Abstract
Background and Aims
Fragmentation of natural habitats can negatively impact plant populations by leading to reduced genetic variation and increased genetic distance as populations become geographically and genetically isolated from one another. To test whether such detrimental effects occur within an urban landscape, the genetic structure of six populations of the perennial herb Viola pubescens was characterized in the metropolitan area of Greater Cincinnati in southwestern Ohio, USA.
Methods
Using three inter-simple sequence repeat (ISSR) markers, 51 loci amplified across all urban populations. For reference, four previously examined agricultural populations in central/northern Ohio and a geographically distant population in Michigan were also included in the analysis.
Key Results
Urban populations retained high levels of genetic variation (percentage of polymorphic loci, Pp = 80·7 %) with similar genetic distances among populations and an absence of unique alleles. Geographic and genetic distances were correlated with one another, and all populations grouped according to region. Individuals from urban populations clustered together and away from individuals from agricultural populations and from the Michigan population in a principle coordinates analysis. Hierarchical analysis of molecular variance (AMOVA) revealed that most of the genetic variability was partitioned within populations (69·1 %) and among groups (22·2 %) of southwestern Ohio, central/northern Ohio and Michigan groups. Mean Fst was 0·308, indicating substantial population differentiation.
Conclusions
It is concluded that urban fragmentation does not appear to impede gene flow in V. pubescens in southwestern Ohio. These results are consistent with life history traits of this species and the possibility of high insect abundance in urban habitats due to diverse floral resources and nesting sites. Combined with the cleistogamous breeding system of this species, pollinator availability in the urban matrix may buffer populations against detrimental effects of habitat fragmentation, at least in larger forest fragments. Consequently, it may be inappropriate to generalize about genetic effects of fragmentation across landscapes or even across plant species with different pollination systems.
Key words: Habitat fragmentation, ISSR, population genetic structure, urban effects, Viola pubescens
INTRODUCTION
Habitat fragmentation is increasing throughout the world and has many important ecological and genetic consequences for endemic plant populations. In general, plant species in fragmented areas may experience reduced habitat and population size as well as potential disturbances in pollinator service and seed dispersal (Jennersten, 1988; Rathcke and Jules, 1993; Kwak et al., 1998; Spira, 2001; Bhattacharya et al., 2003). Over time, this can lead to increased inbreeding (Ellstrand and Elam, 1993), lower reproductive success (Steffan-Dewenter and Tscharntke, 1999) and disrupted gene flow. Consequently, isolated plant populations can experience loss of genetic variation, increased population differentiation and genetic drift (Young et al., 1996; Mills and Tallmon, 1999). Over the long term, fragmentation may also reduce the ability of populations to adapt to changing environments (Fisher, 1930; Mills and Tallmon, 1999), thereby increasing local extinction events. In some cases, however, fragmentation may not be detrimental (Young et al., 1996) and may even promote gene flow (Young and Merriam, 1994; Aldrich and Hamrick, 1998; White et al., 2002). Although genetic effects of fragmentation in plants have been examined in a variety of landscapes, including grasslands (Young et al., 1999), woodlands (Prober et al., 1998), deciduous forests (Foré et al., 1992; Young et al., 1993; Young and Merriam, 1994; Cruzan, 2001) and agricultural areas (Berg et al., 1998; Culley and Grubb, 2003), urban areas have remained relatively unexplored.
Urban habitat fragmentation results in a mixture of natural areas interspersed with residential and commercial developments and often dissected by major transportation corridors. Consequently, the matrix of an urban landscape can be much more complex than the matrices of other landscapes, such as agricultural areas where crop monocultures typically separate fragments. This complexity of the urban matrix may potentially impose a more substantial barrier to gene flow than other matrices, especially if movement of pollinators and seed dispersers is negatively impacted by urbanization. On the other hand, the urban matrix may potentially promote gene flow if pollinators and seed dispersers persist and flourish within the urban matrix.
An ideal location in which to examine the genetic effects of urban fragmentation is southwestern Ohio. As with many other areas of the USA, this locality has experienced increased urbanization as the city of Cincinnati has expanded into outlying areas. The city was one of the first settlements in the Ohio Valley circa 1778 when mixed mesophytic and beech–maple forests were common (Braun, 1950; Gordon, 1969). It is estimated that > 95 % of Ohio was originally covered with forests (Griffith et al., 1993), which were gradually cleared for agriculture and settlement, until by 1910 only 10 % of the state was forested. Following passage of laws encouraging forest development as well as abandonment of farms as people migrated to urban locations, forest cover in Ohio gradually increased to > 30 % by 1991 (Griffith et al., 1993). Today, > 13 % of southwestern Ohio is forested (Griffith et al., 1993), a portion of which is contained within urban parks in the Greater Cincinnati metropolitan area. This area is now home to > 2 million people, occupying 13 counties in Ohio, Kentucky and Indiana, and was considered the 24th largest metropolitan area in the country in 2000 (US Bureau of the Census, 2003).
Within urban parks in Greater Cincinnati can be found Viola pubescens var. scabriuscula, the Smooth Yellow Violet. This native perennial is ideal for study because it is partially dependent upon insect pollinators for seed set (Culley, 2002), it contains substantial amounts of genetic variation relative to other species (Culley and Wolfe, 2001) and its population genetic structure was previously characterized within an agricultural landscape (Culley and Grubb, 2003). In this case, populations in small forest fragments exhibited reduced genetic variation and increased population differentiation relative to populations in larger fragments (Culley and Grubb, 2003). The goal of the current study was to quantify the genetic structure of urban populations of V. pubescens to answer the following questions. (a) Do urban populations exhibit lower levels of genetic variation relative to non-urban populations? (b) Are urban populations more genetically distant and differentiated from one another compared with non-urban populations? (c) Is gene flow impeded by the urban matrix? To our knowledge, this is the first study of genetic effects of habitat fragmentation in a plant species within an urban environment.
MATERIALS AND METHODS
The study organism
Viola pubescens var. scabriuscula is a stemmed, non-clonal violet that is a common resident of deciduous forests in eastern North America (Ballard, 1994). Foliage is largely glabrous, which distinguishes this variety from the densely pubescent V. pubescens var. pubescens (Ballard, 1994); the varieties also differ in number of stems, basal leaves and teeth on leaf margins (Lévesque and Dansereau, 1966; Cain, 1967; Ballard, 1994). The species itself exhibits dimorphic cleistogamy (Culley and Klooster, 2007), producing two types of flowers at different times: yellow chasmogamous flowers are produced first in the early spring and inconspicuous cleistogamous flowers subsequently appear after the forest canopy forms in late spring (Culley, 2002). Individuals continue to produce automatically self-pollinated cleistogamous flowers until plant senescence in early autumn. Chasmogamous flowers are visited by a variety of generalist pollinators, including bumblebees, carpenter bees, butterflies, halictid bees and beeflies (Culley, 2002). Outcrossing rates vary substantially between years, ranging from 0·40 to 0·73 (Culley, 2002). Seeds are dispersed both ballistically up to 4 m away from the maternal plant and also by ants (Culley, 2002), so long-distance gene flow probably only occurs via pollen. Individuals can live at least 5 years (Culley, 2002) and possibly longer, as in Viola sororia which has an individual life expectancy > 10 years (Solbrig et al., 1980).
Sampling
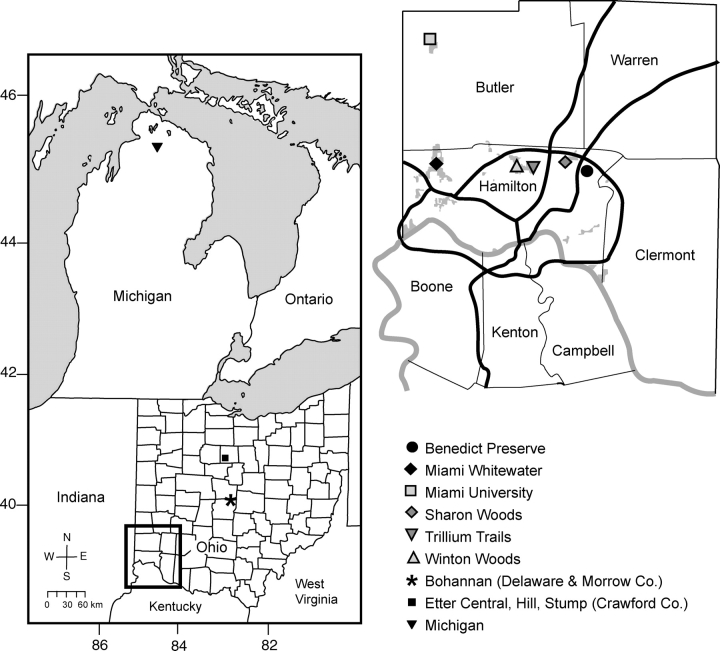
Fresh leaf samples were obtained during Spring, 2003 from 39–49 individuals in each of six populations of V. pubescens var. scabriuscula scattered throughout the Greater Cincinnati area in southwestern Ohio (Fig. 1; Table 1). These populations are found in secondary growth forest that formed after agriculture was abandoned in the region and most probably originated from relict populations inhabiting smaller farm woodlots in the area. Four of the populations (Miami Whitewater, Sharon Woods, Trillium Trails and Winton Woods) are located in urban parks owned by the Hamilton County Park District in Hamilton County, Ohio. The remaining two populations inhabit preserves owned by the University of Cincinnati in Hamilton County, Ohio (The Harris M. Benedict Botanical Preserve) and Miami University in neighbouring Butler County, Ohio (Miami University Natural Areas). Within each site, samples were collected from individual plants located at least 2 m apart to prevent sampling the same plant twice. Tissue samples were placed on ice for transport back to the University of Cincinnati, where they were stored at − 75 °C.
Fig. 1.
Locations of Viola pubescens populations examined in the current study. Shown in the left panel are reference populations in central Ohio, northern Ohio and Michigan (numerals along the border represent latitude and longitude, respectively). The right panel represents the enlarged area in southwestern Ohio that shows the location of six populations in the Greater Cincinnati metropolitan area. County lines are indicated in both panels for Ohio or Greater Cincinnati, and major roadways (dark lines) and Hamilton County parks (grey areas) are indicated in the right panel.
Table 1.
Locations of populations of Viola pubescens var. scabriuscula in Ohio and Michigan
| Site | County | Area (ha) | Npop | Nsampled | Latitude | Longitude |
|---|---|---|---|---|---|---|
| Southwestern Ohio (Urban) | ||||||
| Benedict (BEN) | Hamilton | 26·30 | <800 | 47 | 39·26577 | −84·35498 |
| Miami Whitewater (MWW) | Hamilton | 779·22 | >900 | 49 | 39·25163 | −84·70099 |
| Sharon Woods (SHW) | Hamilton | 192·01 | >800 | 39 | 39·27797 | −84·40200 |
| Trillium Trails (TT) | Hamilton | 9·30 | <500 | 40 | 39·25668 | −84·47898 |
| Winton Woods (WW) | Hamilton | 548·63 | >800 | 43 | 39·26173 | −84·51552 |
| Miami University (MIU) | Butler | 10·50 | >600 | 46 | 39·52912 | −84·70796 |
| Central/Northern Ohio (Agricultural) | ||||||
| Etter Central (ETT) | Crawford | 21·13 | <800 | 37 | 40·74555 | −82·94684 |
| Hill (HLL) | Crawford | 25·09 | <800 | 35 | 40·71079 | −83·02582 |
| Stump (STP) | Crawford | 31·30 | <1000 | 34 | 40·75498 | −82·92736 |
| Bohannan (BOH) | Delaware/Morrow | 40·50 | >1000 | 33 | 40·35049 | −82·92864 |
| Outgroup | ||||||
| Michigan (MI) | Emmet, MI | >500 | >1000 | 18 | 45·54386 | −84·85573 |
Listed are the approximate area of each site that is forested, an estimate of the total number of individuals in the population (Npop) and the subset of individuals that were sampled (Nsampled).
These six southwestern Ohio populations were also compared with four different non-urban Ohio (hereafter referred to as ‘central/northern Ohio populations’) and Michigan populations of V. pubescens var. scabriuscula and one Michigan population of V. pubescens var. pubescens that had been previously examined (Culley and Wolfe, 2001). Bohannan, a central Ohio population in Delaware and Morrow Counties, is located in Bohannan Woods, a scientific preserve owned by Ohio Wesleyan University. Three sites in northern Ohio (Etter Central, Hill and Stump) are privately owned woodlots in Crawford County, Ohio; populations in these sites inhabit isolated wooded areas within an agricultural landscape that was historically part of the Sandusky Plains (Culley and Grubb, 2003). Finally, a distant site in Emmet County, Michigan contained intermingled populations of V. pubescens var. scabriuscula and pubescens individuals. Samples from these populations were originally analysed at The Ohio State University (Culley and Wolfe, 2001) using laboratory techniques as described below.
Generation of genetic data
DNA from the southwestern Ohio samples was extracted using a modified mini-prep cetyltrimethyl ammonium bromide (CTAB) technique of Doyle and Doyle (1987) and then stored at − 20 °C until further analysis. Dominant inter-simple sequence repeat (ISSR) markers were used to quantify the genetic structure of all populations. ISSR markers are similar to randomly amplified polymorphic DNA (RAPD) markers (Wolfe and Liston, 1998) except that ISSR primers are typically longer nucleotide sequences, resulting in a higher primer annealing temperature that gives greater band reproducibility than RAPD markers. Three simple sequence repeats (SSRs) were used as primers to generate 51 bands in single-primer reactions. Primer Mao [(CTC)4RC] yielded 15 bands, primer 17898 [(CA)6RY] produced 22 bands and primer 844 [(CT)8RG] generated 14 bands.
Polymerase chain reaction (PCR) was conducted individually for each primer with either 15 µL (for primers Mao and 17898) or 25 µL (for primer 844) reaction volumes using optimized conditions as follows: 0·2 mm dNTPs, 3 mm (for Mao and 17898) or 2 mm (for 844) MgCl2, 0·4 µm primer, 1 × Taq DNA polymerase buffer, 0·25 U (for Mao and 844) or 0·50 U (for 17898) of Taq (Gibco/BRL; Invitrogen) and 0·3 µL (for Mao and 844) or 0·5 µL (for 844) of DNA. A larger reaction volume was used for primer 844 because it exhibited inconsistent band amplifications at lower reaction volumes. The brand of Taq polymerase and its buffer was found to be important because different brands (e.g. Promega Corp., Madison, WI, USA) may amplify only a sub-set of loci. Amplifications were performed in an Eppendorf Mastercycler using the following conditions: initial denaturation at 94 °C for 1·5 min; 35 cycles each of 94 °C for 45 s, 45 °C (for Mao and 17898) or 46 °C (for 844) for 45 s, and 72 °C for 1·5 min; a final cycle consisted of 94 °C for 45 s, 45 °C or 46 °C for 45 s, and 72 °C for 5 min.
Following PCR, blue/orange loading dye (Promega Corp.) was added to each reaction and samples were loaded onto a 1·2 % agarose gel in 1 × TAE buffer. Additionally, 1 kb ladders (Promega Corp.) and negative and positive controls were loaded onto each gel, which were then run at constant voltage (145 V). Each gel was stained with ethidium bromide and digitized under UV light using the Kodak 1D software package and a Kodak DC290 camera (Eastman Kodak, Rochester, NY, USA). The images were analysed using the same software, which assigns a fragment size to each band using an algorithm based on the 1 kb ladder. These fragment sizes were used to assign loci for each primer, and bands for each assigned locus were scored as diallelic (1 = band present; 0 = band absent).
Data analysis
To avoid common problems associated with the analysis of dominant data (Culley and Wolfe, 2001), analyses only used band presence or absence data and did not involve Hardy–Weinberg equilibrium. This is necessary because populations of V. pubescens are known to deviate from Hardy–Weinberg equilibrium with co-dominant allozyme markers (Culley and Grubb, 2003). It was assumed that there was no co-migration of alleles from different loci, alleles shared by two individuals descend from a common ancestor and each locus consisted of only two alleles that segregate in Mendelian inheritance. Genetic variation was characterized for all 12 populations by calculating the number of shared and unique bands, the percentage of polymorphic loci (Pp) and the percentage of loci that were fixed for a single band. Shannon's index of diversity (H′) was also estimated for each population by calculating the proportion of bands observed per locus across all individuals. Differences between southwestern Ohio and central/northern populations in Pp, H′ and the percentage of loci with fixed bands were analysed with individual Student's t-tests in SAS version 9·1 (SAS Institute, Cary, NC, USA). To test for separate relationships between Pp or H′ with fragment area on both regional and local scales, Spearman's coefficient of rank correlation was calculated separately in SAS for all V. pubescens var. scabriuscula populations and for the sub-sets of southwestern Ohio and central/northern Ohio populations.
Genetic distance was quantified using the Nei and Li (1979) coefficient to compare the number of alleles shared between individuals or populations, excluding shared band absences. This coefficient was first calculated as a distance measure for each pair of individuals in MSVP version 3·11 h (Kovach Computing Services, Anglesey, UK) and used in a principle coordinates analysis to determine whether individuals would cluster according to their designated population. The Nei and Li similarity coefficient was calculated for pairs of populations using !WAVSIML (V. Ford, unpublished; see Crawford et al., 1998 for formulae), and genetic distances were computed for each pair of populations as (1 – similarity). Distance values range from 0 (no difference among groups) to 1 (complete difference among groups). Distances were used to construct a population-level UPGMA tree in NTSYSpc version 2·11a (Rohlf, 1998).
To determine if a relationship existed between genetic and geographic distances of pairs of populations, a Mantel test was conducted using the TFPGA software package (Miller, 1997). Geographic distances were calculated as straight line distances between pairs of sites using latitude and longitude of each location.
Population differentiation was examined with a hierarchical analysis of molecular variance (AMOVA) for populations of V. pubescens var. scabriuscula using Arlequin version 3·1 (Excoffier, 2005). This method treats dominant data as haplotypes and partitions the total variance into covariance components associated with differences among individuals within populations, among individuals in different populations within groups and among groups. These regional groups consisted of: (a) urban populations in southwestern Ohio; (b) agricultural populations in central/northern Ohio; and (c) the Michigan population. Overall Fst was calculated and significance tests were performed in Arlequin (Excoffier et al., 1992) using the approach of Weir and Cockerham (1984) with 1023 permutations. AMOVA was also conducted separately for the southwestern OH populations and for the central/northern OH populations to generate FST values for these regions.
RESULTS
Genetic variation
High levels of variation were observed in urban populations of V. pubescens in Greater Cincinnati (Table 2). The percentage of loci that were polymorphic (Pp) ranged from 78·4 % in the Benedict and Winton Woods populations to 84·3 % in the Miami University population. Overall, the mean proportion of polymorphic loci in the Greater Cincinnati populations was 80·7 %, not significantly different from the 79·9 % observed in central/northern Ohio populations for the same loci (t = 0·21, d.f. = 8, P = 0·809). Similarly, the mean Shannon diversity index did not differ significantly (t = 1·05, d.f. = 8, P = 0·323) between populations in southwestern Ohio (H′ = 3·83) and in central/northern Ohio (4·22). Values of Pp were not correlated with fragment area, either for all populations of V. pubescens var. scabriuscula (r = 0·032, P = 0·925) or for the subset of southwestern Ohio populations (r = − 0·294, P = 0·571), but Pp was substantially correlated with fragment area for the central/northern Ohio populations (r = 0·949, P = 0·051). There were also no significant correlations between fragment area and H′ for all populations of V. pubescens var. scabriuscula (r = 0·042, P = 0·907) or for the southwestern Ohio populations (r = − 0·371, P = 0·469). However, H′ was significantly correlated with fragment area in the central/northern Ohio populations (r = 1·00, P < 0·0001).
Table 2.
Descriptive statistics based on ISSR data for southwestern Ohio, central/northern Ohio and Michigan populations of Viola pubescens
| Population | Pp | H′ | Fixed |
|---|---|---|---|
| Southwestern Ohio (Urban) | |||
| Benedict | 78·4 | 3·43 | 5·9 |
| Miami University | 84·3 | 3·99 | 3·9 |
| Miami Whitewater | 80·4 | 3·63 | 3·9 |
| Sharon Woods | 82·4 | 4·14 | 5·9 |
| Trillium Trails | 80·4 | 4·12 | 9·8 |
| Winton Woods | 78·4 | 3·67 | 3·9 |
| Mean | 80·7 | 3·83 | 5·6 |
| Central/northern Ohio (Agricultural) | |||
| Bohannan | 86·3 | 5·28 | 0·0 |
| Etter Central | 70·6 | 3·36 | 3·9 |
| Hill | 76·5 | 3·73 | 3·9 |
| Stump | 86·3 | 4·50 | 2·0 |
| Mean | 79·9 | 4·22 | 2·4 |
| Michigan | |||
| var. scabriuscula | 76·5 | 4·57 | 0·0 |
| var. pubescens | 82·3 | 4·95 | 2·0 |
Shown are the percentage of polymorphic loci (Pp), Shannon's Index (H′) and the percentage of loci that were fixed for the same band (Fixed), averaged over 51 loci. Data from the central Ohio, northern Ohio and Michigan populations represent a sub-set of the data reported in Culley and Wolfe (2001). All populations consisted of V. pubescens var. scabriuscula except for the Michigan var. pubescens population.
Unique alleles were not detected in any of the southwestern Ohio populations, but 3·9–9·8 % of loci within each population were completely fixed for a single allele. The average percentage of fixed alleles in the southwestern Ohio populations (5·6 %) was slightly higher than in central/northern Ohio populations (2·4 %; t = 2·24, d.f. = 8, P = 0·055). Of the 51 loci, 32 (63 %) had alleles that were found in at least some individuals of all six southwestern Ohio populations. Comparing across all populations, 49 of the 51 loci in southwestern Ohio populations were also detected in the north/central Ohio and Michigan populations and two loci were only detected in southwestern populations. An additional 34 bands that had previously been observed at low rates in the central/northern Ohio populations (Culley and Wolfe, 2001) could not be reliably detected in the current study and therefore were not included in the analysis.
Genetic distance
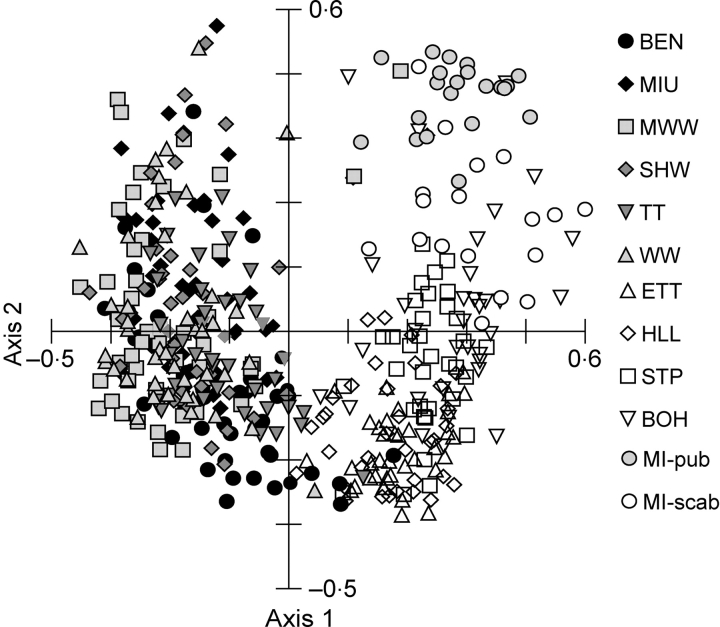
As calculated from the ISSR data using the Nei and Li (1979) distance coefficient, individuals tended to cluster with others from the same population (Fig. 2). In general, there were three main clusters of individuals with minimal overlap representing: (a) urban populations in southwestern Ohio; (b) agricultural populations in central and northern Ohio; and (c) populations from northern Michigan. On a population level, Nei and Li genetic distances for all pairwise comparisons of the six southwestern Ohio populations were quite similar to one another, ranging from 0·300 to 0·366 (mean = 0·327; Table 3). Distance measures between the north/central Ohio populations were more variable, ranging from 0·287 to 0·482 (mean = 0·399). Genetic distances between the southwestern Ohio and north/central groups of populations were higher, ranging from 0·339 to 0·587 (mean = 0·453; Table 3). Finally, southwestern populations were most distant from the Michigan populations of either the same variety (var. scabriuscula; mean distance = 0·611) or var. pubescens (0·648; Table 3).
Fig. 2.
Principle coordinate analysis of Viola pubescens populations based on Nei and Li genetic distances among individual plants from urban and agricultural populations in Ohio, and populations in Michigan. All populations consist of V. pubescens var. scabriuscula except the noted var. pubescens population in Michigan (MI-pub). Axes 1 and 2 explained 25·6 and 16·5 % of the variation, respectively. Population abbreviations are given in Table 1.
Table 3.
Genetic distances (below diagonal) and geographic distances (above diagonal; km) between pairs of populations
| Population | BEN | MIU | MWW | SHW | TT | WW | BOH | ETT | HLL | STP | MI var. scabriuscula | MI var. pubescens |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BEN | – | 42·2 | 29·9 | 4·3 | 10·7 | 13·8 | 171·6 | 203·8 | 196·8 | 205·7 | 205·7 | 205·7 |
| MIU | 0·337 | – | 30·9 | 38·4 | 36·2 | 34·1 | 177·3 | 202·0 | 194·4 | 203·9 | 669·7 | 669·7 |
| MWW | 0·308 | 0·337 | – | 25·9 | 19·1 | 19·1 | 194·8 | 223·7 | 216·3 | 225·6 | 700·6 | 700·6 |
| SHW | 0·303 | 0·339 | 0·313 | – | 7·0 | 9·9 | 173·6 | 205·1 | 198·0 | 207·0 | 698·5 | 698·5 |
| TT | 0·317 | 0·366 | 0·346 | 0·323 | – | 3·2 | 180·0 | 211·0 | 203·9 | 212·9 | 700·6 | 700·6 |
| WW | 0·300 | 0·356 | 0·311 | 0·311 | 0·338 | – | 181·9 | 212·5 | 205·3 | 214·4 | 699·9 | 699·9 |
| BOH | 0·517 | 0·572 | 0·587 | 0·561 | 0·546 | 0·573 | – | 44·0 | 40·9 | 45·0 | 599·0 | 599·0 |
| ETT | 0·347 | 0·433 | 0·419 | 0·410 | 0·388 | 0·416 | 0·468 | – | 7·7 | 1·9 | 556·1 | 556·1 |
| HLL | 0·339 | 0·422 | 0·404 | 0·397 | 0·383 | 0·404 | 0·462 | 0·287 | – | 9·7 | 558·1 | 558·1 |
| STP | 0·416 | 0·486 | 0·478 | 0·457 | 0·443 | 0·467 | 0·482 | 0·355 | 0·342 | – | 555·6 | 555·6 |
| MI var. pubescens | 0·583 | 0·604 | 0·628 | 0·612 | 0·609 | 0·629 | 0·597 | 0·535 | 0·551 | 0·556 | – | |
| MI var. pubescens | 0·637 | 0·642 | 0·665 | 0·635 | 0·646 | 0·665 | 0·657 | 0·636 | 0·601 | 0·611 | 0·617 | – |
Genetic distances were calculated using 51 ISSR loci, based on (1-similarity) using the Nei and Li (1979) coefficient. Population abbreviations are given in Table 1.
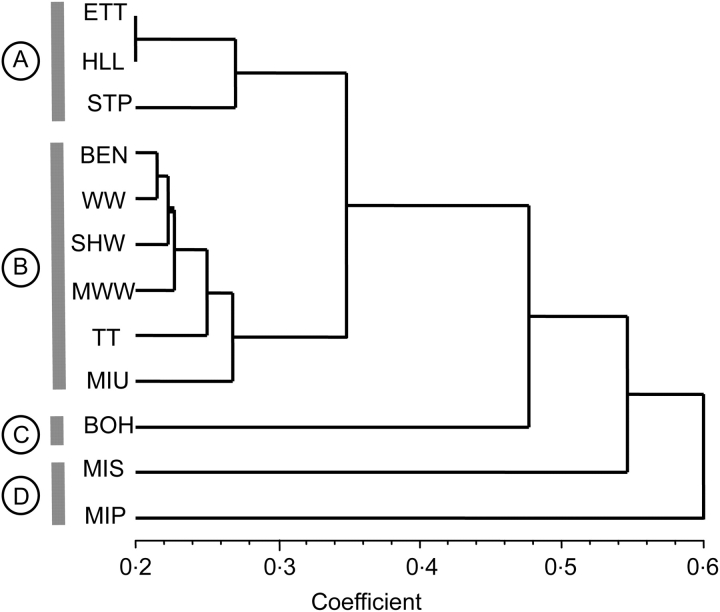
The UPGMA phenogram indicated that populations generally clustered according to their geographic location (Fig. 3). All populations in southwestern Ohio formed a grouping, composed of populations in Hamilton County that clustered together before they grouped with the Miami University population in neighbouring Butler County. This southwestern Ohio group then clustered with a group of agricultural populations in Crawford County, before clustering with the agricultural population in Delaware and Morrow Counties (Bohannan) and then the geographically distant population in Michigan. Finally, the population of V. pubescens var. pubescens in Michigan was the most distantly associated with the other populations of var. scabriuscula. There was a significant relationship between genetic and geographic distances for all 11 populations of V. pubescens var. scabriuscula (Mantel test; r = 0·8103, P = 0·001) but this was most probably due to a significant isolation × distance effect because a lower coefficient was detected when the single Michigan population was removed (r = 0·615, P = 0·005). There was no significant relationship between genetic and geographic distances for the six southwestern Ohio populations (Mantel test; r = 0·545, P = 0·140).
Fig. 3.
UPGMA phenogram based on Nei and Li coefficients showing clustering relationships of populations of Viola pubescens. Shown are groups of populations in (A) northern Ohio in Crawford County; (B) southwestern Ohio; (C) central Ohio in Delaware/Morrow Counties; and (D) northern Michigan. Population abbreviations are given in Table 1; except for MIS (MI var. scabriuscula) and MIP (MI var. pubescens).
Population differentiation
Hierarchical AMOVA of three population groups (southwestern Ohio, central/northern Ohio and Michigan) revealed that genetic variability was partitioned 69·1 % within populations, 8·7 % among populations within groups and 22·2 % among groups (Table 4). Values of overall FST for individual loci ranged from − 0·004 to 0·695, with a mean of 0·308 that was significantly different from zero (P < 0·00001). When analysed separately, urban populations within southwestern Ohio and agricultural populations in central/northern Ohio had FST values of 0·111 and 0·113, respectively; both values were significantly greater than zero (P < 0·00001).
Table 4.
Table of the hierarchical AMOVA examining differences among and within groups of urban and agricultural populations of Viola pubescens var. scabriuscula
| Source of variation | d.f. | Sum of squares | Variance component | Percentage variation |
|---|---|---|---|---|
| Among groups | 2 | 438·12 | 1·846 | 22·17 |
| Among populations within groups | 8 | 279·86 | 0·722 | 8·67 |
| Within population | 410 | 2360·90 | 5·758 | 69·16 |
| Total | 420 | 3078·88 | 8·326 |
Three groups consisted of populations in southwestern Ohio, north/central Ohio and Michigan. Values are based on 51 ISSR loci.
DISCUSSION
Urban effects of habitat fragmentation have received little attention to date, although fragmentation in general often lowers genetic variation and increases genetic differentiation of plant populations (Young et al., 1996; Mills and Tallmon, 1999). Such effects may also occur in urban areas if the matrix imposes substantial barriers to gene flow (Ledig, 1992), leading to increased isolation and genetic drift of populations. However, in the urban landscape of southwestern Ohio, populations of V. pubescens exhibited high genetic variation and substantial gene flow, as indicated for example by the low amount of variation that was attributed among populations within groups (8·7 %). In general, genetic variation in urban populations of V. pubescens was substantially higher than for other long-lived perennials with mixed-mating systems (Hamrick and Godt, 1996), a result consistent with previous investigations of this species in agricultural areas (Culley and Wolfe, 2001; Culley and Grubb, 2003). Although urban populations were genetically differentiated from one another, similar levels of differentiation were also observed for non-urban populations of the same species. Combined with the absence of unique bands, these results indicate that habitat fragmentation in urban areas does not substantially impede gene flow in V. pubescens.
This study is distinctive in that it is one of the first reports of whether the genetic structure of a plant species is affected by habitat fragmentation in an urban environment. Within animals, high levels of genetic variation have also been found in urban populations of butterflies (Kronforst and Fleming, 2001) and woodland beetles (Desender et al., 2005), but not in toads (Hitchings and Beebee, 1998). The fact that largely deleterious effects of fragmentation were not detected in V. pubescens is not completely unusual in plant populations, as negligible and even positive effects of fragmentation have been found in non-urban habitats. For example, pollen dispersal increased in Swietenia humilis in dry forest fragments (White et al., 2002), and isolated individuals of Symphonia globulifera were responsible for most seedlings in nearby remnant tropical forest (Aldrich and Hamrick, 1998). In temperate forest, fragmented populations of Acer saccharum did not exhibit reduced genetic variation (Young et al., 1993), although genetic structure was affected (Young and Merriam, 1994). Interestingly, all these species are long-lived trees while V. pubescens, as a herbaceous perennial with a relatively shorter generation time, would be expected to be more susceptible to deleterious effects of fragmentation.
In V. pubescens, a scarcity of deleterious effects of urban fragmentation could in part be explained by characteristics of the species itself and of the urban environment. First, reproductive traits of V. pubescens promote occasional long-distance gene flow and resistance to population extinction, thus preventing loss of genetic diversity in fragments. For example, dual production of chasmogamous and cleistogamous flowers (Culley, 2002) creates opportunities for outcrossing by generalist pollinators but also guarantees seed production independent of pollinator activity. Chasmogamous flowers also have the added benefit of delayed selfing if left unvisited (Culley, 2002). Viola pubescens is a long-lived perennial with a seed bank, factors that enhance population persistence. Overall, these traits may protect populations against short-term effects of habitat fragmentation, but may not be as effective in the long-term if populations continue to lose pollinators and habitat (Culley and Grubb, 2003). Other plant species may be more detrimentally affected by habitat fragmentation, especially if they are annual, obligate outcrossers with specific habitat requirements or require specialized pollinators.
Genetic variation and gene flow in V. pubescens may also be maintained in urban areas because insect pollinators are not as detrimentally affected by fragmentation as originally thought (Rathcke and Jules, 1993; Didham et al., 1996; Kearns et al., 1998; Kwak et al., 1998; Spira, 2001). Many previous investigations focused on non-urban landscapes in which the intervening matrix is relatively inhospitable for pollinators (e.g. Jennersten, 1988; Steffan-Dewenter and Tscharntke, 1999). In the urban landscape, the matrix may be more permeable because it contains residential yards, undeveloped land and transportation corridors that sustain pollinators with food and possibly nesting sites as they move between fragments. Urban areas enhance insect abundance but not diversity (McIntyre and Hostetler, 2001; Tommasi et al., 2004), even though both are expected to decline in fragments (Rathcke and Jules, 1993). Compared with natural areas, certain bee species are more abundant in metropolitan areas (McIntyre and Hostetler, 2001; Tommasi et al., 2004), and many insects that pollinate V. pubescens (Culley, 2002) have been observed in residential areas of southwestern Ohio (T. Culley, pers. obs.), including bumblebees known to fly far from urban nesting sites (Chapman et al., 2003; but see Bhattacharya et al., 2003). Although busy roadways could potentially restrict pollinator movement and hence gene flow in urban locations, this does not seem to occur in populations of V. pubescens. For example, the Benedict and Sharon Woods populations are separated by US Interstate 71, a busy six-lane roadway, but they exhibited one of the lowest pairwise genetic distances of all populations. Such pollinator activity in urban areas would buffer populations against expected declines in genetic variation associated with fragmentation (Young et al., 1996; Mills and Tallmon, 1999), in part by counteracting genetic isolation that has been observed in small agricultural populations (Culley and Grubb, 2003).
Substantial levels of genetic variation detected in this study may also reflect the relatively large size of urban habitat fragments (25–779 ha), many of which are large public parks. In contrast, fragments in agricultural areas are typically only 0·5–40 ha (Culley and Grubb, 2003). Larger urban fragments could buffer populations against loss of genetic variation by minimizing edge effects, maximizing population size and promoting pollinator persistence within the fragments (Rathcke and Jules, 1993; Buchmann and Nabhan, 1996; Spira, 2001). Not surprisingly, levels of genetic variation of V. pubescens were not correlated with fragment size in the urban landscape as they were in the agricultural area (see also Culley and Grubb, 2003). Although this could be caused in part by different glacial histories of the two regions, both areas were almost completely deforested in the early 20th century and thus it is more likely that the genetic results are due to fragmentation rather than to glaciation. Fragment size, however, is not completely responsible for genetic patterns because the smallest urban populations contained more genetic variation than the closest matching sized agricultural population. For example, urban populations at Miami University and Trillium Trails were smaller (<11 ha) than the smallest agricultural population, Etter Central (21 ha), but yet contained a higher percentage of polymorphic loci (84·3 and 80·4 %, vs. 70·6 %). This suggests that other factors, such as those described previously, are also involved in the genetic response of urban populations to fragmentation.
CONCLUSIONS
Populations of V. pubescens that inhabit urban forest fragments in southwestern Ohio are less prone to deleterious genetic effects of habitat fragmentation than previously thought. It now appears that at least some of our basic understanding regarding genetic effects of habitat fragmentation is in need of revision, especially for urban areas. Because different urban environments may not all have the same effect on plant populations, the impact of various urban settings also needs to be explored. This study suggests that it may not be appropriate to generalize about fragmentation effects across landscapes because processes affecting gene flow may differ depending upon the surrounding environmental matrix as well as the plant species itself. To appreciate more fully the genetic implications of fragmentation in the urban landscape for plants, additional investigations are needed using a variety of plant species in different urban areas. In this way, it will finally be possible to understand the genetic consequences and ecological effects of urban habitat fragmentation, an ever-increasing problem in the world today.
ACKNOWLEDGEMENTS
Permission to collect samples at the various sites was obtained from the Hamilton County Park District and Miami University in Oxford, Ohio. T. Grubb Jr and P. Doherty assisted in collecting samples from the populations in Crawford County, Ohio. We thank M. Klooster, N. Hardiman, R. Stokes and three anonymous reviewers for valuable comments on the manuscript.
LITERATURE CITED
- Aldrich PR, Hamrick JL. Reproductive dominance of pasture trees in a fragmented tropical forest mosaic. Science. 1998;281:103–105. doi: 10.1126/science.281.5373.103. [DOI] [PubMed] [Google Scholar]
- Ballard HE. Violets of Michigan. Michigan Botanist. 1994;33:131–199. [Google Scholar]
- Berg G, Nordal I, Hestmark G. The effect of breeding systems and pollination vectors on the genetic variation of small plant populations within an agricultural landscape. Oikos. 1998;81:17–29. [Google Scholar]
- Bhattacharya M, Primack RB, Gerwein J. Are roads and railroads barriers to bumblebee movement in a temperate suburban conservation area? Biological Conservation. 2003;109:37–45. [Google Scholar]
- Braun EL. Deciduous forests of eastern North America. Philadelphia: The Blakiston Company; 1950. [Google Scholar]
- Buchmann SL, Nabhan GP. The forgotten pollinators. Washington, DC: Island Press; 1996. [Google Scholar]
- Cain SA. Studies on the stemmed yellow violets of eastern North America. II. Mass-collections of Viola pubescens and V. eriocarpa in the Michigan area. Naturaliste Canadien. 1967;94:79–129. [Google Scholar]
- Chapman RE, Wang J, Bourke AFG. Genetic analysis of spatial foraging patterns and resource sharing in bumble bee pollinators. Molecular Ecology. 2003;12:2801–2808. doi: 10.1046/j.1365-294x.2003.01957.x. [DOI] [PubMed] [Google Scholar]
- Crawford DJ, Esselman EJ, Windus JL, Pabin CS. Genetic variation in running buffalo clover (Trifolium stoloniferum: Fabaceae) using random amplified polymorphic DNA markers (RAPDs) Annals of the Missouri Botanical Garden. 1998;85:81–89. [Google Scholar]
- Cruzan MB. Population size and fragmentation thresholds for the maintenance of genetic diversity in the herbaceous endemic Scutellaria montana (Lamiaceae) Evolution. 2001;55:1569–1580. doi: 10.1111/j.0014-3820.2001.tb00676.x. [DOI] [PubMed] [Google Scholar]
- Culley TM. Reproductive biology and delayed selfing in Viola pubescens (Violaceae), an understory herb with chasmogamous and cleistogamous flowers. International Journal of Plant Sciences. 2002;163:113–122. [Google Scholar]
- Culley TM, Grubb TC. Genetic effects of habitat fragmentation in Viola pubescens (Violaceae), a perennial herb with chasmogamous and cleistogamous flowers. Molecular Ecology. 2003;12:2919–2930. doi: 10.1046/j.1365-294x.2003.01971.x. [DOI] [PubMed] [Google Scholar]
- Culley TM, Klooster MR. Botanical Review. 2007. The cleistogamous breeding system: a review of its frequency, evolution and ecology in angiosperms. in press. [Google Scholar]
- Culley TM, Wolfe AD. Population genetic structure of the cleistogamous plant species Viola pubescens, as indicated by isozyme and ISSR molecular markers. Heredity. 2001;86:545–556. doi: 10.1046/j.1365-2540.2001.00875.x. [DOI] [PubMed] [Google Scholar]
- Desender K, Small E, Gaublomme E, Verdyck P. Rural–urban gradients and the population genetic structure of woodland ground beetles. Conservation Genetics. 2005;6:51–62. [Google Scholar]
- Didham RK, Ghazoul J, Stork NE, Davis AJ. Insects in fragmented forests: a functional approach. Trends in Ecology and Evolution. 1996;11:255–260. doi: 10.1016/0169-5347(96)20047-3. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bulletin. 1987;19:11–15. [Google Scholar]
- Ellstrand NC, Elam DR. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics. 1993;24:217–242. [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3·0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. London: Oxford University Press; 1930. [Google Scholar]
- Foré SA, Hickey RJ, Vankat JL, Guttman SI, Schaefer RL. Genetic structure after forest fragmentation: a landscape ecology perspective on Acer saccharum. Canadian Journal of Botany. 1992;70:1659–1668. [Google Scholar]
- Gordon RM. The natural vegetation of Ohio in pioneer days. Bulletin of the Ohio Biological Survey. 1969;3:1–109. [Google Scholar]
- Griffith DM, DiGiovanni D, Witzel TL, Wharton EH. Forest statistics for Ohio, 1991. Radnor, PA: US Department of Agriculture, Forest Service, and Northeastern Forest Experiment Station; 1993. Resource Bulletin NE-128. [Google Scholar]
- Hamrick JL, Godt MJW. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society B: Biological Sciences. 1996;351:1291–1298. [Google Scholar]
- Hitchings SP, Beebee TJC. Loss of genetic diversity and fitness in Common Toad (Bufo bufo) populations isolated by inimical habitat. Journal of Evolutionary Biology. 1998;11:269–283. [Google Scholar]
- Jennersten O. Pollination in Dianthus deltoides (Caryophyllaceae): effects of habitat fragmentation on visitation and seed set. Conservation Biology. 1988;2:359–366. [Google Scholar]
- Kearns CA, Inouye DW, Waser NM. Endangered mutualisms: the conservation of plant–pollinator interactions. Annual Review of Ecology and Systematics. 1998;29:83–112. [Google Scholar]
- Kronforst MR, Fleming TH. Lack of genetic differentiation among widely spaced subpopulations of a butterfly with home range behavior. Heredity. 2001;86:243–250. doi: 10.1046/j.1365-2540.2001.00830.x. [DOI] [PubMed] [Google Scholar]
- Kwak MM, Velterop O, van Andel J. Pollen and gene flow in fragmented habitats. Applied Vegetation Science. 1998;1:37–54. [Google Scholar]
- Ledig FT. Human impacts on genetic diversity in forest ecosystems. Oikos. 1992;63:87–108. [Google Scholar]
- Levesque FL, Dansereau P. Études sur les violettes jaunes caulescentes de l'est de l'amérique du nord. I. Taxonomie, nomenclature, synonymie et bibliographie. Naturaliste Canadien. 1996;93:489–569. [Google Scholar]
- McIntyre NE, Hostetler ME. Effects of urban land use on pollinator (Hymenoptera: Apoidea) communities in a desert metropolis. Basic and Applied Ecology. 2001;2:209–218. [Google Scholar]
- Miller MP. Tools for population genetic analyses (TFPGA) 1·3: a Windows program for the analysis of allozyme and molecular population genetic data. 1997. Computer software distributed by the author. [Google Scholar]
- Mills LS, Tallmon DA. The role of genetics in understanding forest fragmentation. In: Rochelle JA, Lehmann LA, Wisniewski J, editors. Forest fragmentation: wildlife and management implications. Boston: Brill; 1999. pp. 171–186. [Google Scholar]
- Nei M, Li W-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proceedings of the National Academy of Sciences of the USA; 1979. pp. 5269–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober SM, Spindler LH, Brown AHD. Conservation of the grassy white box woodlands: effects of remnant population size on genetic diversity in the allotetraploid herb Microseris lanceolata. Conservation Biology. 1998;12:1279–1290. [Google Scholar]
- Rathcke BJ, Jules ES. Habitat fragmentation and plant–pollinator interactions. Current Science. 1993;65:273–277. [Google Scholar]
- Rohlf FJ. NTSYSpc: numerical taxonomy and multivariate analysis system. Setauket, New York: Exeter Software; 1998. Version 2·02 h. [Google Scholar]
- Solbrig OT, Newell SJ, Kincaid DT. The population biology of the genus Viola. I. The demography of Viola sororia. Journal of Ecology. 1980;68:521–546. [Google Scholar]
- Spira TP. Plant–pollinator interactions: a threatened mutualism with implications for the ecology and management of rare plants. Natural Areas Journal. 2001;21:78–88. [Google Scholar]
- Steffan-Dewenter I, Tscharntke T. Effects of habitat isolation on pollinator communities and seed set. Oecologica. 1999;121:432–440. doi: 10.1007/s004420050949. [DOI] [PubMed] [Google Scholar]
- Tommasi D, Miro A, Higo HA, Winston ML. Bee diversity and abundance in an urban setting. Canadian Entomology. 2004;136:851–869. [Google Scholar]
- US Bureau of the Census. Population in metropolitan and metropolitan statistical areas ranked by 2000 population for the United States and Puerto Rico: 1990 and 2000. 2003. Available online at: http://www.census.gov/population/www/cen2000/phc-t29.html . [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- White GM, Boshier DH, Powell W. Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. Proceedings of the National Academy of Sciences of the USA; 2002. pp. 2038–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AD, Liston A. Contributions of PCR-based methods to plant systematics and evolutionary biology. In: Soltis DE, Soltis PS, Doyle JJ, editors. Plant molecular systematics II. Dordrecht: Kluwer Academic Publishers; 1998. pp. 43–86. [Google Scholar]
- Young AG, Merriam HG. Effects of forest fragmentation on the spatial genetic structure of Acer saccharum Marsh. (sugar maple) populations. Heredity. 1994;72:201–208. [Google Scholar]
- Young AG, Merriam HG, Warwick SI. The effects of forest fragmentation on genetic variation in Acer saccharum Marsh. (sugar maple) populations. Heredity. 1993;71:277–289. [Google Scholar]
- Young A, Boyle T, Brown T. The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]
- Young AG, Brown AHD, Zich FA. Genetic structure of fragmented populations of the endangered daisy Rutidosis leptorrhynchoides. Conservation Biology. 1999;13:256–265. [Google Scholar]