Abstract
A recessive nonsense mutation in the zebrafish recombination activating gene 1 (rag1) gene results in defective V(D)J recombination; however, animals homozygous for this mutation (rag1-/-) are reportedly viable and fertile in standard, nonsterile aquarium conditions but display increased mortality after intraperitoneal injection with mycobacteria. Based on their survival in nonsterile environments, we hypothesized that the rag1-/- zebrafish may possess an “enhanced” innate immune response to compensate for the lack of an adaptive immune system. To test this hypothesis, microarray analyses were used to compare the expression profiles of the intestines and hematopoietic kidneys of rag1 deficient zebrafish to the expression profiles of control (heterozygous) siblings. The expression levels of 12 genes were significantly altered in the rag1-/- kidney including the up regulation of a putative interferon stimulated gene, and the down regulation of genes encoding fatty acid binding protein 10, keratin 5 and multiple heat shock proteins. The expression levels of 87 genes were shown to be significantly altered in the rag1-/- intestine; the majority of these differences reflect increased expression of innate immune genes, including those of the coagulation and complement pathways. Subsequent analyses of orthologous coagulation and complement genes in Rag1-/- mice indicate increased transcription of the complement C4 gene in the Rag1-/-intestine.
Keywords: zebrafish, mouse, rag1, innate immunity, SCID
1. Introduction
Severe combined immunodeficiency (SCID) defines a group of genetic conditions in which individuals lack a functional adaptive immune system. In humans SCID can prove lethal when patients are exposed to a number of common pathogens as well as to a wide range of opportunistic microorganisms. SCID can be caused by mutations in multiple genes, including the recombination activating gene 1 (RAG1) which mediates V(D)J recombination. RAG1-deficient (RAG1-/-) humans represent ∼20% of SCID patients, and present with a characteristic absence or reduction of lymphatic organs and tonsils. RAG1-/- SCID is associated with a loss of B and T cells but is associated with a normal level of natural killer (NK) cells resulting in a T-B-NK+ phenotype. The phenotype of the Rag1-/- mouse model is characterized by an increased susceptibility to infection, and requires specific pathogen-free (SPF) conditions.
The zebrafish (Danio rerio), which has been a powerful model species for embryological studies, is emerging as a model for immunological studies (Trede et al., 2001; Yoder et al., 2002; Traver et al., 2003; Trede et al., 2004; Van Der Sar et al., 2004; Phelps and Neely, 2005; Meeker and Trede, 2008; Sullivan and Kim, 2008). Zebrafish encode genes representative of both the innate and adaptive immune response. The adaptive immune responses of fish and mammals share many similarities, and thus, zebrafish, with its unique developmental and in vivo imaging advantages, is becoming increasingly appreciated as a model for vertebrate immune function. Although efficient technologies are lacking for targeted disruption of genes by homologous recombination in zebrafish, target induced local lesions in genomes (TILLING) methodology is being employed routinely for generating “knock-out” zebrafish (Deiters and Yoder, 2006). A disruption of the rag1 gene by an ENU-induced point mutation that creates a premature stop codon in the rag1t26683 allele (herein referred to the rag1- allele) has been identified using TILLING; this allele encodes a truncated Rag1 protein (Wienholds et al., 2002). Although zebrafish homozygous for this mutation (rag1-/-) are more susceptible to an injected dose of Mycobacterium marinum and their immunoglobulin genes fail to undergo V(D)J recombination, these immune-compromised animals appear to survive in nonsterile aquarium facilities, albeit with reduced fertility (Wienholds et al., 2002; Swaim et al., 2006). Based on these observations, we hypothesized that the rag1-/- zebrafish may possess an “enhanced” innate immune response to compensate for the lack of an adaptive immune system. This hypothesis is supported by earlier observations that Rag1-/- and other immune compromised mice can possess elevated complement and NK cell activity (Shultz et al., 1995; Shultz et al., 2000). In order to better understand the impact of this rag1 mutation on the transcriptome of zebrafish, we compared the transcriptional state of the kidneys and intestines from these animals to normal (heterozygous) siblings and extended studies to the Rag1-/- mouse model. Our results establish that the expression of 12 genes are altered in the rag1-/- zebrafish kidney and 87 genes are altered in the rag1-/- zebrafish intestine. Within the rag1-/- zebrafish intestine numerous innate immune genes, including multiple complement and coagulation factors, are expressed at elevated levels. Furthermore, transcriptional analyses of murine complement and coagulation genes reveal that the complement C4 gene is expressed at elevated levels in the intestine of Rag1-/- mice.
2. Methods
2.1. Animals
Adult zebrafish were maintained at 28 °C in a recirculating aquarium facility (Aquatic Habitats, Apopka, FL) and fed twice daily. For removal of organs, zebrafish were euthanized in a buffered solution of 0.02% Tricaine methanesulfonate (Finquel MS-222; Argent Chemical Laboratories, Redmond WA). The abdominal wall was opened and the intestine was removed. After all intra-abdominal organs were removed the kidney was visualized ventral to the spinal canal and removed.
Female 7-8 month old Rag1-/- (RAG1-B6.12957) and congenic (C57BL/6J) mice were purchased from Jackson Laboratories (Bar Harbor, ME) and euthanized by CO2 asphyxiation. Tissues were collected and snap frozen in liquid nitrogen.
2.2. Zebrafish genotyping
Zebrafish homozygous for a mutant allele (rag1t26683) of the rag1 gene (Wienholds et al., 2002) and heterozygous siblings were collected from matings of heterozygous animals. Genotyping of adult animals was performed on sperm for males and fertilized eggs for females to avoid tail clip-induced injury that could lead to lethal infection in rag1 deficient zebrafish (Swaim et al., 2006). DNA was obtained using the HotSHOT method as described previously (Meeker et al., 2007) and genotyping was performed with a derived cleaved-amplified polymorphic sequence (dCAPS) methodology (Neff et al., 2002). In brief, a partial sequence of the rag1 gene was amplified by PCR using the forward primer Rag1-dCAPs-F-Kpn (GAGTCAGCAGACGAACTGCGGTAC) and the reverse primer Rag1-dCAPS-R2 (ATTCAGTCGCATTGCCAATATCACAG). Two non-complementary bases at the 3′ end of the Rag1-dCAPs-F-Kpn primer (underlined) create a KpnI site in the amplicons from the wild-type allele whereas amplicons from the mutant allele lack this KpnI site. Amplicons were generated using Titanium™ Taq polymerase (BD Bioscience), 1 ng/μl genomic DNA and 35 PCR cycles annealing at 60 °C degrees. The resultant PCR products were digested to completion with KpnI and resolved on a 3% NuSieve (3:1) agarose gel. The interpretation of the genotype was straightforward for males. Females were crossed to wild-type males and eight resulting embryos were genotyped as described above on day 1. Interpretation of the maternal genotype was: 100% of embryos heterozygous (rag1+/-) = homozygous for the mutant allele; 100% of embryos homozygous wild-type = homozygous wild-type; >0% but <100% of embryos heterozygous = heterozygous. The rate of mistaking a heterozygous female for a homozygous mutant under these conditions is ≤0.4% using the binomial distribution (where p=0.5, k=0, n=8). In addition, genotypes of males and females were verified post mortem.
2.3. RNA
Tissues from genotyped adult zebrafish were collected directly into Trizol (Invitrogen, Carlsbad, CA), homogenized using a Tissuemiser Homogenizer (Fisher Scientific, Pittsburgh, PA), stored at -80 °C, thawed within 2 months and total RNA was purified as described by the manufacturer. Frozen mouse tissues were thawed, homogenized in Trizol and total RNA was purified. The quality and quantity of the resulting RNA was assessed using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA.)
2.4. Microarray Analyses
One μg of total zebrafish RNA was converted into labeled cRNA with nucleotides coupled to the fluorescent dye (Cy3 or Cy5) using the Low RNA Input Linear Amplification Kit (Agilent Technologies) following the manufacturer's protocol. The quality and quantity of the resulting labeled cRNA was assessed using a NanoDrop ND-1000 spectrophotometer and an Agilent 2100 Bioanalyzer. Equal amounts of Cy3 and Cy5-labeled cRNA (750 ng) from two different samples were hybridized to zebrafish microarrays (Agilent Zebrafish Oligo Microarrays, G2518A option 001) for 17 hours at 60 °C. Two biological replicates were completed for the following 4 dual hybridizations: 1) Cy3-rag1+/- kidney cRNA and Cy5-rag1-/- kidney cRNA, 2) Cy5-rag1+/- kidney cRNA and Cy3-rag1-/- kidney cRNA, 3) Cy3-rag1+/- intestine cRNA and Cy5-rag1-/- intestine cRNA, and 4) Cy5-rag1+/- intestine cRNA and Cy3-rag1-/- intestine cRNA. The hybridized arrays were then washed and scanned using an Agilent G2565BA scanner.
Data were extracted from the scanned image using Feature Extraction version 8.5 (Agilent Technologies) and imported to GeneSpring GX software version 7.3.1 (Silicon Genetics, Redwood City, CA) using enhanced Agilent FE plug-in for further analysis. The default normalization in GeneSpring was changed to the FE Normalization scenario in order to normalize imported data. To account for dye-swap-specific variation between arrays, a data transformation was executed and the signal channel and control channel measurements for dye-swapped replicates were reversed. An additional data transformation was applied to reset the row expression intensity values which were less than 0.01 to 0.01. Per Spot and Per Chip Intensity-dependent normalization was applied to rescale all genes to the same normalized value range. Finally, per gene normalization to median was completed by dividing the signal of each gene by the median of its measurements in all samples. Cross gene-error model was active based on the replicate. In order to differentiate groups of replicates, parameters were defined (kidney = K and intestine = I) and the same replicate value (K-/- vs. K+/- and I-/- vs. I+/-) was assigned to replicate samples in the same parameter. Comparisons of gene expression data were made between K-/- and K+/- as well as I-/- and I+/- separately.
“Filter on flag” was used to create quality control (QC) gene lists for each category based on flag value (present or marginal) assigned by feature extraction (FE) software during data import in at least 2 out of 4 samples using 21549 all gene list. In addition, “filter on expression” was used to create lists of genes, which display a ≥1.5-fold difference in the rag1-/- samples as compared to the rag1+/- samples. For identification of differentially expressed genes, filter on confidence was used with t-test P value cutoff of 0.05 and fold change for the gene list was generated using filter on Volcano plot.
2.5. Gene List Annotation and Gene Ontology analyses
The nucleotide sequence of the Target Identity (ID) for each Agilent probe (GenBank, TIGR or ENSEMBL ID) was used to assign a transcript/gene Sequence Description and Gene Symbol for each probe. When certain TIGR or ENSEMBL IDs failed to identify a specific transcript/gene nucleotide sequence conclusively, the nucleotide sequence of the Agilent probe was used in a BLASTn search to define the Sequence Description and Gene Symbol. Probes failing to identify any transcript/gene sequence via BLAST analyses were removed from the Gene List.
DAVID software (Huang et al., 2007) was used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Zebrafish Information Network (ZFIN) IDs for each zebrafish sequence were uploaded to DAVID, converted to DAVID IDs and analyzed. As a comparative approach, the ZFIN IDs were converted to human Ensemble Gene IDs using BIOMART software (Durinck et al., 2005) and uploaded to DAVID for analyses.
2.6. Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR)
Total RNA (2 μg) was annealed to oligo dT primers, reverse transcribed with Superscript III (Invitrogen), diluted 5-fold with dH2O and utilized for quantitative PCR. Primers for qRT-PCR of zebrafish transcripts which were designed to span 70 to 100 bp and at least one intron are listed in Table 1. Optimal annealing temperatures were determined by assessing the quantity and purity of zebrafish amplicons when annealed at 55, 60, 65 and 70 °C and visualized by agarose gel electrophoresis. Primers for qRT-PCR of mouse transcripts were selected from the Primer Bank database (http://pga.mgh.harvard.edu/primerbank: Wang and Seed, 2003) and are listed in Table 1. All PCRs were performed using 1 μl of diluted cDNA, in a 25 μl reaction volume (iQ SYBR Green Supermix, BIO-RAD). All reactions were performed and monitored with a BioRad iCycler and iCycler iQ Optical System Software, version 3.0a. Thermal cycling parameters included an initial denaturing at 94 °C for 2 minutes followed by 45 cycles of: denaturing at 94 °C for 30 seconds, annealing at 60 °C (zebrafish primers) or 56 °C (mouse primers) for 30 seconds and extension at 72 °C for 30 seconds. Reactions from individual biological replicates were completed in triplicate and the average relative levels of expression were calculated by normalizing to levels of β actin as described (Livak and Schmittgen, 2001). When multiple biological replicates were analyzed, the average relative level of expression from each replicate was considered as a single data point and the mean (and standard error) calculated. A negative control (no cDNA) for each primer set was utilized to exclude sample contamination.
Table 1. Oligonucleotide Primer Sequences.
| Species | Sequence Description | ZFIN/GenBank Identifiera | Forward Primer | Reverse Primer |
|---|---|---|---|---|
| zebrafish | complement component 9 (c9) | ZDB-GENE-050522-442 | CCGAGATCCTAATTCAATGG | TGTCGATGAGGTCTCCCTAG |
| zebrafish | fibrinogen gamma (fgg) | ZDB-GENE-040426-1998 | TGGACGTGGATGGACTGTAC | GTCATCGGGTGAGAGGTAAC |
| zebrafish | complement component bfb | ZDB-GENE-990415-34 | ATGGGAAGTGGAATGGAAGC | GGTACGTGACTTCATCATCA |
| zebrafish | LOC79635 similar to complement C4-2 | XM 001334604 | TGGTTTGATGCTCGCAGCAG | CCATTCCAGAGAGCTTCCTC |
| zebrafish | vitronectin b (vtnb) | ZDB-GENE-041116-1 | CTTCACACGATACACAGATC | TGATGCCAATCCAGTCTTTG |
| zebrafish | alpha-1-microglobulin/bikunin precursor, like (ambpl) | ZDB-GENE-040426-1608 | GTGACGACATGCCTATGTTC | ACATCTGACAGCTCATGATG |
| zebrafish | si:ch211-240l19.7 | ZDB-GENE-041210-329 | CGGAACCCGGAACAACTGAG | ACAGCACAACAGCGGCTTTG |
| zebrafish | zgc:171537 | ZDB-GENE-i080204-13 | TCATCAACATTGGTAACGCC | GTTCCATGAGAAGTCGAACA |
| zebrafish | β-actin2 | ZDB-GENE-000329-3 | AGAGCTACGAGCTGCCTGAC | TCTCGTGGATACCGCAAGAC |
| mouse | Coagulation factor X (F10) | NM 007972 | GAGGGACACCTACGACTATGAT | GCCCAGTCTTTCTGAGGCA |
| mouse | Fibrinogen alpha (Fga) | NM 010196 | GACCTGAGGCGCAGAATTGA | CTCCAGGCGCTTCATGTCTAT |
| mouse | Fibrinogen beta (Fgb) | NM 181849 | AGTGTTGTGTCCTACGGGATG | CTGAGGAGGTATCGGAAACAGA |
| mouse | Fibrinogen gamma (Fgg) | NM 133862 | ACCAGAGATAACTGTTGCATCCT | CCACGTCGGTTTGGTAAGAAG |
| mouse | Serpina1 | BC012874 | GAGCATTGGCACAGCGTTTG | AAGCGATGGTTGGATGTCAGC |
| mouse | Complement C3 | NM 009778 | CCAGCTCCCCATTAGCTCTG | GCACTTGCCTCTTTAGGAAGTC |
| mouse | complement C4 | NM 009780 | TGCCAATGAAGACTACGAAGA | TGCCATTTCGCCAGATACACA |
| mouse | Complement C9 | NM 013485 | TGTGGAAATGACTTTCAGTGTGA | GCTCTGATTCTTCCGCTACTC |
| mouse | Complement factor b (Bf) | NM 008198 | GGAGTACCTATGTCCCTCTGG | CAATCTTTTGGTCTCGGGTCTG |
| mouse | β-actin | NM 007393 | TGTTTGAGACCTTCAACACC | TAGGAGCCAGAGCAGTAATC |
GenBank accession numbers are listed for sequences not annotated by ZFIN.
2.7. Histopathology
Selected rag1+/- and rag1-/- zebrafish were euthanized with Tricaine methanesulfonate (see above) and a small incision was made on the ventral body wall from the anus to the anterior extent of the main body cavity. Fish were fixed in 10% neutral buffered formalin and processed routinely. Specimens were embedded in paraffin in left lateral recumbency, and multiple step sections were cut at 5 microns. Sections were stained with hematoxylin and eosin (H&E), and analyzed via light microscopy by a single pathologist.
3. Results
3.1. Microarray analyses of rag1-/- zebrafish
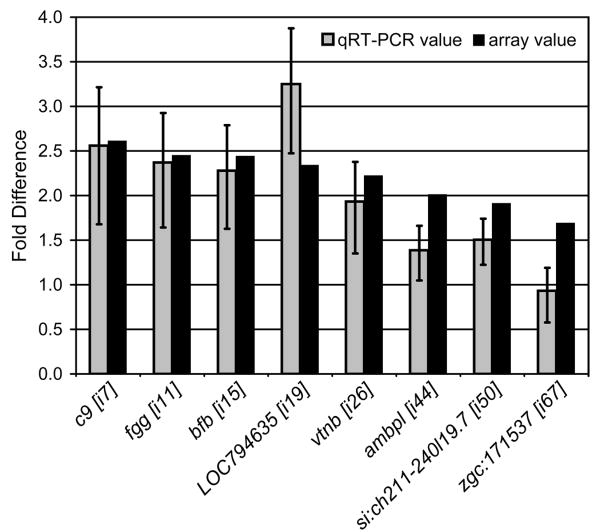
In order to assess the impact of the rag1 mutation on gene expression, we performed transcriptional profiling in the hematopoietic kidney (functional equivalent to bone marrow) and the intestine (major site for microbial challenge) of rag1-/- zebrafish and control heterozygous siblings. Microarray analyses identified 87 genes exhibiting altered transcription at 95% confidence in the rag1-/- intestine; twelve genes with altered transcription (95% confidence) were noted in the rag1-/- kidney (Table 2). Each of the genes was assigned a unique Sequence ID (“Seq. ID” in Table 2), which includes a letter designating source (e.g., i for intestine; k for kidney). Sequence IDs are indicated by brackets throughout the text (e.g. [i1]). The observed differences in transcription levels range between a ∼4.2-fold increase in the intestine and a ∼3.3-fold decrease in the kidney. Increased expression was confirmed by qRT-PCR for 7 of 8 selected transcripts (Figure 1).
Table 2. Zebrafish genes with altered expression in rag1-/- intestine and kidney.
| Seq. ID |
Tissue | Agilent Probe | Sequence Description | Gene Symbol | ZFIN Identifier | GenBank Identifier |
Fold Change |
|---|---|---|---|---|---|---|---|
| i1 | Intestine | A_15_P115601 | LOC555289 similar to microfibrillar-associated protein 4 | LOC555289 | n.d. | XM 677760 | 4.20 |
| i2 | Intestine | A 15 P105559; A 15 P108046; A_15_P104191 | hemopexin | hpx | ZDB-GENE-030131-5773 | NM 001111147 | 3.36 |
| i3 | Intestine | A 15 P110618; A 15 P103668; A_15_P105258 | heat shock cognate 70-kd protein | hsp70 | ZDB-GENE-990415-91 | NM 131397 | 3.35 |
| i4 | Intestine | A_15_P112162 | zgc:63663 | zgc:63663 | ZDB-GENE-040426-1221 | NM 200614 | 3.05 |
| i5 | Intestine | A_15_P101682 | zgc:55406 | zgc:55406 | ZDB-GENE-040426-2891 | NM 213213 | 2.85 |
| i6 | Intestine | A_15_P107518 | complement factor B | cfb | ZDB-GENE-980526-487 | NM 131338 | 2.66 |
| i7 | Intestine | A 15 P112489; A 15 P110271; A_15_P103802 | complement component C9 | c9 | ZDB-GENE-050522-442 | NM 001024435 | 2.62 |
| i8 | Intestine | A 15 P113576; A_15_P104301 | zgc:92882 | zgc:92882 | ZDB-GENE-040718-176 | NM 001002474 | 2.56 |
| i9 | Intestine | A_15_P106533 | transferrin-a | tfa | ZDB-GENE-980526-35 | NM 001015057 | 2.49 |
| i10 | Intestine | A 15 P105661; A 15 P115510; A_15_P109239 | keratin 5 | krt5 | ZDB-GENE-991110-23 | NM 131156 | 2.48 |
| i11 | Intestine | A_15_P121055 | fibrinogen, gamma polypeptide | fgg | ZDB-GENE-040426-1998 | NM 213054 | 2.46 |
| i12 | Intestine | A_15_P102047 | LOC555334 similar to intelectin | LOC555334 | n.d. | XM 677825 | 2.44 |
| i13 | Intestine | A_15_P115458 | inter-alpha (globulin) inhibitor H2 | itih2 | ZDB-GENE-040426-1942 | NM 213007 | 2.44 |
| i14 | Intestine | A 15 P103556; A_15_P112538 | fibrinogen, B beta polypeptide | fgb | ZDB-GENE-030131-9261 | NM 212774 | 2.44 |
| i15 | Intestine | A_15_P106657 | complement component bfb | bfb | ZDB-GENE-990415-34 | NM 131241 | 2.43 |
| i16 | Intestine | A_15_P117565 | apolipoprotein A-IV | apoa4 | ZDB-GENE-030131-1263 | NM 001079861 | 2.42 |
| i17 | Intestine | A_15_P111514 | angiotensinogen | agt | ZDB-GENE-030131-1205 | NM 198063 | 2.40 |
| i18 | Intestine | A_15_P101542 | serine (or cysteine) proteinase inhibitor, clade C (antithrombin), member 1 | serpinc1 | ZDB-GENE-030131-264 | NM 182863 | 2.38 |
| i19 | Intestine | A_15_P109004 | LOC794635 similar to complement C4-2 | LOC794635 | n.d. | XM 001334604 | 2.34 |
| i20 | Intestine | A 15 P110547; A 15 P105980; A_15_P119726 | serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 | serpina1 | ZDB-GENE-030131-1421 | NM 001077758 | 2.33 |
| i21 | Intestine | A_15_P109234 | complement component c3c | c3c | ZDB-GENE-990415-37 | NM 001037236 | 2.32 |
| i22 | Intestine | A 15 P120995; A_15_P111775 | fibronectin 1b | fn1b | ZDB-GENE-030131-6545 | NM 001013261 | 2.30 |
| i23 | Intestine | A_15_P116122 | si:ch211-219i10.1 | si:ch211-219i10.1 | ZDB-GENE-030131-9732 | NM 001030062 | 2.29 |
| i24 | Intestine | A_15_P100047 | LOC557557 similar to complement control protein factor I-B | LOC557557 | n.d. | BC129471 | 2.27 |
| i25 | Intestine | A_15_P105778 | insulin-like growth factor binding protein 1 | igfbp1 | ZDB-GENE-021231-1 | NM 173283 | 2.25 |
| i26 | Intestine | A_15_P119259 | vitronectin b | vtnb | ZDB-GENE-041116-1 | BC055570 | 2.22 |
| i27 | Intestine | A_15_P101717 | alpha-2-HS-glycoprotein | ahsg | ZDB-GENE-041114-50 | NM 212622 | 2.22 |
| i28 | Intestine | A_15_P116686 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 | serpina7 | ZDB-GENE-041010-47 | NM 001005972 | 2.21 |
| i29 | Intestine | A_15_P120815 | carboxyl ester lipase, tandem duplicate 2 | cel.2 | ZDB-GENE-061110-10 | BC076049 | 2.19 |
| i30 | Intestine | A_15_P107918 | glucose-6-phosphatase, catalytic | g6pc | ZDB-GENE-031001-4 | NM 001003512 | 2.17 |
| i31 | Intestine | A_15_P104121 | fibrinogen alpha chain | fga | ZDB-GENE-031010-21 | NM 001002039 | 2.16 |
| i32 | Intestine | A_15_P118096 | zgc:92137 | zgc:92137 | ZDB-GENE-040801-179 | NM 001003729 | 2.15 |
| i33 | Intestine | A_15_P108361 | SHC SH2-domain binding protein 1 | shcbp1 | ZDB-GENE-030131-5865 | NM 199843 | 2.15 |
| i34 | Intestine | A_15_P113038 | si:ch73-252g14.4 | si:ch73-252g14.4 | ZDB-GENE-030131-1133 | NM 001100029 | 2.14 |
| i35 | Intestine | A_15_P113080 | urate oxidase | uox | ZDB-GENE-030826-24 | NM 001002332 | 2.12 |
| i36 | Intestine | A_15_P102721 | zgc:55941 | zgc:55941 | ZDB-GENE-040426-2014 | NM 213249 | 2.12 |
| i37 | Intestine | A 15 P109892; A_15_P117633 | zgc:77778 | zgc:77778 | ZDB-GENE-040426-2281 | NM 213203 | 2.12 |
| i38 | Intestine | A_15_P105655 | si:dkey-38l12.3 | si:dkey-38l12.3 | ZDB-GENE-030131-9563 | BC091470 | 2.09 |
| i39 | Intestine | A_15_P120671; A_15_P100607 | LOC563048 similar to hyaluronic acid binding protein 2 | LOC563048 | n.d. | BC122335 | 2.08 |
| i40 | Intestine | A_15_P113509 | LOC100006895 similar to LOC567732 protein | LOC100006895 | n.d. | XM 001923641 | 2.07 |
| i41 | Intestine | A_15_P111167 | zgc:158628 | zgc:158628 | ZDB-GENE-070112-2012 | NM 001080698 | 2.06 |
| i42 | Intestine | A_15_P117436 | yippee-like 3 | ypel3 | ZDB-GENE-030516-4 | NM 212790 | 2.03 |
| i43 | Intestine | A_15_P117580 | catechol-O-methyltransferase domain containing 1 | comtd1 | ZDB-GENE-030131-1072 | AI477552 | 2.01 |
| i44 | Intestine | A_15_P106260 | alpha-1-microglobulin/bikunin precursor, like | ambpl | ZDB-GENE-040426-1608 | NM 201118 | 2.01 |
| i45 | Intestine | A_15_P109389 | wu:fb20e08 | wu:fb20e08 | ZDB-GENE-030131-300 | AI522512 | 1.99 |
| i46 | Intestine | A 15 P111115; A_15_P107308 | ceruloplasmin | cp | ZDB-GENE-010522-1 | NM 131802 | 1.97 |
| i47 | Intestine | A_15_P100784 | zgc:77825 | zgc:77825 | ZDB-GENE-040426-1887 | NM 205643 | 1.93 |
| i48 | Intestine | A_15_P102952 | retinol binding protein 2b, cellular | rbp2b | ZDB-GENE-040715-7 | NM 001002307 | 1.93 |
| i49 | Intestine | A_15_P110951 | C100002040 similar to myosin heavy chain fast skeletal type 2 | LOC100002040 | n.d. | XM 001339170 | 1.91 |
| i50 | Intestine | A_15_P101461 | si:ch211-240l19.7 | si:ch211-240l19.7 | ZDB-GENE-041210-329 | NM 001082929 | 1.91 |
| i51 | Intestine | A_15_P118717 | mannan-binding lectin serine peptidase 2 | masp2 | ZDB-GENE-060130-154 | BC046065 | 1.90 |
| i52 | Intestine | A_15_P101796 | apolipoprotein A-I | apoa1 | ZDB-GENE-990415-14 | NM 131128 | 1.88 |
| i53 | Intestine | A_15_P100991 | zgc:55398 | zgc:55398 | ZDB-GENE-040426-705 | NM 201048 | 1.84 |
| i54 | Intestine | A_15_P109600 | complement component 6 | c6 | ZDB-GENE-040426-1358 | NM 200638 | 1.81 |
| i55 | Intestine | A_15_P110687 | solute carrier family 38, member 4 | slc38a4 | ZDB-GENE-041010-14 | NM 001005944 | 1.80 |
| i56 | Intestine | A_15_P101041 | zgc:112493 | zgc:112493 | ZDB-GENE-050522-259 | NM 001020703 | 1.78 |
| i57 | Intestine | A_15_P107335 | carboxypeptidase B1 (tissue) | cpb1 | ZDB-GENE-030131-1132 | NM 001110021 | 1.77 |
| i58 | Intestine | A_15_P108307 | LOC100150154 similar to complement component 1, q subcomponent-like 4 | LOC100150154 | n.d. | XM 001921998 | 1.76 |
| i59 | Intestine | A_15_P109564 | zgc:152809 | zgc:152809 | ZDB-GENE-060901-6 | NM 001045860 | 1.75 |
| i60 | Intestine | A_15_P112264 | zgc:92753 | zgc:92753 | ZDB-GENE-040718-307 | NM 001002568 | 1.74 |
| i61 | Intestine | A_15_P107235 | complement component 8, gamma polypeptide | c8g | ZDB-GENE-040426-1898 | NM 200863 | 1.73 |
| i62 | Intestine | A_15_P118422 | heat shock protein 90-alpha 2 | hsp90a.2 | ZDB-GENE-031001-3 | NM 001045073 | 1.72 |
| i63 | Intestine | A_15_P111237 | fatty acid desaturase 2 | fads2 | ZDB-GENE-011212-1 | NM 131645 | 1.72 |
| i64 | Intestine | A_15_P103131 | phospholipase A2, group XIIB | pla2g12b | ZDB-GENE-040426-2771 | NM 213430 | 1.70 |
| i65 | Intestine | A 15 P107233; A_15_P116880 | zgc:77439 | zgc:77439 | ZDB-GENE-031010-24 | NM 212801 | 1.70 |
| i66 | Intestine | A_15_P112820 | apolipoprotein C-II | apoc2 | ZDB-GENE-030131-2168 | AL916586 | 1.70 |
| i67 | Intestine | A_15_P113156 | zgc:171537 | zgc:171537 | ZDB-GENE-i080204-13 | NM 001110109 | 1.69 |
| i68 | Intestine | A_15_P107222 | lipase, hepatic | lipc | ZDB-GENE-040426-1361 | NM 201022 | 1.68 |
| i69 | Intestine | A_15_P104732 | si:ch211-93f2.1 | si:ch211-93f2.1 | ZDB-GENE-041014-96 | BC155642 | 1.68 |
| i70 | Intestine | A_15_P104772 | 3-oxoacid CoA transferase 1b | oxct1b | ZDB-GENE-060929-212 | NM 001077150 | 1.68 |
| i71 | Intestine | A_15_P114341 | zgc:56326 | zgc:56326 | ZDB-GENE-040426-986 | NM 201167 | 1.67 |
| i72 | Intestine | A_15_P107695 | fatty acid binding protein 10, liver basic | fabp10 | ZDB-GENE-020318-1 | NM 152960 | 1.66 |
| i73 | Intestine | A_15_P103003 | zgc:56053 | zgc:56053 | ZDB-GENE-040426-1994 | NM 213050 | 1.64 |
| i74 | Intestine | A_15_P109129 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 3 | cited3 | ZDB-GENE-030425-5 | NM 200078 | 1.61 |
| i75 | Intestine | A 15 P102267; A_15_P119634 | zgc:92630 | zgc:92630 | ZDB-GENE-040718-449 | NM 001002696 | 1.61 |
| i76 | Intestine | A_15_P114906 | secreted immunoglobulin domain 4 | sid4 | ZDB-GENE-050726-3 | NM 001034182 | 1.61 |
| i77 | Intestine | A_15_P106615 | coagulation factor X | f10 | ZDB-GENE-021206-9 | NM 201462 | 1.60 |
| i78 | Intestine | A_15_P111997 | zgc:73355 | zgc:73355 | ZDB-GENE-040426-1775 | NM 200803 | 1.60 |
| i79 | Intestine | A_15_P118819 | zgc:92061 | zgc:92061 | ZDB-GENE-040718-78 | NM 001002383 | 1.59 |
| i80 | Intestine | A_15_P107520 | zgc:92406 | zgc:92406 | ZDB-GENE-040808-53 | NM 001003736 | 1.55 |
| i81 | Intestine | A_15_P107195 | apolipoprotein Eb | apoeb | ZDB-GENE-980526-368 | NM 131098 | 1.55 |
| i82 | Intestine | A_15_P114005 | cyp2k19 cytochrome P450 monooxygenase | cyp2k19 | n.d. | BC134885 | 1.55 |
| i83 | Intestine | A_15_P112103 | si:rp71-1g18.11 (C1ql4) | si:rp71-1g18.11 | ZDB-GENE-040724-34 | NM 001030139 | 1.55 |
| i84 | Intestine | A_15_P116013 | chymotrypsinogen B1 | ctrb1 | ZDB-GENE-030131-1171 | NM 212618 | 1.52 |
| i85 | Intestine | A_15_P109273 | zgc:73262 | zgc:73262 | ZDB-GENE-040426-1735 | NM 200765 | -1.53 |
| i86 | Intestine | A_15_P109192 | solute carrier family 44, member 4 | slc44a4 | ZDB-GENE-040426-1371 | NM 200413 | -1.58 |
| i87 | Intestine | A_15_P103533 | DKEY-202E17.1 novel protein similar to vertebrate core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1(C1GALT1) | LOC555344 | n.d. | NM 001128331 | -1.63 |
| k88 | Kidney | A_15_P113023 | zgc:77651 | zgc:77651 | ZDB-GENE-040426-2381 | NM 213247 | 1.89 |
| k89 | Kidney | A_15_P107584 | chromosome 14 open reading frame 159 | c14orf159 | ZDB-GENE-070725-6 | BC155801 | 1.89 |
| k90 | Kidney | A_15_P105717 | putative interferon stimulated gene 12 (ISG12-2) | zgc:152791 | ZDB-GENE-060901-4 | NM 001007133 | 1.53 |
| k91 | Kidney | A_15_P107483 | zgc:63563 | zgc:63563 | ZDB-GENE-030131-8928 | NM 200044 | 1.52 |
| k92 | Kidney | A_15_P105778 | insulin-like growth factor binding protein 1 | igfbp1 | ZDB-GENE-021231-1 | NM 173283 | -1.55 |
| k93 | Kidney | A_15_P109004 | LOC794635 similar to complement C4-2 | LOC794635 | n.d. | XM 001334604 | -1.64 |
| k94 | Kidney | A_15_P106235 | zgc:92533 | zgc:92533 | ZDB-GENE-040801-181 | NM 001003445 | -1.92 |
| k95 | Kidney | A_15_P112150 | heat shock protein 90-alpha 1 | hsp90a.1 | ZDB-GENE-990415-94 | NM 131328 | -2.47 |
| k96 | Kidney | A 15 P103668; A_15_P110618 | heat shock cognate 70-kd protein | hsp70 | ZDB-GENE-990415-91 | NM 131397 | -2.58 |
| k97 | Kidney | A 15 P118357; A_15_P118422 | heat shock protein 90-alpha 2 | hsp90a.2 | ZDB-GENE-031001-3 | NM 001045073 | -2.69 |
| k98 | Kidney | A_15_P107695 | fatty acid binding protein 10, liver basic | fabp10 | ZDB-GENE-020318-1 | NM 152960 | -3.28 |
| k99 | Kidney | A_15_P105661 | keratin 5 | krt5 | ZDB-GENE-991110-23 | NM 131156 | -3.34 |
Figure 1. Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR) of select zebrafish genes.
Eight genes identified as having an increased expression in the rag1-/- zebrafish intestine (X axis) were analyzed by qRT-PCR. CT values were normalized to β actin2 and the relative fold-difference in expression was calculated (2-ΔΔCT) as described (Livak et al., 2001). Values (gray bars) represent normalized expression levels in rag1-/- zebrafish relative to expression in rag1+/- zebrafish: error bars indicate standard error of the mean. Fold difference values from microarray analyses are shown (black bars) for comparison (numbers in brackets cross-reference to Table 2).
3.2. Transcriptional differences between kidneys of rag1-/- and rag1+/- zebrafish
In the rag1-/- kidney, increased transcription was detected for 4 genes [k88-k91], only one of which, (putative interferon stimulated gene 12; [k90]) has a predictable function. In contrast, decreased levels of transcription are noted for 8 genes [k92-k99] including keratin 5 (krt5), fatty acid binding protein 10 (fabp10), insulin-like growth factor binding protein 1 (igfbp1), heat shock protein 90a2 (hsp90a2), hsp70 and hsp90a and a gene similar to complement C4-2.
3.3. Transcriptional differences between intestines of rag1-/- and rag1+/- zebrafish
In the rag1-/- intestine, decreased transcription was detected for 3 genes [i85-i87], including solute carrier family 44, member 4 (slc44a4) and a gene similar to core 1 synthase, glycoprotein-N-acetylgalactosamine 3-β-galactosyltransferase, 1 (C1GALT1). Of the 84 genes, which are up regulated in rag1-/- zebrafish intestines [i1-i84], many are associated with an immune response including multiple complement factors (discussed below), antimicrobial peptides [i2], apolipoproteins [i16, i52, i66] and chitinases which are likely targeted against chitin from parasites ([i5] and [i36] are similar to chitinases from other species: Elias et al., 2005).
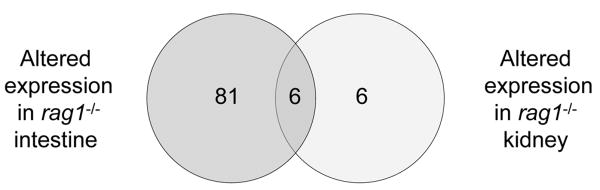
3.4. Six genes are transcriptionally altered in both rag1-/- kidney and intestine
Interestingly, 6 of the 8 genes that are down regulated in the rag1-/- zebrafish kidney were found to be up regulated in the rag1-/- zebrafish intestine (Figure 2). These genes are krt5, fabp10, hsp90a2, hsp70, a gene similar to complement C4-2 and igfbp1. None of the genes that are up regulated in the rag1-/- zebrafish kidney were found to be transcriptionally altered in the rag1-/- zebrafish intestine. The significance of the opposing transcriptional response for these 6 genes in the kidney and intestine is unknown.
Figure 2. Comparison of altered gene expression in intestine and kidney of rag1-/- zebrafish.
Venn diagram showing the number of genes altered in rag1-/- zebrafish intestine and rag1-/- zebrafish kidney (as compared to tissues from rag1+/- zebrafish.) Overlapping genes demonstrated altered expression in both tissues: note that all 6 of these genes demonstrated an increased expression in rag1-/- zebrafish intestine and decreased expression in rag1-/- zebrafish kidney.
3.5. Gene ontology analyses
Gene ontology analyses was performed using the ZFIN ID assigned to each gene in Table 2 and DAVID software (Huang et al., 2007). Of the genes with ZFIN IDs, 77 had been assigned GO Terms and were identified by DAVID software. By restricting the results to GO Terms with six or more associated genes (from Table 2) and for which P<0.01, 11 different GO Terms were identified (6 Biological Processes and 5 Molecular Functions), including Response to Wounding, Response to External Stimulus, and Protease Inhibitor Activity (Table 3).
Table 3. Genes with altered expression in rag1-/- zebrafish that are associated with Gene Ontology (GO) termsa.
| GO Term (P value)b | Ontologyc | DAVID IDd | Gene Name (DAVID) | ZFIN ID | Seq IDe | ||
|---|---|---|---|---|---|---|---|
| Response to Wounding (1.3E-5); Response to External Stimulus (2.1E-4) | BP | 2636106 | fibrinogen, b beta polypeptide | ZDB-GENE-030131-9261 | i14 | ||
| 2610274 | fibrinogen, gamma polypeptide | ZDB-GENE-040426-1998 | i11 | ||||
| 2611966 | complement component c3c | ZDB-GENE-990415-37 | i21 | ||||
| 2609369 | coagulation factor x | ZDB-GENE-021206-9 | i77 | ||||
| 2614980 | complement factor b | ZDB-GENE-980526-487 | i6 | ||||
| 2610608 | fibrinogen alpha chain | ZDB-GENE-031010-21 | i31 | ||||
| Response to Stress (4.4E-5) | BP | 2636106 | fibrinogen, b beta polypeptide | ZDB-GENE-030131-9261 | i14 | ||
| 2610274 | fibrinogen, gamma polypeptide | ZDB-GENE-040426-1998 | i11 | ||||
| 2609392 | insulin-like growth factor binding protein 1 | ZDB-GENE-021231-1 | k92 | ||||
| 2611966 | complement component c3c | ZDB-GENE-990415-37 | i21 | ||||
| 2609369 | coagulation factor x | ZDB-GENE-021206-9 | i77 | ||||
| 2614980 | complement factor b | ZDB-GENE-980526-487 | i6 | ||||
| 2610608 | fibrinogen alpha chain | ZDB-GENE-031010-21 | i31 | ||||
| 2609941 | heat shock cognate 70-kd protein | ZDB-GENE-990415-91 | i3, k96 | ||||
| 2637387 | heat shock protein 90-alpha | ZDB-GENE-990415-94 | k95 | ||||
| Regulation of Biological Quality (8.0E-5) | BP | 2636106 | fibrinogen, b beta polypeptide | ZDB-GENE-030131-9261 | i14 | ||
| 2610274 | fibrinogen, gamma polypeptide | ZDB-GENE-040426-1998 | i11 | ||||
| 2613279 | zgc:92406 | ZDB-GENE-040808-53 | i80 | ||||
| 2610892 | transferrin-a | ZDB-GENE-980526-35 | i9 | ||||
| 2609392 | insulin-like growth factor binding protein 1 | ZDB-GENE-021231-1 | k92 | ||||
| 2610130 | zgc:55398 | ZDB-GENE-040426-705 | i53 | ||||
| 2637785 | zgc:56326 | ZDB-GENE-040426-986 | i71 | ||||
| 2609369 | coagulation factor x | ZDB-GENE-021206-9 | i77 | ||||
| 2610625 | transferrin | ZDB-GENE-980526-35 | i9 | ||||
| 2610608 | fibrinogen alpha chain | ZDB-GENE-031010-21 | i31 | ||||
| Carbohydrate Metabolic Process (2.5E-3) | BP | 2613213 | zgc:92137 | ZDB-GENE-040801-179 | i32 | ||
| 2637785 | zgc:56326 | ZDB-GENE-040426-986 | i71 | ||||
| 2609504 | inter-alpha (globulin) inhibitor h2 | ZDB-GENE-040426-1942 | i13 | ||||
| 2613123 | zgc:55941 | ZDB-GENE-040426-2014 | i36 | ||||
| 2606698 | zgc:92882 | ZDB-GENE-040718-176 | i8 | ||||
| 2610186 | zgc:56053 | ZDB-GENE-040426-1994 | i73 | ||||
| 2638086 | zgc:77912 | ZDB-GENE-040426-2891 | i5 | ||||
| Nitrogen Compound Metabolic Process (5.0E-3) | BP | 2637785 | zgc:56326 | ZDB-GENE-040426-986 | i71 | ||
| 2609504 | inter-alpha (globulin) inhibitor h2 | ZDB-GENE-040426-1942 | i13 | ||||
| 2613123 | zgc:55941 | ZDB-GENE-040426-2014 | i36 | ||||
| 2610186 | zgc:56053 | ZDB-GENE-040426-1994 | i73 | ||||
| 2638086 | zgc:77912 | ZDB-GENE-040426-2891 | i5 | ||||
| 2637387 | heat shock protein 90-alpha | ZDB-GENE-990415-94 | k95 | ||||
| Serine-type Endopeptidase Inhibitor Activity (7.8E-6) | MF | 2642993 | serine (or cysteine) proteinase inhibitor, clade c (antithrombin), member 1 | ZDB-GENE-030131-264 | i18 | ||
| 2608876 | angiotensinogen | ZDB-GENE-030131-1205 | i17 | ||||
| 2612650 | serpin peptidase inhibitor, clade a (alpha-1 antiproteinase, antitrypsin), member 7 | ZDB-GENE-041010-47 | i28 | ||||
| 2609504 | inter-alpha (globulin) inhibitor h2 | ZDB-GENE-040426-1942 | i13 | ||||
| 2611808 | zgc:66321 | ZDB-GENE-040426-1608 | i44 | ||||
| 2620612 | serine (or cysteine) proteinase inhibitor, clade a (alpha-1 antiproteinase, antitrypsin), member 1 | ZDB-GENE-030131-1421 | i20 | ||||
| Endopeptidase Inhibitor Activity (2.6E-5); Protease Inhibitor Activity (3.4E-5); Enzyme Inhibitor Activity (6.4E-5) | MF | 2642993 | serine (or cysteine) proteinase inhibitor, clade c (antithrombin), member 1 | ZDB-GENE-030131-264 | i18 | ||
| 2608876 | angiotensinogen | ZDB-GENE-030131-1205 | i17 | ||||
| 2612650 | serpin peptidase inhibitor, clade a (alpha-1 antiproteinase, antitrypsin), member 7 | ZDB-GENE-041010-47 | i28 | ||||
| 2609504 | inter-alpha (globulin) inhibitor h2 | ZDB-GENE-040426-1942 | i13 | ||||
| 2611966 | complement component c3c | ZDB-GENE-990415-37 | i21 | ||||
| 2609369 | coagulation factor x | ZDB-GENE-021206-9 | i77 | ||||
| 2612404 | chymotrypsinogen b1 | ZDB-GENE-030131-1171 | i84 | ||||
| 2607012 | alpha-2-hs-glycoprotein | ZDB-GENE-041114-50 | i27 | ||||
| 2611808 | zgc:66321 | ZDB-GENE-040426-1608 | i44 | ||||
| 2620612 | serine (or cysteine) proteinase inhibitor, clade a (alpha-1 antiproteinase, antitrypsin), member 1 | ZDB-GENE-030131-1421 | i20 | ||||
| Enzyme Regulator Activity (2.5E-4) | MF | 2642993 | serine (or cysteine) proteinase inhibitor, clade c (antithrombin), member 1 | ZDB-GENE-030131-264 | i18 | ||
| 2608876 | angiotensinogen | ZDB-GENE-030131-1205 | i17 | ||||
| 2612650 | serpin peptidase inhibitor, clade a (alpha-1 antiproteinase, antitrypsin), member 7 | ZDB-GENE-041010-47 | i28 | ||||
| 2609504 | inter-alpha (globulin) inhibitor h2 | ZDB-GENE-040426-1942 | i13 | ||||
| 2611966 | complement component c3c | ZDB-GENE-990415-37 | i21 | ||||
| 2609369 | coagulation factor x | ZDB-GENE-021206-9 | i77 | ||||
| 2612404 | chymotrypsinogen b1 | ZDB-GENE-030131-1171 | i84 | ||||
| 2607012 | alpha-2-hs-glycoprotein | ZDB-GENE-041114-50 | i27 | ||||
| 2611808 | zgc:66321 | ZDB-GENE-040426-1608 | i44 | ||||
| 2620612 | serine (or cysteine) proteinase inhibitor, clade a (alpha-1 antiproteinase, antitrypsin), member 1 | ZDB-GENE-030131-1421 | i20 | ||||
| 2637387 | heat shock protein 90-alpha | ZDB-GENE-990415-94 | k95 | ||||
GO Terms and genes were defined by DAVID software using “Functional Annotation Chart” and selecting “Gene Ontology”. In order for a GO term to be listed in this table, at least 6 genes from Table 2 must be associated with the term with a P value # 0.01.
GO Identity refers to Gene Ontology http://www.geneontology.org/.
Ontologies were restricted to Biological Processes (BP) and Molecular Function (MF).
DAVID identity refers to DAVID software, http://david.abcc.ncifcrf.gov/home.jsp.
Zebrafish Sequence ID refers to Table 2.
3.6. Enhanced transcription of mediators of the complement and coagulation pathways in rag1-/- zebrafish intestine
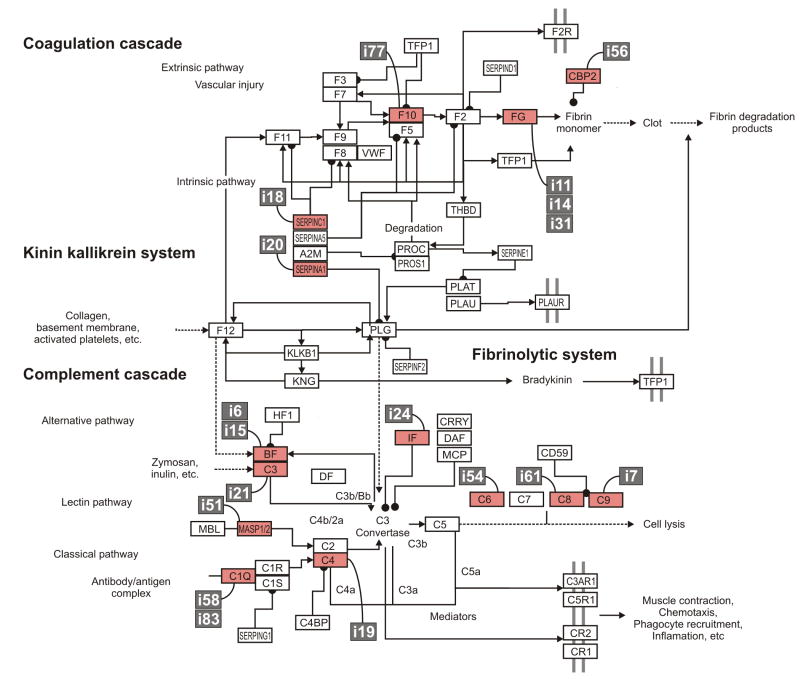
In order to further evaluate the functional relationships of the genes with altered expression in rag1-/- zebrafish, their ZFIN IDs were converted to Ensembl IDs for the corresponding human gene and uploaded into DAVID software. This conversion resulted in DAVID recognizing 50 human orthologs of the 93 different zebrafish sequences shown in Table 2. Using these human sequences, DAVID software identified a single KEGG pathway (P<0.01) which identified 16% of the genes to be involved in the complement and coagulation pathway (P = 2.2E-8). These genes are carboxypeptiodase b2, sepinc1, fibrinogen α, β and γ as well as complement components C3, C6, C9. The significance of this association is strengthened by two additional observations: (1) four additional zebrafish genes from our gene list (Table 2) are associated with this pathway (f10, serpina1, masp2, and c8g) and (2) six additional genes on our gene list are similar to genes involved in this pathway and likely play a role in the complement pathway in zebrafish (Figure 3: [i6] and [i15] are similar to complement factor B; [i24] is similar to complement factor I; [i19] is similar to C4; and [i58] and [i83] are similar to C1Q). It is noted that although [i58] and [i83] are similar to C1Q, it is possible that they may not function in immunity as seen for other C1Q-like genes (Mei et al., 2008b). In summary, 18 of the 84 (∼20%) genes with increased expression in the rag1-/- zebrafish intestine are involved or likely involved in the coagulation and complement pathways.
Figure 3. Increased expression of complement and coagulation factors in rag1-/- intestine.
Of the 84 genes up regulated in the intestine of rag1-/- zebrafish, ∼20% are associated with the complement or coagulation cascades. The mammalian complement/coagulation cascade pathway is shown. Pathway information is adapted from the KEGG database (Kanehisa et al., 2006). Orthologous or similar zebrafish genes are indicated by their Sequence ID (from Table 2; white text on gray background). Proteins with corresponding zebrafish genes which were identified by microarray analyses are highlighted in red. Complement genes with elevated transcription in the rag1-/- zebrafish intestine include c3, c6, c8g, c9, and masp2. Additional genes with elevated transcription in the rag1-/- zebrafish intestine include genes similar to C1Q, C4, complement factor B (BF) and complement factor I (IF). Coagulation genes exhibit elevated transcription in the rag1-/- zebrafish intestine include serpina1, serpinc1, carboxypeptidase B2 (CBP2), coagulation factor X (F10), and fibrinogen α, β and γ (FG).
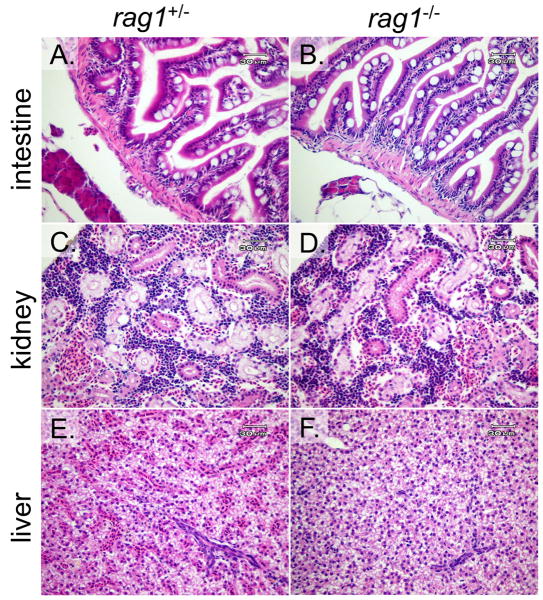
3.7. Histopathology: Tissues from rag1-/- zebrafish had no remarkable abnormalities
Standard histopathology was utilized to compare the morphology and organization of the internal organs of homozygous rag1-/- (Wienholds et al., 2002) and heterozygous rag1+/- mutants. Microscopic examination of the kidneys from rag1-/- fish revealed normal tubules and hematopoietic cellular components but otherwise was unremarkable. Intestines in both rag1-/- and rag1+/- animals also had no remarkable lesions, with a normal population of lymphocytes within the lamina propria. Livers in animals from both groups had normal to moderately vacuolated hepatocytes; the biliary tree was considered normal (Figure 4).
Figure 4. Comparative histopathology of rag1+/- and rag1-/- zebrafish tissues.
Hematoxylin and eosin (H&E) stained tissue sections of rag1+/- (A, C and E) and rag1-/- animals (B, D and F). Tissues include intestine (A and B), kidney (C and D) and liver (E and F). No remarkable microscopic abnormalities were found in either the heterozygous or homozygous mutant zebrafish.
3.8. Complement C4 is elevated in Rag1-/- mouse intestine
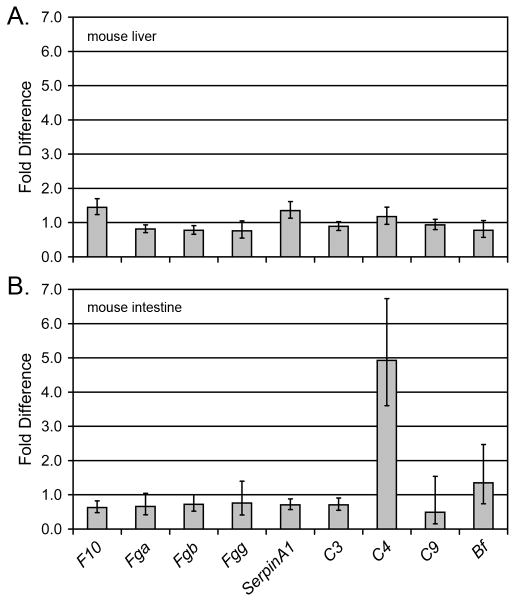
In order to determine if the elevated levels of transcription of complement and coagulation pathway genes observed in the zebrafish extend to other vertebrate species, qRT-PCR for the corresponding genes was carried out with Rag1-/- and wild type (congenic) mice. No significant differences in transcription of these genes were observed in liver, the primary site of complement production in mammals (Figure 5a); however, a significant increase in transcription of complement C4 was observed in the intestine of Rag1-/- mice (Figure 5b).
Figure 5. The transcription of complement C4 is elevated in the Rag1-/- mouse intestine.
Nine complement and coagulation genes (X axis) were analyzed by quantitative RT-PCR from (a) liver and (b) intestine of 3 Rag1-/- and 3 congenic mice. Values were normalized to β actin and the relative fold-difference in expression was calculated (2-ΔΔCT) as described (Livak et al., 2001). Values (Y axis) represent normalized expression levels in rag1-/- mice relative to expression in congenic rag1+/- mice.
4. Discussion
Severe combined immunodeficiency (SCID) defines a group of genetic conditions, including mutations in purine metabolism, cytokine receptor subunits, T cell receptor associated protein kinases, CD45 and RAG proteins (reviewed by Buckley, 2004). RAG1 and RAG2 mediate V(D)J recombination (Schwarz et al., 1991; Buckley, 2004) and RAG1-deficient (RAG1-/-) humans and mice present with a loss of B and T lymphocytes, yet maintain the presence of natural killer (NK) cells resulting in a T-B-NK+ phenotype (Mombaerts et al., 1992; Grundy and Sentman, 2006; Sobacchi et al., 2006). In humans SCID is lethal when patients are exposed to standard environmental pathogens. Isolation in a sterile environment represents one means to manage such individuals. Maternal bone marrow transplants, when feasible, have become a standard curative approach for SCID patients (Buckley, 2004). Similarly, the Rag1-/- mouse model has an increased susceptibility to infections and these animals are housed in SPF facilities. Although rag1-/- zebrafish originally were reported to be viable in standard, nonsterile aquaria (Wienholds et al., 2002), it has been shown that they are more susceptible to mycobacterial infection when infected by intraperitoneal injection (Swaim et al., 2006). It has not been reported if rag1-/- zebrafish are more susceptible to infection by immersion (rather than by injection), leaving open the possibility that lymphocyte-independent zebrafish possess a formidable barrier to environmental-based infection.
Based on observations that Rag1-/- mice can have elevated levels of NK cell activity depending on the background strain (Shultz et al., 2000), we hypothesized that rag1-/- zebrafish may possess an “enhanced” innate immune response to compensate for the lack of an adaptive immune system. This hypothesis was also supported by functional assays using the “scid” mouse which harbors a genetic disruption of Prkdc, a second murine model for SCID (Bosma et al., 1983; Shultz et al., 2000). Sera from scid mice can possess elevated levels of complement activity depending on the background strain (Shultz et al., 1995).
The microarray data presented here demonstrates that the transcriptional status of multiple innate immune genes is elevated in the intestine, but not the kidney of the rag1-/- zebrafish and includes multiple genes involved in the complement and coagulation pathway. Genes representative of both the lectin and alternative pathways of the complement cascade were shown to have elevated transcription in the rag1-/- intestine. For example, masp2 and c3, which are transcriptionally elevated in the rag1-/- intestine, are essential for activating the lectin and alternative pathways, respectively. Transcriptional changes of the classical pathway in the rag1-/- intestine are not as compelling as bony fish encode up to 52 genes with C1q domains (Mei and Gui, 2008) and the zebrafish C1q-like genes with elevated transcription in the rag1-/- intestine may not be directly involved in the complement cascade (Mei et al., 2008a). Nevertheless, the increased expression of c4 within rag1-/- intestine (of both zebrafish and mice) may be informative as C4 plays an early role in both the classical and lectin pathways. In addition, not all zebrafish genes involved in these pathways were represented on the Agilent arrays leaving the possibility that additional complement and coagulation genes may be up regulated in the rag1-/- intestine. For example, there are 4 zebrafish genes, hbl1, hbl2, hbl3 and hbl4, that are closely related to the gene encoding the mammalian mannose binding lectin (MBL in Figure 3; Jackson et al., 2007). In mammals, MBL binds carbohydrates on the surface of a wide range of pathogens and complexes with MASPs which activate the lectin pathway of the complement cascade. Although hbl3, which is the most similar to human MBL, was represented on the Agilent arrays and showed no significant transcriptional alteration in the rag1-/- intestine, it is possible that the expression of other hbl genes may be elevated in these animals.
The presented qRT-PCR data demonstrate an increased transcription of complement C4 in the intestine, but not in the liver of Rag1-/- mice. It is interesting to note that the background strain for our murine Rag1-/- studies was C57BL/6J, which also was the background stain for the scid mice exhibiting a heightened complement activity (Shultz et al., 1995) and the background strain for gnotobiotic Rag1-/- studies demonstrating an increased transcription of multiple innate immune response genes (including complement genes) within the intestine after the introduction of bacterial species into germ free mice (Peterson et al., 2007). As the background strain of mice can influence the innate immune response in SCID models, it is possible that crossing the rag1- allele onto different zebrafish lines may produce animals with varying levels of innate immune function.
Multiple segments of both zebrafish T cell receptor (TCR) and immunoglobulin (Ig) genes were incorporated in the Agilent gene chips used in these studies, but no significant differences in transcriptional levels were observed between rag1-/- and rag1+/- tissues. It is possible that although V(D)J recombination is deficient in the rag1-/- zebrafish, transcription of TCR and Ig genes occurs normally, which could result in a preponderance of sterile transcripts. Alternatively, it may be that expression levels of these genes within the tissues studied are too low to be effectively monitored by microarray analyses. In our experiments, the majority of ∼50 TCR and Ig gene sequences on these arrays were flagged as “absent” and the probes consistently producing a signal showed no significant difference between rag1-/- and rag1+/- tissues.
Three general models can be envisioned to explain the increased transcription of innate immune genes in rag1 deficient animals and why differences are observed between zebrafish and mice. The first model invokes the genetic differences between zebrafish and mice. The second and third models rely on the microbiota of the intestine to shape the genetic response of the host.
The first model is based on whole genome sequencing efforts which indicate that after the divergence of bony fish and mammals, there was a fish-specific whole genome duplication (Hufton et al., 2008). This hypothesis is supported by the presence of 17 Toll-like receptors in zebrafish (Meijer et al., 2004). In addition, there may have been selective pressure to maintain a robust innate immune response since in certain fish, (demonstrable) antibody production can take at least 4-6 weeks, which is much longer than the time leading to host mortalilty for many pathogens (at LD50) and places an increased burden on the innate immune response (Ellis, 2001). It follows that fish potentially possess a more complex, and perhaps, more effective innate immune system than mammals. In this model, the robust zebrafish innate immune system is able to detect the absence of adaptive immunity via unknown mechanisms, and respond by amplifying distinct components of the innate response. The observed complexity of the intestinal versus the kidney response might be due to the substantial microbial burden within the intestinal lumen. In contrast, the less robust innate response in mice is only slightly altered in the absence of Rag1 function.
The second model is founded on the observation that the intestines of zebrafish and mouse are exposed to very different populations of commensal microorganisms (Rawls et al., 2006): the zebrafish intestinal microbiota is numerically dominated by members of the Proteobacteria and Fusobacteria phyla, while the mouse intestinal microbiota is dominated by the Firmicutes and Bacteroidetes phyla. In this model it is proposed that in the absence of Rag1 function, the zebrafish intestinal microbial community is inherently more immunogenic than the mouse intestinal microbial community, thus inducing the transcriptional response observed in the rag1-/- zebrafish intestine.
In the third model, it is proposed that the absence of Rag1 function results in the establishment of an intestinal microbial community with altered composition and/or activity compared to controls. This altered population of commensal microorganisms could then elicit a different set of innate immune responses than is typically evoked by the wild-type microbiota. This is supported by the observation that the expression of multiple innate immune response genes, including at least one complement gene, is altered in the intestines of gnotobiotic zebrafish when raised in the presence of different bacterial species (Rawls et al., 2004). Moreover, loss of the transcription factor T-bet in mice results in population shifts in the intestinal microbial community which are associated with spontaneous and communicable colitis (Garrett et al., 2007), suggesting that host genotype can have profound influence on the composition and activity of the intestinal microbial community. With this model, the different transcriptional responses between the intestines of rag1-/- zebrafish and Rag1-/- mice are explained by relatively larger changes in microbial community composition as a function of rag1 genotype in fish as compared to mammals – therefore fish display a larger transcriptional response in proportion to changes in the intestinal microbiota.
It is possible that all three models may contribute to the observed transcriptional differences between the intestines of rag1-/- zebrafish and Rag1-/- mice. It will be of interest to evaluate the expression of the complement and coagulation genes in rag1-/- zebrafish after exposure to antibiotics or in gnotobiotic rag1-/- animals after controlled exposures to select microorganisms.
The observation that animal models of SCID can possess an “enhanced” innate immune response has now been documented in both mouse and zebrafish; however, the cellular and molecular mechanisms which lead to an alteration in innate immunity are unresolved. Along these lines, it is not known why the kidney of rag1-/- zebrafish, which is the functional counterpart to bone marrow, shows no detectable (by microscopy) lack of lymphocytes when Rag1-/- mice present with a severe reduction in bone marrow lymphocytes (Mombaerts et al., 1992). Although B and T cell function and the process of V(D)J recombination are strikingly similar between fish and mammals, major differences in lymphoid compartmentalization and likely specialization are noted throughout the vertebrate radiations. Furthermore, the lack of reagents with specificity for defined lymphoid markers within zebrafish confounds efforts to draw analogies between the distribution and developmental staging of lymphocytes in zebrafish versus mouse. These deficiencies preclude a more thorough assessment of the rag1-/- lymphocyte phenotype in zebrafish at this time.
Acknowledgments
The authors are indebted to Elwood Linney and Ed Lobenhofer for helpful discussions on microarray studies, John Rawls for insightful discussions on host-microbe interactions, John Sleasman for helpful discussions on immunodeficiencies and Barb Pryor for editorial assistance. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE12655 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12655). This work was supported by NIH grant R01 AI057559 (to GWL) and by funding from the All Children's Hospital Foundation (to GWL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Bosma GC, Custer RP, Bosma MJ. A severe combined immunodeficiency mutation in the mouse. Nature. 1983;301:527–530. doi: 10.1038/301527a0. [DOI] [PubMed] [Google Scholar]
- Buckley RH. Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Annu Rev Immunol. 2004;22:625–655. doi: 10.1146/annurev.immunol.22.012703.104614. [DOI] [PubMed] [Google Scholar]
- Deiters A, Yoder JA. Conditional Transgene and Gene Targeting Methodologies in Zebrafish. Zebrafish. 2006;3:415–429. doi: 10.1089/zeb.2006.3.415. [DOI] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- Elias JA, Homer RJ, Hamid Q, Lee CG. Chitinases and chitinase-like proteins in T(H)2 inflammation and asthma. J Allergy Clin Immunol. 2005;116:497–500. doi: 10.1016/j.jaci.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Ellis AE. Innate host defense mechanism of fish against viruses and bacteria. Dev Comp Immunol. 2001;25:827–839. doi: 10.1016/s0145-305x(01)00038-6. [DOI] [PubMed] [Google Scholar]
- Garrett WS, Lord GM, Punit S, Lugo-Villarino G, Mazmanian SK, Ito S, Glickman JN, Glimcher LH. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy MA, Sentman CL. Immunodeficient mice have elevated numbers of NK cells in non-lymphoid tissues. Exp Cell Res. 2006;312:3920–3926. doi: 10.1016/j.yexcr.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Huang dW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton AL, Groth D, Vingron M, Lehrach H, Poustka AJ, Panopoulou G. Early vertebrate whole genome duplications were predated by a period of intense genome rearrangement. Genome Res. 2008;18:1582–1591. doi: 10.1101/gr.080119.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AN, McLure CA, Dawkins RL, Keating PJ. Mannose binding lectin (MBL) copy number polymorphism in Zebrafish (D. rerio) and identification of haplotypes resistant to L. anguillarum. Immunogenetics. 2007;59:861–872. doi: 10.1007/s00251-007-0251-5. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meeker ND, Hutchinson SA, Ho L, Trede NS. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. Biotechniques. 2007;43:610, 612, 614. doi: 10.2144/000112619. [DOI] [PubMed] [Google Scholar]
- Meeker ND, Trede NS. Immunology and zebrafish: Spawning new models of human disease. Dev Comp Immunol. 2008;32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Mei J, Chen B, Yue H, Gui JF. Identification of a C1q family member associated with cortical granules and follicular cell apoptosis in Carassius auratus gibelio. Mol Cell Endocrinol. 2008a;289:67–76. doi: 10.1016/j.mce.2008.02.016. [DOI] [PubMed] [Google Scholar]
- Mei J, Gui J. Bioinformatic identification of genes encoding C1q-domain-containing proteins in zebrafish. J Genet Genomics. 2008;35:17–24. doi: 10.1016/S1673-8527(08)60003-X. [DOI] [PubMed] [Google Scholar]
- Mei J, Zhang QY, Li Z, Lin S, Gui JF. C1q-like inhibits p53-mediated apoptosis and controls normal hematopoiesis during zebrafish embryogenesis. Dev Biol. 2008b;319:273–284. doi: 10.1016/j.ydbio.2008.04.022. [DOI] [PubMed] [Google Scholar]
- Meijer AH, Gabby Krens SF, Medina RI, He S, Bitter W, Ewa Snaar-Jagalska B, Spaink HP. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol. 2004;40:773–783. doi: 10.1016/j.molimm.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Iocomini J, Johnson RS, Herrup J, Tonegawa S. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18:613–615. doi: 10.1016/s0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Phelps HA, Neely MN. Evolution of the Zebrafish Model: From Development to Immunity and Infectious Disease. Zebrafish. 2005;2:87–103. doi: 10.1089/zeb.2005.2.87. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K, Hansen-Hagge TE, Knobloch C, Friedrich W, Kleihauer E, Bartram CR. Severe combined immunodeficiency (SCID) in man: B cell-negative (B-) SCID patients exhibit an irregular recombination pattern at the JH locus. J Exp Med. 1991;174:1039–1048. doi: 10.1084/jem.174.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz LD, Lang PA, Christianson SW, Gott B, Lyons B, Umeda S, Leiter E, Hesselton R, Wagar EJ, Leif JH, Kollet O, Lapidot T, Greiner DL. NOD/LtSz-Rag1null mice: an immunodeficient and radioresistant model for engraftment of human hematolymphoid cells, HIV infection, and adoptive transfer of NOD mouse diabetogenic T cells. J Immunol. 2000;164:2496–2507. doi: 10.4049/jimmunol.164.5.2496. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Schweitzer PA, Christianson SW, Gott B, Schweitzer IB, Tennent B, McKenna S, Mobraaten L, Rajan TV, Greiner DL. Multiple defects in innate and adaptive immunologic function in NOD/LtSz-scid mice. J Immunol. 1995;154:180–191. [PubMed] [Google Scholar]
- Sobacchi C, Marrella V, Rucci F, Vezzoni P, Villa A. RAG-dependent primary immunodeficiencies. Hum Mutat. 2006;27:1174–1184. doi: 10.1002/humu.20408. [DOI] [PubMed] [Google Scholar]
- Sullivan C, Kim CH. Innate Immune System of the Zebrafish, Danio rerio. Nucleic Acids and Molecular Biology. 2008;21:113–133. [Google Scholar]
- Swaim LE, Connolly LE, Volkman HE, Humbert O, Born DE, Ramakrishnan L. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun. 2006;74:6108–6117. doi: 10.1128/IAI.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Herbomel P, Patton EE, Murphey RD, Yoder JA, Litman GW, Catic A, Amemiya CT, Zon LI, Trede NS. The zebrafish as a model organism to study development of the immune system. Adv Immunol. 2003;81:253–330. [PubMed] [Google Scholar]
- Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- Trede NS, Zapata A, Zon LI. Fishing for lymphoid genes. Trends Immunol. 2001;22:302–307. doi: 10.1016/s1471-4906(01)01939-1. [DOI] [PubMed] [Google Scholar]
- Van Der Sar AM, Appelmelk BJ, Vandenbroucke-Grauls CM, Bitter W. A star with stripes: zebrafish as an infection model. Trends Microbiol. 2004;12:451–457. doi: 10.1016/j.tim.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Wang X, Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Schulte-Merker S, Walderich B, Plasterk RHA. Target-selected inactivation of the zebrafish rag1 gene. Science. 2002;297:99–102. doi: 10.1126/science.1071762. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Nielsen ME, Amemiya CT, Litman GW. Zebrafish as an immunological model system. Microbes and Infection. 2002;4:1469–1478. doi: 10.1016/s1286-4579(02)00029-1. [DOI] [PubMed] [Google Scholar]