Abstract
Neuroendocrine signaling pathways play a central role in modulating animal body size in response to environmental signals. Little is known, however, regarding how these neuroendocrine circuits are controlled. An important advance in this area is reported in this issue of Genes & Development by Kaplan and colleagues (pp. 1877–1893), who show that serotonergic neurons regulate the growth of peripheral tissues in Drosophila through the insulin/IGF pathway.
Keywords: Nucleostemin, serotonin, insulin, neurons, growth, size
How animals grow to a specific final body size remains one of the great puzzles of biology. Although it is clear that body size is determined by both genetic and environmental components, the relative contribution of these factors varies between species (Visscher et al. 2008). In humans, for example, ∼20% of variation in body height is due to environmental factors (Silventoinen 2003), while the size of genetically identical fruit flies can differ by several fold depending on dietary conditions and population density (Beadle et al. 1938). In spite of numerous observations of this phenomenon, however, the molecular mechanisms by which genetic and environmental factors are integrated to control growth remain poorly understood.
Studies in higher organisms have defined a central role for neuroendocrine signaling in controlling growth and coordinating this response with varying environmental conditions. One of the best characterized systems is the mammalian hypothalamic–pituitary growth hormone (GH) axis. External stimuli, such as exercise, stress, and nutritional status, modulate the release of two neuropeptides from the hypothalamus, growth hormone-releasing hormone (GHRH) and somatostatin. These factors, in turn, control GH secretion by neuroendocrine cells of the anterior pituitary gland. GH has systemic effects on growth by regulating the synthesis and release of insulin-like growth factors (IGFs) in peripheral tissues. Many factors affect GHRH and somatostatin release, including central neurotransmitters such as dopamine, adrenaline, serotonin, and acetylcholine (Reichlin 1998). The complexity of the mammalian brain, however, makes it difficult to understand how these neuroendocrine circuits regulate growth at a cellular and molecular level. In contrast, studies of neuroendocrine and insulin/IGF signaling in the fruit fly, Drosophila, provide a simpler genetic system to characterize growth control. In the fly, Drosophila insulin-like peptides (DILPs) are produced by small clusters of neuroendocrine cells in the brain and act to regulate cellular growth in peripheral tissues (Geminard et al. 2006). Although it is likely that, similar to mammals, neuronal factors modulate DILP production, the nature of these upstream regulators remains poorly understood. A study by Kaplan et al. (2008) in this issue of Genes & Development provides exciting evidence that these neuronal signals do indeed exist. They show that serotonergic neurons control adult Drosophila body size by regulating the insulin/IGF pathway in peripheral tissues, paving the way for a better understanding of how animals coordinate their growth and development with varying environmental conditions.
An unexpected role for a nucleostemin family GTPase in growth control
Kaplan et al. (2008) were drawn into studies of growth regulation through their functional characterization of a GTPase family that is collectively referred to as YRG (Ylqf-related GTPase) (Reynaud et al. 2005) or MMR1/HSR1 GTPases (Vernet et al. 1994). There are at least four YRG family members in the budding yeast Saccharomyces cerevisiae, six in Drosophila, and seven in mammals (Reynaud et al. 2005). To date, all members of this family studied at a molecular level appear to function in ribosome biogenesis (Reynaud et al. 2005). Lsg1p regulates 60S ribosomal subunit biogenesis (Kallstrom et al. 2003). Nog2p is required for late 60S maturation steps (Saveanu et al. 2001), while Nug1p is involved in the export of preribosomal particles from the nucleolus to the nucleoplasm (Bassler et al. 2001). Based largely on work in yeast and mammalian cell culture, YRG proteins have been implicated in growth (Grn1p) (Du et al. 2006), proliferation (Nucleostemin) (Tsai and McKay 2002), and cell survival (Nug1p, Nog2p, Mtg1p, and Lsg1p) (Reynaud et al. 2005). In contrast to these cellular studies, however, relatively little is known regarding how YRG family members function in animal models, with two reports showing that nucleostemin can regulate cell proliferation in mice and Xenopus embryos (Beekman et al. 2006; Zhu et al. 2006).
In an effort to expand our understanding of YRG proteins in multicellular organisms, Kaplan et al. (2008) examine the function of four YRG genes in Drosophila. They show that two of them, ns1 and ns2 (orthologs of NUG1 and NOG2, respectively), are essential for embryonic viability, while a P-element insertion disrupting ns4 (the ortholog of GNL1) has no apparent effect. Interestingly, however, loss-of-function mutants for the LSG1 ortholog ns3 exhibit a significant developmental delay with many animals dying as late larvae or pupae. Surviving ns3 adults appear normal and have a normal life span, but are, on average, 60% the size of wild-type flies and have fewer and smaller cells. Given that YRG proteins are implicated in ribosome biogenesis, one simple explanation for this effect on growth could be that ribosomal function is altered in ns3 mutants. Partial loss-of-function mutations in ribosomal protein genes, as is found in heterozygous Minute mutants, inhibit cell growth (Lambertsson 1998). Indeed, ns3 is expressed widely in the animal, and levels of the ribosomal protein RPS6 are significantly reduced in extracts from ns3 mutant larvae, supporting a role in cellular ribosomal activity. In sharp contrast to this model, however, specific expression of wild-type NS3 in ns3 mutant wings has no effect on cell size or number, indicating that ns3 must act outside these cells to control growth. Moreover, clones of ns3 mutant cells in the wing are identical in size to those of adjoining wild-type cells, indicating that this gene regulates growth in a non-cell-autonomous manner. In addition, no difference was detected in the levels of RPS6 protein in ns3 mutant clones compared to their wild-type neighbors, suggesting that NS3 does not play a direct role in ribosome biogenesis in peripheral cells. These remarkable results establish a foundation for the rest of the paper—defining how ns3 regulates systemic growth in a non-cell-autonomous manner.
NS3 regulates insulin signaling
Mutations in the Drosophila insulin/IGF pathway lead to phenotypes that resemble those of ns3 mutants, suggesting that ns3 exerts its effects on growth through insulin/IGF signaling. Mutants for the single fly homolog of the mammalian insulin and IGF receptor, InR, display severe developmental delays and lethality at early stages of development (Chen et al. 1996). Surviving adults appear normal, but are significantly smaller than wild-type flies due to a reduction in both cell size and cell number. Mutants that disrupt signaling components downstream from InR, including the Akt1 kinase, also display similar phenotypes (Garofalo 2002; Edgar 2006). Based on this, Kaplan et al. (2008) examined the effects of ns3 mutations on the insulin/IGF pathway and show that Akt1 phosphorylation, a hallmark of active insulin signaling, is significantly reduced in ns3 mutants. Moreover, expression of a constitutively active form of Akt1 in the adult eye is sufficient to rescue the reduced size of this tissue in ns3 mutants as well as restore the appropriate number and size of cells in a cell-autonomous manner. This result provides strong evidence that ns3 exerts its effects on the growth of peripheral tissues through the insulin/IGF pathway (Fig. 1). The absence of effects on growth or Akt1 phosphorylation in ns3 mutant clones, however, indicate that ns3 acts outside of these cells to impart its effects on insulin/IGF signaling.
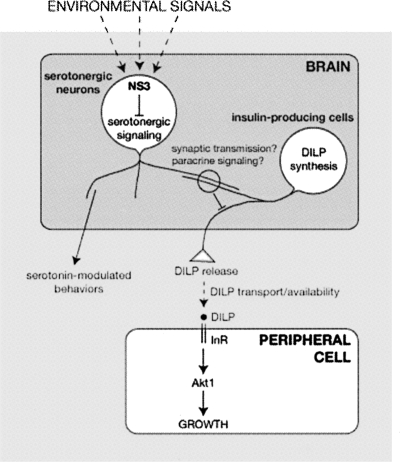
Figure 1.
A model for the regulation of body size by Drosophila NS3. NS3 acts through serotonergic neurons to regulate DILP release by the insulin producing cells. The reduced level of released DILPs in ns3 mutants leads to decreased peripheral insulin signaling through the insulin receptor (InR) and downstream effector Akt1, resulting in reduced overall growth. Close apposition of serotonergic neuron and IPC projections suggest that these cells communicate through synaptic or paracrine signaling to coordinate growth with serotonin-modulated behaviors and the environment.
A simple explanation for how ns3 might regulate peripheral insulin/IGF signaling would be through the insulin-producing cells (IPCs), the prominent DILP expressing neuroendocrine cells of the brain (Brogiolo et al. 2001; Rulifson et al. 2002). Similar to ns3 mutants, IPC ablation results in developmental delays and reduced growth (Rulifson et al. 2002; Broughton et al. 2005). IPCs extend processes that terminate on the heart, suggesting that they exert their systemic effects on growth through direct release of DILPs into the circulatory system (Rulifson et al. 2002). Using a dilp2-GAL4 driver to express NS3 specifically in the IPCs of ns3 mutant animals, however, has no effect on ns3 mutant phenotypes, indicating that this gene acts outside of the IPCs to affect peripheral insulin/IGF signaling. Accordingly, Kaplan et al. (2008) tested whether ns3 acts through any of three key signaling centers that could impact insulin/IGF activity during development: the larval fat body, ring gland, or brain. The fat body can regulate body size through secretion of dALS, the Drosophila ortholog of the mammalian IGF-binding protein, acid-labile subunit. dALS regulates both growth and metabolism, apparently by forming a complex with DILPs and modulating their activity in the hemolymph (Arquier et al. 2008). Basal ecdysone production in the ring gland can also control organismal growth through effects on insulin signaling (Colombani et al. 2005). Finally, given the extensive data in mammalian systems for neuronal regulation of growth through the GH/IGF pathway, it remains possible that the central nervous system could modulate insulin/IGF signaling in Drosophila. Tissue-specific expression of NS3 in the fat body or ring gland had no effect on ns3 mutant phenotypes. NS3 expression in neurons, however, rescued the size and developmental timing of ns3 mutant animals. This exciting result indicates that neurons outside of the IPCs are critical for ns3 function, and provides evidence that the Drosophila central nervous system can regulate body size through effects on insulin/IGF signaling.
A role for serotonergic neurons in regulating growth
The investigators go on to conduct an elegant series of rescue experiments using GAL4 drivers to express NS3 in different neuronal populations, and show that NS3 expression in dopaminergic and serotonergic neurons, but not in dopaminergic neurons alone, is sufficient to rescue the size and developmental timing of ns3 mutants. Moreover, specific disruption of ns3 function in dopaminergic and serotonergic neurons results in a developmental delay and reduced adult body size, similar to that seen in ns3 mutants. These results imply that ns3 exerts its systemic effects on timing and growth through the serotonergic neurons of the brain (Fig. 1). The biogenic monoamine serotonin is an ancient signaling molecule found in all phyla that possess nervous systems (Weiger 1997). Serotonin acts as a classical neurotransmitter, as a neuromodulator, or as a hormone that controls important behaviors, such as feeding, sexual behavior, and aggression. A null mutation has been reported in the neuronal form of Drosophila TPH (DTRHn), which encodes tryptophan hydroxylase, and catalyzes the first and rate-limiting step of serotonin biosynthesis (Neckameyer et al. 2007). DTRHn mutants have slightly reduced feeding rates, mild developmental delays, and display some lethality during larval and pupal stages, although no size defects were reported.
Kaplan et al. (2008) show that serotonin levels are significantly elevated in the heads of ns3 mutant flies. If this compound is released from serotonergic neurons, then these increased levels should lead to increased serotonin signaling. The investigators test this model by feeding flies 5-HTP (5-hydroxy-L-tryptamine), the immediate precursor of serotonin. They show that this treatment results in a developmental delay in wild-type animals, reminiscent of the ns3 mutant phenotype, and that ns3 mutants are more sensitive to 5-HTP than their wild-type counterparts, suggesting that they already have increased serotonergic signaling. It would be interesting to try the converse experiment—to ask whether ns3 mutant phenotypes can be rescued by the DTRHn mutation. This would provide genetic evidence in support of a functional role for increased serotonin levels in developmental timing and growth.
Given the central role for serotonergic signaling in feeding behavior, one explanation for the reduced size of ns3 mutants could be that they simply feed less, and therefore are deprived of essential nutrients throughout development. The observation that ns3 mutants consume the same amount of food as control animals during early larval stages, however, argues against this possibility (Kaplan et al. 2008). Rather, serotonergic signaling appears to coordinate feeding behavior with growth at the organismal level. Consistent with this possibility, reduction of brain serotonin signaling by injection of a serotonergic-specific neurotoxin induces hyperphagia and increased growth in rats (Saller and Stricker 1976). The mechanisms that underlie these effects, however, remain elusive.
Serotonin neurotransmission may control DILP release
Finally, Kaplan et al. (2008) perform experiments to determine how serotonergic neurons regulate insulin/IGF signaling in Drosophila. Serotonin has been shown to enhance insulin synthesis, release, and target tissue sensitivity in mammals (for review, see Lam and Heisler 2007). Similarly, mutations in the Caenorhabiditis elegans insulin signaling pathway and the single C. elegans tryptophan hydroxylase gene, tph-1, result in overlapping phenotypes (Sze et al. 2000; Liang et al. 2006). Many tph-1 mutant defects can also be suppressed by mutations in the C. elegans FOXO homolog daf-16, a prominent downstream insulin effector in the worm (Sze et al. 2000; Liang et al. 2006). However, the complexity of serotonergic signaling in mammals and insulin signaling in C. elegans (which possess 37 insulin-like ligands) (Pierce et al. 2001) complicates our understanding of how these pathways might be linked at the cellular or molecular level. This study by Kaplan et al. (2008) assembles the first pieces of this puzzle.
The investigators focus their attention on DILP-2, a prominent DILP produced by the IPCs during larval stages. Although DILP expression can be regulated at both the transcriptional and post-transcriptional level (Brogiolo et al. 2001; Ikeya et al. 2002), little is known regarding the control of DILP processing or DILP release from IPCs. The investigators show that dilp-2 transcript levels do not change in ns3 mutants, while levels of DILP-2 protein increase by sevenfold in the IPCs of these animals. This abundant DILP-2 is apparently not released from the IPCs because peripheral insulin signaling is reduced, rather than increased, in ns3 mutants. A test of this hypothesis requires direct measurement of hemolymph DILP titers, a methodology that remains to be developed in Drosophila. Nonetheless, these results by Kaplan et al. (2008) provide some of the first evidence supporting a role for DILP release in contributing to insulin/IGF regulation in the fly. This observation also provides a foundation for future genetic studies aimed at defining the mechanisms that modulate DILP release from the IPCs.
In an effort to address the remaining question of how serotonergic neurons exert their effects on DILP levels, Kaplan et al. (2008) show that the axonal projections of serotonergic neurons are closely apposed to those of the IPCs (Fig. 1). This observation raises the exciting possibility that serotonergic neurons might synapse directly on the IPCs to control DILP release. Four serotonergic receptors have been identified in Drosophila, all of which are G protein-coupled receptors that share considerable sequence similarity with their mammalian homologs (Witz et al. 1990; Saudou et al. 1992). Mutants exist for at least three of them (serotonin1A, serotonin1B, 5HT2), and these could be tested for their effects on body size and/or their ability to suppress ns3 mutant defects (Yuan et al. 2006). In addition, given that the spatial distribution of serotonergic receptors remains poorly characterized in the fly brain, it would be interesting to know if any of these receptors are clustered at postsynaptic sites on the IPCs in front of active presynaptic sites, thus defining a serotonergic synapse.
Roles for the brain in regulating body size
Little is known about how the brain coordinates growth with environmental signals. Sensory neurons have been shown to play a role in body size determination in C. elegans, independently of the ability of the animal to locate food (Fujiwara et al. 2002). This response is mediated, at least in part, through egl-4, which encodes a cGMP-dependent kinase that is expressed specifically in sensory neurons. More recently, ablation of neurons that express prothoracicotropic hormone in Drosophila was shown to result in increased adult body size, although this can be attributed to a prolonged larval feeding stage as a consequence of reduced ecdysone titers (McBrayer et al. 2007). In a study more similar to that reported here, Lee et al. (2008) demonstrated a role for the Drosophila short neuropeptide F (sNPF), which is the ortholog of mammalian neuropeptide Y, in body size determination. sNPF is expressed in multiple neurons of the brain, some of which appear to directly contact the IPCs (Lee et al. 2004, 2008). In agreement with this observation, sNPF expression in a subset of sensory neurons in the brain can modulate dilp2 mRNA levels in the IPCs and control insulin signaling in peripheral tissues, providing a mechanism for its effects on body size. This paper by Kaplan et al. (2008) extends our understanding of roles for the brain in body size determination in an important way—showing that in addition to its roles in modulating feeding and behavior, serotonin is a determinant of the size of peripheral adult tissues. This report paves the way for further studies aimed at dissecting the serotonergic neuronal circuits coordinating growth with behavior in the fly, and provides a simple genetic model for determining how this level of growth control might be achieved in other animals.
Acknowledgments
We thank M. Horner, M. Metzstein, and J. Tennessen for critical comments on the manuscript. Research in our lab on growth and metabolism is supported by NIH grant R01 DK075607.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1700708.
References
- Arquier N., Geminard C., Bourouis M., Jarretou G., Honegger B., Paix A., Leopold P. Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 2008;7:333–338. doi: 10.1016/j.cmet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Bassler J., Grandi P., Gadal O., Lessmann T., Petfalski E., Tollervey D., Lechner J., Hurt E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Beadle G., Tatum E., Clancy C. Food levels in relation to rate of development and eye pigmentation in Drosophila melanogaster. Biol. Bull. 1938;75:447–462. [Google Scholar]
- Beekman C., Nichane M., De Clercq S., Maetens M., Floss T., Wurst W., Bellefroid E., Marine J.C. Evolutionarily conserved role of nucleostemin: Controlling proliferation of stem/progenitor cells during early vertebrate development. Mol. Cell. Biol. 2006;26:9291–9301. doi: 10.1128/MCB.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R., Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 2001;11:213–221. doi: 10.1016/s0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Broughton S.J., Piper M.D., Ikeya T., Bass T.M., Jacobson J., Driege Y., Martinez P., Hafen E., Withers D.J., Leevers S.J., et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc. Natl. Acad. Sci. 2005;102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Jack J., Garofalo R.S. The Drosophila insulin receptor is required for normal growth. Endocrinology. 1996;137:846–856. doi: 10.1210/endo.137.3.8603594. [DOI] [PubMed] [Google Scholar]
- Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C., Antoniewski C., Carre C., Noselli S., Leopold P. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science. 2005;310:667–670. doi: 10.1126/science.1119432. [DOI] [PubMed] [Google Scholar]
- Du X., Rao M.R., Chen X.Q., Wu W., Mahalingam S., Balasundaram D. The homologous putative GTPases Grn1p from fission yeast and the human GNL3L are required for growth and play a role in processing of nucleolar pre-rRNA. Mol. Biol. Cell. 2006;17:460–474. doi: 10.1091/mbc.E05-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A. How flies get their size: Genetics meets physiology. Nat. Rev. Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- Fujiwara M., Sengupta P., McIntire S.L. Regulation of body size and behavioral state of C. elegans by sensory perception and the EGL-4 cGMP-dependent protein kinase. Neuron. 2002;36:1091–1102. doi: 10.1016/s0896-6273(02)01093-0. [DOI] [PubMed] [Google Scholar]
- Garofalo R.S. Genetic analysis of insulin signaling in Drosophila. Trends Endocrinol. Metab. 2002;13:156–162. doi: 10.1016/s1043-2760(01)00548-3. [DOI] [PubMed] [Google Scholar]
- Geminard C., Arquier N., Layalle S., Bourouis M., Slaidina M., Delanoue R., Bjordal M., Ohanna M., May M., Colombani J., et al. Control of metabolism and growth through insulin-like peptides in Drosophila. Diabetes. 2006;55:85–88. [Google Scholar]
- Ikeya T., Galic M., Belawat P., Nairz K., Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 2002;12:1293–1300. doi: 10.1016/s0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- Kallstrom G., Hedges J., Johnson A. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 2003;23:4344–4355. doi: 10.1128/MCB.23.12.4344-4355.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.D., Zimmermann G., Suyama K., Meyer T., Scott M.P.2008A nucleostemin family GTPase, NS3, acts in serotonergic neurons to regulate insulin signaling and control body size Genes & Dev. doi: 10.1101/gad.1670508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam D.D., Heisler L.K. Serotonin and energy balance: Molecular mechanisms and implications for type 2 diabetes. Expert Rev. Mol. Med. 2007;9:1–24. doi: 10.1017/S1462399407000245. [DOI] [PubMed] [Google Scholar]
- Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv. Genet. 1998;38:69–134. doi: 10.1016/s0065-2660(08)60142-x. [DOI] [PubMed] [Google Scholar]
- Lee K.S., You K.H., Choo J.K., Han Y.M., Yu K. Drosophila short neuropeptide F regulates food intake and body size. J. Biol. Chem. 2004;279:50781–50789. doi: 10.1074/jbc.M407842200. [DOI] [PubMed] [Google Scholar]
- Lee K.S., Kwon O.Y., Lee J.H., Kwon K., Min K.J., Jung S.A., Kim A.K., You K.H., Tatar M., Yu K. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat. Cell Biol. 2008;10:468–475. doi: 10.1038/ncb1710. [DOI] [PubMed] [Google Scholar]
- Liang B., Moussaif M., Kuan C.J., Gargus J.J., Sze J.Y. Serotonin targets the DAF-16/FOXO signaling pathway to modulate stress responses. Cell Metab. 2006;4:429–440. doi: 10.1016/j.cmet.2006.11.004. [DOI] [PubMed] [Google Scholar]
- McBrayer Z., Ono H., Shimell M., Parvy J. P., Beckstead R.B., Warren J.T., Thummel C.S., Dauphin-Villemant C., Gilbert L.I., O’Connor M.R. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell. 2007;13:857–871. doi: 10.1016/j.devcel.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckameyer W.S., Coleman C.M., Eadie S., Goodwin S.F. Compartmentalization of neuronal and peripheral serotonin synthesis in Drosophila melanogaster. Genes Brain Behav. 2007;6:756–769. doi: 10.1111/j.1601-183X.2007.00307.x. [DOI] [PubMed] [Google Scholar]
- Pierce S.B., Costa M., Wisotzkey R., Devadhar S., Hamburger S.A., Buchman A.R., Heller J., Platt D.M., Pasquinelli A.A., et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes & Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichlin S. Neuroendocrinology. In: Foster D.W., Kronenberg H.M., et al., editors. Williams textbook of endocrinology. W.B. Saunders Company; Philadelphia, PA: 1998. pp. 165–248. [Google Scholar]
- Reynaud E.G., Andrade M.A., Bonneau F., Ly T.B., Knop M., Scheffzek K., Pepperkok R. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 2005;3:21. doi: 10.1186/1741-7007-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E.J., Kim S.K., Nusse R. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Saller C.F., Stricker E.M. Hyperphagia and increased growth in rats after intraventricular injection of 5,7-dihydroxytryptamine. Science. 1976;192:385–387. doi: 10.1126/science.1257774. [DOI] [PubMed] [Google Scholar]
- Saudou F., Boschert U., Amlaiky N., Plassat J.L., Hen R. A family of Drosophila serotonin receptors with distinct intracellular signalling properties and expression patterns. EMBO J. 1992;11:7–17. doi: 10.1002/j.1460-2075.1992.tb05021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C., Bienvenu D., Namane A., Gleizes P.E., Gas N., Jacquier A., Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 2001;20:6475–6484. doi: 10.1093/emboj/20.22.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silventoinen K. Determinants of variation in adult body height. J. Biosoc. Sci. 2003;35:263–285. doi: 10.1017/s0021932003002633. [DOI] [PubMed] [Google Scholar]
- Sze J.Y., Victor M., Loer C., Shi Y., Ruvkun G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature. 2000;403:560–564. doi: 10.1038/35000609. [DOI] [PubMed] [Google Scholar]
- Tsai R.Y., McKay R.D. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes & Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet C., Ribouchon M.T., Chimini G., Pontarotti P. Structure and evolution of a member of a new subfamily of GTP-binding proteins mapping to the human MHC class I region. Mamm. Genome. 1994;5:100–105. doi: 10.1007/BF00292335. [DOI] [PubMed] [Google Scholar]
- Visscher P.M., Hill W.G., Wray N.R. Heritability in the genomics era—Concepts and misconceptions. Nat. Rev. Genet. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- Weiger W.A. Serotonergic modulation of behaviour: A phylogenetic overview. Biol. Rev. Camb. Philos. Soc. 1997;72:61–95. doi: 10.1017/s0006323196004975. [DOI] [PubMed] [Google Scholar]
- Witz P., Amlaiky N., Plassat J.L., Maroteaux L., Borrelli E., Hen R. Cloning and characterization of a Drosophila serotonin receptor that activates adenylate cyclase. Proc. Natl. Acad. Sci. 1990;87:8940–8944. doi: 10.1073/pnas.87.22.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Q., Joiner W.J., Sehgal A. A sleep-promoting role for the Drosophilaserotonin receptor 1A. Curr. Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Zhu Q., Yasumoto H., Tsai R.Y. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol. Cell. Biol. 2006;26:9279–9290. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]