Abstract
Background and Aims
The endemic tree Nothofagus alessandrii (Fagaceae) has been historically restricted to the coastal range of Region VII of central Chile, and its forests have been increasingly destroyed and fragmented since the end of the 19th century. In this study, the patterns of within- and among-population genetic diversity in seven fragments of this endangered narrowly endemic tree were examined.
Methods
Allozyme electrophoresis of seven loci of N. alessandrii was used to estimate genetic diversity, genetic structure and gene flow.
Key Results
High levels of genetic diversity were found as shown by mean expected heterozygosity (He = 0·182 ± 0·034), percentage of polymorphic loci (Pp = 61·2 %), mean number of alleles per locus (A = 1·8) and mean number of alleles per polymorphic locus (Ap = 2·3). Genetic differentiation was also high (GST = 0·257 and Nm = 0·7). These values are high compared with more widespread congeneric species.
Conclusions
Despite its endemic status and restricted geographical range N. alessandrii showed high levels of genetic diversity. The observed patterns of diversity are explained in part by historical processes and more recent human fragmentation.
Key words: Genetic diversity, endangered species, Nothofagus alessandrii, narrow endemic, conservation
INTRODUCTION
The geographic range of a species has been proposed as a good predictor of its genetic diversity in natural populations (Karron, 1987; Hamrick and Godt, 1989, 1996a; Gitzendanner and Soltis, 2000; Cole, 2003). Usually, species with narrow distributional ranges posses lower levels of genetic diversity than their widespread congeners, because they are associated with historically small and less continuously distributed populations (Hamrick et al., 1979). Therefore, they are more exposed to genetic drift, inbreeding and low levels of gene flow than widespread species (Hamrick and Godt, 1989). While some studies support this hypothesis (e.g. Ledig and Conkle, 1983; Ayres and Ryan, 1999; Wolf et al., 2000), others have shown some endemic species with very narrow distributional ranges to have moderate to high levels of genetic variation (Karron et al., 1988; Karron, 1991; Linhart and Premoli, 1993; Smith and Pham, 1996; Cardoso et al., 1998; Delgado et al., 1999; Gitzendanner and Soltis, 2000, González-Astorga and Núñez-Farfán, 2001; Premoli et al., 2001; González-Astorga and Castillo-Campos, 2004).
Life history traits such as breeding-system, seed and pollen dispersal and longevity have also been considered as good predictors of the distribution of genetic diversity within and among populations (Hamrick and Godt, 1989, 1996a, b). In general, wind-pollinated and/or wind-dispersed outcrossing species show a higher proportion of genetic diversity within populations with low levels of genetic differentiation (Hamrick and Godt, 1996b).
Central Chile is considered as one of the ‘hotspots’ of biodiversity (Myers et al., 2000). While the most characteristic vegetation of central Chile is the sclerophyllous matorral, the wetter conditions found on the west-facing slopes of the Coastal Range permit the development of forests (San Martín and Donoso, 1996). The maulino forest (sensu San Martin and Donoso, 1996) of Central Chile is of a mesic forest type, dominated by two long-lived and broadleaved caducifolious species: Nothofagus alessandrii and Nothofagus glauca. This forest type is restricted to the south-west coastal range of central Chile from 150 to 450 m a.s.l. within the extremely narrow latitudinal range of 35–36°S.
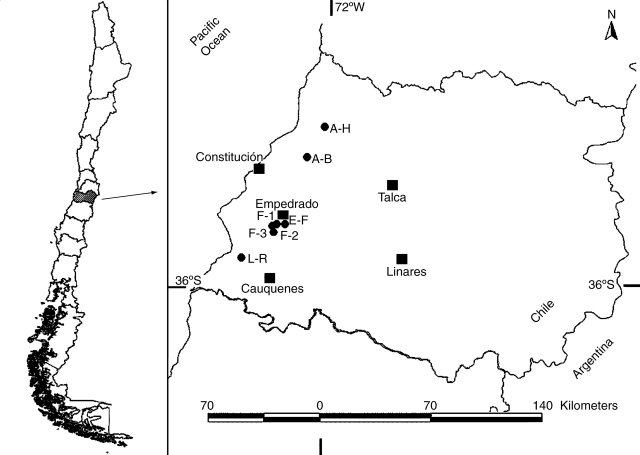
Nothofagus alessandrii (Fagaceae), commonly known as ruil, is an endemic species that has been historically restricted to the Coastal Range of Talca and Cauquenes provinces (Fig. 1), in Region VII of central Chile (Donoso and Landaeta, 1983). Unlike southern Chilean temperate forests (39–43°S), there is a lack of palynological records for the maulino forest zone. Thus, the pleistocene history of N. alessandrii is less well understood than that of their south-American congeners. However, given its high levels of endemism, this region has been suggested as a potential glacial refuge (Villagrán and Hinojosa, 1997; Villagrán et al., 1998).
Fig. 1.
Location of the seven populations of Nothofagus alessandrii sampled along the coastal range of Region VII of central Chile. Squares are cities and dots are the populations sampled.
Based on morphological evidence, early studies postulated that Nothofagus alessandrii is a primitive species within the genus (Humpries, 1981). More recently, morphological and molecular studies have confirmed the early branching position of N. alessandrii within the genus (Hill and Jordan, 1993; Martin and Dowd, 1993; Manos, 1997), and a molecular clock analysis performed by Knapp et al. (2005) showed that the origin of N. alessandrii may be as ancient as 42–61 Myr. This is an important reason to conserve this species. Despite of its biological significance, N. alessandrii forests have been increasingly destroyed and fragmented from the end of the 19th century on. Currently, extensive commercial plantations of introduced species such as Pinus radiata and Eucalyptus spp., surround the native remnant forests dominated by N. alessandrii. Between 1981 and 1991, levels of perturbation in this ecosystem were among the highest in Chile, with disappearance rates of 8·15 % per year (Bustamante and Castor, 1998). Due to this high rate of disturbance, N. alessandrii is considered to be one of the most endangered species in Chile (Benoit, 1989). To date, only 12% (0·42 km2) of its distributional range is under protection in the National System of Protected Areas (SNASPE) of Chile (Bustamante and Castor, 1998). Only two populations are considered within these protected areas: ‘El Fin Empedrado Reserve’ with 0·25 km2 and ‘Los Ruiles Reserve’ with 0·45 km2. Most of the adult trees found within the protected areas are stump re-sprouts from trees cut or burned during the exploitation period of this species (Donoso and Landaeta, 1983; Rodríguez et al., 1983; San Martin and Donoso, 1996; C. Torres-Díaz, pers. obs.). Although N. alessandrii clearly has the ability to resprout, vegetative propagation has not been documented, and sexual reproduction seems to be very limited, as it is difficult to find seedlings and juveniles in the field (San Martín and Donoso, 1996).
Although data regarding the longevity of N. alessandrii are unavailable, individuals of some species of this genus are expected to live for >200 years (e.g. N. obliqua) (Rodríguez et al., 1983). Nothofagus alessandrii reaches reproductive maturity after 10 years of age (Rodriguez et al., 1995). Considering that the beginning of the fragmentation process is recent compared with the longevity of this species, even if the forest of N. alessandrii is highly fragmented at present, the surviving adult trees probably represent a good sample of the genetic diversity and structure of the natural populations present prior to the fragmentation process. In this study, allozyme analyses were used to examine the genetic diversity of N. alessandrii throughout its distributional range. Our main aims were (a) to determine the genetic diversity within and among remnant populations of N. alessandrii; (b) to compare the levels of genetic diversity within and among populations of N. alessandrii with its widespread and endemic South-American congeners; and (c) to discuss the relevance of this information for future strategies of conservation of this narrowly distributed endemic and endangered tree species of central Chile.
MATERIALS AND METHODS
Study site
Bustamante and Castor (1998) estimated that all of the remnant forest fragments of N. allesandrii were distributed in nine localities across the coastal range of the Río Maule Region. During the austral summers of 2001 and 2002, seven populations of Nothofagus alessandrii (approx. 90 ha) were sampled along the entire distributional range of the species. Two of them were located within the protected areas (El Fin Empedrado National Reserve and Los Ruiles National Reserve). The other five, all representing small unprotected fragments of varying size, were sampled on account of their size and accessibility. Geographical co-ordinates, altitude, area and the number of individuals sampled in each population are shown in Table 1. All populations are fragmented and isolated by commercial plantations of Pinus radiata and Eucalyptus globulus.
Table 1.
Names and codes of populations, geographical location (°S/°W), altitude, surface area, approximate population size, exact sample sizes and conservation status (CS) for all fragments sampled
| Population | (°S/°W) | Altitude (m) | Surface (km2) | Population size (approx. no. of trees) | Sample size | CS |
|---|---|---|---|---|---|---|
| 1. Fragment 1 (F-1) | 35°37/72°18′ | 424 | 0·015 | <100 | 35 | PP |
| 2. Fragment 2 (F-2) | 35°39/72°20′ | 455 | 0·030 | <100 | 25 | PP |
| 3. Fragment 3 (F-3) | 35°40/72°20′ | 360 | 0·015 | <100 | 15 | PP |
| 4. Alto Huelón (A-H) | 35°06/72°02′ | 300 | 0·088 | >500 | 45 | PP |
| 5. Agua Buena (A-B) | 35°16/72°08′ | 300 | 0·048 | 200–300 | 58 | PP |
| 6. El Fin Empedrado (E-F) | 35°37/72°18′ | 441 | 0·250 | 500 | 90 | NR |
| 7. Los Ruiles (L-R) | 35°49/72°30′ | 206 | 0·450 | >500 | 40 | NR |
NR, National Reserve (El Fin Empedrado and Reserva Los Ruiles); PP (private property, Fragments 1, 2, 3; Alto Huelón and Agua Buena).
Electrophoretic analyses
Young leaves of each individual of Nothofagus alessandrii were collected in each population. Standard methods of starch gel electrophoresis were followed: the isozyme extraction buffer consisted of 0·1 m of Tris–HCl (pH 7·5), 14 mm 2-mercaptoethanol, 1 mm EDTA (tetrasodium salt), 10 mm MgCl2, 10 mm KCl and 5–10 mg polyvinylpyrrolidone per 0·5 mL buffer (Gottlieb, 1981). Two buffer systems were used with 12 % starch gels. (1) An electrode buffer of 0·5 m Tris, 0·65 m boric acid, 10 mm EDTA, pH 8·0, was diluted 1:9 for gel buffer. The isozymes resolved with this buffer were glucose-6-phosphate isomerase (GPI, E.C.5·3·1·9), phosphoglucomutase (PGM, E.C. 5·4·2·2) and malate dehydrogenase (MDH, E.C. 1·1·1·37). (2) An electrode buffer of 40 mm citric acid titrated to pH 6·1 with N-(3-aminopropyl)-morpholine was diluted 1:19 for gel buffer. The isozymes resolved with this buffer were phosphogloconate dehydrogenase (PGD; EC 1·1·1·44). The staining protocols of Wendel and Weeden (1989) were used, and genetic variability was inferred by observing the banding patterns within and among populations, based on the knowledge of the enzymes active subunit compositions (Wendel and Weeden, 1989).
Data analyses
The allelic frequencies for each locus, and the proportion of polymorphic loci (Pp, sensu stricto, where one locus is considered polymorphic if it has more than one allele) were calculated. To analyse deviations from expected genotypic frequencies under Hardy–Weinberg equilibrium Markov chain exact tests (Raymond and Rousset, 1995) and χ2-tests (Snedecor and Cochran, 1967) were performed. These analyses were carried out for each locus and each population, as well as for the entire gene pool. The Hardy–Weinberg expected heterozygosities (He, genetic diversity), observed heterozygosities (Ho), mean number of alleles per locus (A) and mean number of alleles per polymorphic locus (Ap) were calculated as well. Statistical genetic analyses were done using the software ‘Tools for Population Genetic Analysis (TFPGA)’ (Miller, 1997). Wright's fixation index or inbreeding coefficient for each population was also calculated as F = 1 – (Ho/He) to determine deviations from random-mating expectations. To test for deviations from zero χ2-tests were used. Gene Stat-PC 3·3 software (Lewis, 1993) was used to determine the apportionment of the total genetic variation within and among populations using Nei's statistics (Nei, 1973). Measures of genetic diversity were estimated for each locus. The total genetic diversity (HT) that is distributed within (HS) and among populations (DST), and GST, which represents the proportion of the total genetic diversity residing among populations, were calculated. The total inter-population gene flow was estimated with the formula Nm = (1 – GST)/(4 GST) (Wright, 1951).
Also the relationship between gene flow among population pairs (M) and geographic distance (d) was examined. F statistics were calculated following the methods of Weir and Cockerham (1984) to assess the isolation by distance model using Slatkin's method (Slatkin, 1993). FST values between all populations pairs were estimated using all loci. Based on these FST values, all pairwise effective rates of migrants were calculated as M = [(1/FST) – 1]/4. A linear regression between log10 M and log10 d (d geographic distance) was performed to determine if there was a linear relationship between both variables. Slatkin (1993) showed that in a stepping stone model, gene flow and geographic distance should be inversely correlated. If a significant linear relationship is found, this could be interpreted as evidence of isolation by distance. To evaluate the significance of this relationship a Mantel test (Mantel, 1967) was performed.
To evaluate the question whether or not both El Fin Empedrado and Reserva los Ruiles (both of them protected by SNASPE) preserve a significant proportion of the genetic diversity the genetic diversity of protected and unprotected areas (private proprieties) were compared. In calculations, the fragments were considered as subpopulations of two big populations. In one population, unprotected fragments were pooled, and in the other, protected fragments were pooled. Differences of the mean expected and observed heterozygosity between groups were tested using a t-test with a jack-knife procedure (Sokal and Rohlf, 1995). Finally, to compare N. alessandrii with its congeners studied by Premoli (1997), comparisons of the mean expected and observed heterozygosity were also performed; the differences were also evaluated with t-tests using jack-knife procedures.
RESULTS
Hardy–Weinberg equilibrium
Seven loci were resolved for the seven fragments of N. alessandrii studied: MDH-2 (two alleles), MDH-3 (two), GPI-1 (four), GPI-2 (two), PGD-1 (three), PGM-1 (three) and PGM-2 (four). Allele frequencies of all fragments are shown in Table 2. Considering the entire gene pool, Chi-squared tests indicated that only PGM-1 was in Hardy–Weinberg equilibrium. In contrast, a Markov chain approach showed that none of the loci were in Hardy–Weinberg equilibrium. There was a deficit of heterozygotes in fragments A and B and L–R (χ2 = 23·9, P = 0·005; and χ2 = 10·7, P = 0·001, respectively), while an excess of heterozygotes was detected for fragment 1 (χ2 = 10·5, P = 0·001). F values for fragments F-2, F-3, A-H and E-F did not differ significantly from zero (Table 3).
Table 2.
Allelic frequencies for all population fragments of Nothofagus alessandrii sampled
| Locus | Populations |
||||||
|---|---|---|---|---|---|---|---|
| F-1 | F-2 | F-3 | A-H | A-B | E-F | L-R | |
| MDH-2 | |||||||
| A | 0·943 | 0·864 | 0·769 | 0·867 | 0·860 | 0·976 | 0·773 |
| B | 0·057 | 0·136 | 0·231 | 0·133 | 0·140 | 0·024 | 0·227 |
| MDH-3 | |||||||
| A | 0·983 | 1·000 | 0·792 | 0·625 | 0·900 | 1·000 | 0·983 |
| B | 0·017 | 0·000 | 0·208 | 0·375 | 0·100 | 0·000 | 0·017 |
| GPI-1 | |||||||
| A | 0·420 | 1·000 | 0·933 | 0·346 | 0·658 | 0·974 | 0·516 |
| B | 0·048 | 0·000 | 0·067 | 0·628 | 0·342 | 0·026 | 0·469 |
| C | 0·500 | 0·000 | 0·000 | 0·026 | 0·000 | 0·000 | 0·015 |
| D | 0·032 | 0·000 | 0·000 | 0·000 | 0·000 | 0·000 | 0·000 |
| GPI-2 | |||||||
| A | 1·000 | 0·952 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 |
| B | 0·000 | 0·048 | 0·000 | 0·000 | 0·000 | 0·000 | 0·000 |
| PGD-1 | |||||||
| A | 0·942 | 0·500 | 1·000 | 1·000 | 1·000 | 1·000 | 1·000 |
| B | 0·058 | 0·125 | 0·000 | 0·000 | 0·000 | 0·000 | 0·000 |
| C | 0·000 | 0·375 | 0·000 | 0·000 | 0·000 | 0·000 | 0·000 |
| PGM-1 | |||||||
| A | 0·500 | 0·900 | 0·875 | 0·735 | 0·958 | 1·000 | 1·000 |
| B | 0·150 | 0·100 | 0·125 | 0·147 | 0·042 | 0·000 | 0·000 |
| C | 0·350 | 0·000 | 0·000 | 0·118 | 0·000 | 0·000 | 0·000 |
| PGM-2 | |||||||
| A | 0·475 | 0·954 | 0·063 | 1·000 | 1·000 | 1·000 | 0·727 |
| B | 0·350 | 0·046 | 0·750 | 0·000 | 0·000 | 0·000 | 0·182 |
| C | 0·175 | 0·000 | 0·063 | 0·000 | 0·000 | 0·000 | 0·091 |
| D | 0·000 | 0·000 | 0·125 | 0·000 | 0·000 | 0·000 | 0·000 |
Table 3.
Genetic diversity statistics for all populations of N. alessandrii sampled
| Populations | n | A | Ap | Pp | He | Ho | F | H-W deviations |
||
|---|---|---|---|---|---|---|---|---|---|---|
| Tests | − | + | ||||||||
| Fragment 1 | 24·7 | 2·4 | 2·7 | 85·7 | 0·299 (0·112) | 0·372 (0·163) | −0·244 | 6 | 0 | 1 |
| Fragment 2 | 18·7 | 1·9 | 2·2 | 71·4 | 0·176 (0·080) | 0·189 (0·088) | −0·080 | 5 | 0 | 0 |
| Fragment 3 | 11·1 | 2·0 | 2·4 | 71·4 | 0·217 (0·068) | 0·198 (0·070) | 0·088 | 5 | 0 | 0 |
| Alto Huelón | 27·9 | 1·9 | 2·5 | 57·1 | 0·234 (0·089) | 0·205 (0·083) | 0·124 | 4 | 0 | 0 |
| Agua Buena | 28·9 | 1·6 | 2·0 | 57·1 | 0·137 (0·064) | 0·083 (0·043) | 0·399 | 4 | 1 | 0 |
| El Fin Empedrado | 51·7 | 1·3 | 2·0 | 42·9 | 0·017 (0·008) | 0·017 (0·008) | −0·013 | 4 | 0 | 0 |
| Reserva Los Ruiles | 30·4 | 1·9 | 2·5 | 57·1 | 0·195 (0·090) | 0·166 (0·088) | 0·149 | 4 | 1 | 0 |
| Population average | 27·4 | 1·8 | 2·3 | 61·2 | 0·182 (0·034) | 0·176 (0·042) | ||||
| Species average | 191·6 | 2·9 | 2·9 | 100 | 0·209 (0·055) | 0·148 (0·031) | ||||
| Parks average | 22·3 | 1·9 | 2·5 | 68·5 | 0·106 (0·089) | 0·091 (0·075) | ||||
| Private property average | 40·6 | 2·9 | 2·9 | 50·0 | 0·213 (0·027) | 0·209 (0·046) | ||||
n = mean number of individuals analyzed per locus; A = mean number of alleles per locus; Ap = mean number of alleles per polymorphic locus; Pp = percentage of polymorphic loci (no frequency criterion); He = mean unbiased expected heterozygosity; Ho = mean observed heterozygosity; the values shown are means (standard errors). F = fixation index or inbreeding coefficient, and results of tests for deviations from Hardy–Weinberg (H-W) equilibrium (tests = number of loci out of a total of seven for which tests could be performed, – = number of loci with a significant deficiency of heterozygotes,+=number of loci with a significant excess of heterozygotes, P < 0·05.
Genetic diversity
The mean number of alleles per locus per population (A) was = 1·8, ranging from 1·3 in E-F to 2·4 in fragment F-1. Surprisingly, four private alleles were found in smaller populations, with two private alleles in F-2 (GPI-2-B and PGD-1-C) and one private allele in F-1 (GPI-1-A) and F-3 (PGM-2-D) (Table 2). The mean expected heretozygosity across all populations was high He = 0·182 (±0·034), ranging from He = 0·017 (±0·008) in population E-F to He = 0·299 (±0·112) in population F-1. The mean value of percentage of polymorphic loci (Pp) was 61·2, ranging from 42·9 in E-F to 85·7 in F-1 populations (Table 3). The fragment size was not associated with any estimator of genetic diversity (data not shown).
Genetic structure and isolation by distance
The apportionment of genetic diversity using Nei's indexes was: total genetic diversity HT = 0·245, within population genetic diversity HS = 0·182, genetic diversity between populations DST = 0·063 and the genetic differentiation GST = 0·257 (Table 4). The number of migrants per generation (Nm) was 0·7.
Table 4.
Comparison of mean genetic diversity and its components: total (HT), within (HS), among populations (DST) and among-population differentiation (GST) (Nei, 1973), number of migrants per generation (Nm) and number of populations sampled for each Nothofagus species
| Species | Range | Ho | HT | HS | DST | GST | Nm | NPOPS | Reference |
|---|---|---|---|---|---|---|---|---|---|
| N. alessandrii | Narrow/endemic | 0·176 | 0·246 | 0·182 | 0·063 | 0·257 | 0·7 | 7 | This study |
| N. nitida | Narrow | 0·036 | 0·156 | 0·141 | 0·015 | 0·047 | 5·0 | 4 | Premoli (1997) |
| N. betuloides | Widespread | 0·098 | 0·301 | 0·236 | 0·065 | 0·120 | 1·8 | 4 | Premoli (1997) |
| N. dombeyi | Widespread | 0·082 | 0·228 | 0·199 | 0·029 | 0·074 | 3·1 | 5 | Premoli (1997) |
| N. pumilio | Widespread | 0·013 | 0·047 | 0·045 | – | 0·013 | 19 | 12 | Premoli (2003) |
| N. alpina | Narrow | 0·170 | – | – | – | – | – | – | Marchelli and Gallo (2001) |
| N. truncata* | Widespread | 0·051 | 0·161 | 0·153 | 0·008 | 0·049 | 4·9 | 30 | Haase (1992) |
* Nothofagus truncata from New Zealand.
There was no correlation between the pairwise effective rate of migrants (M) and the geographic distance matrixes among population pairs (correlation between matrices r = 0·434; Mantel Z = −5·07, P = 0·08), suggesting the absence of isolation-by-distance in the populations studied.
Comparison between protected and unprotected areas and with related congeners
Expected heterozygosity was higher for unprotected than for protected areas (He = 0·294 and He = 0·090; respectively t = 10·9, P < 0·001). The same occurred for the observed heterozygosity (Ho = 0·229 and Ho = 0·067; t = 19·7, P < 0·001).
The comparisons of the levels of genetic diversity (mean expected and observed heterozygosity within populations) between N. alessandrii and its widespread and narrow Nothofagus congeners sampled by Premoli (1997) showed that N. alessandrii has significantly higher genetic diversity (t-test, P < 0·001 for all comparisons).
DISCUSSION
Although the geographic range of species has been suggested to be a good predictor of genetic diversity of natural populations (Karron, 1987; Hamrick and Godt, 1989, 1996a, Gitzendanner and Soltis, 2000; Cole, 2003) where narrowly distributed or endemic species attain lower genetic diversity levels than their widespread congeners, new examples of narrow endemics with high genetic diversity are regularly added to the literature (Cardoso et al., 1998; Delgado et al., 1999; González-Astorga and Castillo-Campos, 2004). According to the present results, this is the case for Nothofagus alessandrii as well. High genetic diversity in narrowly endemic species is commonly associated with unique history such as recent origin from widespread congeners, hybridization, maintenance of genetic diversity within refugial populations and ecological traits such as the ability to survive in a range of different habitats (Smith and Pham, 1996; Godt and Hamrick, 1998; Gitzendanner and Soltis, 2000).
Genetic diversity and comparisons with related congeners
Given that species with restricted geographic distributions usually occur in small and isolated populations, population genetic theory predicts that these populations will be prone to the loss of genetic diversity due to genetic drift (Wright, 1931). Considering that N. alessandrii has been historically restricted to a narrow latitudinal range on south-facing slopes of the coastal mountains of central Chile (Donoso and Landaeta, 1983), low levels of genetic diversity compared with its widespread congeners were expected. In contrast, appreciable levels of total genetic diversity for the narrow endemic N. alessandrii were found, which suggests that this species has not been strongly affected by genetic drift and/or inbreeding processes, maintaining relatively high levels of mean expected heterozygosity within populations (He = 0·182).
This high level of genetic diversity is congruent with glacial refugia. According to the present results, N. alessandrii showed levels of total genetic diversity (HT = 0·246) similar to those reported by Premoli (1997) for its more related widespread congeners N. dombeyi (HT = 0·228) and N. betuloides (HT = 0·301), and similar values to those reported by Hamrick and Godt (1996b) for the Fagaceae species (He = 0·198). Further, total genetic diversity in N. alessandrii was higher than those reported for the Chilean endemic N. nitida (HT = 0·156; Premoli, 1997), and the South American widespread N. pumilio (HT = 0·047; Premoli, 2003), and N. truncata from New Zealand (HT = 0·161; Haase, 1992) (Table 4).
At the population level, the genetic diversity of N. alessandrii was higher (Pp = 61·2 %; A = 1·8, Ap = 2·3 and He = 0·182) than the mean values reported by Hamrick and Godt (1989) for long-lived woody perennials (Pp = 49·3 %; Ap = 1·76 and He = 0·148). If N. alessandrii is compared with the three congener species studied by Premoli (1997), the mean percentage of polymorphic loci (Pp) was higher, but this may be explained by the fact that this author found several monomorphic loci (Tables 3 and 5). However, the mean number of alleles per polymorphic locus (Ap) in N. alessandrii was lower than that of its congeners (Tables 3 and 5). Additionally, higher levels of mean observed and expected heterozygosity were also found in N. alessandrii compared with its widespread congeners (t-test, P < 0·001 for all comparisons). Therefore, the present results do not support the idea that endemic plant species harbour less total allozyme variation than more widespread species (Hamrick and Godt, 1989, 1996b; Cole, 2003).
Table 5.
Abstract of genetic variation for Nothofagus species found in the literature
| Species | Pp | A | Ap | He | Reference |
|---|---|---|---|---|---|
| N. nitida | 10·0* | 1·3 | 2·7 | 0·045 | Premoli (1997) |
| N. betuloides | 26·7* | 1·5 | 2·8 | 0·116 | Premoli (1997) |
| N. dombeyi | 29·0* | 1·6 | 2·9 | 0·093 | Premoli (1997) |
| N. pumilio | 12·3* | 1·2 | – | 0·025 | Premoli (2003) |
| N. alpina | 75·0 | – | – | – | Marchelli and Gallo (2001) |
| N. truncata | 20·0** | 1·8 | – | 0·054 | Haase (1992) |
Pp is the percentage of polymorphic loci estimated under (* 95% criteria or ** 99%), mean number of alleles per locus (A), mean number of alleles per polymorphic locus (Ap) and mean expected heterozygosity (He).
Currently, an increasing number of studies indicate that the expectation of endemic plant species harbouring lower genetic variation than widespread species was an overgeneralization. For instance, Gitzendanner and Soltis (2000) found significant, but small, differences between rare and widespread congeneric species for a series of estimators of the genetic diversity such as the percentages of polymorphic loci, mean number of alleles per locus and observed heterozygosity. However, they failed to find differences for HT. Other examples of endemic species with narrow geographical range showing high genetic diversity are Cupressus macrocarpa (Kafton, 1976), Adenophorus periens (Ranker, 1994), Daviesia suaveolens (Young and Brown, 1996), Caesalpinia echinata (Cardoso et al., 1998), Pinus rzedowskii (Delgado et al., 1999), Brongniartia vazquezii (González-Astorga and Nuñez-Farfán, 2001) and Antirhea aromatica (González-Astorga and Castillo-Campos, 2004). Therefore, the present results constitute a possible new example of endemic species with a very narrow distributional range exhibiting high genetic diversity compared with its widespread congeners. It is noted that the small number of alleles segregating at isozyme loci imposes a limit on the resolution of the present comparisons. Highly polymorphic codominant DNA markers may provide a more detailed account of levels and patterns of diversity in Nothofagus spp. in the future.
Genetic structure of the fragments
Although nothing is known about the distance of pollen and seed dispersal in N. alessandrii, both pollen and seeds have obvious adaptations for wind-dispersal (Rodríguez et al., 1983). Other wind-dispersed and wind-pollinated species of Nothofagus (N. alpina, N. betuloides and N. dombeyi) have been reported to be highly self-incompatible (Riveros et al., 1995). Premoli (1996), using protein electrophoresis from progeny arrays of N. nitida, N. betuloides and N. dombeyi, determined that only 13 % of the seeds are produced by self-fertilization. Thus, considering the evidence for the most closely related species to N. alessandrii (Manos, 1997), it seems likely that this species is predominantly outcrossing as well. Hamrick and Godt (1996b) showed that wind-pollinated outcrossing species usually have low values of GST (approx. 0·094), while endemic long-lived perennials tend to have values of GST near 0·150. They also reported a mean GST value of 0·085 for 27 species of the Fagaceae family. However, it was found that a high proportion of the total genetic diversity resides among populations (GST = 0·257; Table 3), with very low levels of mean historical gene flow (Nm = 0·7), which contrasts with our expectations considering the breeding system and longevity of this species. Despite this, genetic differentiation in N. alessandrii seems to be more similar to that reported by Hamrick and Godt (1996b) for endemic species with wind-dispersed seeds (GST = 0·259).
Nothofagus alessandrii is located in a zone which has been proposed as a glacial refuge (Villagrán and Hinojosa, 1997; Villagrán et al., 1998). This suggests that differentiation among populations was already high before human fragmentation, as a consequence of glacial fragmentation. However, the lack of specific palynological records for N. alessandrii makes the reconstruction of the glacial history of this species difficult. Hence, the present result could be the reflection of both historical and human-mediated processes.
Human-mediated fragmentation can drastically affect natural populations by modifying their genetic diversity and genetic structure (Frankel and Soulé, 1981; Ellstrand and Elam, 1993; Frankham et al., 2002). The high level of among-population differentiation found here could be explained, in part, by the presence of private alleles in fragments that are geographically very close (F-1, F-2 and F-3; see Fig. 1). Although it is known that the onset of the genetic effects of fragmentation is frequently delayed in long-lived woody species, such high differentiation at a local scale could be interpreted to be a consequence of human-mediated fragmentation. In this context, fragments would probably represent a small and biased sample of a more continuous forest that due to fragmentation now represents a random sample of the original forest. Thus, due to fragmentation, genetic bottlenecks could have increased the population differentiation that already existed prior to fragmentation. Furthermore, it is possible that the Pinus radiata and Eucalyptus globulus matrix is acting as a barrier, probably limiting the gene flow between fragments, either via seeds and/or pollen, hence, increasing their isolation. This can be particularly important if N. alessandrii, as well as all South American Nothofagus species, is predominantly outcrossing.
Not only genetic diversity is expected to be different between narrow and widespread congeners, but the apportionment of genetic diversity is predicted to be higher in narrowly distributed species as well, compared with widespread congeners (Cole, 2003). However, Hamrick and Godt (1989), Gitzendanner and Soltis (2000) and Cole (2003) did not find significant differences between widespread and narrowly distributed congeners in how genetic diversity is partitioned within and among populations (GST and/or FST). In contrast, in the present study, higher inter-population differentiation and lower levels of historical gene flow were found in N. alessandrii compared with its Nothofagus congeners studied by Premoli (1997) (Table 4). Nothofagus nitida, N. betuloides and N. bombeyi showed moderate and low genetic differentiation with low GST and high levels of gene flow (Nm >1), which is in contrast to the high GST and low Nm found in N. alessandrii (Table 4).
Implications for the conservation of N. alessandrii
Higher levels of genetic diversity (observed and expected heterozygosity) were found in unprotected than in protected fragments. Further, the inbreeding coefficient FIS was positive in the protected population Reserva Los Ruiles, showing a significant deficiency of heterozygotes. These results suggest that protected populations are preserving only a small fraction of the species genetic diversity. Moreover, the presence of rare alleles in small unprotected fragments suggests that they can serve as reservoirs of rare or private alleles. This finding is important because it emphasizes the importance of conserving small remnant fragments that can harbour rare allelic variants, and hence preserve species genetic variation. It has been suggested that, when genetic differentiation between populations is high, large numbers of natural protected areas will be necessary for preserving the genetic diversity of a threatened species (Frankel and Soulé, 1981; Eguiarte and Piñero, 1990). Considering that genetic differentiation in N. alessandrii was high, future conservation efforts must aim at preserving more fragments surrounding the El Fin Empedrado area and the northern limit of its distribution. This is particularly important because this forest type has an extremely high rate of disappearance (Bustamante and Castor, 1998). Currently, there are many small unprotected fragments within private properties (almost 88 % of the species distribution) and most of these are restricted to deep ravines in south-facing slopes. This is the case because Chilean legislation forbids the cutting of forests when they are growing on steep slopes. Therefore, forestry companies and private land-owners must be included in future efforts for effective conservation of this highly endangered endemic tree species.
There are no specific studies about the regeneration dynamics of N. alessandrii. Although reproductively mature individuals of N. alessandrii in population fragments F-2, A-H and E-F were observed, no seedlings were found. Therefore, a comprehensive conservation strategy for this species should include data on regeneration ecology, breeding systems, and the effects of the Pinus/Eucalyptus matrix on gene flow between fragments and its consequences on fitness.
ACKNOWLEDGEMENTS
This work was funded by University of Concepción, Project GIA No. 201·111·025-1·4, Fundación Andes Proyect No. C-14055, Chilean Millennium Science Initiative Grant No. P02-051 and the BBVA-2004 Prize in Conservation Biology. We thank the CONAF (VII Region, Chile) and Forestal CELCO SA for permitting collection of plant material. Cristian Torres-Díaz held a Doctoral Fellowship supported through project MECESUP UCO 9906. For research assistance and sample collection we thank Maritza Mihoč and Marco Molina.
LITERATURE CITED
- Ayres DR, Ryan FJ. Genetic diversity and structure of the narrow endemic Wyethia reticulata and its congeners W. bolanderi (Asteraceae) using RAPD and allozyme techniques. American Journal of Botany. 1999;86:344–353. [PubMed] [Google Scholar]
- Benoit I. Libro rojo de la flora terrestre de Chile. Santiago: Corporación Nacional Forestal, Ministerio de Agricultura; 1989. [Google Scholar]
- Bustamante RO, Castor C. The decline of an endangered temperate ecosystem: the ruil (Nothofagus alessandrii) forest in central Chile. Biodiversity and Conservation. 1998;7:1607–1626. [Google Scholar]
- Cardoso MA, Provan J, Powell W, Ferreira PCG, de Oliveira DE. High genetic differentiation among remnant populations of the endangered Caesalpinia echinata Lam. (Leguminoseae-Caesalpinioideae) Molecular Ecology. 1998;7:601–608. [Google Scholar]
- Cole CT. Genetic variation in rare and common plants. Annual Review of Ecology and Systematics. 2003;34:213–237. [Google Scholar]
- Delgado P, Piñero D, Chaos A, Pérez-Nasser N, Alvarez-Buylla ER. High population differentiation and genetic variation in the endangered Mexican pine Pinus rzedowskii (Pinaceae) American Journal of Botany. 1999;86:669–676. [PubMed] [Google Scholar]
- Donoso C, Landaeta E. Ruil (Nothofagus alessandrii), a threatened Chilean tree species. Environmental Conservation. 1983;10:159–162. [Google Scholar]
- Eguiarte LE, Piñero D. Genética de la conservación: leones vemos, genes no sabemos. Ciencias. 1990;4:34–47. Número especial. [Google Scholar]
- Ellstrand NC, Elam DR. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics. 1993;24:217–242. [Google Scholar]
- Frankel HO, Soulé ME. Conservation and evolution. Cambridge: Cambridge University Press; 1981. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- Gitzendanner MA, Soltis PS. Patterns of genetic variation in rare and widespread plant congeners. American Journal of Botany. 2000;87:783–792. [PubMed] [Google Scholar]
- Godt MJW, Hamrick JL. Allozyme diversity in the endangered pitcher plant Sarracenia rubra spp. alabamensis (Sarraceniaceae) and its close relative S. rubra spp. rubra. American Journal of Botany. 1998;85(802):802–810. [PubMed] [Google Scholar]
- González-Astorga J, Castillo-Campos G. Genetic variability of the narrow endemic species tree Antirhea aromatica (Rubiaceae, Guettardeae) in a tropical forest of México. Annals of Botany. 2004;93:521–528. doi: 10.1093/aob/mch070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Astorga J, Núñez-Farfán J. Effects of habitat fragmentation on the genetic structure of the narrow endemics Brongniartia vazquezii. Evolutionary Ecology Research. 2001;3:861–872. [Google Scholar]
- Gottlieb DL. Electrophoretic evidence and plant populations. Progress in Phytochemistry. 1981;7:1–46. [Google Scholar]
- Haase P. Isozyme variability and biogeography of Nothofagus truncata (Fagaceae) New Zealand Journal of Botany. 1992;30:315–328. [Google Scholar]
- Hamrick JL, Godt MJ. Allozyme diversity in plant species. In: Brown A, Clegg M, Khaler A, Weir B, editors. Plant population genetics, breeding and genetic resources. Sunderland, MA: Sinauer Press; 1989. pp. 43–63. [Google Scholar]
- Hamrick JL, Godt MJ. Conservation genetics of endemic plant species. In: Avise JC, Hamrick JL, editors. Conservation genetics case histories from nature. New York, NY: Chapman and Hall; 1996a. pp. 281–304. [Google Scholar]
- Hamrick JL, Godt MJ. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London. 1996b;351:1291–1298. [Google Scholar]
- Hamrick JL, Linhart YB, Mitton JB. Relationships between life history characteristics and electrophoretically-detectable genetic variation in plants. Annual Review of Ecology and Systematics. 1979;10:173–200. [Google Scholar]
- Hill RS, Jordan GJ. The evolutionary history of Nothofagus (Nothofagaceae) Australian Systematic Botany. 1993;6:111–126. [Google Scholar]
- Humphries TM. Biogeographical methods and the southern beeches (Fagaceae: Nothofagus) In: Funks VA, Brooks DR, editors. Advances in cladistics. Bronx, NY: New York Botanical Garden; 1981. pp. 177–207. [Google Scholar]
- Kafton DL. Isozyme variability and reproductive phenology in Monterrey cypress. Berkeley, CA: University of California; 1976. PhD Thesis. [Google Scholar]
- Karron JD. A comparison of levels of genetic polimorphism and self-compatibility in geographically restricted and widespread plant congeners. Evolutionary Ecology. 1987;1:47–58. [Google Scholar]
- Karron JD. Patterns of genetic variation and breeding systems in rare plant species. In: Falk DA, Holsinger KE, editors. Genetics and conservation of rare plants. New York, NY: Oxford University Press; 1991. pp. 87–98. [Google Scholar]
- Karron JD, Linhart YB, Chaulk CA, Robertson CA. Genetic structure of populations of geographically restricted and widespread species of Astragalus (Fabaceae) American Journal of Botany. 1988;75:1114–1119. [Google Scholar]
- Knapp M, Stöckler K, Havell D, Delsuc F, Sebastiani F, Lockhart PJ. Relaxed molecular clock provides evidence for long-distance dispersal of Nothofagus (southern beech) PLoS Biology. 2005 doi: 10.1371/journal.pbio.0030014. e-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledig FT, Conkle MT. Gene diversity and genetic structure in a narrow endemic, Torrey pine (Pinus torreyana Parry ex Carr.) Evolution. 1983;37::79–85. doi: 10.1111/j.1558-5646.1983.tb05515.x. [DOI] [PubMed] [Google Scholar]
- Linhart YB, Premoli AC. Comparison of the genetic variability in Aletes humilis, a rare plant species, and its common relative Aletes acaulis in Colorado. American Journal of Botany. 1993;80:598–605. doi: 10.1002/j.1537-2197.1993.tb13846.x. [DOI] [PubMed] [Google Scholar]
- Lewis PO. Genestat-pc 3·3. Raleigh, NC: Department of Statistics, North Carolina State University; 1993. [Google Scholar]
- Manos PS. Systematics of Nothofagus (Nothofagaceae) based on RDNA spacer sequences (ITS) – taxonomic congruence with morphology and plastid sequences. American Journal of Botany. 1997;84:1137–1155. [PubMed] [Google Scholar]
- Mantel N. The detection of disease clustering and generalized regression approach. Cancer Research. 1967;27:209–220. [PubMed] [Google Scholar]
- Martin PG, Dowd JM. Using sequences of rbcL to study phylogeny and biogeography of Nothofagus species. Australian Journal of Botany. 1993;6:441–447. [Google Scholar]
- Marchelli P, Gallo LA. Genetic diversity and differentiation in a southern beech subjected to introgresive hybridization. Heredity. 2001;87:284–293. doi: 10.1046/j.1365-2540.2001.00882.x. [DOI] [PubMed] [Google Scholar]
- Miller MP. A Windows program for the analysis of allozyme and molecular population genetic data. Logan, UT: Utah State University; 1997. Tools for population genetic analyses (TFPGA), ver. 1·3. [Google Scholar]
- Myers N, Mittermeier RA, Fonseca GAB. Kent J. Biodiversity hotspot for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- Nei M. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences of the USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premoli AC. Allozyme polymorphisms, outcrossing rates and hybridization of South American Nothofagus. Genética. 1996;97:55–64. [Google Scholar]
- Premoli AC. Genetic variation in a geographically restricted and two widespread species of South American Nothofagus. Journal of Biogeography. 1997;24:883–892. [Google Scholar]
- Premoli AC. Isozyme polymorphisms provide evidence of clinal variation with elevation in Nothofagus pumilio. Journal of Heredity. 2003;94:218–226. doi: 10.1093/jhered/esg052. [DOI] [PubMed] [Google Scholar]
- Premoli AC, Souto CP, Allnutt TR, Newton AC. Effects of population disjunction on isozyme variation in the widespread Pilgerodendron uviferum. Heredity. 2001;8:337–343. doi: 10.1046/j.1365-2540.2001.00906.x. [DOI] [PubMed] [Google Scholar]
- Ranker TA. Evolution of high genetic variability in the rare Hawaiian fern Adenophorus pariens and implications for conservation management. Biological Conservation. 1994;70:19–24. [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1·2): population genetics software for exact tests and ecumenicism. Journal of Heredity. 1995;86:248–249. [Google Scholar]
- Riveros M, Paredes MA, Rosas MT, Cardenas E, Armesto J, Arroyo MTK, et al. Reproductive biology in species of the genus Nothofagus. Environmental and Experimental Botany. 1995;35:519–524. [Google Scholar]
- Rodríguez G, Rodríguez R, Barrales HL. Concepción, Chile: Editorial Anibal Pinto; 1995. Chilean ornamental plants. [Google Scholar]
- Rodriguez R, Matthei O, Quezada M. Concepción, Chile: Universidad de Concepción; 1983. Flora arbórea de Chile. [Google Scholar]
- San Martín J, Donoso C. Estructura florística e impacto antrópico en el bosque maulino de Chile. In: Armesto JJ, Villagrán C, Arroyo MK, editors. Ecología de los bosques nativos chilenos. Santiago de Chile: Editorial Universitaria; 1996. pp. 153–167. [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Smith JF, Pham TV. Genetic diversity of the narrow endemic Allium aaseae (Alliaceae) American Journal of Botany. 1996;83:717–726. [Google Scholar]
- Snedecor GW, Cochran G. Statistical methods. 6th edn. Ames, IA: Iowa State University Press; 1967. [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: the principles of statistics in biological research. New York, NY: WH Freeman and Co; 1995. [Google Scholar]
- Villagrán C, Hinojosa LF. Historia de los bosques del sur de Sudamérica. II. Análisis fitogeográfico. Revista Chilena de Historia Natural. 1997;70:241–267. [Google Scholar]
- Villagrán C, Le-Quesne C, Aravena JC, Jiménez H, Hinojosa LF. El rol de los cambios de clima del Cuaternario en la distribución actual de la vegetación de Chile central-sur. Bamberg Geographische. 1998;15:227–242. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of populations structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Weeden NF. Visualization and interpretation of plants isozymes. In: Soltis DE, Soltis PS, editors. Isozymes in plant biology. Portland, OR: Discorides Press; 1989. pp. 5–45. [Google Scholar]
- Wolf AT, Howe RW, Hamrick JL. Genetic diversity and population structure of the serpentine endemic Calystegia collina (Convolvulaceae) in Northern California. American Journal of Botany. 2000;87:1138–1146. [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Young AB, Brown ADH. Comparative population genetic structure of the rare shrub Daviesia suaveolens and its common congener D. mimosoides. Conservation Biology. 1996;10:1220–1228. [Google Scholar]