Abstract
The epithelium of the adult mammalian intestine is in a constant dialog with its underlying mesenchyme to direct progenitor proliferation, lineage commitment, terminal differentiation, and, ultimately, cell death. The epithelium is shaped into spatially distinct compartments that are dedicated to each of these events. While the intestinal epithelium represents the most vigorously renewing adult tissue in mammals, the stem cells that fuel this self-renewal process have been identified only recently. The unique epithelial anatomy makes the intestinal crypt one of the most accessible models for the study of adult stem cell biology. This review attempts to provide a comprehensive overview of four decades of research on crypt stem cells.
Keywords: Crypt base columnar cell, intestine, stem cell
Adult tissue stem cells can be defined by two essential features. First, a population of tissue stem cells should be maintained over long periods of time, often the entire lifetime of the organism (“longevity”). And second, these long-lived stem cell populations should be able to generate the differentiated cell types of the tissue. Most adult stem cells can generate multiple cell types (“multipotency”), yet examples also exist in which they only generate offspring of a single lineage. Two experimental strategies can unveil these combined features in candidate stem cell populations. In this review, these are arbitrarily termed the transplantation strategy and the genetic marking strategy. The transplantation strategy utilizes molecular markers to enrich putative stem cell populations, followed by in vitro culture and/or transplantation into recipient animals. This approach has been highly successful in the identification of the hematopoietic stem cell from bone marrow (Spangrude et al. 1988) and—more recently—of cancer stem cells in leukemias (Bonnet and Dick 1997) and in solid tumors (Singh et al. 2004; Dalerba et al. 2007; O’Brien et al. 2007; Prince et al. 2007). In an elegant example, Visvader and colleagues regrew an entire mammary gland (Shackleton et al. 2006) from a single isolated stem cell. Similarly, clonal analyses have shown that cells derived from a single follicle stem cell can give rise to epidermis, sebaceous gland, and hair follicles (Blanpain et al. 2004; Claudinot et al. 2005). Of note, multipotency can only be definitively demonstrated when transplantation can be performed with a single cell, which is rarely possible. As an alternative strategy, candidate stem cells are genetically marked in situ, after which the introduced marker allows the visualization of the modified stem cell and its clonal offspring over time. As an example of the latter approach, a progesterone-responsive version of the Cre recombinase enzyme was specifically expressed in cells residing in the bulge region of hair follicles using a transgenic Keratin-15 promoter (Morris et al. 2004). Activation of the Cre enzyme by progesterone irreversibly activated the genetic marker R26R-LacZ in the bulge cells. A simple blue stain for LacZ activity revealed that over time the entire hair follicle and its associated hair derived from the marked bulge stem cells, thus establishing that the bulge cells are endowed with longevity and pluripotency.
Genetic marking is often technically challenging, as one needs to be able to genetically modify individual stem cells. Sometimes a combination of the two approaches is applied, particularly when single cell transplantation is not feasible and in situ marking of individual cells cannot be achieved. In such cases, one can genetically mark individual cells in enriched stem cell populations; for instance, by retroviral integration (Jordan and Lemischka 1990) or by titration of individual putative stem cells into a stem cell suspension of a different genetic background prior to transplantation (Smith et al. 1991).
Quiescence and asymmetric cell division
In addition to longevity and multipotency, stem cells are often tacitly assumed to be endowed with two other characteristics. Stem cells are commonly believed to divide very infrequently (“quiescence”), and when this occurs, to generate one rapidly cycling daughter cell, while the other daughter replaces the parent stem cell (“asymmetric cell division”). The rapidly cycling daughter cells, also called transit-amplifying (TA) cells, are responsible for building tissue mass. TA cells typically undergo a limited number of cell divisions, after which they terminally differentiate.
While good examples exist of quiescent stem cells (e.g., in the bulge of the hair follicle) (Tumbar et al. 2004), there is actually no a priori reason why stem cells should be quiescent. The best studied animal stem cells, the germ cells of Drosophila, are actively cycling (Spradling et al. 2001). Furthermore, while the size of stem cell populations should be stable over time, there is no logical reason why this should be accomplished at the single stem cell level, i.e., by obligatory asymmetric cell division (Kiel et al. 2007, and refs therein). Physically defined stem cell niches could maintain stable stem cell populations, while allowing individual stem cells to take any of three courses in mitosis: the generation of two stem cells, of two daughter cells, or of one of each. Of note, the champion of all stem cells, the Embryonic Stem (ES) cell, is a very rapidly cycling cell that never undergoes asymmetric cell divisions, at least when kept under appropriate culture conditions.
Surrogate markers for stemness: DNA label retention
In many tissues, the technology is not available to isolate/transplant or to genetically mark adult stem cells. In such cases, markers known to identify stem cells in unrelated tissues, such as CD34, cKit, or the Hoechst dye-defined Side Population may be used as surrogate stem cell markers (Natarajan and FitzGerald 2007). It is immediately apparent that this approach should be applied with great caution. Long-term retention of DNA represents another commonly used surrogate marker of “stemness” (Kiel et al. 2007). An elegant variation of this strategy involves the use of Histone-GFP marking (Tumbar et al. 2004). Stem cells are commonly believed to be unique in their ability to retain DNA labels such as BrdU incorporated into their genome during periods of mitotic activity stimulated, for example, by severe tissue injury. This is based on the assumption that stem cells tend to be quiescent, while their direct descendants, the vigorously proliferating TA cells, rapidly dilute out any incorporated DNA label.
As an alternative mechanism behind DNA label retention, John Cairns formulated the “immortal strand hypothesis” 35 years ago (Cairns 1975). Stem cells may selectively retain their old DNA strands, while donating the newly synthesized DNA strands to their TA daughters. To this day, the immortal strand hypothesis has been the subject of controversy (Lansdorp 2007; Rando 2007). Few convincing examples have been reported, while its molecular machinery remains unidentified. In fact, it was shown very recently that one of the best characterized adult stem cells, the hematopoietic stem cell, does not asymmetrically segregate its DNA (Kiel et al. 2007).
Independent of its mechanism, it is important to realize that DNA label retention is only a surrogate marker of stemness. It is to be used with great caution, because terminally differentiated cells (or for instance invading lymphocytes or tissue macrophages) may retain DNA labels even better than stem cells, since by definition they do not divide.
Anatomy of the adult small and large intestine
The intestinal tract is essentially a tube, whose wall is composed of three tissue layers arrayed in a concentric structure. The outer layer consists of several sheets of smooth muscle that, together with the intramural enteric nervous system, execute the rhythmic peristaltic movements of the intestine. The space between the outer muscle and the inner epithelial layer is filled by connective tissue (“stroma”) that contains numerous blood and lymph vessels, nerve fibers, and various cells of the immune system. On the inside, the luminal surface consists of a simple epithelium; a single-cell layer termed the mucosa. The mucosa is responsible for the processing and absorption of nutrients as well as for the compaction of the stool (Sancho et al. 2003).
The intestinal tract can be anatomically divided into two well-defined segments: the small intestine and the large intestine or colon. The small intestine is subdivided into three proximal-distal segments: the duodenum, jejunum, and ileum. The absorptive surface area of the small intestine is dramatically increased by numerous finger-like protrusions that point toward the lumen, the so-called villi, and invaginations into the submucosa known as the crypts of Lieberkühn. The mucosa of the large intestine lacks villi; crypts invaginate deep into the submucosa.
Four differentiated cell types mediate the functions of the intestinal epithelium (Sancho et al. 2003): absorptive, enteroendocrine, mucosecreting, and Paneth cells. The relative abundance of each of these three cell types varies markedly within different segments of the intestine. Absorptive cells (also called enterocytes) are the more abundant cell type in the small intestine and are responsible for the absorption of nutrients from the food and for the secretion of a cocktail of hydrolytic enzymes into the lumen. Numbers of mucosecreting cells (also called goblet cells) increase from proximal (small intestine) to distal (colon and rectum) as the stool becomes increasingly compacted. Enteroendocrine cells represent a small proportion (<1%) of the cells in the epithelium. They control gut physiology by secreting a variety of hormones including serotonine, substance P, and secretin. Multiple subtypes can be defined by the specific intestinal hormones that are produced (Schonhoff et al. 2004). Paneth cells reside at the bottom-most positions of the crypts of the small intestine. They secrete antimicrobial agents such as cryptidins (termed defensins in humans) and lysozyme, which play an essential role in the control of the microbial environment of the intestine (Porter et al. 2002). Finally, some lesser-known cell types should be mentioned, such as the M cells that cover the lymphoid Peyer’s patches (Gebert et al. 1996), brush/tuft/caveolated cells (Nabeyama and Leblond 1974), and cup cells (Madara 1982). However, little insight exists into their lineage relationships.
Around birth, epithelial proliferation is confined to shallow pockets residing between the villi of the small intestine of mice. Mature small intestinal crypts appear in the first weeks after birth, by a process in which the intervillus pockets invade the wall of the small intestine. Similarly, colon crypts become progressively deeper in early postnatal life. Intervillus pockets are initially polyclonal, but rapidly become monoclonal through a poorly understood process of refinement (Schmidt et al. 1988). To accommodate the growth of the organ into adulthood, the number of crypt units steadily increases by crypt fission, a process in which new crypts form by branching off from existing crypts (Totafurno et al. 1987).
Cell renewal and stem cells in the intestinal epithelium
In the murine small intestine, the epithelium renews every 5 d. Vigorous proliferation occurs within the crypt compartment. This process is fueled by stem cells that have long remained elusive, but were believed to reside near the crypt bottom. The readily distinguishable TA crypt progenitors divide every 12–16 h, generating some 300 cells per crypt every day (Marshman et al. 2002). When the committed TA cells reach the crypt–villus junction, they rapidly and irreversibly differentiate. The proliferation is balanced by apoptosis and cell shedding at the other end of the epithelial conveyor belt, the tip of the villus. Thus, the epithelial sheet is in a continuous upward movement. Epithelial cells produced in the lower part of the crypt migrate up onto an adjacent villus in a coherent column (Heath 1996; Marshman et al. 2002). Six or more independent crypts surround a single villus, resulting in an equal number of parallel columns of epithelial cells running toward the villus tip. Only Paneth cells escape this flow; they have a residence time of 3–6 wk at the crypt base. Newly formed TA cells reside within crypts for ∼48–72 h, undergoing up to six rounds of cell division (Marshman et al. 2002), while migrating upward. During this period, the TA cells become committed to specific cell fates, a process best described by a set of elegant experiments utilizing the murine Dlb-1 locus. The product of this locus generates an intestinal binding site for the lectin Dolichus biflorus agglutinin. SWR mice are Dlb-1−/−, while C57BL/6 mice are Dlb-1+/+. In F1 mice, mutagens readily induce somatic mutations in individual cells, resulting in unstained clones on a stained background of heterozygous cells. Winton and Ponder (1990) originally used this approach to formally prove the existence of long-lived, pluripotent stem cells, by showing that a single mutated cell can give rise to all epithelial cell types. Unfortunately, since mutagenesis occurs randomly in this approach, no conclusion can be drawn regarding the nature or location of the stem cells. Bjerknes and Cheng refined this strategy by showing that mutagen treatment of Dlb-1−/− SWR mice results in stained mutant cells on a background of unstained wild-type cells (Bjerknes and Cheng 1999). The isolation of crypt–villus units from such mice allows entire clones to be visualized at single cell resolution. This study revealed the existence of multiple short-lived crypt progenitors that have the ability to give rise to one or more lineages, most likely through consecutive binary decisions. Short-lived multipotent “Mix” progenitors presumably derive directly from multipotent stem cells. Mix in turn generates committed progenitors for the different epithelial lineages; e.g., C1 for (columnar) enterocytes and M1 for mucus-producing goblet cells.
Other genetic clonal marking strategies exploit mutation of the X-linked glucose-6-phosphate dehydrogenase gene in male mice (Griffiths et al. 1988; Park et al. 1995), of a hypothetical enzyme involved in O-acetylation (Campbell et al. 1996), or of mitochondrial cytochrome c oxidase (Taylor et al. 2003; Greaves et al. 2006) in human colon. Another elegant tracing strategy follows epigenetic changes in gene methylation patterns as a lineage marker (Yatabe et al. 2001; Shibata 2008). These studies in essence confirm that all epithelial cell types derive from a single stem cell, that crypts contain multiple stem cells, and that crypts can multiply through lateral fission.
Stem cells: two schools of thought
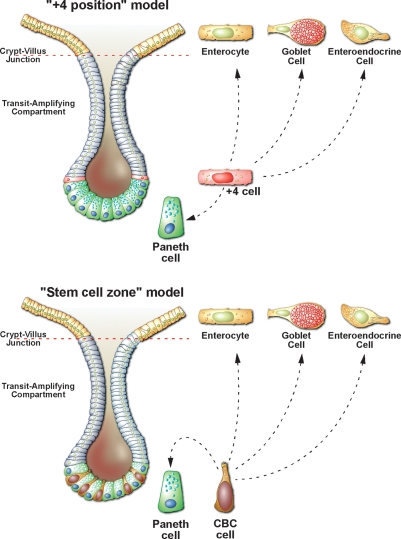
As indicated above, the crypts of the small intestine and colon have long been known to harbor a functional stem cell compartment (Marshman et al. 2002; Bjerknes and Cheng 2006), yet a paucity of unique molecular markers has hampered the definitive identification of the stem cells proper. While there has been general agreement that every crypt contains four to six independent stem cells, two schools of thought exist as to their exact identity, the “+4 position” model and the “stem cell zone” model (Fig. 1). For more than a century, the small intestinal crypt has been viewed as a tube of proliferating cells bounded from below by Paneth cells. Since the late 1950s, the “+4 position” model has therefore placed the stem cells at position +4 relative to the crypt bottom, with the first three positions being occupied by the terminally differentiated Paneth cells. Chris Potten and colleagues (Potten et al. 1974) have provided experimental support for the +4 stem cell model. They have reported the existence of label-retaining cells residing specifically at this position (Potten et al. 1974). Moreover, they have observed that the +4 cells are extremely radiation sensitive, a property that would functionally protect the stem cell compartment from genetic damage (Potten 1977). In the proposed model, damaged stem cells are replaced by the first two to three generations of TA cells, which would have a much better repair capacity, and which would fall back into the +4 position while regaining stem cell properties. The +4 cells are actively cycling. Label retention by the +4 cell has been proposed to result from asymmetric segregation of old and new DNA strands (Potten 1977; Potten et al. 2002). As outlined in the introduction, definitive proof of stemness requires that putative stem cells can be experimentally linked to their progeny. The current literature gives no insight into the nature of the cellular progeny of +4 cells. Therefore, the position of the +4 cells in the epithelial hierarchy remains uncertain.
Figure 1.
The exact identity of the intestinal stem cells has proven controversial over the last 30 years, with two opposing models dominating the literature. (Top panel) In the“+4 position”model proposed in the late 1950s, it was assumed that that the crypt base is exclusively populated by terminally differentiated Paneth Cells and the stem cells must therefore be located just above the Paneth cells at the +4 position. This model, largely championed by Chris Potten and colleagues (Marshman et al. 2002) predicts that the enterocytes, goblet cells, and enteroendocrine cells are derived from +4 cell progeny that differentiate as they migrate out of the crypts onto the villi. In contrast, the Paneth cells differentiate as they migrate down from the +4 position toward the crypt base. (Bottom panel) A more recent, but less well accepted model, the “stem cell zone” model proposed by Leblond and colleagues in the early 1970s (Cheng and Leblond 1974a, b) states that small, undifferentiated, cycling cells (termed crypt base columnar cells) intermingled with the Paneth cells are likely to be the true intestinal stem cells. Definitive proof for either model has proven elusive due to the lack of specific markers for these cells
The second school of thought has been based on the identification over 30 yr ago of Crypt Base Columnar (CBC) cells; small, undifferentiated, cycling cells hidden between the Paneth cells (Cheng and Leblond 1974a, b; Bjerknes and Cheng 1981a, b, 1999; Stappenbeck et al. 2003). Originally based on morphological considerations, but more recently also on the Dlb-1-based clonal marking techniques, Leblond, Cheng and Bjerknes have proposed that the CBC cells may represent the true stem cells. In the stem cell zone model, Mix cells are proposed to represent the direct offspring of the CBC cells. Mix cells would occupy the positions directly above the Paneth cells, the “common origin of diffentiation”. At the “common origin”, the cells commit to one of the various fates. Maturing Paneth cells precursors will migrate downward, with the oldest Paneth cells residing at the very base of the crypt. All other cell types migrate upward, as described above.
While these studies have revealed important aspects and parameters of the self-renewing intestinal epithelium, a paucity of unique stem cell markers has hampered the definitive identification of intestinal stem cells. Musashi-1 (Kayahara et al. 2003; Potten et al. 2003; Asai et al. 2005) and β1-integrin (Fujimoto et al. 2002) have been suggested to mark stem cells. While activity of these genes is high in the bottom third of crypts, their expression appears too broad to mark stem cells specifically. Several other markers have been identified for the +4 cells, i.e., phospho-PTEN and phospho-AKT (He et al. 2004), sFRP5 (Gregorieff et al. 2005), Sox4 (Van der Flier et al. 2007), and Dcamkl1 (Giannakis et al. 2006). All of these markers are specific for the +4 cells. As argued above, however, positional information is insufficient evidence to define molecules as stem cell markers.
Wnt target genes as candidate stem cell markers
A rational approach to the identification of stem cells and their molecular markers exploits the insights into the molecular machinery controlling self-renewal of the intestinal epithelium. A large series of recent studies indicate that the Wnt signaling pathway has a unique and central role in the (patho-) physiology of the intestine. Multiple secreted Wnt factors are produced by the epithelial cells at the crypt bottom (Gregorieff et al. 2005), potentially generating a morphogen-like gradient of Wnt signals along the crypt–villus axis. Several lines of in vivo evidence indicate that proliferation of the TA cells in the crypt is strictly dependent on continuous stimulation of the Wnt pathway. First, progenitors located at the bottom of the crypts accumulate nuclear β-catenin, implying that these cells respond to Wnt stimulation (van de Wetering et al. 2002). Second, removal of Tcf4, β-catenin, or overexpression of the Wnt inhibitor Dkk-1 all result in a complete loss of proliferation (Korinek et al. 1998; Pinto et al. 2003; Kuhnert et al. 2004). Third, mutations in the negative regulator of Wnt signaling APC, or expression of oncogenic forms of β-catenin result in hyperproliferation of the epithelium (Harada et al. 1999; Romagnolo et al. 1999; Smits et al. 1999; Sansom et al. 2004; Andreu et al. 2005). Of note, these activational Wnt pathway mutations are causative to colon cancer in man (Korinek et al. 1997; Morin et al. 1997).
In addition to their mitogenic activity, Wnt signals perform at least two other functions in crypts. First, terminal differentiation of Paneth cells at the crypt bottom paradoxically requires Wnt signals (Andreu et al. 2005; van Es et al. 2005). And second, the Wnt gradient drives a graded expression of the cell-sorting receptors EphB2 and EphB3 (Batlle et al. 2002), which in turn is responsible for sorting cells along the crypt–villus axis. In particular, Wnt-driven expression of EphB3 guides Paneth cells to crypt bottoms against the general flow of epithelial cells, a notion that fits well with the “stem cell zone” hypothesis.
In an attempt to determine the genetic program that is inappropriately activated in APC-mutant human colon cancer cells, we found that the same genetic program is physiologically expressed in proliferative cells in healthy crypts (van de Wetering et al. 2002). The program consists of a core of ∼80 Wnt target genes (Van der Flier et al. 2007). We reasoned that the possibility existed that a subset of these genes could be uniquely expressed in crypt stem cells. To identify such markers, we performed histological expression studies for each of the 80 Wnt target genes.
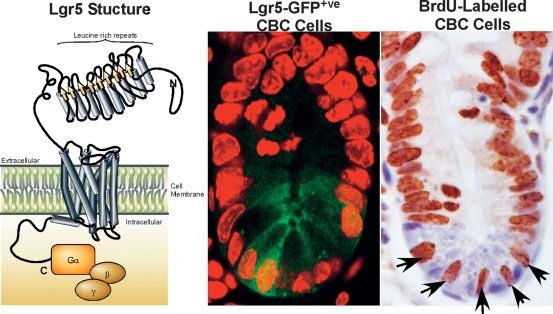
Although the overwhelming majority of the genes were expressed throughout the proliferative crypt compartment (van de Wetering et al. 2002; Van der Flier et al. 2007) or in mature Paneth cells (van Es et al. 2005), we detected several Wnt target genes with a much more restricted expression within crypts. One of these, the Lgr5/Gpr49 gene, was expressed in a particularly unique fashion. The Lgr5 gene encodes an orphan G protein-coupled receptor, characterized by a large leucine-rich extracellular domain (Fig. 2). It is closely related to receptors with glycoprotein hormone ligands, such as the TSH, FSH, and LH receptors. Lgr5 is highly expressed in stem cells of another Wnt-driven self-renewing structure, the hair follicle (Morris et al. 2004). In situ hybridization on small intestinal tissue revealed highly restricted expression at the crypt bottom. This expression pattern clearly differed from that obtained with any of the other 80 genes in the Wnt signature. The Lgr5-marked cells were reminiscent of the cycling CBC cells, described by Leblond and colleagues (Cheng and Leblond 1974b).
Figure 2.
(Left panel) Lgr5 is predicted to encode a 7-transmembrane protein with a large extracellular domain for ligand binding and a short cytoplasmic tail for coupling to G-proteins. (Middle panel) The Lgr5 protein is exclusively expressed on the Crypt-Base-Columnar (CBC) cells interspersed between the Paneth cells at the base of the intestinal crypts (visualized here by confocal microscopy for EGFP fluorescence in crypts from the Lgr5-EGFP KI mouse). (Right panel) The Lgr5+ve CBC cells are actively cycling, with the entire population staining positive for BrdU after 24 h of continuous labeling.
While our study was in progress, Hsueh and colleagues (Morita et al. 2004) published the Lgr5−/− phenotype. A developmental abnormality of the tongue and lower jaw causes newborns to swallow large amounts of air, leading to their postnatal demise. A knock-in allele, in which LacZ was integrated just N-terminal to the first transmembrane domain of Lgr5, confirmed this phenotype (Barker et al. 2007) and allowed a detailed expression analysis from which the following could be concluded: Lgr5 displays a complex expression pattern during embryogenesis, yet expression in most tissues subsides around birth. In adult mice, expression is restricted to rare, scattered cells in multiple tissues including the eye, the olfactory bulb of the brain, hair follicles, mammary glands, adrenal gland, and the epithelium of the stomach and intestinal tract. Importantly, Lgr5 expression in the intestine is indeed restricted to the CBC cells that are squeezed in between the Paneth cells (Fig. 2). By morphology, the slender Lgr5+ve CBC cells with their scant cytoplasm and flat, triangular nuclei are readily distinguishable from the adjacent Paneth cells. The CBC cells invariably express the Ki67 cell cycle marker, which provides a simple means of their identification amongst the noncycling Paneth cells. BrdU labeling studies indicate that the average cycling time of CBC cells is in the order of 1 d, ruling out that they are quiescent (Fig. 2), while TA cells cycle every 12 h (Marshman et al. 2002). In the colon, Lgr5 expression occurs in cells of similar shape at the crypt base.
In order to visualize live CBC cells and to follow their potential progeny, we generated another knock-in allele by integrating a cassette encoding Enhanced Green Fluorescent Protein (EGFP) and a tamoxifen-inducible version of the Cre recombinase enzyme into the first exon. The EGFP pattern observed in adult tissues faithfully recapitulated the Lgr5-LacZ expression pattern. Confocal imaging captured live EGFP+ve cells in small intestine (Fig. 2, middle panel) and colon. By Immuno-Electron Microscopy, the unique ultrastructural anatomy of the GFP+ve cells was found to be identical to that of the CBC cells described by Cheng and LeBlond (1974a, b). Typically, the CBC cells are broad at their base. Their flat wedge-shaped nucleus makes up an estimated 80% of the volume of the cell. A slender extension of apical cytoplasm reaches between the neighboring Paneth cells to the crypt lumen and carries some microvilli.
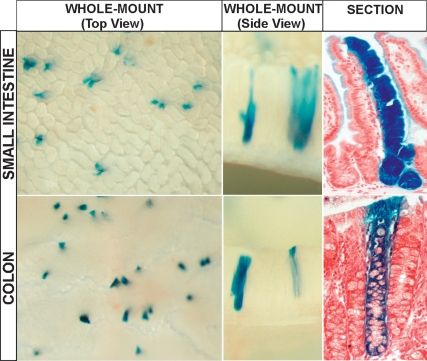
Following the genetic marking strategy described in the introduction, the knock-in allele was crossed with the Cre-activatable R26R-LacZ reporter. Injection of tamoxifen should activate the CreERT2 fusion enzyme uniquely in the CBC cells, irreversibly marking these cells by Cre-mediated excision of the roadblock sequence in the R26R-LacZ reporter. The activated LacZ reporter then acts as a genetic mark in the CBC cells and their potential progeny. The lineage tracing strategy worked surprisingly well. Adult mice were injected with a single, low-dose tamoxifen pulse to activate the R26R-LacZ reporter stochastically and at low frequency in CBC cells, hitting one cell per 10 crypts on average. The mice were sacrificed from 1 d to 6 mo post-induction. On day 1 post-injection, occasional CBC cells in the crypts of small intestine and colon expressed the LacZ marker. At later time points, parallel ribbons of blue cells emanated from these marked CBC cells and ran up to the tips of adjacent villi. These ribbons started reaching the villus tips 5 d post-injection. The CBC cells were capable of long-term maintenance of the self-renewing epithelium, since 2 mo (Barker et al. 2007) and even 6–12 mo after induction, during which time the intestinal epithelium will have been renewed at least 10–60 times, respectively, the appearance and frequency of blue crypts and ribbons remained unchanged (Fig. 3).
Figure 3.
Whole-mount photograph of small intestine (top series) and colon (bottom series) from Lgr5-EGFP-IresCreERT2/Rosa26-LacZ mice induced 6 mo previously with low-dose Tamoxifen. Villi containing ribbons of blue cells originating from the Lgr5+ve CBC cells are readily visible in the small intestine. (Top right panel) The various cell-types of the villus epithelium are all present within each of these blue ribbons, proving that the Lgr5+ve CBC cells are multipotent as well as being long-lived. (Bottom rightpanel) Similar observations are made in the colon, demonstrating that the Lgr5+ve cells are the stem cells of both the small intestine and colon.
Within the clonal blue ribbons of 60-d induced intestine, the four epithelial cell types (enterocytes, goblet cells, Paneth cells, and enteroendocrine cells) occurred at normal ratios. As described above, Bjerknes and Cheng (1999) have reported the existence of different types of long-lived epithelial clones; i.e., columnar (enterocyte) clones, mucous (goblet) clones, and mixed clones (Bjerknes and Cheng 1999). The clones observed in our study were exclusively of the mixed variety. In colon, blue clones emanating from the crypt bottom contained colonocytes as well as goblet cells.
Thus, the Lgr5+ve cells in small intestine and colon cells fulfill the definition of stemness in displaying longevity and multipotency. The observations confirm earlier estimates that each crypt contains approximately six independent, long-lived stem cells. Counter-intuitively, although CBC cells are stem cells, they appear never to be quiescent. Rather, the CBC cells complete a cell cycle every day. As they likely persist for the entire life of a mouse, the CBC cells must go through many hundreds of cell divisions. The logistical consequences in terms of maintenance of telomeres and genomic integrity, and of protection against cellular transformation must be colossal.
Sangiorgio and Capecchi (2008) very recently described a comparable lineage-tracing experiment utilizing a newly generated Bmi-Cre-ER knock-in allele. The Bmi-1 gene encodes a component of a Polycomb Repressing Complex 1, shown to play an essential role in maintenance of chromatin silencing (Valk-Lingbeek et al. 2004; Widschwendter et al. 2007). The Bmi1 gene has recently attracted significant attention because of its role in self-renewal of neuronal, hematopoietic, and leukemic cells (Lessard and Sauvage 2003; Molofsky et al. 2003; Leung et al. 2004). Using the Cre reporter strategy described above, these authors report that the first cells in which the activated Cre reporter can be observed, 20 h after induction, were predominantly located at the presumed position +4 directly above the Paneth cells. Over the next few days, the marked cells produced labeled offspring at a rate that appears similar to what is seen in Lgr5-based tracing experiments. No cell cycle kinetic analysis is reported for the “Bmi-1 cells”, yet the rapid appearance of labeled offspring would argue against a quiescent state for these cells. Like CBC cells, the Bmi-labeled clones generated ribbons emanating from crypts onto associated villi. Within these ribbons, all cell types were present, indicative of multipotency. The number of labeled clones only slightly decreased over a 1-year period, implying longevity of the underlying Bmi+ stem cells.
CBC cells and Bmi-1 cells therefore share important functional characteristics. They both produce offspring within days, yet persist for at least a year and both are multipotent. At first glance, there are also some significant differences between the two studies. (1) The Bmi-labeled cells only occur in ∼10% of the proximal part of the small intestine and are not seen elsewhere in the intestinal tract. (2) Only one to two Bmi-labeled cells are observed per positive crypt. (3) The Bmi-labeled cells are first seen predominantly at position +4. The first two differences may be due to the relatively weak/variegated activity of the BmiCreER allele as observed by Sangiorgio and Capecchi (2008) and to the indirect way by which Bmi-1 expression is visualized. The apparent difference in location of CBC cells and Bmi cells is less easily reconciled. Are the Bmi cells the same +4 cells as the ones observed by Potten and colleagues? If so, they should be DNA label retaining and highly radiation sensitive, neither of which has currently been assessed. Or alternatively, do the Bmi cells and the CBC cells represent overlapping or even identical populations of stem cells? Direct visualization of Bmi1 expression in comparison with Lgr5, to Paneth markers, and to markers of the +4 cells should resolve these differences.
The anatomy of the intestinal crypt is uniquely suited to study adult stem cells in their niche. With the Lgr5- and Bmi-based genetic tools in hand, it is now possible to visualize, isolate, and genetically modify the stem cells of the adult intestine at will. It is anticipated that this will facilitate rapid progress in our understanding of the biology of the intestinal stem cell niche over the next few years.
Acknowledgments
We are grateful to Eduardo Batlle and Esther Verheyen for critical reading of the manuscript and Hugo Snippert for help with preparation of the figures.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1674008.
References
- Andreu P., Colnot S., Godard C., Gad S., Chafey P., Niwa-Kawakita M., Laurent-Puig P., Kahn A., Robine S., Perret C., et al. Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development. 2005;132:1443–1451. doi: 10.1242/dev.01700. [DOI] [PubMed] [Google Scholar]
- Asai R., Okano H., Yasugi S. Correlation between Musashi-1 and c-hairy-1 expression and cell proliferation activity in the developing intestine and stomach of both chicken and mouse. Dev. Growth Differ. 2005;47:501–510. doi: 10.1111/j.1440-169X.2005.00825.x. [DOI] [PubMed] [Google Scholar]
- Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Batlle E., Henderson J.T., Beghtel H., van den Born M.M., Sancho E., Huls G., Meeldijk J., Robertson J., de van Wetering M., Pawson T., et al. β-Catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- Bjerknes M., Cheng H. The stem-cell zone of the small intestinal epithelium. I. Evidence from Paneth cells in the adult mouse. Am. J. Anat. 1981a;160:51–63. doi: 10.1002/aja.1001600105. [DOI] [PubMed] [Google Scholar]
- Bjerknes M., Cheng H. The stem-cell zone of the small intestinal epithelium. III. Evidence from columnar, enteroendocrine, and mucous cells in the adult mouse. Am. J. Anat. 1981b;160:77–91. doi: 10.1002/aja.1001600107. [DOI] [PubMed] [Google Scholar]
- Bjerknes M., Cheng H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology. 1999;116:7–14. doi: 10.1016/s0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- Bjerknes M., Cheng H. Intestinal epithelial stem cells and progenitors. Methods Enzymol. 2006;419:337–383. doi: 10.1016/S0076-6879(06)19014-X. [DOI] [PubMed] [Google Scholar]
- Blanpain C., Lowry W.E., Geoghegan A., Polak L., Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Bonnet D., Dick J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Cairns J. Mutation selection and the natural history of cancer. Nature. 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- Campbell F., Williams G.T., Appleton M.A., Dixon M.F., Harris M., Williams E.D. Post-irradiation somatic mutation and clonal stabilization time in the human colon. Gut. 1996;39:569–573. doi: 10.1136/gut.39.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Leblond C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am. J. Anat. 1974a;141:461–479. doi: 10.1002/aja.1001410403. [DOI] [PubMed] [Google Scholar]
- Cheng H., Leblond C.P. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am. J. Anat. 1974b;141:537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- Claudinot S., Nicolas M., Oshima H., Rochat A., Barrandon Y. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc. Natl. Acad. Sci. 2005;102:14677–14682. doi: 10.1073/pnas.0507250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Dylla S.J., Park I.K., Liu R., Wang X., Cho R.W., Hoey T., Gurney A., Huang E.H., Simeone D.M., et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K., Beauchamp R.D., Whitehead R.H. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123:1941–1948. doi: 10.1053/gast.2002.37065. [DOI] [PubMed] [Google Scholar]
- Gebert A., Rothkotter H.J., Pabst R. M cells in Peyer’s patches of the intestine. Int. Rev. Cytol. 1996;167:91–159. doi: 10.1016/s0074-7696(08)61346-7. [DOI] [PubMed] [Google Scholar]
- Giannakis M., Stappenbeck T.S., Mills J.C., Leip D.G., Lovett M., Clifton S.W., Ippolito J.E., Glasscock J.I., Arumugam M., Brent M.R., et al. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J. Biol. Chem. 2006;281:11292–11300. doi: 10.1074/jbc.M512118200. [DOI] [PubMed] [Google Scholar]
- Greaves L.C., Preston S.L., Tadrous P.J., Taylor R.W., Barron M.J., Oukrif D., Leedham S.J., Deheragoda M., Sasieni P., Novelli M.R., et al. Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc. Natl. Acad. Sci. 2006;103:714–719. doi: 10.1073/pnas.0505903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A., Pinto D., Begthel H., Destree O., Kielman M., Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Griffiths D.F., Davies S.J., Williams D., Williams G.T., Williams E.D. Demonstration of somatic mutation and colonic crypt clonality by X-linked enzyme histochemistry. Nature. 1988;333:461–463. doi: 10.1038/333461a0. [DOI] [PubMed] [Google Scholar]
- Harada N., Tamai Y., Ishikawa T., Sauer B., Takaku K., Oshima M., Taketo M.M. Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.C., Zhang J., Tong W.G., Tawfik O., Ross J., Scoville D.H., Tian Q., Zeng X., He X., Wiedemann L.M., et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-β-catenin signaling. Nat. Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- Heath J.P. Epithelial cell migration in the intestine. Cell Biol. Int. 1996;20:139–146. doi: 10.1006/cbir.1996.0018. [DOI] [PubMed] [Google Scholar]
- Jordan C.T., Lemischka I.R. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes & Dev. 1990;4:220–232. doi: 10.1101/gad.4.2.220. [DOI] [PubMed] [Google Scholar]
- Kayahara T., Sawada M., Takaishi S., Fukui H., Seno H., Fukuzawa H., Suzuki K., Hiai H., Kageyama R., Okano H., et al. Candidate markers for stem and early progenitor cells, Musashi-1 and Hes1, are expressed in crypt base columnar cells of mouse small intestine. FEBS Lett. 2003;535:131–135. doi: 10.1016/s0014-5793(02)03896-6. [DOI] [PubMed] [Google Scholar]
- Kiel M.J., He S., Ashkenazi R., Gentry S.N., Teta M., Kushner J.A., Jackson T.L., Morrison S.J. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 2007;449:238–242. doi: 10.1038/nature06115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V., Barker N., Morin P.J., van Wichen D., de Weger R., Kinzler K.W., Vogelstein B., Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P.J., Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Kuhnert F., Davis C.R., Wang H.T., Chu P., Lee M., Yuan J., Nusse R., Kuo C.J. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. 2004;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp P.M. Immortal strands? Give me a break. Cell. 2007;129:1244–1247. doi: 10.1016/j.cell.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Lessard J., Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Leung C., Lingbeek M., Shakhova O., Liu J., Tanger E., Saremaslani P., Van Lohuizen M., Marino S. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- Madara J.L. Cup cells: Structure and distribution of a unique class of epithelial cells in guinea pig, rabbit, and monkey small intestine. Gastroenterology. 1982;83:981–994. [PubMed] [Google Scholar]
- Marshman E., Booth C., Potten C.S. The intestinal epithelial stem cell. Bioessays. 2002;24:91–98. doi: 10.1002/bies.10028. [DOI] [PubMed] [Google Scholar]
- Molofsky A.V., Pardal R., Iwashita T., Park I.K., Clarke M.F., Morrison S.J. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin P.J., Sparks A.B., Korinek V., Barker N., Clevers H., Vogelstein B., Kinzler K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- Morita H., Mazerbourg S., Bouley D.M., Luo C.W., Kawamura K., Kuwabara Y., Baribault H., Tian H., Hsueh A.J. Neonatal lethality of LGR5 null mice is associated with ankyloglossia and gastrointestinal distension. Mol. Cell. Biol. 2004;24:9736–9743. doi: 10.1128/MCB.24.22.9736-9743.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R.J., Liu Y., Marles L., Yang Z., Trempus C., Li S., Lin J.S., Sawicki J.A., Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Nabeyama A., Leblond C.P. “Caveolated cells” characterized by deep surface invaginations and abundant filaments in mouse gastro-intestinal epithelia. Am. J. Anat. 1974;140:147–165. doi: 10.1002/aja.1001400203. [DOI] [PubMed] [Google Scholar]
- Natarajan T.G., FitzGerald K.T. Markers in normal and cancer stem cells. Cancer Biomark. 2007;3:211–231. doi: 10.3233/cbm-2007-34-506. [DOI] [PubMed] [Google Scholar]
- O’Brien C.A., Pollett A., Gallinger S., Dick J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- Park H.S., Goodlad R.A., Wright N.A. Crypt fission in the small intestine and colon. A mechanism for the emergence of G6PD locus-mutated crypts after treatment with mutagens. Am. J. Pathol. 1995;147:1416–1427. [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Gregorieff A., Begthel H., Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes & Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter E.M., Bevins C.L., Ghosh D., Ganz T. The multifaceted Paneth cell. Cell. Mol. Life Sci. 2002;59:156–170. doi: 10.1007/s00018-002-8412-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potten C.S. Extreme sensitivity of some intestinal crypt cells to X and γ irradiation. Nature. 1977;269:518–521. doi: 10.1038/269518a0. [DOI] [PubMed] [Google Scholar]
- Potten C.S., Kovacs L., Hamilton E. Continuous labelling studies on mouse skin and intestine. Cell Tissue Kinet. 1974;7:271–283. doi: 10.1111/j.1365-2184.1974.tb00907.x. [DOI] [PubMed] [Google Scholar]
- Potten C.S., Owen G., Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J. Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- Potten C.S., Booth C., Tudor G.L., Booth D., Brady G., Hurley P., Ashton G., Clarke R., Sakakibara S., Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- Prince M.E., Sivanandan R., Kaczorowski A., Wolf G.T., Kaplan M.J., Dalerba P., Weissman I.L., Clarke M.F., Ailles L.E. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando T.A. The immortal strand hypothesis: Segregation and reconstruction. Cell. 2007;129:1239–1243. doi: 10.1016/j.cell.2007.06.019. [DOI] [PubMed] [Google Scholar]
- Romagnolo B., Berrebi D., Saadi-Keddoucci S., Porteu A., Peuchmaur M., Vandewalle A., Kahn A., Perret C. Intestinal dysplasia and adenoma in transgenic mice after overexpression of an activated β-catenin. Cancer Res. 1999;59:3875–3879. [PubMed] [Google Scholar]
- Sancho E., Batlle E., Clevers H. Live and let die in the intestinal epithelium. Curr. Opin. Cell Biol. 2003;15:763–770. doi: 10.1016/j.ceb.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Sangiorgio E., Capecchi M.R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008 doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom O.J., Reed K.R., Hayes A.J., Ireland H., Brinkmann H., Newton I.P., Batlle E., Simon-Assmann P., Clevers H., Nathke I.S., et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes & Dev. 2004;18:1385–1390. doi: 10.1101/gad.287404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt G.H., Winton D.J., Ponder B.A. Development of the pattern of cell renewal in the crypt-villus unit of chimaeric mouse small intestine. Development. 1988;103:785–790. doi: 10.1242/dev.103.4.785. [DOI] [PubMed] [Google Scholar]
- Schonhoff S.E., Giel-Moloney M., Leiter A.B. Minireview: Development and differentiation of gut endocrine cells. Endocrinology. 2004;145:2639–2644. doi: 10.1210/en.2004-0051. [DOI] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Shibata D. Stem cells as common ancestors in a colorectal cancer ancestral tree. Curr. Opin. Gastroenterol. 2008;24:59–63. doi: 10.1097/MOG.0b013e3282f2a2e9. [DOI] [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Smith L.G., Weissman I.L., Heimfeld S. Clonal analysis of hematopoietic stem-cell differentiation in vivo. Proc. Natl. Acad. Sci. 1991;88:2788–2792. doi: 10.1073/pnas.88.7.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits R., Kielman M.F., Breukel C., Zurcher C., Neufeld K., Jagmohan-Changur S., Hofland N., van Dijk J., White R., Edelmann W., et al. Apc1638T: A mouse model delineating critical domains of the adenomatous polyposis coli protein involved in tumorigenesis and development. Genes & Dev. 1999;13:1309–1321. doi: 10.1101/gad.13.10.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude G.J., Heimfeld S., Weissman I.L. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Spradling A., Drummond-Barbosa D., Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- Stappenbeck T.S., Mills J.C., Gordon J.I. Molecular features of adult mouse small intestinal epithelial progenitors. Proc. Natl. Acad. Sci. 2003;100:1004–1009. doi: 10.1073/pnas.242735899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.W., Barron M.J., Borthwick G.M., Gospel A., Chinnery P.F., Samuels D.C., Taylor G.A., Plusa S.M., Needham S.J., Greaves L.C., et al. Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 2003;112:1351–1360. doi: 10.1172/JCI19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totafurno J., Bjerknes M., Cheng H. The crypt cycle. Crypt and villus production in the adult intestinal epithelium. Biophys. J. 1987;52:279–294. doi: 10.1016/S0006-3495(87)83215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W.E., Rendl M., Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek M.E., Bruggeman S.W.M., van Lohuizen M. Stem cells and cancer: The polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- de van Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A.P., et al. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/s0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- Van der Flier L.G., Sabates-Bellver J., Oving I., Haegebarth A., De Palo M., Anti M., Van Gijn M.E., Suijkerbuijk S., de Van Wetering M., Marra G., et al. The Intestinal Wnt/TCF Signature. Gastroenterology. 2007;132:628–632. doi: 10.1053/j.gastro.2006.08.039. [DOI] [PubMed] [Google Scholar]
- van Es J.H., Jay P., Gregorieff A., van Gijn M.E., Jonkheer S., Hatzis P., Thiele A., van den Born M., Begthel H., Brabletz T., et al. Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat. Cell Biol. 2005;7:381–386. doi: 10.1038/ncb1240. [DOI] [PubMed] [Google Scholar]
- Widschwendter M., Fiegl H., Egle D., Mueller-Holzner E., Spizzo G., Marth C., Weisenberger D.J., Campan M., Young J., Jacobs I., et al. Epigenetic stem cell signature in cancer. Nat. Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Winton D.J., Ponder B.A. Stem-cell organization in mouse small intestine. Proc. Biol. Sci. 1990;241:13–18. doi: 10.1098/rspb.1990.0059. [DOI] [PubMed] [Google Scholar]
- Yatabe Y., Tavare S., Shibata D. Investigating stem cells in human colon by using methylation patterns. Proc. Natl. Acad. Sci. 2001;98:10839–10844. doi: 10.1073/pnas.191225998. [DOI] [PMC free article] [PubMed] [Google Scholar]