≈ 0.25–0.3) conformations. These results demonstrate how single-molecule techniques can be used to eliminate the fluorescence background arising from heterogeneity observed in Figure 4b.
Figure 5.
GAAA tetraloop-receptor docking measured by single-molecule FRET. Time-resolved single-molecule FRET traces measured at 0.35 mM Mg2+ are shown for (a) the A7 construct, (b) the U7 construct, and (c) the A14 construct. Cy3 donor fluorescence is plotted in green and the Cy5 acceptor fluorescence in red, with the corresponding EFRET plotted in blue. The molecules fluctuate between the docked state (EFRET ≈ 0.75) and the undocked state (EFRET ≈ 0.25–0.30).
Values for Kdock for the three RNA constructs were determined from the time-resolved FRET traces by calculating the fraction of the time individual molecules spend in the docked and undocked states and then averaging these fractions over many molecules (Table 3). Individual molecules were typically measured for 2–30 s prior to photobleaching of one of the fluorescence dyes. Comparison of data for the U7 or A14 with the A7 construct show that the length and sequence of the linker have a small effect on the free energy of docking (ΔΔGodock < 0.4 kcal/mol, Table 3).
The time-resolved fluorescence FRET trajectories in Figure 5 were also used to determine the docking and undocking kinetics for the three RNA constructs. The values determined for kdock and kundock at 0.35 mM Mg2+ are summarized in Table 3. These rate constants were used to calculate Kdock for each of the constructs (Table 3). These Kdock values are within error of those calculated from the populations of the docked and undocked states (Table 3).
Inhibition of Tetraloop-Receptor Docking by Complementary Oligonucleotides
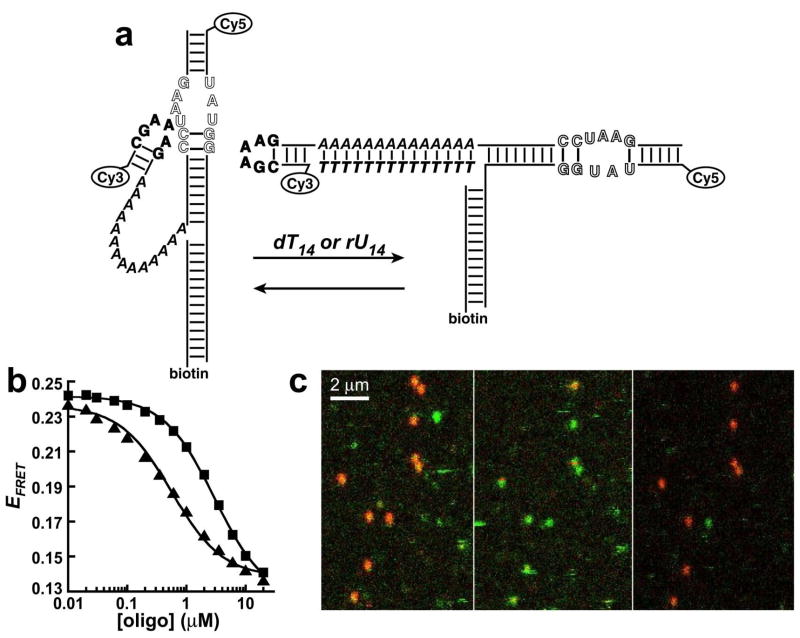
The single-stranded linker in this system represents a specific target for inhibition of docking. Hybridizing the linker with a complementary oligonucleotide results in a rigid double helix, which shifts the equilibrium towards the undocked state (Figure 6a). The thermodynamics of helix formation for RNA-DNA and RNA-RNA helices predict weak association of an A7 sequence with a complementary oligonucleotide (KD > 0.5 mM) (33, 34). Consistent with this prediction, no change in EFRET was observed in ensemble experiments when the A7 construct was titrated with dT7 or rU7 (up to 220 μM) under conditions favorable for docking (10 mM Mg2+) (data not shown). However, titration of the A14 construct with dT14 or rU14 caused a clear decrease in EFRET, demonstrating inhibition of docking (Figure 6b). This inhibition was easily reversed by addition of excess dA14 or rA14 to sequester the inhibitor, which restored the EFRET to its initial value (data not shown). The FRET data for the inhibitors fit well to binding isotherms (Equation 2), yielding apparent KD values of 0.51 ± 0.09 and 3.1 ± 0.3 μM for the dT14 and rU14, respectively (Figure 6b). Tighter association of dT14 relative to the rU14 is predicted from the thermodynamics of helix formation (33, 34). The KD values observed here for helix formation are 2–3 orders of magnitude weaker than the predicted values of 1–10 nM, reflecting the free energy cost of disrupting the tetraloop-receptor interaction.
Figure 6.
Specific inhibition of GAAA tetraloop-receptor docking for the A14 construct. (a) Schematic of inhibition of docking by a dT14 or rU14 oligonucleotide complementary to the single-stranded linker. (b) Ensemble FRET measurements of titration of the A14 construct with dT14 (triangles) and rU14 (squares). Error bars are approximately within the size of the symbols. The solid lines represent fits to a binding isotherm (Equation 2). (c) Single-molecule raster-scanned images of the A14 construct at 10 mM Mg2+, plotted in false color with donor fluorescence in green and acceptor fluorescence in red. The same RNA molecules are being observed in each image. The RNAs are primarily docked in the absence of inhibitor (left), undocked after flushing with 10 μM dT14 (center), and docked again after flushing with 10 μM dA14 (right).
Raster-scanned fluorescence experiments on the A14 construct were used to directly visualize docking inhibition of multiple single molecules. These experiments measure donor (green) and acceptor (red) fluorescence intensities, which are plotted as a function of position on the microscope cover slip to yield images that show the relative EFRET of each molecule (Figure 6c). A flow cell apparatus was used to image the same RNA molecules under various solution conditions. The A14 construct is highly docked (red) in the absence of inhibitor (Figure 6c, left panel), undocked after flushing with 10 μM dT14 (middle panel), and returns to highly docked after flushing with 10 μM dA14 (right panel). Not every molecule in Figure 6c survives for detection in all three panels due to dye photobleaching or dissociation of the RNAs from the surface.
DISCUSSION
Tertiary folding in RNAs involves formation of stabilizing interactions between two or more highly negatively charged domains. Thus, charge-charge repulsion represents a significant barrier to tertiary structure formation. The association of mono- or divalent cations with the RNA reduces this repulsion, stabilizing the folded structure (4). Tertiary structure formation typically requires divalent cations, where Mg2+ is normally the physiologically relevant metal (4, 35, 36). The interplay between Mg2+ binding and RNA folding is complex, with RNAs being stabilized by both specifically and non-specifically bound Mg2+ ions (37, 38). In some cases specifically bound Mg2+ ions directly coordinate to the RNA, whereas in other instances Mg2+ makes water-mediated contacts to the RNA (35). In many RNAs, a variety of multivalent cations (such as Ca2+, Mn2+, or Co(NH3)63+) or monovalent cations (such as K+ and Na+) can substitute for Mg2+ in promoting folding.
FRET is a common structural probe for biological macromolecules (39), and this method has been applied in a variety of ensemble (40–42) and single-molecule (38, 43–46) studies of RNA structure and folding. Ensemble FRET experiments were used here to probe the ability of various cations to stabilize GAAA tetraloop-receptor docking. All of the cations tested are able to promote docking. The K1/2 values for the divalent ions Ca2+, Mg2+, and Mn2+ are similar for the GAAA tetraloop-receptor (Table 1). RNAs such as RNase P and the group I intron also showed little variation in their K1/2 values for folding with these divalent cations (47–49). In our tetraloop-receptor system, Co(NH3)63+ promotes docking much more effectively than the divalent ions (≈ 50 times tighter K1/2, Table 1), whereas Na+ and K+ are much less effective than the divalents (≈ 250 times weaker K1/2). Therefore, charge density appears to be an important factor in the ability of a cation to stabilize docking in this tetraloop-receptor system.
The Co(NH3)63+ experiments were also used to test whether GAAA tetraloop-receptor docking requires direct coordination between the RNA and a metal ion(s). Mg2+ can interact with RNA as either a fully hydrated ion complex (Mg(H2O)62+) or with one or more of the coordinated H2O ligands replaced by a ligand from the RNA (35, 36). By contrast, RNA can only make outer-sphere contacts to Co(NH3)63+ (32). Our data show that Co(NH3)63+ effectively promotes GAAA tetraloop-receptor docking (K1/2 = 0.012 mM, Table 1). Thus, no inner sphere metal ion-RNA interactions are required for GAAA tetraloop-receptor docking.
The K1/2 values for these various cations in the GAAA tetraloop-receptor system are similar to what was previously observed for folding of the Tetrahymena ribozyme, which had K1/2 values of 0.017 mM for Co(NH3)63+, 0.27 mM for Mg2+, and 460–640 mM for Na+ and K+ (50). Notably, in the tetraloop-receptor system the K1/2 values for monovalents versus Mg2+ differ by only ≈ 250-fold, whereas these values differ by ≈ 2400-fold for the Tetrahymena ribozyme. The hairpin ribozyme and the S15 binding domain of the 16S rRNA also show relatively low K1/2 values for Na+ (200 and 350 mM, respectively) (44, 51). However, these K1/2 values for Na+ are considerably weaker than the K1/2 values for Mg2+ (≈ 6000- and ≈ 2500-fold, respectively). Thus, the requirement for Mg2+ over monovalent ions is less stringent for the GAAA tetraloop-receptor interaction than for many other RNA tertiary interactions.
The cooperativity of metal ion binding will also influence RNA folding. The multivalent cations Ca2+, Mg2+, Mn2+, and Co(NH3)63+ all yielded Hill coefficients of ≈ 1 (Table 1). Hill coefficients for different metal ions vary considerably in other RNA systems. For example, Mg2+-induced folding of a natural hammerhead ribozyme is noncooperative (42), whereas high cooperativity is observed for the full Tetrahymena ribozyme (n ≈ 20 under conditions of 24 mM Na+, 50 mM HEPES pH 7.5, 4–10 °C) and its isolated P4-P6 subdomain (n ≈ 3–5 under conditions of 10 mM sodium cacodylate pH 7.5, 42 °C) (50, 52). It is important to note that the concentration of monovalent cations can have a large effect on the observed Hill coefficient. Furthermore, Hill coefficients are determined indirectly from the Mg2+ dependence of native structure formation and thus only measure the cooperativity of the weakest binding Mg2+ ion(s) necessary for folding. Therefore, a Hill coefficient represents a lower limit for the number of Mg2+ ions required for folding.
One interpretation of the results here is that the Hill coefficient of ≈ 1 for the divalent ions represents binding to the divalent site observed in the receptor domain in the crystal structure of tetraloop-receptor complex (18). Previous EPR data for the bimolecular association of a GAAA tetraloop and tetraloop receptor as a function of Mg2+ also showed a Hill coefficient of ≈ 1 (24). In addition, gel shift studies yielded a Hill coefficient of ≈ 2 for the Mg2+ dependence of formation of a homodimer RNA complex containing two GAAA tetraloop-receptor interactions (25). All these results are consistent with one metal binding site for each GAAA tetraloop-receptor interaction.
In contrast, cooperativity was observed for both K+- and Na+- induced docking (Hill coefficients ≈ 2, Table 1). Different cooperativities for monovalent and divalent ions have been observed previously in other RNA systems. For example, Mg2+ was more cooperative than Na+ in the folding of the Tetrahymena ribozyme (50), whereas Na+ was more cooperative than Mg2+ for folding of a 2-way junction hairpin ribozyme (38), and the same cooperativity was observed for Na+ and Mg2+ in the 4-way junction hairpin ribozyme (51). If the Hill coefficient of 1 measured here for the divalents represents one ion binding to a specific site in the tetraloop-receptor, then a Hill coefficient of 2 could simply reflect the need for this site to bind two monovalent ions to achieve charge neutralization. Alternatively, electrostatic non-linear Poisson-Boltzmann calculations have been used to model diffusely bound ions in tRNAs, and this study found an average of 1.9 monovalent ions are required to replace each Mg2+ ion (37). Thus, the two-fold higher Hill coefficient for monovalents observed here is also consistent with stabilization of the tetraloop-receptor docking by only non-specific, diffusely bound ions.
It is also possible that the difference in Hill coefficients between monovalent and divalent cations results from the specific K+ binding site in the GAAA tetraloop-receptor interaction previously observed by X-ray crystallography (53). NMR studies have shown that the free receptor has a significantly different conformation than the tetraloop-bound receptor (17), so this specific monovalent binding site likely is not formed in the undocked state. The n ≈ 2 observed for K+- and Na+-dependent docking could result from cooperative binding of two monovalent ions to form the docked state. However, because the solution contains 100 mM NaCl, cooperativity would not be observed for the multivalent cations if the specific monovalent site can simply bind Na+ once the docked state forms in the presence of the multivalent ion.
The Ea of 12.7 kcal/mol for the GAAA tetraloop-receptor is much smaller than what has been observed for folding of several larger RNAs such as the Bacillus subtilis RNase P (50 kcal/mol) (54) and the P4-P6 (26 kcal/mol) (55) and P3-P7 (23 kcal/mol) (56) domains of the Tetrahymena ribozyme. Large Ea values such as these can result from breaking stable interactions in unfolded, intermediate, or misfolded states in the folding pathway (54–57). Other RNAs have much smaller Ea values for folding; for example, the 2-way junction and 4-way junction hairpin ribozymes (1–5 kcal/mol) (58) or the catalytic core of the bI5 group I intron (0.5 kcal/mol) (59). Conformational searches have been proposed to dominate the folding rates of these RNAs (58, 60). The significant Ea value for the GAAA tetraloop-receptor system suggests that the docking rate is dominated by breaking stable interactions in the undocked state rather than by a conformational search. As noted earlier in the discussion, the receptor has significantly different conformations in the free- and in the receptor-bound states (17, 18). Since the docked-state conformation contains a Mg2+ binding site, Mg2+ would be expected stabilize this conformation even in the absence of the tetraloop. However, EPR studies of a spin-labeled receptor domain have shown that the docked-state conformation is not significantly populated in the free receptor, even at very high Mg2+ (125 mM) (61). Thus, the free-state conformation of the receptor appears to be quite stable, and a significant portion of the observed Ea may simply represent the cost of breaking the favorable interactions in the free-state to form the docked-state conformation.
To test the effect of the A7 single-stranded linker on docking in our GAAA tetraloop-receptor system, control experiments with U7 and A14 linkers were performed. The Kdock values measured from single-molecule FRET experiments for U7 and A14 are within a factor of two of A7 (Table 3), demonstrating the docking equilibrium is not significantly affected by the length or composition of the linker. The Kdock for the A14 construct is slightly smaller than for A7, which may simply reflect a somewhat larger entropic cost for docking in the A14 construct. Weaker base stacking for an oligo U compared to oligo A sequence makes the U7 linker more flexible than the A7, leading to the slightly larger Kdock for the U7 construct than for the A7. These results are confirmed by the ensemble FRET measurements, which show similar K1/2 values and Hill coefficients for the three linkers (Table 3).
Another estimate for Kdock can be obtained from the inhibitor studies on the A14 construct. If we assume the reaction in Figure 6a consists of two steps, disruption of the docking interaction and formation of an A14-U14 duplex, then the ΔGo for the overall reaction is the sum of the ΔGos for the two steps. For the U14 inhibitor, the ΔGo for the overall reaction determined here is −7.52 kcal/mol (KD = 3.1 μM at 10 mM Mg2+). The ΔGo for formation of an A14-U14 duplex is −9.82 kcal/mol, as calculated by summing nearest neighbor parameters, a penalty for duplex initiation, and a penalty for two terminal A-U base-pairs for an RNA-RNA duplex (34). The 2.30 kcal/mol difference between these values is the apparent ΔGo for undocking (Kdock = 49), which agrees reasonably well with the Kdock of 19 measured by single-molecule experiments under these Mg2+ conditions (A7 linker, (27)). For the dT14 inhibitor, the ΔGo for the overall reaction was measured as −8.58 kcal/mol (KD = 0.51 μM at 10 mM Mg2+). A ΔGo of −12.3 kcal/mol was calculated for formation of an A14-dT14 duplex using RNA-DNA nearest neighbor parameters, a penalty for initiation of an RNA-DNA duplex (33), and the same penalty as above for the two terminal A-dT base-pairs. This yields a Kdock of 530, which is significantly different from the Kdock = 19. Thus, this method for calculating Kdock overestimates free energy for docking, indicating that other factors besides the energy of duplex formation need to be taken into account to accurately model binding of the inhibitor.
Regulation of GAAA tetraloop-receptor docking with these complementary oligos suggests an intriguing application, that of an RNA-based nanomolecular machine. Binding of the oligos results in a significant linear movement of the tetraloop relative to the receptor, and this motion is reversed by removal of the inhibitor. This motion represents a two-stroke cycle of extension and contraction driven by oligonucleotide “fuel.” Previous studies have explored conformational changes in nucleic acids as potential nanodevices, such as DNA systems that switch between: i) a G-quadruplex and an extended duplex, ii) a B-form helix and a Z-form helix, or iii) a “tweezer” system that switches between closed and open states (62). Similar to the G-quadruplex system, the tetraloop-receptor device here employs hybridization of the complementary oligo to switch between compact and extended states (63). In the extended state of the tetraloop-receptor device both the tetraloop and the receptor are available to interact with other molecules. This property could be exploited to design a system that reversibly switches between unimolecular and bimolecular states. For example, the homodimer tetraloop-receptor system previously described by Jaeger et al. (25) could be modified to function in this way by replacing the double-stranded region between the tetraloop and the receptor with a single-stranded region. In the absence of the complementary oligo this system would form an intramolecular tertiary interaction, and in the presence of the oligo the system would switch to the dimer state.
The single-molecule kinetic data for the A7, U7, and A14 constructs yield nearly identical kundock values, with small differences for the kdock values (Table 3). Thus, disruption of interactions between the tetraloop and receptor associated with the undocking reaction does not appear to be affected by the linker length or composition. However, the linker may have a small effect on the docking reaction by changing the rate of collision between the two domains. The rate constant for collision between two RNA domains joined by a short flexible tether (such as a single-stranded domain) has been estimated to be ≈ 106 s−1 (45). This value is orders of magnitude greater than the kdock values measured here for the tetraloop-receptor (≈ 101 s−1). A simple interpretation for this large difference is that only a small fraction of the collisions result in docking, as has been proposed for other RNAs (45). Thus, the rate constant for conversion of the free receptor conformation to the bound receptor conformation is likely much smaller than the rate constant for collision of the tetraloop and receptor domains.
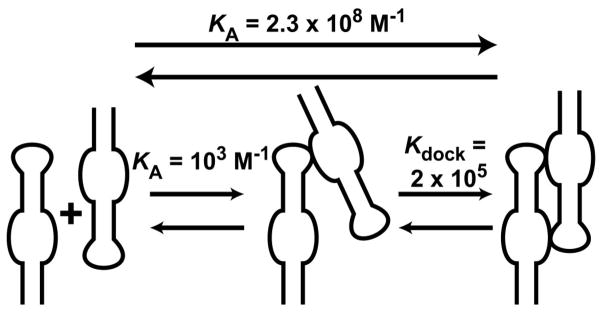
The similarities in the kinetic and equilibrium data for the different linkers (Table 3) indicate that the single-stranded linker acts primarily as a passive tether between the GAAA tetraloop and receptor. Thus, is it instructive to compare the results with the homodimer tetraloop-receptor system discussed above, where the system was designed to position optimally a tetraloop for docking to its receptor (25). This homodimer system (which is stabilized by two GAAA tetraloop-receptor contacts) has a KA of 2.3 × 108 M−1 (KD of 4.3 nM) at 15 mM Mg2+. Formation of the homodimer can be described by the two-step reaction illustrated in Scheme 1. The KA for the first step can be estimated as the KA for bimolecular binding of a single GAAA tetraloop and receptor, which has been previously measured as ≈ 1 × 103 M−1 at 15 mM Mg2+ (24) (KD ≈ 3 mM, which is divided by 4 here to account for the self-complementary homodimer construct). Since the equilibrium constants for both the overall reaction and first step are known, Kdock for the second step is calculated as 2 × 105. The second step is more favorable because the first step includes a high entropic cost for bringing the two molecules together, and thus the second step is effectively an intramolecular docking event. This Kdock of 2 × 105 can then be compared with the intramolecular A7 system, where Kdock = 19 (at 10 mM Mg2+) (27). This 104-fold difference in Kdock for these two systems is quite striking. To try to understand the origin of this difference, a simple model was used to estimate Kdock for a completely flexible linker. This model assumes that the only function of the linker is to restrict the accessible volume for the GAAA tetraloop relative to the receptor. For the A7 linker, the accessible volume is modeled as a sphere with a radius ≈ 38 Å (the maximum combined length of the linker and the tetraloop domain). The effective concentration of the tetraloop domain in this volume is 14 mM (assuming 50% of the volume is excluded by steric hindrance). The previously measured KD of ≈ 4 mM (at 10 mM Mg2+) for the bimolecular association of a GAAA tetraloop and receptor (24) predicts 80% occupancy of the docked state at a 14 mM effective tetraloop concentration, which corresponds to a Kdock of 4 for an intramolecular system. This prediction agrees reasonably well with the previously measured Kdock of 19 for the A7 construct (27). These results confirm that the single-stranded linker is well modeled by a passive flexible tether. Thus, the 104-fold difference in (intramolecular) Kdock between a flexible linker versus the optimized homodimer system should not be unexpected and emphasizes the critical effect that optimal positioning can have in stabilizing RNA tertiary interactions.
Scheme 1.
Supplementary Material
Figure S1 shows fluorescence-imaged native polyacrylamide gels on the A7 construct and various combinations of its constituent single oligonucleotide strands. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank Joseph Warner for assistance with the stopped-flow instrument and Drs. Robert Batey and Thomas Perkins for helpful discussions.
Footnotes
This work was supported in part by grants from: NIH (AI 33098), NSF, NIST and the W. M. Keck Foundation initiative in RNA science at the University of Colorado, Boulder. CDD was also supported in part by Biophysics Training Grant NIH (GM 65103).
Abbreviations: NMR, nuclear magnetic resonance; FRET, fluorescence resonance energy transfer; HPLC, high-performance liquid chromatography; HEPES, N-(2-hydroxyethyl)piperazine-N′-2-ethanesulfonic acid; EDTA, ethylenediaminetetraacetate; Tris, tris(hydroxymethyl)aminomethane; THE, Tris HEPES EDTA; EPR, electron paramagnetic resonance.
Mention of commercial products is for information only; it does not imply NIST recommendation or endorsement, nor does it imply that the products mentioned are necessarily the best available for the purpose.
References
- 1.Cech TR. The ribosome is a ribozyme. Science. 2000;289:878–879. doi: 10.1126/science.289.5481.878. [DOI] [PubMed] [Google Scholar]
- 2.Moore PB, Steitz TA. The involvement of RNA in ribosome function. Nature. 2002;418:229–235. doi: 10.1038/418229a. [DOI] [PubMed] [Google Scholar]
- 3.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 4.Tinoco I, Jr, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 5.Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- 6.Rupert PB, Ferre-D’Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 7.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci U S A. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puglisi JD, Wyatt JR, Tinoco I., Jr Conformation of an RNA pseudoknot. J Mol Biol. 1990;214:437–453. doi: 10.1016/0022-2836(90)90192-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore PB. Structural motifs in RNA. Ann Rev Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- 10.Batey RT, Rambo RP, Doudna JA. Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 11.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy FL, Cech TR. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J Mol Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 13.Tanner MA, Cech TR. An important RNA tertiary interaction of group I and group II introns is implicated in gram-positive RNase P RNAs. RNA. 1995;1:349–350. [PMC free article] [PubMed] [Google Scholar]
- 14.Massire C, Jaeger L, Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J Mol Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- 15.Heus HA, Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- 16.Jucker FM, Pardi A. GNRA tetraloops make a U-turn. RNA. 1995;1:219–222. [PMC free article] [PubMed] [Google Scholar]
- 17.Butcher SE, Dieckmann T, Feigon J. Solution structure of a GAAA tetraloop receptor RNA. EMBO J. 1997;16:7490–7499. doi: 10.1093/emboj/16.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 19.Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- 20.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J Mol Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 21.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Young BT, Silverman SK. The GAAA tetraloop-receptor interaction contributes differentially to folding thermodynamics and kinetics for the P4-P6 RNA domain. Biochemistry. 2002;41:12271–12276. doi: 10.1021/bi0264869. [DOI] [PubMed] [Google Scholar]
- 23.Abramovitz DL, Pyle AM. Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction. J Mol Biol. 1997;266:493–506. doi: 10.1006/jmbi.1996.0810. [DOI] [PubMed] [Google Scholar]
- 24.Qin PZ, Butcher SE, Feigon J, Hubbell WL. Quantitative analysis of the isolated GAAA tetraloop/receptor interaction in solution: a site-directed spin labeling study. Biochemistry. 2001;40:6929–6936. doi: 10.1021/bi010294g. [DOI] [PubMed] [Google Scholar]
- 25.Jaeger L, Leontis N. Tecto-RNA: One-dimensional self-assembly through tertiary interactions. Angew Chem Int Ed Engl. 2000;39:2521–2524. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 26.Jaeger L, Westhof E, Leontis NB. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hodak JH, Downey CD, Fiore JL, Pardi A, Nesbitt DJ. Docking kinetics and equilibrium of a GAAA tetraloop-receptor motif probed by single-molecule FRET. Proc Natl Acad Sci U S A. 2005;102:10505–10510. doi: 10.1073/pnas.0408645102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grosshans CA, Cech TR. Metal ion requirements for sequence-specific endoribonuclease activity of the Tetrahymena ribozyme. Biochemistry. 1989;28:6888–6894. doi: 10.1021/bi00443a017. [DOI] [PubMed] [Google Scholar]
- 29.Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- 30.Harada Y, Sakurada K, Aoki T, Thomas DD, Yanagida T. Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J Mol Biol. 1990;216:49–68. doi: 10.1016/S0022-2836(05)80060-9. [DOI] [PubMed] [Google Scholar]
- 31.Jou RW, Cowan JA. Ribonuclease-H activation by inert transition-metal complexes - mechanistic probes for metallocofactors - insights on the metallobiochemistry of divalent magnesium-ion. J Am Chem Soc. 1991;113:6685–6686. [Google Scholar]
- 32.Young K, Gill F, Grasby J. Metal ions play a passive role in the hairpin ribozyme catalysed reaction. Nucleic Acids Res. 1997;25:3760–3766. doi: 10.1093/nar/25.19.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugimoto N, Nakano S, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 34.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 35.Pyle A. Metal ions in the structure and function of RNA. J Biol Inorg Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 36.Draper DE, Grilley D, Soto AM. Ions and RNA folding. Annu Rev Biophys Biomol Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 37.Misra VK, Draper DE. The linkage between magnesium binding and RNA folding. J Mol Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 38.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc Natl Acad Sci U S A. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2. Kluwer Academic/Plenum Publishers; New York: 1999. [Google Scholar]
- 40.Tuschl T, Gohlke C, Jovin TM, Westhof E, Eckstein F. A three-dimensional model for the hammerhead ribozyme based on fluorescence measurements. Science. 1994;266:785–789. doi: 10.1126/science.7973630. [DOI] [PubMed] [Google Scholar]
- 41.Walter NG, Harris DA, Pereira MJ, Rueda D. In the fluorescent spotlight: global and local conformational changes of small catalytic RNAs. Biopolymers. 2001;61:224–242. doi: 10.1002/bip.10144. [DOI] [PubMed] [Google Scholar]
- 42.Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DM. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 44.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartley LE, Zhuang X, Das R, Chu S, Herschlag D. Exploration of the transition state for tertiary structure formation between an RNA helix and a large structured RNA. J Mol Biol. 2003;328:1011–1026. doi: 10.1016/s0022-2836(03)00272-9. [DOI] [PubMed] [Google Scholar]
- 46.Nahas MK, Wilson TJ, Hohng S, Jarvie K, Lilley DM, Ha T. Observation of internal cleavage and ligation reactions of a ribozyme. Nat Struct Mol Biol. 2004;11:1107–1113. doi: 10.1038/nsmb842. [DOI] [PubMed] [Google Scholar]
- 47.Brannvall M, Mikkelsen NE, Kirsebom LA. Monitoring the structure of Escherichia coli RNase P RNA in the presence of various divalent metal ions. Nucleic Acids Res. 2001;29:1426–1432. doi: 10.1093/nar/29.7.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Streicher B, von Ahsen U, Schroeder R. Lead cleavage sites in the core structure of group I intron-RNA. Nucleic Acids Res. 1993;21:311–317. doi: 10.1093/nar/21.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith D, Burgin A, Haas E, Pace N. Influence of metal ions on the ribonuclease P reaction. Distinguishing substrate binding from catalysis. J Biol Chem. 1992;267:2429–2436. [PubMed] [Google Scholar]
- 50.Heilman-Miller SL, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme. I. Equilibrium stabilization by cations. J Mol Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 51.Wilson TJ, Lilley DM. Metal ion binding and the folding of the hairpin ribozyme. RNA. 2002;8:587–600. doi: 10.1017/s1355838202020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deras ML, Brenowitz M, Ralston CY, Chance MR, Woodson SA. Folding mechanism of the Tetrahymena ribozyme P4-P6 domain. Biochemistry. 2000;39:10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- 53.Basu S, Rambo RP, Strauss-Soukup J, Cate JH, Ferre-D’Amare AR, Strobel SA, Doudna JA. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nat Struct Biol. 1998;5:986–992. doi: 10.1038/2960. [DOI] [PubMed] [Google Scholar]
- 54.Pan T, Sosnick TR. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat Struct Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- 55.Silverman SK, Deras ML, Woodson SA, Scaringe SA, Cech TR. Multiple folding pathways for the P4-P6 RNA domain. Biochemistry. 2000;39:12465–12475. doi: 10.1021/bi000828y. [DOI] [PubMed] [Google Scholar]
- 56.Rook MS, Treiber DK, Williamson JR. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J Mol Biol. 1998;281:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 57.Silverman SK, Cech TR. An early transition state for folding of the P4-P6 RNA domain. RNA. 2001;7:161–166. doi: 10.1017/s1355838201001716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pljevaljcic G, Klostermeier D, Millar DP. The tertiary structure of the hairpin ribozyme is formed through a slow conformational search. Biochemistry. 2005;44:4870–4876. doi: 10.1021/bi047772i. [DOI] [PubMed] [Google Scholar]
- 59.Buchmueller KL, Webb AE, Richardson DA, Weeks KM. A collapsed non-native RNA folding state. Nat Struct Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- 60.Treiber DK, Williamson JR. Beyond kinetic traps in RNA folding. Curr Opin Struct Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 61.Qin PZ, Feigon J, Hubbell WL. Site-directed spin labeling studies reveal solution conformational changes in a GAAA tetraloop receptor upon Mg2+-dependent docking of a GAAA tetraloop. J Mol Biol. 2005;351:1–8. doi: 10.1016/j.jmb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 62.Seeman NC. From genes to machines: DNA nanomechanical devices. Trends Biochem Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alberti P, Mergny JL. DNA duplex-quadruplex exchange as the basis for a nanomolecular machine. Proc Natl Acad Sci U S A. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 shows fluorescence-imaged native polyacrylamide gels on the A7 construct and various combinations of its constituent single oligonucleotide strands. This material is available free of charge via the Internet at http://pubs.acs.org.