SUMMARY
Melanoma and other cancers harbor oncogenic mutations in the protein kinase B-Raf, which leads to constitutive activation and dysregulation of MAP kinase signaling. In order to elucidate molecular determinants responsible for B-Raf control of cancer phenotypes, we present a method for phosphoprotein profiling, using negative ionization mass spectrometry to detect phosphopeptides based on their fragment ion signature caused by release of PO3−. The method provides an alternative strategy for phosphoproteomics, circumventing affinity enrichment of phosphopeptides and isotopic labeling of samples. Ninety phosphorylation events were regulated by oncogenic B-Raf signaling, based on their responses to treating melanoma cells with MKK1/2 inhibitor. Regulated phosphoproteins included known signaling effectors and cytoskeletal regulators. We investigated MINERVA/FAM129B, a target belonging to a protein family with unknown category and function, and established the importance of this protein and its MAP kinase-dependent phosphorylation in controlling melanoma cell invasion into 3-dimensional collagen matrix.
INTRODUCTION
Approximately two-thirds of sporadic melanomas and cell lines derived from human melanomas contain mutations in B-Raf which cause constitutive activation and dysregulation of phosphorylation by the MAP kinase pathway involving the MAP kinase kinases, MKK1 and MKK2, and the MAP kinases, ERK1 and ERK2 (Gray-Schopfer et al., 2007). Many B-Raf mutations yield forms with constitutively elevated catalytic activity, ~80% of which give rise to a single amino acid substitution, V600E (Brose et al., 2002; Davies et al., 2002). Others do not activate B-Raf itself, but rather elevate the activity of Raf-1 through mechanisms that are still undefined (Wan et al., 2004). Melanomas with wild-type B-Raf also show elevated MAP kinase signaling which can be ascribed to oncogenic mutations in N-Ras and c-Kit, as well as autocrine signaling through growth factors and their receptors (Curtin et al., 2006; Gorden et al., 2003; Satyamoorthy et al., 2003). Given the prevalence of dysregulated MAP kinase signaling in melanoma, identifying pathway targets is key to understanding the molecular determinants responsible for cancer progression.
The importance of MAP kinase pathway activation in melanoma has been demonstrated by inhibiting mutant B-Raf with RNAi, inactivating MKK1/2 with anthrax lethal factor, and inhibiting B-Raf or MKK1/2 with small molecule inhibitors (Gray-Schopfer et al., 2007). Such treatments block cell proliferation, survival, and anchorage independent growth, while enhancing melanogenesis. Transcriptional targets regulated by constitutive B-Raf/MKK/ERK signaling have been reported to control events in tumor formation. These include cyclin D1, Brn2 and microphthalmia-associated transcription factor (MITF) which modulate cell proliferation (Bhatt et al., 2005; Goodall et al., 2004; Wellbrock and Marais, 2005), and HIF1a, Bim1 and PUMA, which regulate cell survival against apoptotic cell stress (Wang et al., 2007; Kumar et al., 2007). In addition, B-Raf promotes melanoma cell invasion and bypass of immune surveillance (Sumimoto et al., 2006). In contrast to targets regulated at the level of gene expression, little is known about proteins regulated post-translationally in response to oncogenic B-Raf signaling in melanoma cells. In particular, identifying cellular targets for phosphorylation is needed to gain a more comprehensive understanding of the responses to MAP kinase pathway dysregulation in melanoma.
Mass spectrometry has emerged as a key technology for screening protein post-translational modifications, allowing simultaneous phosphorylation site mapping and quantitation in a single experiment. A commonly used strategy is to prepare proteolytic digests of phosphoproteins, from which phosphopeptides can be identified by m/z and sequenced by MS/MS. In large scale studies of complex samples, phosphopeptides are enriched by immunoaffinity, metal chelating, or phosphate adsorbing resins, followed by peptide HPLC coupled to MS/MS and MS3 sequencing (Beausoleil et al., 2004; Olsen et al., 2006). Enrichment is needed due to the problem of inadequately sampling phosphopeptides against larger numbers of unphosphorylated peptides in complex samples, a problem compounded by low phosphorylation stoichiometries in phosphoproteins. To quantify changes in phosphorylation, metabolic labeling with isotopically distinguishable amino acids is a prevailing method (Ong and Mann, 2005). When metabolic labeling is not possible chemical labeling approaches such as ITRAQ and “tandem mass tags” may be used (Zhang et al., 2005; Thompson et al., 2003). Samples are mixed, and relative changes in abundance are determined from the ratio of intensities between differentially labeled phosphopeptides. This circumvents problems in variable recovery from phosphopeptide affinity resins.
Alternative strategies to identify and quantify phosphopeptides are needed, because not all biological samples can be readily labeled isotopically. Furthermore, different enrichment methods show different preferences in phosphopeptide recovery, governed by chemical differences that are not completely defined (Bodenmiller et al., 2007). Therefore, we wished to explore methods for profiling phosphopeptides that would not depend on enrichment, and could be performed quantitatively in a label-free manner. One approach uses a precursor ion scanning method to detect phosphopeptides by their signature fragment ions of −79 m/z, due to loss of PO3− (Carr et al., 1996). In order to detect this signature, peptides must be negatively charged during ionization. This has the advantage of increased sensitivity for phosphopeptides, which often compete poorly against unphosphorylated peptides for positive charging (Janek et al., 2001). In addition, this method detects phosphotyrosine (pY) as well as phosphoserine (pS) and phosphothreonine (pT) containing peptides, unlike neutral loss methods which primarily detect pS and pT or 216 m/z precursor ion scanning which detects pY (Steen et al., 2001). The method has been effective in mapping phosphorylation sites on single proteins or simple protein mixtures (Williamson et al., 2006), but has never been extended to samples as complex as cell lysates, in part because negative mode mass spectrometry is less well developed than positive mode detection of peptide analytes.
Here we report the application of −79 Da precursor ion scanning mass spectrometry towards a large scale study of phosphoproteins targeted by constitutive B-Raf/MKK/ERK signaling in melanoma cells. Our results demonstrate successful selection and sequencing of phosphopeptides in proteolytic digests without affinity enrichment, as well as label-free quantitation of regulated protein phosphorylation events. Targets of B-Raf include known enzymes in the MAP kinase cascade, as well as proteins phosphorylated at motifs consistent with substrates for MAP kinases and MAP kinase-activated protein (MAPKAP) kinases. We also demonstrate pathway-dependent phosphorylation of FAM129B, a member of an uncharacterized gene family, and establish its importance in controlling melanoma cell invasion. Our study reveals that phosphoprotein profiling can be achieved in complex samples without phosphopeptide enrichment or stable isotope labeling, and identifies a potential cancer target regulated by phosphorylation in response to oncogenic B-Raf signaling.
RESULTS
Phosphoproteome profiling by −79 Da precursor ion scanning
The WM115 cell line is derived from a primary human melanoma tumor harboring the oncogenic B-Raf-V600E mutation (Hsu et al., 2002). As a result, signaling through the B-Raf/MKK/ERK pathway is dysregulated, reflected by constitutive phosphorylation and activation of the MAP kinases, ERK1 and ERK2. Constitutive signaling is efficiently suppressed by treating cells with U0126, a cell permeable inhibitor of MKK1/2, which binds the kinase N-terminal domain and disrupts ATP binding (Fig. S1). In our screen, cells containing active B-Raf were treated in the presence or absence of U0126 for 4 h, and proteins were extracted from each sample and analyzed in parallel (Fig. 1A). Cytosolic proteins were separated by quaternary anion exchange (QAE) chromatography, and proteins in each fraction were proteolyzed with trypsin and cysteines alkylated. Separation of proteins into QAE fractions simplified the resulting peptide mixtures, reducing matrix suppression and improving phosphopeptide identifications.
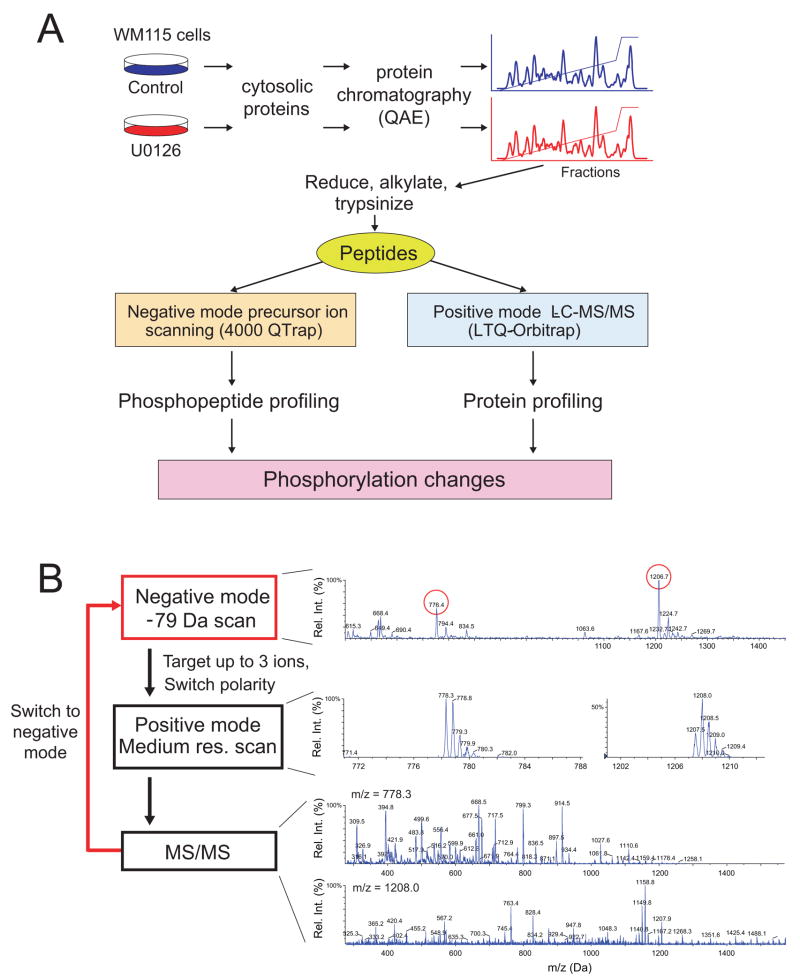
Figure 1. Label-free profiling of phosphoproteins responsive to B-Raf/MKK/ERK in human melanoma cells.
(A) Cytosolic proteins were extracted from WM115 melanoma cells treated with carrier (DMSO, 4 h) or MKK1/2 inhibitor (10 μM U0126, 4 h), then separated by anion exchange chromatography (Mono Q FPLC). Peptides resulting from trypsin proteolysis of each fraction are analyzed by negative mode precursor ion scanning to identify phosphopeptides in each sample, and by positive mode LC-MS/MS to identify proteins and quantify relative abundances by spectral counting. (B) Schematic of the precursor ion scanning experiment and examples of spectra collected in each segment of one scan cycle. Shown are the −79 Da precursor ion scan ([M-2H]2− = −776.4, −1206.7), the positive mode medium resolution scan, and positive mode MS/MS of two phosphopeptides (MH22+ = 778.3, 1208.0).
Phosphopeptide profiling was carried out in negative ionization mode using a triple quadrupole linear ion trap mass spectrometer. Collision-activated dissociation (CAD) of negatively charged phosphopeptides releases PO3−, which can be used as a signature fragment ion for phosphopeptides (Carr et al., 1996). By using the third quadrupole as a mass filter for the −79 Da fragment ion, and scanning across a wide mass range in the first quadrupole, phosphopeptides can be selectively distinguished from a background of unphosphorylated peptides, and their negative m/z measured. Following detection of the negatively charged precursor ion, the instrument polarity is switched to positive mode to acquire medium-resolution linear ion trap MS and MS/MS scans within the same scan cycle (Fig. 1B). This allows MS/MS sequencing of phosphopeptides. Factors that were critical for analyzing highly complex samples using this negative ionization approach are described in Discussion and Suppl. Methods.
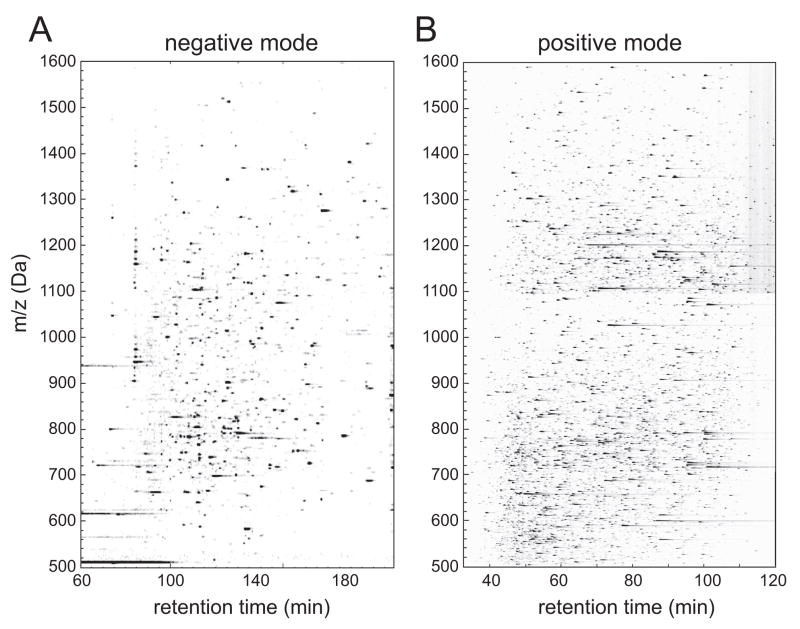
Figure 2A shows a contour map of the −79 Da precursor ion scanning experiment applied to a proteolytic digest of one QAE fraction. Phosphopeptide candidates were revealed by peaks of −79 Da signal intensity, plotted by negative precursor ion m/z vs. HPLC retention time. When peptides were ionized in positive mode, many more ions were detected in the same sample (Fig. 2B). Thus, the lower complexity of the −79 Da precursor ion experiment reflects the selective observation of phosphopeptide candidates in non-affinity enriched samples.
Figure 2. Selective detection of phosphopeptide candidates by −79 Da precursor ion scanning.
Proteins in one QAE fraction (DMSO-33) were proteolyzed with trypsin and analyzed in parallel by negative mode and positive mode mass spectrometry. (A) Contour map showing candidate phosphopeptides revealed by their −79 Da signal intensities, plotted vs HPLC retention time (x-axis) and precursor ion m/z (y-axis). (B) Contour map showing all peptides in the same sample, detected by positive LC-MS/MS. The precursor ion scanning method enables phosphopeptide candidates to be selectively detected within complex mixtures. Differences in timescales are due to different LC gradients used in each experiment.
Measurement of −79 Da signal intensity enabled quantitative comparisons between samples to be made without stable isotope labeling. Replicate analyses of a mixture of 60 synthetic phosphopeptides showed more than 80% of −79 Da precursor ions matched across six technical replicates, with a mean coefficient of variation equaling 0.20 (Fig. S2). In addition, phosphopeptides from QAE fractions showed good agreement in signal intensity between replicate runs. Log2 ratios of intensities, measured for peaks matched between replicate runs, showed standard deviation equal to 0.4 (1.3-fold, Fig. S3). Analysis of peptide serial dilutions showed linearity of signal ranging between 50–1000 fmol (Fig. S4).
We used −79 Da precursor ion scanning to estimate phosphopeptide recovery after purification by Fe3+-immobilized metal affinity chromatography (Fe3+-IMAC). Fig. S5 shows phosphopeptide candidates in a tryptic digest of one QAE fraction, before and after Fe3+-IMAC purification. Manual inspection indicated 71 peaks were detected before IMAC and 24 peaks after IMAC. Thus, only ~1/3 of peptides were recovered after purification, where few peaks showed more than 50% recovery of signal intensity. This suggests that −79 Da precursor ion scanning might be less biased in phosphopeptide detection, and illustrates its utility in assessing the degree of phosphopeptide recovery by affinity-based separations.
In total, 820 unique (i.e., chemically distinct) phosphopeptides (876 phosphorylation sites) were observed in our experiments, reporting only those assignments validated by manual inspection of MS/MS spectra (Table S1). Of these, 568 phosphorylation sites were localized to specific residues, evaluated using an Ascore index (Beausoleil et al., 2006), which calculates the probability of localization based on the presence of site-determining fragment ions in MS/MS spectra. Distributions of phosphoserine (79%), phosphothreonine (17%), and phosphotyrosine (4%) sites were similar to those reported in other large scale studies (Olsen et al., 2006). Not all −79 Da precursor ions corresponded to phosphopeptides. In two cases, peptides were instead modified by glycerylphosphorylethanolamine adducts of glutamic acid, generating −79 Da fragments by internal cleavage (Fig. S6). Both peptides were derived from eukaryotic elongation factor 1α (EEF1α), where adducts of 197 Da at Glu301 and Glu374 were consistent with previous reports of glycerylphosphorylethanolamine modifications at these residues in rabbit EEF1α (Dever et al., 1989). Cysteic acid and methionine sulfone have been reported to yield fragments of −80 Da and −79 Da, due to loss of SO3− and SO2CH3− respectively, which can be detected by −79 Da precursor ion scanning (Williamson et al., 2006). A separate search for cysteic acid and methionine sulfone as variable modifications yielded no matches (not shown). Thus, while −79 Da ion signals could be attributed to unphosphorylated peptides, those we observed were generally specific for phosphopeptides. Overall, 349 (61%) of the localized phosphorylation sites we observed were present in the PhosphoSite database, the largest compilation of human sites (Hornbeck et al., 2004), while 219 (39%) appeared to be novel.
Profiling responses to B-Raf/MKK/ERK signaling
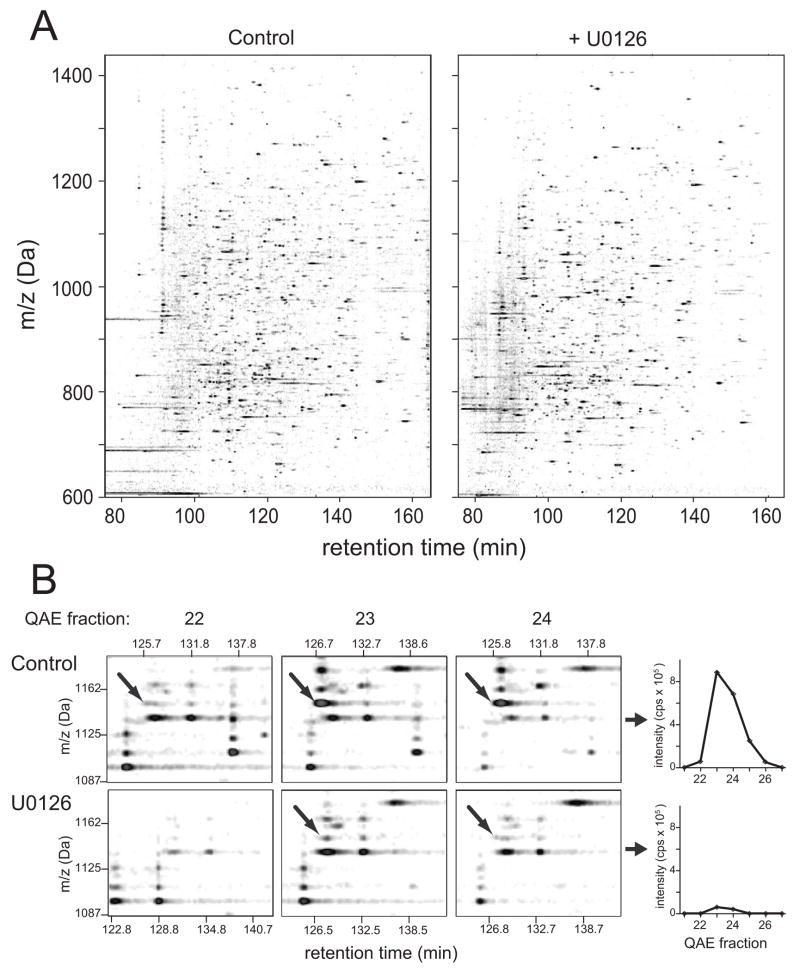
In order to identify phosphoproteins regulated by B-Raf/MKK/ERK signaling, −79 Da ion signals were used to quantify phosphopeptides in melanoma cells, surveying changes in intensity that were responsive to the MKK1/2 inhibitor, U0126. Fig. 3A shows phosphopeptide maps for matched fractions of the control and U0126-treated cells, illustrating that most phosphopeptides were unaltered in response to pathway inhibition. Profiles were generated for each phosphopeptide, tracking the elution of the parent protein across QAE fractions (Fig. 3B). Proteins often separated across multiple QAE fractions and shifted elution in response to U0126. This required matching phosphopeptide peaks across different datasets corresponding to adjacent fractions. Software was developed to automate this task, recombining signal intensities to calculate phosphopeptide profiles and intensity ratios between control vs drug-treated conditions (Fig. 3B, Table S2).
Figure 3. Profiling responses to B-Raf/MKK/ERK signaling.
(A) Contour maps of −79 Da signal intensities, from cells treated for 4 h under control conditions (DMSO carrier, left) or with 10 μM U0126 (right). Panels show the same fraction (DMSO-22, U0126-22) from each QAE separation. (B) Left: Illustration of quantitation method. Phosphopeptides from proteins eluting in more than one fraction were detected by matching m/z and retention times between separate runs, quantifying intensities in each fraction. Computational methods developed for feature identification and quantitation are described in Supplementary Methods. Right: Feature intensity profiles plotted vs QAE fraction for one phosphopeptide ion, which was inhibited in cells treated with U0126. The peak represents the [M-2H]2− ion of a peptide from ERK2 containing two regulatory phosphorylation sites (VADPDHDHTGFLpTEpYVATR).
Ninety phosphosites showed significant changes in intensity between control and drug-treated cells, deviating by more than 1.7-fold (2σ from the mean, Fig. S3). These included sixty phosphorylation sites that decreased significantly in cells treated with inhibitor (Table 1A), indicating positive regulation of protein phosphorylation by oncogenic B-Raf/MKK/ERK signaling. In addition, 30 phosphorylation sites increased upon drug treatment (Table 1B), revealing mechanisms for suppressing phosphorylation by this pathway. Regulated sites were reproducible between biological replicate experiments (Table S4).
Table 1.
Phosphorylation sites responsive to MKK1/2 inhibitor
| Gene | Name | Swiss- Prot |
Sequencea | Residueb | Ratioc | DBd | EGF Stim.e |
Kinasef | Referencesg |
|---|---|---|---|---|---|---|---|---|---|
|
A. Phosphorylation repressed by U0126 | |||||||||
| MAP Kinase Pathway |
|||||||||
| MAPK1 | MAP kinase, ERK2 | P28482 | TGFLpTEYVA | T185 | 0.2 | a,b | + | MEK1, JAK2, Cot | (Dang et al., 1998; Luciano et al., 2004; Walters et al., 2006; Wang et al., 2002) |
| P28482 | TGFLpTEpYVA | T185;Y187 | 0.1 | a,b | + | MEK1,JAK2, Cot | ibid | ||
| MAPK3 | MAP kinase, ERK1 | P27361 | TGFLpTEYVA | T202 | 0.0 | a,b | + | MEK1/2, Cot | (Luciano et al., 2004; Wang et al., 2002) |
| TGFLpTEpYVA | T202;Y204 | 0.2 | a,b | + | MEK1/2, Cot | ibid | |||
| MAPK14 | MAP kinase, P38α | Q16539 | DDEMpTGYVA | T180 | 0.5 | a,b | ND | MKK3/6 | (Wang et al., 2002) |
| DDEMpTGpYVA | T180;Y182 | 0.5 | a,b | + | MKK3/6 | ibid | |||
|
| |||||||||
| Pro-directed |
|||||||||
| ARHGAP17 | Rho GTPase-activating protein 17 | Q68EM7 | PPTR[pS]PSPP | [S674] | 0.0 | x | 0 | ||
| BAG3 | Bag family molecular chaperone regulator 3 | O95817 | PpSPGPSAVPS[pS]P | S377;[S386] | 0.0 | a,bh | + | ||
| CTTN | Cortactin | Q14247 | TQpTPPVpSPA | T401;S405 | 0.3 | a,b | +,0i | ERK(T405) | (Martinez-Quiles et al., 2004) |
| RLPSpSPVYE | S418 | 0.3 | a,b | 0 | ERK | ibid | |||
| FAM129B | MINERVA/FAM129Bj | Q96TA1 | PPPApSPDGV | S633 | 0.1 | x | + | ||
| ASPEpSPPPApSPDGV | S628;S633 | 0.0 | x | + | |||||
| PEASpSPPAS | S679 | 0.4 | a,b | + | |||||
| ASpSPPApSPL | S679;S683 | 0.1 | a,b | + | |||||
| GTPBP1 | GTP binding protein 1 | O00178 | PEPS[pS]PGAA | [S25] | 0.5 | a,b | 0 | ||
| HDGF | Hepatoma-derived growth factor | P51858 | LLEDpSPKRP | S165 | 0.1 | a,b | 0 | ||
| HNRPK | Heterogeneous nuclear ribonucleoprotein K | P61978 | LQLPpSPTAT | S116 | 0.5 | a,b | + | ||
| MARCKS | Myristoylated alanine-rich C-kinase substrate | P29966 | EAGApSPVEK | S101 | 0.0 | a,b | 0 | ||
| AEPGpSPTAA | S118 | 0.0 | a,b | + | MAPKk | ||||
| PCBP2 | Poly(rC)-binding protein 2 | Q15366 | IESSpSPEVK | S272 | 0.3 | a,b | ND | ||
| RPS6KA1 | Ribosomal protein S6 kinase α1 | Q15418 | KD[pS]PGIPPpSAG | [S363];S369 | 0.0 | a,b | ND | ERK, auto | (Dalby et al., 1998; Jensen et al., 1999) |
| GLLMpTPCYT | T577l | 0.0 | a,b | ND | ERK | ibid | |||
| SMC4 | Structural maintenance of chromosomes protein 4 | Q9NTJ3 | GRTE[pS]PATA | [S41] | 0.3 | a,b | ND | ||
| STAT3 | Signal transducer and activator of transcription 3 | P40763 | DLPMpSPRTL | S727 | 0.2 | a,b | ND | p38α,ERK | (Debidda et al., 2005; Plaza-Menacho et al., 2007; Xu et al., 2003) |
| STK10 | Lymphocyte-oriented kinase | O94804 | PNPSpTPSKA | T952 | 0.0 | a | ND | ||
| STMN1 | Stathmin | P16949 | ELILpSPRSK | S25 | 0.3 | a,b | + | CDC2, MAPK | (Marklund et al., 1993) |
| ApSGQAFELILpSP | S16;S25 | 0.5 | a,b | + | CaMK(S16) | (Marklund et al., 1994) | |||
| TPR | Nucleoprotein TPR | P12270 | EAIHpSPQVA | T2141 | 0.0 | a,b | ND | ||
|
| |||||||||
| Basophilic |
|||||||||
| AKT1S1 | Akt1 substrate 1 | Q96B36 | QYAKpSLPVS | S183 | 0.3 | a,b | - | mTOR | (Oshiro et al., 2007; Wang et al., 2008) |
| ANKRD15 | Ankyrin repeat domain- containing protein 15 | Q14678 | KRSYpSAGNA | S325 | 0.2 | x | ND | ||
| DBNL | Drebrin-like protein | Q9UJU6 | ERAMpSTTSI | S269 | 0.4 | a,b | ND | ||
| RAMpSTTpSISS | S269;S272 | 0.1 | a,b | ND | |||||
| EIF4B | Eukaryotic translation initiation factor 4B | P23588 | RLPKpSPPYT | S93 | 0.2 | a,b | 0 | ||
| GSK3B | Glycogen synthase kinase-3 beta | P49841 | PRTTpSFAES | S9 | 0.7m | a,b | ND | RSK2 | (Sutherland et al., 1993) |
| NEDD4L | E3 ubiquitin-protein ligase nedd4-like protein | Q96PU5 | PRSL[pS]SPTV | [S448] | 0.5 | a,b | 0 | SGK | (Debonneville et al., 2001; Fouladkou et al., 2004) |
| PAK4 | p21-activated kinase 4 | O96013 | KRPLpSGPDV | S181 | 0.0 | b | ND | ||
| PCBP1 | Poly(rC)-binding protein 1 | Q15365 | ARQQpSHFAM | S246 | 0.4 | b | 0 | ||
| PPFIBP1 | Liprin-beta-1 | Q86W92 | KLRRpSQSTT | S594n | 0.5 | (b)o | ND | ||
| KRSApSAPTL | S534n | 0.6 | x | ND | |||||
| RDBP | Negative elongation factor E | P18615 | KRSL[pS]EQPV | [S51] | 0.6 | a,b | ND | ||
| RPS6KA1/3 | Ribosomal protein S6 kinase α1/3 | Q15418 | KKAYpSFCGT | S227p | 0.2 | a,b | ND | PDK1 | (Jensen et al., 1999; Merienne et al., 2000) |
| SASH1 | SAM and SH3 domain- containing protein 1 | O94885 | GRTCpSFGGF | S407 | 0.4 | a | ND | ||
| UFD1L | Ubiquitin fusion degradation protein 1 homolog | Q92890 | GEGQpSLRKK | S299 | 0.3 | a,b | ND | ||
| VIM | Vimentin | P08670 | STRTYpSLGS | S39 | 0.5 | a,b | ND | CaMK, PAK | (Goto et al., 2002; Stefanovic et al., 2005) |
| ZYX | Zyxin | Q15942 | REKV[pS]SIDL | [S142] | 0.0 | a,b | + | AKT | (Chan et al., 2007) |
|
| |||||||||
| Acidophilic |
|||||||||
| CLNS1A | Methylosome Subunit plCln | P54105 | EEEDpSDDDV | S102 | 0.2 | a,b | 0 | ||
| CTTN | Cortactin | Q14247 | EPVYpSMEAA | S447 | 0.6 | (a,b) o | ND | ||
| HDGF | Hepatoma-derived growth factor | P51858 | AEGSpSDEEG | S133 | 0.0 | a,b | 0 | ||
| NAEGpSpSDEEG | S132;S133 | 0.2 | a,b | 0 | |||||
| PDIA6 | Protein disulfide- isomerase A6 precursor | Q15084 | DIDLpSDVEL | S428 | 0.1 | a,b | ND | CK2-A1 | (Janson et al., 1997) |
| PPP1R2 | Protein phosphatase inhibitor 2 | P41236 | EDACpSDTEA | S87 | 0.4 | a,b | ND | CK2 | (Sakashita et al., 2003) |
| EQE[pSpS]GEE | [S120;S121] | 0.0 | a,b | 0 | |||||
| RPLP1 | 60S acidic ribosomal protein P1 | P05386 | KEEpSEEpSDDD | S101;S104 | 0.4 | a,b | 0 | ||
| SAPS3 | SAPS domain family member 3 | Q5H9R7 | DDGGpSDEED | S617 | 0.5 | a,b | 0 | ||
| YWHAZ | 14-3-3 protein zeta/delta | P63104 | LDTLpSEESY | S206 | 0.5 | (a)o | ND | ||
|
B. Phosphorylation stimulated by U0126 | |||||||||
| Proline-directed |
|||||||||
| ARS2 | Arsenite-resistance protein 2 | Q9BXP5 | SEPGpTPPLP | S544 | 1.9 | a,b | ND | ||
| CALD1 | Caldesmon | Q05682 | FS[pS]PTAAGpTPN | [S724];T730 | 2.0 | a,b | - | CDC2 | (Yamashiro et al., 1995) |
| C11orf68 | Conserved hypothetical protein | Q9H3H3 | EEEGpSPGGR | S52q | 3.7 | x | ND | ||
| CARHSP1 | Ca++-regulated heat stable protein 1 | Q9Y2V2 | NVVPpSPLPT | S41 | 11.3 | a,b | + | ||
| DNM1L | Dynamin-1-like protein | O00429 | IMPApSPQKG | S616 | Inf | a,b | ND | ||
| EEF1D | Elongation factor 1-delta | P29692 | TQHVpSPMRQ | S133 | 5.3 | a,b | 0 | CDC2 | (Kawaguchi et al., 2003) |
| FOXK1 | Forkhead box protein K1 | P85037 | EAPApSPLRP | S213 | 2.3 | a,b | 0 | ||
| ApSPLRPLYPQIpSP | S213;S223 | 1.7 | a,b | 0 | |||||
| NCK2 | Cytoplasmic protein Nck2 | O43639 | ARDA[pS]PTPS | [S89] | Inf | a,(b)o | ND | ||
| NES | Nestin | P48681 | GDRGpSPFQE | S1576 | Inf | x | ND | ||
| NUCKS1 | Nuclear ubiquitous casein and cyclin-dep.kinase substrate | Q9H1E3 | EEPEpSPPEK | S214 | 8.0 | a,b | 0 | ||
| PCBP2 | Poly(rC)-binding protein 2 | Q15366 | KPSS[pS]PVIF | [S189] | 4.9 | a,b | - | ||
| SMAD2 | Mothers against decapentaplegic homolog 2 | Q15796 | ILPFpTPPVV | T8 | 4.9 | a,b | ND | ERK1 | (Funaba et al., 2002) |
| SRRM2 | Serine/arginine repetitive matrix protein 2 | Q9UQ35 | T[pS]PQANEQSVpTP | [S857];T866 | 3.5 | a,b | ND | ||
|
| |||||||||
| pTyr |
|||||||||
| GSK3B | Glycogen synthase kinase- 3 beta | P49841 | PNVSpYICSR | Y216 | 3.5 | a,b | 0 | auto, MEK1 | (Cole et al., 2004; Takahashi-Yanaga et al., 2004) |
|
| |||||||||
| Basophilic |
|||||||||
| ANXA2 | Annexin A2 | P07355 | LCKLpSLEGD | S12 | 3.3 | a | ND | ||
| ARFGEF2 | Brefeldin A-inhibited guanine nucleotide- exchange protein 2 | Q9Y6D5 | RERG[pS]SLSG | [S276] | 2.6 | x | ND | ||
| HN1 | Hematological and neurological expressed 1 protein | Q9UK76 | RRNSpSEASS | S88 | 3.3 | a,b | + | ||
| NES | Nestin | P48681 | QRRRpSLGEQ | S768 | 3.5 | b | 0 | ||
| PEA15 | Astrocytic phosphoprotein PEA-15 | Q15121 | IRQPpSEEEI | S116 | 7.0r | a,b | - | AKT1 | (Trencia et al., 2003) |
| PAK2 | p21-activated kinase 2 | Q13177 | QKYLpSFTPP | S141 | 2.3 | a,b | 0 | auto | (Jung and Traugh, 2005) |
| PRKAR2A | cAMP-dependent protein kinase RIIα | P13861 | NRRVpSVCAE | S99 | 3.0 | a,b | 0 | PKA | (Takio et al., 1980) |
| TPD52L2 | Tumor protein D54 | O43399 | ATFKpSFEDR | S166 | 5.7 | a,b | - | ||
|
| |||||||||
| Acidophilic |
|||||||||
| EIF5B | Eukaryotic translation initiation factor 5B | O60841 | PNIEpSGNED | S214 | 8.0 | a,b | 0 | ||
| HSP90AA1 | Heat shock protein Hsp 90α2 | P07900 | EDVGpSDEEE | S263 | 2.3 | a,b | -,0i | ||
| PCNP | PEST proteolytic signal- containing nuclear protein | Q8WW12 | EDEDpSEPEE | S119 | 3.0 | x | ND | ||
| SEPT2 | Septin-2 | Q15019 | PDAEpSDEDE | S218 | 3.7 | a,b | 0 | CK2-A1 | (Huang et al., 2006) |
| SSB | Lupus La protein | P05455 | TKFApSDDEH | S366 | Inf | a,b | 0 | CK2-A1 | (Schwartz et al., 2004) |
Phosphorylated residue in bold with flanking sequence. Multiple phosphorylation sites within the same sequence indicate a multiply phosphorylated peptide. Brackets indicate ambiguous site localization after manual inspection.
Swissprot entry residue numbering.
Ratios of U0126/Control for −79 signals summed over all charge states. Where proteolytic variants of the same peptide were quantified, the highest intensity form was reported. “Inf” and “0” indicate that the phosphopeptide was not quantified, respectively in the Control or U0126 condition.
Site reported in (a) Phosphosite (version from 5/25/07, Hornbeck et al., 2004) or (b) Swissprot Release 13.5/38.5, or (x) absent in either (as of 5/31/08).
Phosphosite observed in HeLa cells (Olsen et al., 2006), indicating (+) EGF-stimulated, (-) EGF-repressed, (0) no change, or (ND) not quantified.
Kinase known to target site, with no distinction between in vivo vs in vitro evidence.
References for known kinase, see Suppl. Materials.
S377 reported in Swissprot and Phosphosite; S386 only in Swissprot.
Direction of phosphorylation change differed depending on proteolytic variants.
MINERVA is our proposed name for FAM129B.
Kinase inferred by substrate similarity as reported in Swissprot entry.
Site numbered for RSK2; the identical RSK1 site (T573) is also phosphorylated (Fig. 4).
Manually quantified for the [M-3H]3− form of fully tryptic variant TTpSFAESCKPVQQPSAFGSMK.
Numbering for alternately spliced isoform 2.
Identified site is unlisted, but database lists a nearby phosphosite (within 1–2 amino acids).
Site numbered for RSK2, based on isoform specific regulation (Fig. 4).
Site identified on two peptides, including a missed cleavage variant containing an N-terminal Arg (not shown). The missed cleavage variant specifies a longer protein form (IPI00027794) than that in Swissprot Q9H3H3, revealing that the Swissprot entry reflects a truncation product.
Manually quantified from [M-H]1− and [M-2H]2− forms of fully tryptic variant QPsEEEIIK, which was the predominant form.
Changes in phosphopeptide abundance may reflect changes in phosphorylation stoichiometry, or may instead reflect differences in protein abundance due to synthesis, degradation, or subcellular translocation. Therefore, in parallel with the −79 Da precursor ion scanning experiment, proteins were profiled by positive mode LC-MS/MS. The resulting dataset yielded identifications and quantitative profiles for proteins in each QAE fraction, against which changes in phosphopeptide abundance could be compared (Table S3). Relative changes in protein abundances were evaluated by spectral counting, summing counts across QAE fractions (Old et al., 2005). Of the quantifiable proteins, only 60 showed significant changes in abundance between control vs drug-treated samples (Fig. S7, p ≤0.01). Most proteins showed no change, which we ascribe to the short treatment time (4 h).
Phosphoproteins targeted by oncogenic B-Raf/MKK/ERK signaling
Phosphorylated residues that were modulated by inhibition of MKK1/2 included known regulatory phosphorylation sites on ERK1, ERK2, RSK1 (p90-RSKα1), and RSK2 (p90-RSKα3) (Table 1). Phosphorylation was also observed at residues known to regulate the activation of MKK1 and MKK2, but these were unaffected by U0126 (Table S2), as expected from prior studies showing inhibition by direct binding and sustained MKK phosphorylation in cells treated with inhibitor (Ahn et al., 2001; Ohren et al., 2004). Thus, regulated phosphorylation was observed on effectors of the MAP kinase cascade that were expected to be activated downstream of B-Raf. Protein kinases other than RSK that are known to be substrates for ERK were observed in the protein profiling experiment (MAPKAP kinases 2 and 3, MST1,3,4, MSK1). However, only MAPKAPK 2 and 3 were observed to be phosphorylated; their phosphopeptides were weak in intensity, despite the fact that their protein spectral counts were similar to RSK1 and RSK2 (not shown). The results suggest that RSKs are predominant among kinase targets of ERK in melanoma cells.
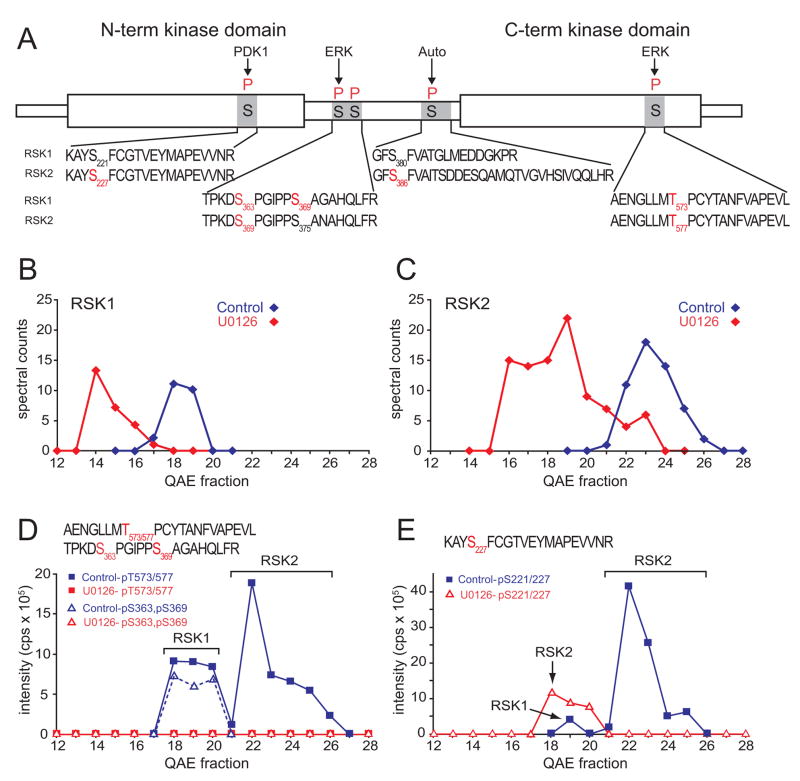
Quantitation of phosphopeptides and protein abundances revealed differential phosphorylation and activation of RSK isoforms, RSK1 and RSK2, which are each comprised of two tandem kinase domains separated by a linker region (Fig. 4A). Previous studies showed that initial phosphorylation of the C-terminal kinase domain and linker region by ERK1/2 promotes subsequent phosphorylation of the N-terminal domain by PDK1, enabling the N-terminal kinase domain to phosphorylate other substrates (Dalby et al., 1998; Frodin et al., 2000). RSK1 and RSK2 eluted earlier by QAE chromatography in cells treated with U0126, consistent with the observed dephosphorylation at several sites (Fig. 4B,C). Because RSK1 and RSK2 resolved from each other, phosphorylation within the C-terminal domain at sites targeted by ERK (Thr573 in RSK1; Thr577 in RSK2) could be verified, despite the fact that the phosphopeptides are identical (Fig. 4D). Phosphorylation at each site was proportional to spectral counts, indicating comparable occupancy in both isoforms, and completely blocked by inhibitor, confirming regulation by B-Raf/MKK/ERK signaling. In contrast, differences in occupancy were observed within the N-terminal domain, at the phosphorylation site targeted by PDK1 (Ser221 in RSK1; Ser227 in RSK2) (Fig. 4E). Phosphorylation of RSK2-Ser227 was significant in controls and suppressed in cells treated with inhibitor, whereas phosphorylation of RSK1-Ser221 was minimal under control conditions. In addition, differential phosphorylation at ERK sites in the linker was suggested, where phosphorylation of RSK1-Ser363/S369 appeared higher than RSK2-S369/S375 (Fig. 4D). These findings illustrate how chromatographic resolution of RSK1 and RSK2, and comparison of phosphopeptide intensities against spectral counts enabled relative occupancy at sites on different isoforms to be estimated. Our results revealed that while the initial C-terminal phosphorylation event is regulated by B-Raf equally on RSK1 and RSK2, RSK2 preferentially undergoes the N-terminal phosphorylation event critically needed for enzyme activation.
Figure 4. Differential activation of RSK isoforms.
(A) Domain structure of RSK isoforms detected in this study, showing observed phosphopeptide sequences (black) and phosphorylated residues (red). (B,C) Spectral counts of isozyme-specific peptides from RSK1 and RSK2, determined by positive mode LC-MS/MS, are plotted vs QAE fraction. Both enzymes eluted earlier when cells were treated with U0126, consistent with their dephosphorylation. (D) −79 Da signal intensities of phosphopeptides derived from RSK1-pT573 or RSK2-pT577, and RSK1-pS363,pS369. Alignment of phosphopeptides and spectral counts shows that T573/577 is phosphorylated in both RSK1 and RSK2 and downregulated by U0126, whereas S363 and S369 are primarily phosphorylated in RSK1. (E) −79 Da signal intensity of a phosphopeptide corresponding to RSK1-pS221 or RSK2-pS227. Alignment with spectral counts shows higher occupancy in RSK2 than RSK1, indicating preferential activation of RSK2.
Other phosphorylation events suppressed by drug treatment, and therefore positively regulated by B-Raf signaling, were catalogued according to primary sequence (Table 1). The majority of sites responsive to B-Raf were either proline-directed or preceded by basic residues, consistent with sequence recognition by MAP kinase (CMGC) family or MAPKAP kinase (AGC) family enzymes. Interestingly, 12 phosphosites showed acidic motifs consistent with the sequence specificity of casein kinases. In addition, sites known to be targeted by mTOR, Akt, and calmodulin kinases were observed. This suggests that other kinase activities are cross-regulated by B-Raf/MKK/ERK and sustained by constitutive signaling in melanoma cells. Of the proteins with phosphorylation sites inhibited by U0126, 12 (23%) were known downstream targets of MAP kinase pathways, and the rest were new candidates for pathway effectors.
Thirty phosphorylation sites were upregulated in response to drug treatment, suggesting protein phosphorylation events that are repressed by oncogenic B-Raf/MKK/ERK signaling (Table 1). An example was GSK3 (Fig. S8A), which is known to be activated through autophosphorylation at the activation lip (Tyr279 in GSK3A, Tyr216 in GSK3B), and repressed by phosphorylation at the N-terminus, which blocks substrate binding (Ser21 in GSK3A; Ser9 in GSK3B) (Cole et al., 2004; Stambolic et al., 1994). Protein profiling revealed the presence of both GSK3A and GSK3B, which partially resolved by QAE chromatography but in this case could not clearly be distinguished (Fig. S8B,C). Phosphorylation of Tyr279/216 was significantly elevated in drug-treated cells, suggesting that one or both forms of GSK3 was derepressed by inhibitor treatment (Fig. S8D). On the other hand, phosphorylation was observed at GSK3B-Ser9 and partly repressed by U0126, consistent with GSK3 inhibition by RSK phosphorylation (Fig. S8E). The results suggest the possibility that oncogenic B-Raf/MKK/ERK signaling might repress GSK3A and/or GSK3B by inhibiting autophosphorylation at a tyrosine residue needed for activity, while enhancing phosphorylation at an inhibitory serine residue on GSK3B.
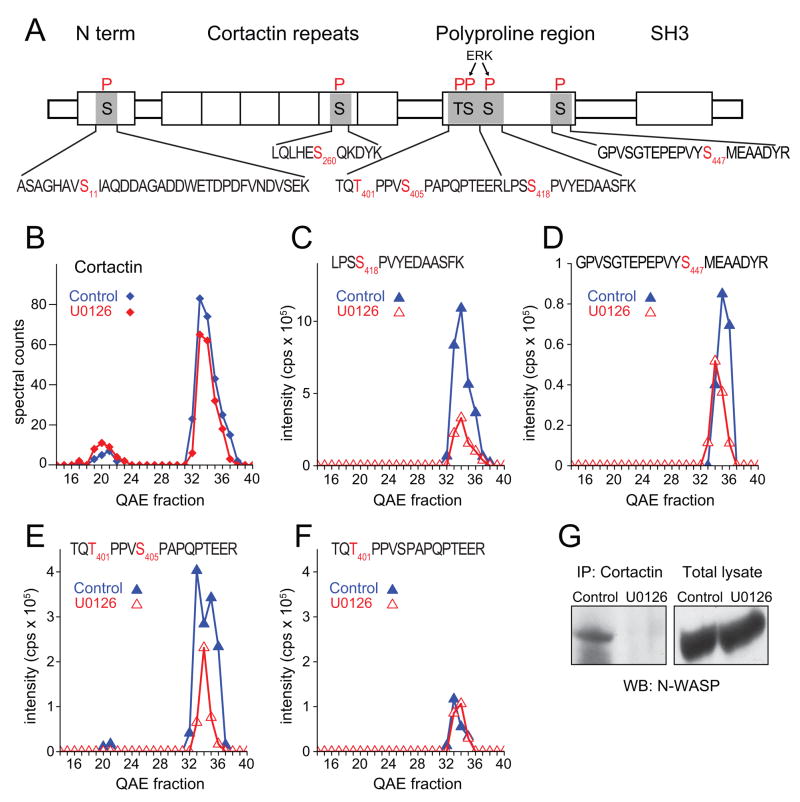
Most phosphoproteins altered in response to U0126 were known effectors of signaling and/or cytoskeletal regulation. An example is cortactin, which promotes cell motility and invasion by controlling actin polymerization at lamellipodia and invadopodia (Weaver, 2008). Cortactin is a well-known Src substrate, and is activated by tyrosine phosphorylation. However, we observed none of the known tyrosine phosphorylation sites on this protein, and instead identified six serine phosphorylation sites (Table 1, Fig. 5A). Spectral counting showed that cortactin resolved into at least two peaks on QAE, an early peak that appeared to be weakly phosphorylated, and a later eluting peak that contained multiple sites of phosphorylation (Fig. 5B). Four of the six sites were significantly repressed by drug treatment, two of which corresponded to proline-directed sites known to be targeted by ERK (Ser405, Ser418), a proline-directed site not previously connected to ERK (Ser401), and a site without clear specificity (Ser447) (Fig. 5C-F). Previous reports have shown that ERK phosphorylation at Ser405 and Ser418 promotes binding of the SH3 domain in cortactin to a proline-rich domain in N-WASP, an allosteric regulator of the Arp2/3 complex (Martinez-Quiles et al., 2004). This is believed to occur through a conformational change that increases accessibility of the cortactin SH3 domain for N-WASP and in turn enables N-WASP to interact with Arp2/3, enhancing nucleation and polymerization of cortical F-actin. Our results predicted that B-Raf-dependent phosphorylation of cortactin would enhance binding to N-WASP. We therefore carried out co-immunoprecipitation assays in melanoma cells, and demonstrated that interactions between cortactin and N-WASP in control cells were significantly disrupted in drug-treated cells (Fig. 5G). Our results suggest that oncogenic B-Raf/MKK/ERK signaling facilitates cortical actin polymerization and membrane ruffling in melanoma cells, by elevating serine phosphorylation at regulatory sites in cortactin.
Figure 5. B-Raf/MKK/ERK promotes cortactin-N-WASP interactions in melanoma.
(A) Domain structure of cortactin, showing observed phosphopeptide sequences (black) and phosphorylated residues (red). (B) Spectral counts indicate two forms which resolve by QAE chromatography. (C,D) Phosphorylation of S418 and S447 is downregulated in cells treated with U0126. (E,F) U0126 downregulates a peptide doubly phosphorylated at T401 and S405. Regulation is attributed to both T401 and S405, because a peptide singly phosphorylated at T401 does not increase in compensation to the decrease in the doubly phosphorylated form, indicating dynamic regulation at both sites. (G) Cortactin interactions with N-WASP, revealed by immunoprecipitating endogenous cortactin from WM115 cell lysates and Western detection of endogenous N-WASP (lane 1). Interactions are disrupted in cells treated with U0126 (lane 2). Similar results were obtained in two independent experiments.
Finally, we examined mechanisms by which the oncogenic B-Raf pathway mediates cancer cell behavior, by investigating phosphorylation targets that have been heretofore uncharacterized. An intriguing case was a protein catalogued as FAM129B (“family with sequence similarity 129, member B”), a 50 kDa polypeptide which belongs to a protein family of unknown category and unknown basic function. Other family members are Niban (FAM129A), a stress-inducible gene found to be upregulated in renal and thyroid cancer models, and B-cell novel protein 1 (BCNP1, FAM129C), a surface membrane protein overexpressed in chronic lymphocytic leukemia (Boyd et al., 2003). FAM129B and Niban/FAM129A share 40% sequence identity, both containing a pleckstrin homology (PH) domain, while FAM129B shares 27% sequence identity with BCNP1/FAM129C. However, little is otherwise known about the function or regulation of these proteins in any organism.
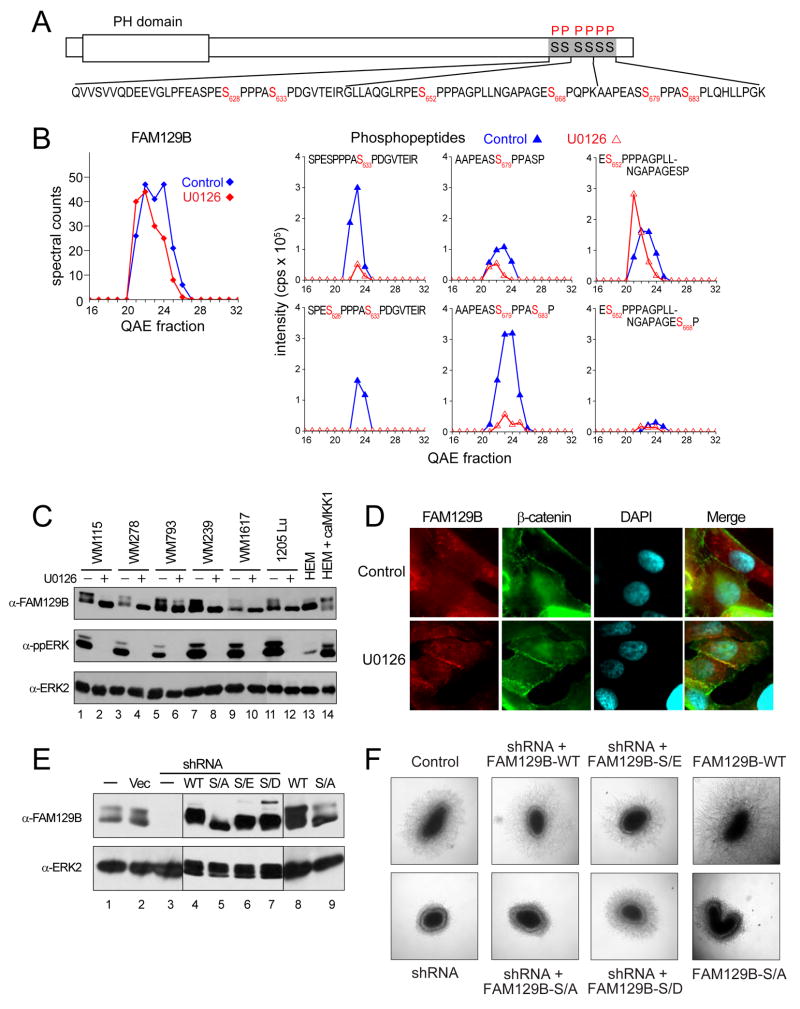
We identified six serine phosphorylation sites on FAM129B, all of which appeared in prior large scale phosphoproteomics studies (Fig. 6A). All six sites showed proline-directed sequences consistent with direct phosphorylation by ERKs, and four sites (Ser628, Ser633, Ser679, S683) were suppressed by more than 2-fold upon pathway inhibition (Fig. 6B). We raised a polyclonal antibody to a unique C-terminal sequence in FAM129B, and showed by Western blotting that the endogenous protein in melanoma cells resolved by gel mobility into multiple forms, all of which converged into a high mobility form upon inhibitor treatment in a manner correlated with dephosphorylation (Fig. 6C, lanes 1,2). Similar gel mobility changes were observed upon inhibitor treatment of five other melanoma cell lines known to harbor oncogenic B-Raf mutations (Fig. 6C, lanes 3–12). In contrast, the gel mobility of FAM129B was rapid in the melanocyte cell line HEM-lp which contains wild type B-Raf, but could be shifted to slower forms in cells stimulated by expression of constitutively active mutant MKK1 (Fig. 6C, lanes 13,14). Likewise, gel mobility retardation was observed in cells from other cancer types harboring activated mutant B-Raf and other oncogenes, as well as in NIH3T3 cells stimulated with PDGF; all shifted to faster mobility upon treatment with U0126 (Fig. S9A,B). Thus, FAM129B appears to be a robust phosphorylation target of B-Raf/MKK/ERK signaling.
Figure 6. FAM129B, a member of a novel gene family, controls melanoma cell invasion.
(A) Domain structure of open reading frame FAM129B, showing observed phosphopeptide sequences (black) and phosphorylated residues (red). (B) Left: Spectral counts show little change in protein abundance with U0126 treatment. Right: Phosphorylation at S628, S633, S679 and S683 is downregulated by U0126, while phosphorylation at S652 and S668 is unaffected. (C) Dephosphorylation of FAM129B by U0126 treatment correlates with faster gel mobility in WM115 and five other melanoma cell lines (lanes 1–12). In contrast, FAM129B shows faster gel mobility in HEM-lp melanocytes (lane 13), and slower mobility when ERK signaling is activated by active mutant MKK1 (lane 14). (D) Dephosphorylation of FAM129B correlates with membrane association. WM115 melanoma cells were treated with or without U0126 for 24 h and immunostained for FAM129B and β-catenin. In control cells, FAM129B reactivity was uniformly dispersed, while β-catenin localized to cell-cell junctions, consistent with cadherin interactions. In U0126-treated cells, FAM129B colocalized with β-catenin at cell-cell junctions. (E) Stable knockdown and rescue of FAM129B. Lanes 1–3: Endogenous FAM129B expression in untreated WM239 cells (lane 1), cells stably expressing vector control (lane 2), or shRNA-FAM129B for knockdown (lane 3). Lanes 4–7: shRNA knockdown cells stably expressing FAM129B-WT or mutant FAM129B-S/A, -S/E, or –S/D substituted at six phosphorylation sites. Lanes 9,10: Naïve WM239 cells stably expressing FAM129B-WT or FAM129B-S/A. (F) FAM129B phosphorylation promotes melanoma cell invasion. Left: Naïve WM239 cells grow into spheroids and invade into 3D collagen matrix after 10 days. Stable knockdown of FAM129B represses invasion but not growth. Middle left: Invasion phenotype of shRNA knockdown is rescued by FAM129B-WT but not FAM129B-S/A. Middle right: Invasion is rescued by FAM129B-S/E and –S/D. Right: Expression of FAM129B-WT in naïve cells increases invasion, while FAM129B–S/A has little effect. Similar results were obtained in two independent experiments (for panels C&D) or three or more independent experiments (for panels E&F).
We asked whether FAM129B localization was modulated in melanoma cells by B-Raf/MKK/ERK signaling, and examined melanoma cells by indirect immunofluorescence. In control cells where B-Raf/MKK/ERK signaling is high, the endogenous FAM129B protein showed a dispersed localization (Fig. 6D, Fig. S10). In contrast, inhibition of B-Raf signaling by drug treatment led to a relocalization of FAM129B into membrane regions where cells contacted other cells, partially overlapping with β-catenin, a marker for cadherin-dependent cell-cell interactions (Fig. 6D, Fig. S10) (Nelson et al., 2004). The results suggest a possible role for FAM129B at the plasma membrane in mediating cell interactions, which is disrupted by C-terminal phosphorylation.
Finally, we investigated the function of FAM129B phosphorylation by B-Raf/MKK/ERK signaling. Melanoma cells were stably transfected with shRNA to achieve efficient knockdown of endogenous FAM129B (Fig. 6E, lanes 1–3), and then rescued by stable transfection with wild type FAM129B or mutants containing Ser→Ala, Ser→Glu, or Ser→Asp substitutions at each of its six phosphorylation sites (Fig. 6E, lanes 4–7). Cells were grown as spheroids embedded in collagen, an assay that mimics 3-dimensional growth and invasion by melanoma cells surrounded by extracellular matrix protein found in human dermis (Smalley et al., 2006). Knockdown of endogenous FAM129B had no effect on spheroid growth rate but significantly blocked cell invasion into collagen (Fig. 6F). Results were not limited to melanoma, as similar responses to FAM129B knockdown were seen in cells from other cancer types (Fig. S9C,D). Expression of FAM129B-WT in the shRNA knockdown cells enhanced cell invasion into matrix, demonstrating that FAM129B promotes melanoma cell invasion. Importantly, the FAM129B-Ser/Ala mutant was unable to rescue the knockdown phenotype, revealing that phosphorylation of this protein is required for invasion (Fig. 6F). By contrast, FAM129B-Ser/Glu and -Ser/Asp both rescued invasion (Fig. 6F). Interestingly, FAM129B-WT enhanced invasion above control levels when expressed in naïve cells expressing endogenous protein, whereas FAM129B-Ser/Ala showed little effect (Fig. 6F). Together, the results suggest that FAM129B enhances invasion when phosphorylated. Based on this new understanding of the properties of FAM129B, we propose renaming this gene to “MINERVA”, to signify its regulation of “Melanoma IN-VAsion by ERK”. Our working model is that MINERVA/FAM129B phosphorylation by B-Raf/MKK/ERK represents an important mechanism for modulating cancer cell behavior.
DISCUSSION
In this study we developed a method to profile phosphoproteins in human cell extracts without requiring phosphopeptide enrichment, by using a negative ionization technique of precursor scanning mass spectrometry to selectively identify phosphopeptides in negative ion mode and isolate them for sequencing. Measurement of −79 Da ion signal intensity allowed us to quantify relative changes in phosphorylation site occupancy, circumventing the need for stable isotope labeling. Thus, the method is applicable to samples which are difficult to examine by isotopic metabolic labeling, such as human tissues and fluids. We then used the method to interrogate targets of B-Raf/MKK/ERK signaling, which is constitutively active in melanoma due to oncogenic genomic mutations in B-Raf. Ninety sites were significantly altered by acute treatment of cells with MKK1/2 inhibitor. Responsive phosphoproteins included ERK and RSK enzymes as well as many signaling effectors and cytoskeletal regulators. Biochemical validation established a function for MINERVA/FAM129B, a member of a novel gene family, in controlling cell invasion into 3-dimensional extracellular matrix in a phosphorylation-dependent manner. Our findings reveal new insight into how oncogenic B-Raf controls properties of cancer cell invasion with potential relevance to metastatic behavior.
The ability to identify phosphopeptides without affinity enrichment represents a significant advance in phosphoproteomics. Peptide selection by LC-MS/MS relies on random sampling weighted by abundance, therefore phosphopeptides in complex, unenriched samples compete poorly against unphosphorylated peptides. This is because phosphopeptide abundances are often low due to substoichiometric phosphorylation, and because phosphopeptide signals are generally weak in positive mode MS, due to their acidity. Use of negative mode MS and detection of the −79 Da signature allows more selective detection and sequencing of phosphopeptide ions against the high background of unphosphorylated peptides. An advantage is that it may at least partly circumvent recovery biases seen with affinity enrichment. The Phosphosite database currently contains more than 25,000 phosphorylation sites in human proteins, most identified after enrichment using Fe3+-IMAC, TiO2, or anti-phosphotyrosine antibodies. As in most large scale studies, the phosphoproteins we observed mainly reflected high abundance proteins where multiple peptides were often detectable. This suggests that some of the 219 localized sites we identified that were not reported previously might be attributable to phosphopeptides that are normally excluded by enrichment methods.
Although the method of precursor ion scanning has been used for many years to identify phosphosites, up to now it has mainly been applied to samples of low complexity, due to limitations in instrument capabilities. A breakthrough was the introduction of the 4000 QTrap mass spectrometer, which can scan for −79 Da fragment ions and then rapidly switch polarity from negative to positive mode for MS/MS. Several advancements allowed us to extend the method to complex samples (Suppl. Methods). A critical improvement was to narrow the isolation window for positive mode scanning. Often in complex samples, the −79 Da phosphopeptide signal is simple but is followed by a more dense pattern of peaks after switching to positive mode. This is due to signals from unphosphorylated peptides alongside phosphopeptides. “Distraction” then occurs, due to preferential MS/MS of coeluting unphosphorylated peptide ions whose intensity or resolution exceeds that of the phosphopeptide ion. Therefore, it was necessary to restrict the mass window to less than 5 Da in the positive scan cycle, in order to partially compensate for this effect. In addition, when switching to positive mode, the instrument selects a positively charged m/z with the same charge state but opposite polarity to the negatively charged precursor ion. However, because the efficiency of charging is not always predictable, the positive ion selected for MS/MS may not represent the charge state with highest intensity. In our analysis, ~5,000 −79 Da precursor ion signals had intensity high enough to match and group peaks between adjacent fractions, and another ~20,000 singletons were observable. Thus, the phosphopeptides we report in this study represent only a small fraction of what we believe can be identified by precursor ion scanning. Software improvements for instrument control are needed to enable sequencing of multiple peaks within the mass window and to more accurately predict positive charge state. With improved software, precursor ion methods should yield greater data capture and representation of the phosphoproteome of complex mixtures.
Of the 90 phosphorylation sites identified in this screen by their sensitivity to U0126, 60 were maintained at an elevated level by constitutive B-Raf signaling. Sensitivity of these phosphosites to acute inhibitor treatment suggests that they are recognized by cellular phosphatases with sufficient basal activity to reverse phosphorylation within the 4 h window. These may be useful intracellular markers of MAP kinase pathway activity state. We compared our findings to a global profiling analysis of EGF-stimulated HeLa cells (Table I, Olsen et al, 2006) which represents the most comprehensive summary of regulated phosphorylation to date. It includes many targets of the MAP kinase pathway, a central mechanism in EGF receptor signaling. Thus, it was not surprising that 32 of the 60 sites we observed to be positively regulated in melanoma were also observed in the HeLa study, of which 16 were also upregulated by EGF stimulation. However, 16 of the phosphosites responsive in melanoma were either unresponsive to EGF or responded in the opposite direction in HeLa cells, suggesting that cells and treatments vary significantly with respect to downstream effectors and susceptibility to dephosphorylation.
The remaining 30 sites were elevated in response to MKK1/2 inhibitor. Thus, one-third of regulated phosphorylation events involved repression of phosphorylation by oncogenic B-Raf signaling, possibly through inhibition of protein kinases or derepression of protein phosphatases. This frequency was higher than that in HeLa cells, where only one-fifth of phosphorylation events decreased in response to EGF treatment (Olsen et al., 2006), and raises the possibility that phosphorylation repression mechanisms may be sustained by B-Raf signaling in cancer in ways that differ from acute growth factor stimulation. For example, cells might induce expression of phosphatases under conditions of sustained signaling, to levels not seen after acute stimulation. Consistent with this we observe that less than 10% of ERK1/2 is activated in melanoma cells and not further enhanced by added mitogen. In contrast, more than 50% ERK1/2 is activated in NIH3T3 cells after acute mitogen stimulation (not shown). This suggests that feedback inhibitory mechanisms exist in melanoma cells, and we speculate that MKK1/2 inhibitor may enhance phosphorylation of many proteins by derepressing these mechanisms.
Although phosphotyrosine comprised 4% of phosphosites in the total dataset, except for ERK1/2 and GSK3, the majority of phosphosites responsive to inhibitor were on Ser/Thr residues. This was expected, based on the phospho-Ser/Thr selectivity of MAP kinase pathway enzymes. Unexpectedly however, acidophilic sites were also observed as regulated targets, suggesting crossregulation of casein kinase enzymes by oncogenic B-Raf/MKK/ERK. In NIH3T3 cells, casein kinase II constitutively associates with the scaffold protein, Kinase Suppressor of Ras (KSR) and contributes to activation of B-Raf and Raf-1 (Ritt et al., 2007). Our findings raise the possibility that casein kinase II may be involved in crossregulatory signaling to additional targets in the MAP kinase pathway besides Raf.
Known ERK1/2 pathway targets identified in this study included RSK1/2, cortactin, stathmin, STAT3, hnRNPK, and MARCKS. The majority were signaling effectors and/or cytoskeletal regulators, which in part reflects the well-known bias of proteomics towards proteins of high abundance. The example of RSK1/2 illustrates the advantage of protein separation before proteolysis in order to reduce sample complexity. Here, phosphorylation of RSK isoforms could be distinguished by combining protein spectral counts with phosphopeptide peak intensity measurements, from which preferential activation of RSK2 could be demonstrated. Differential phosphorylation of protein isoforms is difficult to observe using phosphopeptide enrichment strategies, because phosphosites shared between isoforms often occur within identical peptide sequences that cannot be distinguished by mass.
Among the ~10,000 protein families in the human genome, >40% are of unknown function and not linked to known biological processes. Thus, demonstrating the biological function of unannotated genes provides an excellent way to validate the results of screening. For this study, we chose MINERVA, which belongs to a gene family of unknown category and unknown basic function, and is found widely expressed across tissues. Seven splice variants are predicted, two of which contain the C-terminal phosphorylation site cluster that we observed. The low invasion phenotype of MINERVA knockdown cells, combined with rescue by WT but not Ser→Ala mutant protein, provides convincing evidence that the protein controls events that promote movement through extracellular matrix, and that hyperphosphorylation in response to oncogenic B-Raf is necessary for this function. We speculate that inhibiting B-Raf/MKK/ERK in melanoma cells represses invasion by recruiting unphosphorylated MINERVA to cell-cell membrane junctions, and that phosphorylated MINERVA not only derepresses invasion by relocalizing away from membranes, but also regulates events which actively promote invasion. Our studies demonstrate the role of a previously unannotated gene family in cancer cell invasion.
EXPERIMENTAL PROCEDURES
Materials
Plasmids
A lentivirus plasmid system (a gift of Dr. Xiao-feng Qin, U. Texas MD Anderson Cancer Center (Qin et al., 2003) was used to generate shRNA for knockdown of FAM129B. A retroviral plasmid (pREX-CD2, a gift of Dr. Xuedong Liu, U. Colorado, Boulder) was used for expression of FAM129B. FAM129B was subcloned from I.M.A.G.E. clones MCG-5613 and 10467452 from Open Biosystems Inc. (Huntsville, AL) into the pCMV-HA vector (Clontech, Mountain View, CA) to product an open reading frame corresponding to splice variant NM_022833 (746 aa). This was modified by site-directed mutagenesis (QuikChange, Stratagene, La Jolla, CA) to introduce silent mutations within nucleotides 1274–1294 in order to bypass shRNA recognition. Mutations were also designed to substitute Ser residues 628, 632, 652, 668, 679, and 683 to Ala, Glu, and Asp residues. The resulting FAM129B-WT and FAM129B-S/A, -S/E, and –S/D mutants were subcloned into pREX-CD2, which directs expression of FAM129B and separately expresses the extracellular domain of CD2. Retrovirus was generated in HEK293 Phoenix cells by cotransfection with pREX-FAM129B-CD2 and packaging plasmid pHCMU-VSUG-RA1962.
Antibodies
Rabbit polyclonal antibodies to FAM129B were raised by contract (Sigma GenoSys, St. Louis, MO). The synthetic peptide immunogen corresponded to the C-terminal 14 amino acids of FAM129B splice variants that contain the phosphorylation sites observed in this study (HTTTEDSAGVQTEF, C terminal sequence of NM_022833 and NM_001035534). Antibodies were immunopurified using resin covalently coupled to peptide, eluted with 75 mM Hepes pH 7.2, 3.5 M MgCl2, 25% ethylene glycol, and stored at 4 C.
Cell culture
Cell growth
Melanoma cell lines (WM115, WM278, WM793, WM239, WM1617, 1205Lu) were a gift of Dr. Meenhard Herlyn (Wistar Institute, Philadelphia, PA). Cells were grown in RPMI (Invitrogen), 10% fetal bovine serum. Human epidermal melanocytes (HEM-lp) were grown in Media 254 supplemented with HMGS (Cascade Biologics, Portland, OR) and untreated or infected with adenovirus for expression of constitutively active mutant hMKK1 (ΔN3/S218E/S222D) (Foschi et al., 1997).
Drug treatment
Melanoma cell lines were treated with U0126 (Promega, Madison, WI) at final concentration 10 μM with 0.1% v/v DMSO carrier. Controls were treated in parallel with 0.1% v/v DMSO. Cells were treated for 4 h for proteomics and Western analyses, and for 24 h for immunofluorescence studies.
Spheroid assays
Assays were carried out as described by Smalley et al. (2006). Melanoma cells were plated on 1.5% Noble agar for 3–5 days. Spheroids were collected and centrifuged to remove media, gently resuspended in 2.5 mg/mL bovine dermal collagen (Pure Col, INAMED, Leimuiden, Netherlands) in RPMI-10% FBS, and overlaid on a pre-solidified layer of the same collagen solution. The collagen/cell suspension was allowed to solidify for 1 h, then overlaid with RPMI-10%FBS. Spheroids were grown for 10 days after implanting into collagen, in order to monitor 3-dimensional growth and invasion into collagen, and images were collected by phase microscopy.
Proteomics sample preparation
Cell extraction
WM115 melanoma cells were grown on 150 mm plastic culture dishes (Fisher Scientific, Hampton, NH) to 80% confluence (~5 × 106 cells/plate). Cells were treated with DMSO or U0126 for 4 h at 37 C, then harvested by washing 3 times with ice-cold phosphate buffered saline and scraping into hypotonic lysis buffer (10 mM Tris, pH 8, 1.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol, 1 μM microcystin, 1 mM Na3VO4, 0.7 mL per dish). Cells were disrupted by passage through a 25 gauge × 5/8 inch needle, and nuclei were pelleted by centrifugation (800 × g for 10 min at 4°C). The non-nuclear fraction was then centrifuged in a tabletop ultracentrifuge (TLA100.3, 60,000 rpm, 30 min at 4°C), and the resulting supernatant was removed to yield the cytosolic extract protein pool.
Protein digestion
Cytosolic extract (5 mg protein) was separated by Mono Q FPLC column, eluted with a gradient of 0–0.6 M NaCl over 60 fractions. Proteins in each fraction were alkylated with 14 mM iodoacetamide followed by 8 mM dithiothreitol, and proteolyzed by addition of porcine trypsin (Wako, Richmond, VA) to 4 μg/mL and CaCl2 to 0.5 mM. Samples were digested at 37 C, then stored at −20 C. Peptides were purified on Oasis HLB extraction cartridges (1cc/10 mg, Waters Corp., Milford, Massachusetts) following manufacturer’s protocols. Details are described in Suppl. Methods.
Mass spectrometry
For phosphorylation analysis, each fraction (10 μL, 25% of sample) was analyzed by precursor ion scanning using a 4000 QTrap mass spectrometer (Applied Biosystems, Foster City, CA) interfaced with a NanoLC-2D HPLC (Eksigent, Dublin, CA) for nanoflow chromatography of peptides, using a 15 cm × 75 μm i.d. PepMap100, C18 analytical column with 3 μm bead size (LC Packings, Sunnyvale, CA). Buffer A was 0.1 % formic acid and buffer B was 80% v/v acetonitrile, 0.1 % formic acid. A polarity switching method was used to alternate between detection and sequencing of the phosphopeptides in the same run (Williamson et al., 2006). For protein profiling, one μL of each sample was analyzed using an LTQ-Orbitrap (Thermo Scientific, Waltham, MA) interfaced with an Eksigent NanoLC-2D HPLC. Details are described in Suppl. Methods.
Computational analyses
MS/MS spectra from precursor ion scanning experiments were searched with MASCOT (v. 2.0, MatrixScience, London) using the Human International Protein Index (IPI, v.3.27) database. Parent mass tolerance was 1.2 Da, MS/MS tolerance was 0.6 Da, with fixed modification carbamidomethyl cysteine, and variable modifications methionine oxidation, N-terminal protein acetylation, pyro-glutamic acid on N-terminal Gln, and phosphorylation on Ser, Thr, and Tyr. MS/MS spectra identifications were manually validated before acceptance into Suppl. Table 1. Phosphorylation sites were localized using an in-house implementation of the Ascore (ambiguity score) algorithm described by Beausoleil et al. (2006) written in Perl. Sites were considered localized if Ascore ≥ 14, equivalent to p < 0.05. MS/MS spectra from LTQ/Orbitrap positive mode LC-MS/MS were searched with SEQUEST (v.27 rev 12) and MASCOT. Parent mass tolerance was 1.2 Da, MS/MS tolerance was 0.5 Da, and fixed modifications were set to carbamidomethyl cysteine. Peptides were filtered using in-house MSPlus and MAE algorithms previously described (Resing et al., 2004; Sun et al., 2007).
Supplementary Material
Acknowledgments
We are indebted to David Tinnermeyer and John Fitzpatrick for their help and advice with ABI instrumentation and optimization of precursor ion scanning, Robert Abernathy for computational support, and Theresa Nahreini for flow cytometry cell sorting. This work was supported by NIH grants R01-CA118972 (NGA) and R01-CA126240 (KAR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn NG, Nahreini TS, Tolwinski NS, Resing KA. Pharmacologic inhibitors of MKK1 and MKK2. Methods Enzymol. 2001;332:417–431. doi: 10.1016/s0076-6879(01)32219-x. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Bhatt KV, Spofford LS, Aram G, McMullen M, Pumiglia K, Aplin AE. Adhesion control of cyclin D1 and p27Kip1 levels is deregulated in melanoma cells through BRAF-MEK-ERK signaling. Oncogene. 2005;24:3459–3471. doi: 10.1038/sj.onc.1208544. [DOI] [PubMed] [Google Scholar]
- Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- Boyd RS, Adam PJ, Patel S, Loader JA, Berry J, Redpath NT, Poyser HR, Fletcher GC, Burgess NA, Stamps AC, et al. Proteomic analysis of the cell-surface membrane in chronic lymphocytic leukemia: identification of two novel proteins, BCNP1 and MIG2B. Leukemia. 2003;17:1605–1612. doi: 10.1038/sj.leu.2402993. [DOI] [PubMed] [Google Scholar]
- Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- Carr SA, Huddleston MJ, Annan RS. Selective detection and sequencing of phosphopeptides at the femtomole level by mass spectrometry. Anal Biochem. 1996;239:180–192. doi: 10.1006/abio.1996.0313. [DOI] [PubMed] [Google Scholar]
- Cole A, Frame S, Cohen P. Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. Biochem J. 2004;377:249–255. doi: 10.1042/BJ20031259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- Dalby KN, Morrice N, Caudwell FB, Avruch J, Cohen P. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J Biol Chem. 1998;273:1496–1505. doi: 10.1074/jbc.273.3.1496. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Dever TE, Costello CE, Owens CL, Rosenberry TL, Merrick WC. Location of seven post-translational modifications in rabbit elongation factor 1 alpha including dimethyllysine, trimethyllysine, and glycerylphosphorylethanolamine. J Biol Chem. 1989;264:20518–20525. [PubMed] [Google Scholar]
- Foschi M, Chari S, Dunn MJ, Sorokin A. Biphasic activation of p21ras by endothelin-1 sequentially activates the ERK cascade and phosphatidylinositol 3-kinase. EMBO J. 1997;16:6439–6451. doi: 10.1093/emboj/16.21.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frodin M, Jensen CJ, Merienne K, Gammeltoft S. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 2000;19:2924–2934. doi: 10.1093/emboj/19.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall J, Wellbrock C, Dexter TJ, Roberts K, Marais R, Goding CR. The Brn-2 transcription factor links activated BRAF to melanoma proliferation. Mol Cell Biol. 2004;24:2923–2931. doi: 10.1128/MCB.24.7.2923-2931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorden A, Osman I, Gai W, He D, Huang W, Davidson A, Houghton AN, Busam K, Polsky D. Analysis of BRAF and N-RAS mutations in metastatic melanoma tissues. Cancer Res. 2003;63:3955–3957. [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- Hsu MY, Elder D, Herlyn M. Melanoma: The Wistar Melanoma (WM) Cell Lines. Human Cell Culture. 2002:259–274. [Google Scholar]
- Janek K, Wenschuh H, Bienert M, Krause E. Phosphopeptide analysis by positive and negative ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun Mass Spectrom. 2001;15:1593–1599. doi: 10.1002/rcm.417. [DOI] [PubMed] [Google Scholar]
- Kumar SM, Yu H, Edwards R, Chen L, Kazianis S, Brafford P, Acs G, Herlyn M, Xu X. Mutant V600E BRAF increases hypoxia inducible factor-1alpha expression in melanoma. Cancer Res. 2007;67:3177–3184. doi: 10.1158/0008-5472.CAN-06-3312. [DOI] [PubMed] [Google Scholar]
- Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol Cell Biol. 2004;24:5269–5280. doi: 10.1128/MCB.24.12.5269-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, Yan C, McConnell P, Spessard C, Banotai C, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of Label-free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol Cell Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Resing KA, Meyer-Arendt K, Mendoza AM, Aveline-Wolf LD, Jonscher KR, Pierce KG, Old WM, Cheung HT, Russell S, Wattawa JL, et al. Improving reproducibility and sensitivity in identifying human proteins by shotgun proteomics. Anal Chem. 2004;76:3556–3568. doi: 10.1021/ac035229m. [DOI] [PubMed] [Google Scholar]
- Ritt DA, Zhou M, Conrads TP, Veenstra TD, Copeland TD, Morrison DK. CK2 Is a component of the KSR1 scaffold complex that contributes to Raf kinase activation. Curr Biol. 2007;17:179–184. doi: 10.1016/j.cub.2006.11.061. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K, Li G, Gerrero MR, Brose MS, Volpe P, Weber BL, Van Belle P, Elder DE, Herlyn M. Constitutive mitogen-activated protein kinase activation in melanoma is mediated by both BRAF mutations and autocrine growth factor stimulation. Cancer Res. 2003;63:756–759. [PubMed] [Google Scholar]
- Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–1144. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- Stambolic V, Woodgett JR. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem J. 1994;303(Pt 3):701–704. doi: 10.1042/bj3030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen H, Kuster B, Fernandez M, Pandey A, Mann M. Detection of tyrosine phosphorylated peptides by precursor ion scanning quadrupole TOF mass spectrometry in positive ion mode. Anal Chem. 2001;73:1440–1448. doi: 10.1021/ac001318c. [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Imabayashi F, Iwata T, Kawakami Y. The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J Exp Med. 2006;203:1651–6. doi: 10.1084/jem.20051848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Meyer-Arendt K, Eichelberger B, Brown R, Yen CY, Old WM, Pierce K, Cios KJ, Ahn NG, Resing KA. Improved validation of peptide MS/MS assignments using spectral intensity prediction. Mol Cell Proteomics. 2007;6:1–17. doi: 10.1074/mcp.M600320-MCP200. [DOI] [PubMed] [Google Scholar]
- Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P. Apoptosis induction in human melanoma cells by inhibition of MEK is caspase-independent and mediated by the Bcl-2 family members PUMA, Bim, and Mcl-1. Clin Cancer Res. 2007;13:4934–4942. doi: 10.1158/1078-0432.CCR-07-0665. [DOI] [PubMed] [Google Scholar]
- Weaver AM. Cortactin in tumor invasiveness. Canc Lett. 2008;265:157–166. doi: 10.1016/j.canlet.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C, Marais R. Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation. J Cell Biol. 2005;170:703–708. doi: 10.1083/jcb.200505059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson BL, Marchese J, Morrice NA. Automated Identification and Quantification of Protein Phosphorylation Sites by LC/MS on a Hybrid Triple Quadrupole Linear Ion Trap Mass Spectrometer. Mol Cell Proteomics. 2006;5:337–346. doi: 10.1074/mcp.M500210-MCP200. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-resolved Mass Spectrometry of Tyrosine Phosphorylation Sites in the Epidermal Growth Factor Receptor Signaling Network Reveals Dynamic Modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.