Abstract
Background and Aims
Witches' broom disease is caused by the hemibiotrophic basidiomycete Moniliophthora perniciosa, and is one of the most important diseases of cacao in the western hemisphere. Because very little is known about the global process of such disease development, expressed sequence tags (ESTs) were used to identify genes expressed during the Theobroma cacao–Moniliophthora perniciosa interaction.
Methods
Two cDNA libraries corresponding to the resistant (RT) and susceptible (SP) cacao–M. perniciosa interactions were constructed from total RNA, using the DB SMART Creator cDNA library kit (Clontech). Clones were randomly selected, sequenced from the 5′ end and analysed using bioinformatics tools including in silico analysis of the differential gene expression.
Key Results
A total of 6884 ESTs were generated from the RT and SP cDNA libraries. These ESTs were composed of 2585 singlets and 341 contigs for a total of 2926 non-redundant sequences. The redundancy of the libraries was low and their specificity high when compared with the few other cacao libraries already published. Sequence analysis allowed the assignment of a putative functional category for 54 % of sequences, whereas approx. 22 % of sequences corresponded to unknown function and approx. 24 % of sequences did not show any significant similarity with other proteins present in the database. Despite the similar overall distribution of the sequences in functional categories between the two libraries, qualitative differences were observed. Genes involved during the defence response to pathogen infection or in programmed cell death were identified, such as pathogenesis related-proteins, trypsin inhibitor or oxalate oxidase, and some of them showed an in silico differential expression between the resistant and the susceptible interactions.
Conclusions
As far as is known this is the first EST resource from the cacao–M. perniciosa interaction and it is believed that it will provide a significant contribution to the understanding of the molecular mechanisms of the resistance and susceptibility of cacao to M. perniciosa, to develop strategies to control witches broom, and as a source of polymorphism for molecular marker development and marker-assisted selection.
Key words: Theobroma cacao, Moniliophthora perniciosa, ESTs, resistance, programmed cell death, witches' broom disease
INTRODUCTION
A central goal of genome analysis is to identify and classify all the genes of a particular species. Functional genomics seeks to understand the precise roles of these genes, including unique and redundant functions. Apart from few species such as arabidopsis, for which the complete genome is already available, gene discovery in most plants is primarily based on sample sequencing of expressed sequence tags (ESTs). The manner in which a number of biological questions can be addressed has profoundly evolved in the last few years with the advent of genomics. Indeed, the possibility to conduct large-scale analysis in functional genomics now opens the way to identification of large sets of co-regulated genes involved in biological processes. It is thus possible not only to identify novel and possibly important molecular events, but also to investigate biological processes at the level of gene networks rather than individual genes. This type of approach is attractive for a better understanding of complex development programmes such as those activated during interactions between plants and microorganisms. In this analysis, there is particular interest in the plant genetic programmes involved during development of witches' broom disease caused in cacao (Theobroma cacao) by the hemibiotrophic fungus Moniliophthora perniciosa (Stahel) Aime & Phillips-Mora [= Crinipellis perniciosa (Stahel) Singer] (Aime and Phillips-Mora, 2005).
Cacao is a tropical sub-canopy tree originally from the rain forest of the Amazon basin. It is cultivated primarily to provide cacao liquor, butter and powder for the chocolate industry, not only for its flavour properties, but also for emerging health benefits (Kris-Etherton and Keen, 2002). Cacao is an important commodity: >20 million people depend directly on cocoa for their livelihood, and approx. 90 % of the production – mainly from the Ivory Cost, Ghana and Indonesia – are exported in the form of beans or semi-manufactured cocoa products to Europe and the USA (Food and Agriculture Organization, http://www.fao.org). Despite its environmental and economical importance, cacao has received little attention with respect to molecular genetics and genomic research (Jones et al., 2002; Verica et al., 2004) and, until now, no molecular study has been carried out to understand the genetic programme of cacao during its infection by pests or diseases. Witches' broom disease, caused by M. perniciosa, is one of the major cacao diseases in South America and the Caribbean Islands, destroying plantations and leading to important economical and social changes in the areas concerned such as the State of Bahia in Brazil (Rocha et al., 1993; Purdy and Schmidt, 1996). Basidiospores infect meristematic tissues (shoots, flower cushions, single flowers and developing fruits), and induce a range of symptoms depending on the organ infected and the developmental stage: (a) infected apical meristems present hypertrophic growth (‘brooms’); (b) infected flower cushions usually lead to vegetative shoot production and pathenocarpic fruits; (c) pod infection can directly result in seed loss due to pod rot. The disease shows two distinct stages: a biotrophic phase and a necrotrophic/saprotrophic phase. In the biotrophic phase, the fungus causes hypertrophy and hyperplasia of the tissues, loss of apical dominance, and proliferation of axillary shoots, which results in the formation of abnormal stems (green broom). In the second stage, the fungus changes to the saprotrophic phase and causes necrosis and death of infected tissues distal from the original infection site, producing a dry broom. Basidiocarp production and spore formation occur on infected necrotic tissue (Silva et al., 2002; Scarpari et al., 2005). To recover cacao plantations, numerous efforts have been made such as the development of new cacao varieties, use of cloned resistant plant material or biological control of the disease (Rudgard et al., 1993). Although these technical procedures have been efficient, they do not represent an adequate method of control of witches' broom. The high genetic variability of the fungus, associated with the high frequency of genetic recombination (Rincones et al., 2006) could break the cacao resistance as observed for some cacao tree hybrids, which contain the Scavina-6 resistant parent (Wheeler and Mepsted, 1988; Rios-Ruiz, 2001). For this reason, the identification of differential represented genes between susceptible and resistant cacao trees to witches' broom disease is essential to understand biological events of the cacao–M. perniciosa interaction.
Here the generation and analysis of the first ESTs of Theobroma cacao–M. perniciosa interaction are reported. The main goal was to obtain a first global and comparative view of the susceptible and resistant interactions by characterizing transcript populations in meristems of two different cacao varieties – one susceptible, the other resistant to witches' broom – inoculated with M. perniciosa spores. Special emphasis was given to cDNA sequences related to resistance and to necrosis and death of infected tissues as probable components of the defence and susceptibility reactions occurring in the cacao tree after inoculation by M. perniciosa.
MATERIALS AND METHODS
Plant material and fungus strains
Plantlets of Theobroma cacao L. varieties Catongo (susceptible to Moniliophthora perniciosa) and TSH1188 (resistant to M. perniciosa) were grown in sterile substrate in the greenhouse at CEPEC/CEPLAC (Centro de Pesquisas da Comissão Executiva do Plano da Lavoura Cacaueira, Ilhéus, Bahia, Brazil) from September 2002 to January 2003, under natural light and 90 % relative humidity. Apical meristems of 154 4-week-old plantlets were inoculated by the spraying method using a 105 mL−1 basidiospore suspension from the M. perniciosa Cp1441 CEPEC/CEPLAC strain. After inoculation, plantlets were acclimated during 24 h at 25 °C ± 2 ºC in a water-saturated atmosphere to allow M. perniciosa spore germination, penetration and consequently infection (Frias et al., 1995). A test of spore viability was made in a humid chamber (25 °C) 24 h after inoculation (89 %) and was compared with spore viability obtained before inoculation (90 %). Fifty-six control plantlets were inoculated with sterile water and submitted to the same growing conditions as the inoculated ones. Expression of susceptibility was estimated 4 weeks after inoculation by detection of the Catongo plants with disease symptoms. Disease development was monitored on the growing plants for a period of 90 d. Inoculated and non-inoculated apical meristems from Catongo and TSH1188 were harvested at 24 h, 48 h, 72 h and then every 5 d until 90 d after inoculation (DAI). Infected and non-infected resistant and susceptible apical meristems were harvested, frozen in liquid nitrogen and stored at − 80 °C.
RNA extraction and cDNA library construction
For each library, total RNA was extracted from frozen tissues as described by Gesteira et al. (2003) and cleaned using the Rneasy Plant Mini Kit as described by the manufacturer (Qiagen). Purity and concentration of the purified RNA from each harvesting period were determined spectrophotometrically at 260 nm (Cary® 100 UV-Visible Spectrophotometer; Varian, Palo Alto, CA, USA). The RNA was separated on 1 % DEPC-treated agarose gel and stained with ethidium bromide to confirm RNA integrity. After quantification, 50 ng of RNA from each plant harvest were pooled to obtain a final total RNA amount of about 1 µg. The two cDNA libraries corresponding to the resistant and susceptible cacao–M. perniciosa interactions were constructed from pooled total RNA, using the DB SMART Creator cDNA library kit as described by the manufacturer (Clontech). cDNA longer than 400 bp were cloned directionally into the pDNR-LIB plasmid. ElectroMAX™DH10B™ cells (Invitrogen) were transformed and colonies picked and grown in 96-well microtitre plates in LB, 40 % glycerol medium containing 30 µg L−1 chloramphemicol and stored at − 80 °C.
Plasmid minipreps and sequencing of cDNA clones
Plasmid DNA was obtained from individual clones using the alkaline lysis procedure (Sambrook et al., 1989) adapted for 96-well plates. Plasmid quality and quantity were checked on 1 % TBE-BET agarose gel. For the RT and SP libraries, 3613 and 3271 clones, respectively, were randomly selected and sequenced from the 5′ end by the DyEnamic ET Dye Terminator kit (MegaBACE, GE Health Care) method, using the M13-F 5′-TAAAACGACGGCCAGT-3′ as forward primer. All sequences were produced by the capillary sequencer MegaBACE 1000 (GE Healthcare).
Data processing and computational methods
From each of the 6884 EST sequences generated (a) the largest sequenced stretch with Phred quality ≥ 10 was extracted (allowing 1 % nucleotide with Phred quality < 10) using a Perl script (Ewing et al., 1998), (b) the plasmid vector sequence with cross-match (-minmatch 20, -minscore 5) was removed, (c) the ‘X’ introduced by cross-match in the insert sequence with original nucleotides was substituted, and (d) the poly(A) tail was removed. After this trimming process, only sequences longer than 90 bp were included in the dataset, i.e. 3172 and 2852 sequences from RT and SP, respectively, were considered for further analysis. Finally, the redundancy was eliminated by contig assembling with CAP3 (Huang and Madan, 1999) and codon distribution was obtained from the remaining sequence pool. Specific genes from each library were determined comparing between themselves the available libraries (RT, SP and cacao ESTs available from GenBank; Jones et al., 2002; Verica et al., 2004) by BLASTX and TBLASTX using an expected value ≤ 1·10−4 as significant. For putative function determination and annotation, sequences were compared with the public sequence database (http://www.ncbi.nih.gov/BLAST/) using BLASTX and TBLASTX. Alignments showing similarity with an expected value ≤ 1·10−4 were considered significant. Additional information about the putative function of the ESTs was obtained using ProDom (Corpet et al., 2000), NRDL3D and Pfam programs. Also the GO software (http://www.geneontology.org/) was used to produce a control vocabulary of the annotations (Harris et al., 2004). EST clusters and associated predicted proteins were manually inspected and annotated as described by Journet et al. (2002). To detect potential expressed sequences from M. perniciosa present in the interaction, EST clusters were also compared with the M. perniciosa genome sequences available in the restrict-access database (http://www.lge.ibi.unicamp.br/vassoura/) from the Genome Project Consortium of Bahia, and with the public sequence database from the basidiomycete Ustilago maydis (Austin et al., 2004) using BLASTX and TBLASTX.
The R-statistic was used to identify the significant differences in EST abundance for contig among the RT and SP libraries involved in the plant–pathogen interaction process (Stekel et al., 2000). To test the credibility of the test, a randomization procedure with 1000 runs of randomized data was carried out as described previously (Stekel et al., 2000).
RESULTS
Library construction and sequencing
Two standard libraries, corresponding to resistant (RT) and susceptible (SP) interactions of cacao–M. perniciosa, were constructed (Table 1). The sequences generated from the two primary libraries had an average size of 600 bp. For the RT and SP libraries 3613 and 3271 randomly selected clones were sequenced from the 5' end (Table 2). In all, 6884 sequences were generated and, after trimming for low quality, shortness (<90 bp) and vector contamination, 3172 and 2852 sequences, for RT and SP, respectively, corresponded to the quality criteria for the present study. The overall sequencing success rate, i.e. useful sequences out of the total sequenced, was 88 % and 87 %, for RT and SP, respectively. The lengths of good quality sequences varied between 90 bp and 1058 bp for both libraries with an average of 341 bp and 354 bp for RT and SP, respectively (Table 2). For further analysis, 9338 cacao ESTs available from GenBank (1614 from Jones et al., 2002; 2113 from Verica et al., 2004) were added to the set of sequences generated in this study.
Table 1.
Characteristics of the cDNA libraries constructed and described in this study
| Cultivar* | Phenotype/condition† | Tissue/phase | Type of library | Designation |
|---|---|---|---|---|
| TSH1188 | R/inoculated | Meristems collected at 24 h, 48 h, 72 h then every 5 d until 90 d after inoculation | Full-length | RT |
| Catongo | S/inoculated | Meristems collected at 24 h, 48 h, 72 h then every 5 d until 90 d after inoculation | Full-length | SP |
* TSH: Trinidad Selected Hybrid.
† S, susceptible; R, resistant.
Table 2.
EST generated from the different cDNA libraries and the complete cacao– M. perniciosa unigene set
| Library | No. of sequences generated | No. of sequences analysed1 | Interaction singleton (%)2 | Interaction TC3 | Unigene size (%)4 | Mean size of the sequences (bp)5 | Redundancy (%)6 | Library specific unigenes (%)7 | Contribution (%)8 |
|---|---|---|---|---|---|---|---|---|---|
| RT | 3613 | 3172 (88) | 1520 (48) | 199 | 1719 (54) | 341 | 46 | 1371 (79·7) | 46·8 |
| SP | 3271 | 2852 (87) | 1065 (37) | 142 | 1207 (42) | 354 | 58 | 859 (71·2) | 29·3 |
| Total | 6884 | 6024 | 2585 | 341 | 2926 | 347·5 | 52 | 2230 |
1 Vector sequences and sequences of low quality or smaller than 90 bp were eliminated.
2 The singletons present in each library independent of other libraries. The percentage was calculated as number of singletons/number of sequences analysed from the library.
3 TC = tentative contig. The contigs present in each library independent of other libraries.
4 The unigene set for each library is the sum of singleton plus contigs for the library.
5 The mean size of the unigene sequences.
6 The redundancy of each library calculated as 1 – (unigene library/number of sequence analysed).
7 The unigenes (singletons + contigs) specific to the library. The percentage was calculated as (library-specific unigenes/unigene total).
8 The percentage contribution of unigenes specific to each library as a percentage of total unigenes.
Global analysis of the cacao–M. perniciosa interaction genes
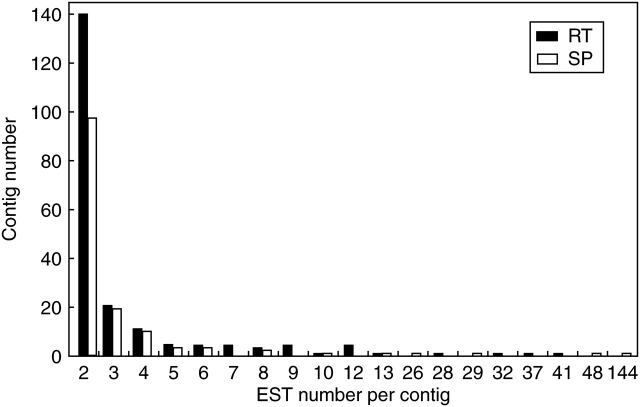
The initial data set of 3172 and 2858 sequences from RT and SP, respectively, was reduced to 1719 and 1207 unique consensus sequences (unigenes). The RT unigene set comprised 199 contigs (1652 sequences) and 1520 singletons, whereas the SP unigene set comprised 142 contigs (1787 sequences) and 1065 singletons (Table 2). The number of ESTs forming each contig varied between two (143 cases) and 41 (one case) for RT and two (99 cases) and 144 (one case) for SP (Fig. 1). The RT and SP libraries showed correct redundancy values (46 % and 58 %, respectively) reflecting the fact that these two libraries were thoroughly sequenced (Table 2). The number of specific unigene sequences for each library was calculated (singletons plus contigs of each library). The percentage of specific unigene sequences, which can be considered as an estimation of the capacity to provide new genes, was high: 79·7 % and 71·2 % for RT and SP, respectively. The contribution of individual libraries to the total unigene set was relatively low, 46·8 % for RT and 29·3 % for SP, indicating that each library contained specific ESTs (Table 2).
Fig. 1.
Histogram showing the distribution of ESTs by contigs. The contig size is the number of EST/contig.
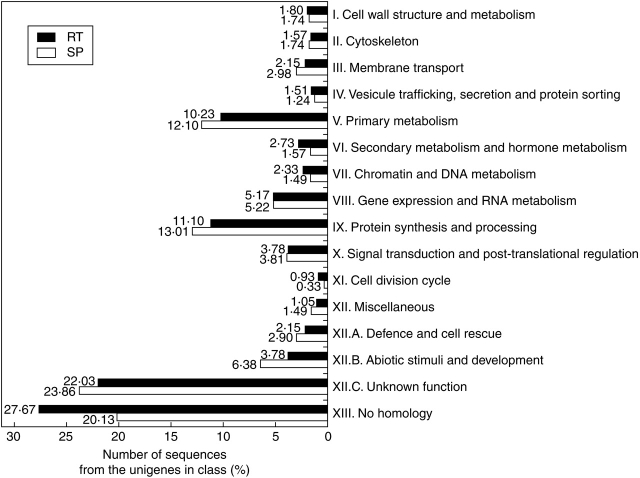
In the process of functional annotation of the putative encoded proteins, clusters were classified into a limited set of 16 broad functional categories, similar to those used in Journet et al. (2002). Figure 2 illustrates the distribution of the 2926 annotated sequences from the RT and SP libraries, respectively. Globally, the EST repartition in functional classes for the RT and SP unigene sets showed only a few differences. For the RT and SP unigene sets, the semi-automatic annotation allowed the placement of 50·3 % and 56·1 %, respectively, in the 14 functional categories sensus stricto, whereas 49·7 % and 43·9 % of the corresponding unigene sets encoded proteins without similarity or with similarity to proteins of unknown function (Fig. 2). Among the 14 functional categories sensu stricto, the most highly represented categories were protein synthesis and processing (11·1 % and 13·01 %, for RT and SP libraries) and the primary metabolism (10·23 % and 12·1 %). In the SP library, the abiotic stimuli and development category was well represented (6·38 %) – compared with 3·78% for the same (abiotic stimuli and development) category from RT – and was larger than the gene expression and RNA metabolism category (5·17 % and 5·22 % for the RT and SP libraries, respectively).
Fig. 2.
Distribution of the 1719 RT and 1207 SP unigene sequences into functional classes. The 16 broad categories that were used for classification during the semi-automatic annotation are indicated, as well as the number of corresponding sequences. Only one class was assigned to each sequence.
To estimate the proportion of fungal ESTs in the libraries used in the present study, the RT and SP sequences were also compared with M. perniciosa and Ustilago maydis databases. The BLAST comparison detected 17 and 15 sequences from RT and SP, respectively, showing homologies with fungal sequences. Most of them (30) were also very similar to plant sequences. Consequently, these 30 ESTs represented highly conserved genes between the plant, animal and fungal kingdoms, and cannot be discriminated as plant or fungal ESTs. Only the remaining two sequences from SP had a high probability of corresponding with a M. perniciosa gene: one which showed similarity with a hydrophobin from Flammulina velutipes (e-value of 1·10−68), the other similarity with a 25-kDa protein elicitor from Pythium aphanidermatum (e-value of 1·10−41).
The genes from the ten most abundant mRNAs in the libraries used in the present study are listed in Table 3. Among them, metallothionein, was highly represented in the total unigene, by two and three contigs in the RT and SP libraries, respectively. Two other genes, the trypsin inhibitor and the pathogenesis-related protein 4b, which were highly represented in the RT library, are related to plant resistance (Chen et al., 1999; van Loon et al., 2006). In the SP library, two contigs of ankyrin-repeat protein were found highly represented. This gene is known to be related to programmed cell death (Dong, 2004; Lu et al., 2005), auxin signalling and pathogen response (Kuhlmann et al., 2003). The abundant expression of these genes reflects the physiological processes of the tissues used to generate the library: resistance of the plant to M. perniciosa for the RT library, and necrosis of the infected tissues – production of necrotic meristems possibly by programmed cell death – for the SP library. Interestingly, two of the most abundant sequences present in the RT library did not show any homology with sequences from the databank, and three sequences from RT and SP libraries presented homology with unknown function sequences from arabidopsis, rice and human.
Table 3.
Putative function of the most abundant sequences present in the RT and SP cDNA libraries
| Library | No. of ESTs in TC | Functional annotation | Species | E value | Size (bp) |
|---|---|---|---|---|---|
| RT | 41 | Unknown function homologue to Orf107a | Arabidopsis thaliana | 2·10−27 | 521 |
| 37*,a | Metallothionein | Betula platyphylla | 6·10−26 | 651 | |
| 32* | Trypsin inhibitor | Theobroma cacao | 2·10−93 | 929 | |
| 28* | Pathogenesis-related protein 4b | Oryza sativa | 1·10−34 | 1058 | |
| 13 | No homology | – | – | 222 | |
| 12 | Ribosomal RNA | Poncirus trifoliata | 0·0 | 893 | |
| 10 | Unknown function | Oryza sativa | 6·10−22 | 386 | |
| 9*,b | Metallothionein | Petunia × hybrida | 2·10−16 | 489 | |
| 9 | No homology | – | – | 829 | |
| 9 | Dehydrin | Vaccinium corymbosum | 2·10−6 | 618 | |
| SP | 144*,a | Metallothionein | Betula platyphylla | 6·10−26 | 669 |
| 49 | Unknown function homologue to Orf107a | Arabidopsis thaliana | 1·10−27 | 266 | |
| 29*,b | Metallothionein | Petunia × hybrida | 3·10−16 | 535 | |
| 26* | Ankyrin repeat protein | Arabidopsis thaliana | 1·10−9 | 625 | |
| 13 | Metallothionein | Gossypium hirsutum | 1·10−24 | 495 | |
| 10* | Ankyrin repeat protein | Arabidopsis thaliana | 7·10−7 | 505 | |
| 8 | Unknown function | Oryza sativa | 7·10−4 | 494 | |
| 8 | Copper chaperone | Arabidopsis thaliana | 2·10−28 | 631 | |
| 6 | Nuclear factor 1 | Mus musculus | 1·10−10 | 447 | |
| 6 | Unknown function | Homo sapiens | 1·10−8 | 497 |
* Sequences expressed differentially as described in the Table 4.
a, b Same letters correspond to sequences homologous between RT and SP libraries.
Comparison of the cacao–M. perniciosa interaction unigene sets with other cacao unigene sets
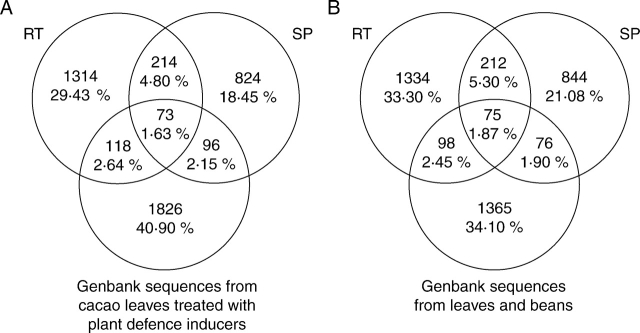
To identify specific cacao–M. perniciosa genes involved in the interaction, the unigene sets were compared with the available ESTs from cacao leaves treated with plant artificial defence inducers (Verica et al., 2004) and from leaves and beans (Jones et al., 2002). When comparing the specific and common sequences among the different cacao libraries available, it was observed that each of them was specific to the cacao physiological stage studied (healthy leaves and beans, cacao treated with defence inducers, and resistant and susceptible interactions with M. perniciosa) (Fig. 3). The RT library shared 287 and 191 sequences with the SP and plant defence inducers-treated leaf libraries, respectively. One hundred and sixty-nine sequences were common to SP and plant defence inducers-treated leaf libraries and only 73 sequences were common to the three libraries (treated, resistant and susceptible to M. perniciosa) (Fig. 3A). In the same way, the RT library shared 287 and 173 sequences with the SP and healthy leaf and bean libraries, respectively. One hundred and fifty-one sequences were common to SP and healthy leaf and bean libraries and only 75 sequences were common to the three libraries (healthy, resistant and susceptible to M. perniciosa) (Fig. 3B). For each comparison (Fig. 3A, B), only small differences were observed between the categories of genes present in each library. Few genes involved in resistance were found in the 191 common genes between RT and the library from plants treated with defence inducers (data not shown).
Fig. 3.
Venn diagram showing the distribution of the sequences present in cacao libraries. (A) From Verica et al. (2004). The percentage values were based on the total sequence number (4465). (B) From Jones et al. (2002). The percentage values were based on the total sequence number (4004).
Characterization of ESTs potentially associated with plant–pathogen interaction
ESTs corresponding to genes encoding proteins potentially related to plant–pathogen interaction were characterized from the cacao–M. perniciosa unigene set collection (113 genes; Tables 4 and 5). RT and SP libraries were sequenced to obtain information on genes whose expression is potentially related to plant–pathogen interaction. Advantage was taken of these two libraries not being normalized to compare the gene expression in silico. Due to the relatively thorough sampling approach, the frequency of ESTs in a given library can be used to obtain information on relative gene expression levels in the tissues from which the library was constructed (Bortoluzzi and Danieli, 1999). A differential analysis was performed using the method reported by Stekel et al. (2000) to study difference in pathogenesis related-gene expression between RT and SP. Table 4 lists the potential pathogenesis related-genes showing differences in the expression pattern between the two interactions. From the nine differentially represented genes six were also found in the most abundant sequence list (Table 3): metallothionein (two contigs), trypsin inhibitor, pathogenesis-related protein and ankyrin repeat protein (two contigs). The other genes corresponded to pathogenesis related-protein 10, NPR1/NIM1-interacting protein and subtilase. Six of the nine differentially represented genes were more highly represented in the SP library (two metallothionein, two ankyrin repeat proteins, PR10 and NPR1/NIM1-interacting protein), the three other differentially represented genes were more highly represented in the RT library (trypsin inhibitor, PR4b and subtilase).
Table 4.
Differentially expressed genes between the two libraries RT and SP in relation to plant–pathogen interaction
| Class1 | Functional annotation | Species | e-value | Size (bp) | R(2) | No. of sequences for RT3 | No. of sequences for SP4 |
|---|---|---|---|---|---|---|---|
| XII.A | Metallothionein | Betula platyphylla | 6·10−26 | 669 | 39·73 | 37* | 144* |
| IX | Trypsin inhibitor | Theobroma cacao | 2·10−93 | 929 | 20·52 | 32* | 0 |
| XII.A | Pathogenesis-related protein 4b | Oryza sativa | 1·10−34 | 1058 | 14·36 | 28* | 1 |
| II | Ankyrin repeat protein | Arabidopsis thaliana | 1·10−9 | 625 | 13·15 | 1 | 26* |
| XII.A | Metallothionein | Petunia × hybrida | 2·10−16 | 489 | 6·66 | 9* | 29* |
| II | Ankyrin repeat protein | Arabidopsis thaliana | 7·10−7 | 505 | 3·81 | 1 | 10* |
| XII.A | Pathogenesis-related protein 10 | Solanum tuberosum | 5·10−48 | 593 | 3·74 | 0 | 5 |
| XII.A | NPR1/NIM1-interacting protein | Arabidopsis thaliana | 1·10−5 | 376 | 2·24 | 0 | 3 |
| IX | Subtilase | Lycopersicon esculentum | 4·10−37 | 538 | 1·92 | 3 | 0 |
1 The class numbers correspond to those described in Fig. 3 and in Journet et al. (2002).
2 R significant to 0·05 calculated as described previously with 1000 permutations.
3 The number of sequences analysed for the RT library was 3172.
4 The number of sequences analysed for the SP library was 2852.
* Sequences listed in the Table 3 as the most expressed in the RT and/or SP libraries.
Table 5.
Other genes potentially related to plant–pathogen interaction present in the RT and SP libraries
| Class* | Functional annotation | Species | e-value | Size (bp) |
|---|---|---|---|---|
| I | Arabinogalactan protein | Gossypium hirsutum | 1·10−40 | 306 |
| I† | Cellulose synthase | Gossypium hirsutum | 1·10−81 | 496 |
| I‡ | Chitinase | Theobroma cacao | 4·10−71 | 382 |
| I | Expansin | Populus tremula × Populus tremuloides | 2·10−70 | 412 |
| I† | Extensin | Populus nigra | 2·10−37 | 718 |
| I | Hydroxyproline-rich glycoprotein | Arabidopsis thaliana | 9·10−36 | 492 |
| I† | O-methyltransferase | Fragaria × ananassa | 1·10−60 | 496 |
| I | Pectate lyase | Fragaria × ananassa | 7·10−47 | 281 |
| I | Pectin methylesterase | Silene latifolia | 9·10−50 | 407 |
| I | Polygalacturonase | Arabidopsis thaliana | 6·10−46 | 413 |
| II | Adhesin | Arabidopsis thaliana | 2·10−17 | 341 |
| II | Hydrophobin | Coprinopsis cinerea | 8·10−24 | 467 |
| II | Kinesin | Arabidopsis thaliana | 6·10−71 | 499 |
| II | Myosin heavy chain | Oryza sativa | 1·10−7 | 421 |
| III | ABC transporter | Arabidopsis thaliana | 3·10−43 | 420 |
| III | Fibre sucrose transporter | Gossypium barbadense | 6·10−58 | 446 |
| III | Multidrug transporter protein | Arabidopsis thaliana | 9·10−35 | 414 |
| IV | Rab gene | Arabidopsis thaliana | 2·10−57 | 378 |
| V‡ | 12-Oxophytodienoate reductase | Catharanthus roseus | 1·10−65 | 504 |
| V | Acyl-coA synthetase | Cicer arietinum | 4·10−20 | 378 |
| V | Asparagine synthetase | Triphysaria versicolor | 9·10−89 | 498 |
| V | ATP citrate-lyase | Arabidopsis thaliana | 3·10−45 | 607 |
| V‡ | Carbonic anhydrase | Gossypium hirsutum | 6·10−11 | 162 |
| V | Germin oxalate oxidase | Gossypium raimondii | 3·10−50 | 491 |
| V† | Glutamine synthetase | Oryza sativa | 5·10−43 | 527 |
| V†,‡ | Glyceraldehyde 3-phosphate dehydrogenase | Oryza sativa | 7·10−46 | 492 |
| V‡ | Hydroxymethyltransferase | Arabidopsis thaliana | 3·10−37 | 248 |
| V† | Lipase | Arabidopsis thaliana | 6·10−49 | 447 |
| V | Methionine sulfoxide reductase | Arabidopsis thaliana | 5·10−44 | 447 |
| V | Monodehydroascorbate reductase | Lycopersicon esculentum | 5·10−52 | 348 |
| V | NADH dehydrogenase | Beta vulgaris | 9·10−52 | 345 |
| V | NADPH oxidase | Arabidopsis thaliana | 7·10−29 | 265 |
| V | Ornithine decarboxylase | Haemonchus contortus | 7·10−16 | 327 |
| V | Polyketide synthase | Oryza sativa | 9·10−9 | 350 |
| V† | S-Adenosyl-l-methionine synthetase | Carica papaya | 4·10−69 | 486 |
| V‡ | Semialdehyde dehydrogenase | Arabidopsis thaliana | 3·10−65 | 434 |
| V†,‡ | Squalene monooxygenase | Medicago truncatula | 5·10−21 | 478 |
| V | Sulfite oxidase | Arabidopsis thaliana | 3·10−71 | 477 |
| V† | Thioredoxin | Ricinus communis | 3·10−54 | 511 |
| VI† | 1-Aminocyclopropane-1-carboxylic acid oxidase | Gossypium barbadense | 4·10−63 | 461 |
| VI | Betaine aldehyde dehydrogenase | Oryza sativa | 6·10−12 | 278 |
| VI | Caffeic acid 3-O-methyltransferase | Prunus dulcis | 2·10−46 | 522 |
| VI | Caffeine synthase | Camellia sinensis | 1·10−16 | 415 |
| VI | Chalcone synthase | Hydrangea macrophylla | 5·10−80 | 522 |
| VI | Cytochrome P450 | Panax ginseng | 3·10−62 | 471 |
| VI | Polyphenol oxidase | Populus tremuloides | 6·10−31 | 355 |
| VII | Acinus L protein | Oryza sativa | 2·10−13 | 442 |
| VIII | bZIP protein | Glycine max | 1·10−55 | 500 |
| VIII | EREBP/AP2-related transcription factor | Mesembryanthemum crystallinum | 2·10−18 | 390 |
| VIII | Glycine-rich RNA binding protein | Pisum sativum | 8·10−32 | 452 |
| VIII | Myb family transcription factor | Arabidopsis thaliana | 2·10−32 | 509 |
| VIII | WRKY transcription factor | Arabidopsis thaliana | 3·10−20 | 448 |
| VIII† | Zinc finger protein family-like | Arabidopsis thaliana | 5·10−39 | 352 |
| IX | 26S proteasome | Oryza sativa | 2·10−73 | 518 |
| IX† | Aspartic proteinase | Theobroma cacao | 9·10−72 | 407 |
| IX | Cyclophilin | Arabidopsis thaliana | 5·10−67 | 445 |
| IX† | Cysteine protease | Phaseolus vulgaris | 8·10−79 | 553 |
| IX | F-box family protein | Arabidopsis thaliana | 2·10−44 | 516 |
| IX‡ | Heat shock protein | Euphorbia esula | 1·10−68 | 407 |
| IX† | Protease inhibitor | Arabidopsis thaliana | 7·10−39 | 470 |
| IX | Proteasome inhibitor | Arabidopsis thaliana | 2·10−41 | 465 |
| IX† | Protein disulfide isomerase (PDI) | Datisca glomerata | 2·10−54 | 474 |
| IX | Serine carboxypeptidase family | Arabidopsis thaliana | 4·10−57 | 495 |
| IX | Ubiquitin family protein | Arabidopsis thaliana | 2·10−48 | 397 |
| IX | U-box domain-containing protein | Arabidopsis thaliana | 5·10−38 | 386 |
| IX‡ | Vacuolar processing enzyme precursor (VPE) | Citrus sinensis | 1·10−57 | 526 |
| X† | Calmodulin-binding family protein | Arabidopsis thaliana | 8·10−80 | 619 |
| X | GTP binding protein | Lycopersicon esculentum | 8·10−70 | 380 |
| X | MAP kinase | Oryza sativa | 0·001 | 466 |
| X | Phosphatase 2A | Oryza sativa | 1·10−48 | 355 |
| X | Protein kinase family protein | Arabidopsis thaliana | 3·10−63 | 376 |
| X | Receptor kinase | Solanum tuberosum | 2·10−53 | 360 |
| XII†,‡ | 14-3-3-like protein | Populus × canescens | 3·10−78 | 464 |
| XII | Beta-1,3-glucanase | Arabidopsis thaliana | 1·10−55 | 423 |
| XII | Cysteine-rich protein | Homo sapiens | 2·10−4 | 307 |
| XII | Glycine-rich protein | Arabidopsis thaliana | 3·10−17 | 319 |
| XII | Lipid transfer protein (LTP) | Arabidopsis thaliana | 2·10−27 | 493 |
| XII | Lipoxygenase | Fragaria × ananassa | 9·10−20 | 431 |
| XII | Peroxidase | Gossypium hirsutum | 2·10−57 | 501 |
| XII.A | Apoptosis inhibitor | Arabidopsis thaliana | 1·10−10 | 275 |
| XII.A‡ | Ascorbate peroxidase | Arabidopsis thaliana | 6·10−4 | 191 |
| XII.A‡ | Avr9/Cf-9 protein | Nicotiana tabacum | 4·10−12 | 412 |
| XII.A | Bax inhibitor | Brassica napus | 4·10−22 | 412 |
| XII.A | Disease resistance-responsive protein | Arabidopsis thaliana | 2·10−56 | 509 |
| XII.A | Glutathione peroxidase | Hevea brasiliensis | 7·10−45 | 524 |
| XII.A | Glutathione S-transferase | Glycine max | 2·10−88 | 715 |
| XII.A | HSR203J like protein | Capsicum chinense | 6·10−21 | 334 |
| XII.A | Lectin | Cicer arietinum | 7·10−16 | 457 |
| XII.A‡ | Leucine-rich repeat family protein | Arabidopsis thaliana | 2·10−58 | 734 |
| XII.A | MLO-like protein 1 | Arabidopsis thaliana | 3·10−23 | 415 |
| XII.A | Pathogenesis-related protein 1 | Vitis vinifera | 4·10−43 | 474 |
| XII.A | SC0A | Lycopersicon esculentum | 7·10−8 | 236 |
| XII.A | Selenium-binding protein | Medicago sativa | 2·10−22 | 275 |
| XII.A | Snakin | Solanum tuberosum | 2·10−20 | 321 |
| XII.A†,‡ | Superoxide dismutase | Fagus sylvatica | 4·10−51 | 372 |
| XII.B | ABA-responsive family protein | Arabidopsis thaliana | 4·10−14 | 264 |
| XII.B | Auxin-responsive family protein | Arabidopsis thaliana | 2·10−26 | 486 |
| XII.B | Brassinosteroid signalling positive regulator | Arabidopsis thaliana | 1·10−13 | 250 |
| XII.B | Ethylene-responsive element binding protein | Gossypium hirsutum | 7·10−7 | 355 |
| XII.B | Jasmonic acid 2 | Lycopersicon esculentum | 8·10−16 | 427 |
| XII.B | Patatin | Arabidopsis thaliana | 5·10−22 | 485 |
| XII.B | Senescence-associated protein | Arabidopsis thaliana | 9·10−50 | 441 |
| XII.B | Syringolide-induced protein | Glycine max | 2·10−22 | 244 |
| XII.B | Thaumatin | Vitis riparia | 1·10−48 | 391 |
* The class numbers correspond to those described in Fig. 3 and in Journet et al. (2002).
† Homologous to sequence from Jones et al. (2002).
‡ Homologous to sequence from Verica et al. (2004).
The genes potentially related to plant–pathogen interaction, but for which it was not possible to detect an in silico differential expression between the RT and SP libraries, are listed in Table 5. Sequences related to resistance to pathogen [Avr9/Cf9, PR (pathogenesis-related) proteins, HSR203J, selenium-binding protein, beta-1,3-glucanase, chitinase, disease-resistant responsive protein and cytochrome P450], and to detoxification (the ABC transporter, multidrug transporter), which play a major role in plant pathogen defence, were encountered. Genes of cell wall biosynthesis and metabolism, such as pectate lyase and cellulose synthase, were also found and may be related to resistance mediated by alteration of plant cell wall composition. Different groups of transcription factors (Myb, WRKY, EREB/AP2, bZIP) involved in regulation of pathogen defence-induced programmes were present in the total unigene from the cacao–M. perniciosa interactions as well as genes which may be related to plant pathogen signal transduction such as receptor kinase, MAP kinase, calmodulin-binding protein and phosphatase 2A. Genes related to programmed cell death and/or senescence such as apoptosis inhibitor, bax and proteasome inhibitors, MLO protein, senescence-associated protein, and genes with function related to oxidative burst (peroxidase, monodehydroascorbate reductase, NADPH oxidase, squalene monoxygenase, sulfite oxidase, semialdehyde dehydrogenase, glutathione peroxidase, germin oxalate oxidase, superoxide dismutase and glutathione S-transferase) were also found. Genes related to protein metabolism were found: 26S proteasome, aspartic proteinase, cysteine protease, serine carboxypeptidase, vacuolar processing enzymes, U-box domain-containing protein, ubiquitin protein, heat shock protein, proteasome inhibitor and protease inhibitor. Different sequences related to hormone pathways were also detected (ethylene-responsive element, ABA-responsive protein, auxin-responsive protein, jasmonic acid 2, 12-oxophytodienoate reductase, lipoxygenase, brassinosteroid signalling positive regulator) and may be related to hormonal modifications through disease development.
DISCUSSION
The EST sequencing approach is of particular interest in organisms for which very little sequence data is available. To date, only a few cacao EST studies have been developed, published and deposited in the dbEST section of Genbank (Jones et al., 2002; Verica et al., 2004). As far as is known, the cacao–M. perniciosa interaction data presented here is therefore the first effort in sequencing of the expressed genome aimed at understanding witches' broom disease. A unigene set of 2926 sequences was generated from 6024 high-quality sequences, and showed a redundancy of about 52 %; a value corresponding to the correct level compared with other published works. Equivalent or lower redundancy levels have been reported (48 % in citrus; Forment et al., 2005), whereas other published libraries showed higher levels of redundancy, such as 72·5 % in Lotus japonicus (Asamizu et al., 2004). The specificity of the RT and SP libraries is high (about 75 %) when compared among themselves, and when compared with the two other published cacao libraries (Jones et al., 2002; Verica et al., 2004). Such high levels were found in other programs comparing ESTs (Lopez et al., 2004; Forment et al., 2005) and showed a high specificity of the sequences obtained in relation to the processes studied, in spite of the restricted number of ESTs present in the total unigene. To reach a high quality of annotation and avoid error propagation (Rouze et al., 1999), the EST clusters were annotated systematically using a semi-automated approach, in which a functional annotation is assigned after human examination of the results of various automated analyses, as described by Journet et al. (2002). Using this classification scheme, a putative functional category could be assigned to about 54 % of the unique sequences. This represents a good level in comparison to other studies using other classification schemes (37 %; Lopez et al., 2004) or using a close semi-automated classification procedure (58 %; Bräutigam et al., 2005). The most prevalent categories were the primary metabolism and protein synthesis and processing, with a slightly higher level in the SP library. This result was not unexpected due to the nature of the plant tissue studied, the apical meristem. Normally, the meristematic cells are involved in division and multiplication, a phenomenon that is amplified in M. perniciosa-infected plants resulting in broom formation (Silva et al., 2002). This could also be correlated to the higher level of sequences related to abiotic stimuli and development preferentially observed in the SP library, and those related to gene expression and RNA metabolism in both libraries.
For the libraries constructed from plants infected by M. perniciosa, in particular for the SP one, pathogen tissue was not separated from host tissue. Therefore, it could be expected that a portion of the sequences derived from this library was of fungal, not cacao, origin. Therefore, the proportion of fungal ESTs in the present libraries was estimated. Only two sequences from the SP library (susceptible cacao–M. perniciosa interaction) demonstrated a high probability of corresponding to M. perniciosa genes, which represents only 0·16 % of the SP sequence set, less than the proportion of ESTs of fungal origin (0·7–9 %) observed in a similar analysis from citrus–Phytophthora interaction (Forment et al., 2005). These data suggest that only a small fraction of the sequences obtained from the pathogen-challenged libraries were derived from the pathogen. In the case of the cacao–M. perniciosa interaction, biochemical and histological analysis had already shown the presence of only few fungal hyphae in stages preceding plant tissue necrosis, with massive fungal invasion of the plant tissues occurring mainly after the death of the broom (Penman et al., 2000; Ceita, 2004; Ceita et al., 2007). Because the present RNA preparation proceeded from tissues harvested from the plant inoculation to the more drastic symptoms, the fungal RNA was highly diluted and corresponding ESTs poorly represented in the SP library. To refine the detection of M. perniciosa sequences within the SP pathogen-challenged library, other methods, such as the automatic codon usage determination, could be used (Ronning et al., 2003).
By examining sequences to the total unigene, it was possible to identify 113 genes related to plant–pathogen interaction, on which an in silico analysis of gene expression was developed based on the R statistic (Stekel et al., 2000). The significant R statistic was R > 1·92 (99·9 % true positive rate) and allowed the detection of nine moderately to highly differentially represented genes. Interestingly, these nine sequences belonged to only three functional categories: cytoskeleton, protein synthesis and processing, and defence and cell rescue. Moreover, six of them also belonged to the most abundant sequence list, pointing out their probable importance to the physiological mechanisms occurring during the cacao–M. perniciosa interactions: plant resistance in the RT plants (Silva et al., 2002) and necrosis process in the SP ones (Ceita, 2004; Ceita et al., 2007). Particular attention was given to genes related to these two distinct physiological processes.
Various genes involved in the resistance process were encountered (e.g. PR proteins, selenium-binding protein, disease resistant-responsive protein) and some of them, such as the trypsin inhibitor and the PR4b genes, were more represented in the RT library. In the case of the PR4b, the in silico analysis was confirmed by sqRT-PCR (semi-quantitative RT-PCR) experiment: the PR4b expression decreased in susceptible inoculated (SI) plants from 0 DAI to 30 DAI and increased in resistant inoculated (RI) plants in the same time course, with a global expression much higher in RI plants than in SI ones (data not shown). It has been shown that trypsin inhibitors presented anti-fungal activity by inhibiting trypsin-like protease from various fungi such as Botrytis cinerea (Chilosi et al., 2000), Aspergillus flavus and Fusarium moniliforme (Chen et al., 1999). In the case of the PR4 protein, different roles have been assigned to this protein sub-family: ribonuclease activity for the PR4 from wheat (Caporale et al., 2004) and chitinase activity for the PR4 from carrot (Kragh et al., 1996; van Loon et al., 2006). In both cases, the demonstrated activity allowed the fungus growth inhibition. A super-expression of these genes in a heterologous system may allow the development of biocontrol methods against the witches' broom disease. More generally, these two genes may constitute a base for more thorough functional studies related to resistance of cacao to M. perniciosa.
The presence of apoptosis and oxidative burst-related genes (apoptosis inhibitor, senescence associated protein, gene related to oxidative burst) strengthens the hypothesis that the susceptible cacao–M. perniciosa interaction involves a programmed cell death process initially occurring in the plant as a defence mechanism which then is diverted by the fungus for its own profit, allowing its sporulation and further propagation (Ceita, 2004; Ceita et al., 2007). Ceita et al. (2007) suggest that oxalate oxidase and ascorbate peroxidase genes play a crucial role in the production of reactive oxygen species and programmed cell death processes by degrading calcium oxalate crystals and producing H2O2. Other genes related to the process of programmed cell death were also found in the cacao–M. perniciosa libraries, such as those encoding protease, in particular plant vacuolar protease (Hatsugai et al., 2004), metallothionein (Mir et al., 2004) and ankyrin-repeat protein (Dong, 2004). One of the metallothionein genes (homologous to the one of Petunia × hybrida; Table 3) showed, by sqRT-PCR analysis, a constant expression in SI plants. In the RI plants, the expression of this metallothionein gene slightly increased from 0 to 60 DAI, even if the global expression in resistant plants remained less than the one observed in susceptible plants (data not shown), as expected in the in silico analysis. Ankyrin contigs were found in the list of the ten most represented genes as well as in the list of the genes more represented in the SP library. Moreover, the PR10 gene whose expression is observed only at 60 DAI in SI plants, confirming the differential expression – higher expression in SI than in RI plants and null expression in RT – obtained by in silico analysis (data not shown), also constitutes a good target for disease mechanism studies because of its function of RNase which could be related to plant defence or to plant RNA degradation in the process of necrosis (Bantignies et al., 2000; Park et al., 2004). Interestingly, the ribonuclease activity of the PR10 family proteins are not considered as being related to the action mechanism of the PR4 previously described (Caporale et al., 2004). It may be supposed that these two kinds of RNase play distinct roles in the resistant and susceptible mechanism of the cacao–M. perniciosa interactions.
Moreover, subsets of the 113 genes had similarity to genes implicated in pathogen detection (Avr9/Cf9 protein, receptor kinase), signal transduction (MAP kinase, calmodulin-binding protein) and gene regulation events (transcription factors). Genes involved in the synthesis pathway of molecules such as alkaloids and tannins were also found in the total unigene, such as caffeine synthase, caffeic acid 3-O-methyltransferase, chalcone syntase, flavonol synthase, flavanone-3-hydroxylase (data not shown), which may be correlated with the alteration of the content of caffeine and tannins (higher in infected plant than in uninfected ones) as observed by Scarpari et al. (2005). Some of the genes encountered hybridized the same cacao BAC (bacterial artificial chromosome) clones (Clement et al., 2004) as observed for the senescence associated protein, calmoduline, PR1, PR10, ABC transporter and auxin repressor genes (data not shown) suggesting a possible organization in clusters as observed in other plant genomes (Gebhart and Valkonen, 2001). Finally, it is important to note that some other genes – with function not related to plant–pathogen interaction, genes of unknown function, or genes without homology – may also be differentially represented and may play a role in the cacao–M. perniciosa interaction, although not analysed here (Rudd, 2003).
Based on the great amount of genes identified in the present study, a large-scale analysis of the expression of candidate genes should be investigated. The development of array technology should allow rapid progress for monitoring changes of gene expression in cacao plants during plant–pathogen interaction. Moreover, because both the RT and SP libraries used in this study were constructed from RNA isolated from multiple time points of the interaction with the pathogen, array experiments will allow assessing temporal expression of these genes in the infection process by the use of probes from individual RNA.
In conclusion, the ESTs generated in this study provide a good tool for more studies to understand the resistant and susceptible interactions of T. cacao and M. perniciosa. It has been shown that oxalate oxidase and ascorbate peroxidase genes participated to the susceptibility process (Ceita et al., 2007), but other genes are under investigation and may contribute to understanding the mechanisms of fungus transition from biotrophic to necrotrophic phase and to relate them to biochemical changes occurring in the green broom (Scarpari et al. 2005) or to in vitro observations (Meinhardt et al., 2006). Even if few fungus genes were found in the present libraries, one of them, the 25-kDa protein elicitor also called NEP (necrosis and ethylene-inducing proteins), was more thoroughly studied and has been shown to be able to induce necrosis and ethylene emission in tobacco and cacao leaves (Garcia et al., 2007). Moreover, some of the genes detected may be used to develop strategies to control witches' broom disease, such as chitinases, glucanases or PR proteins with RNase activity. The sequences may also be a source of single nucleotide polymorphism or simple sequence repeats for molecular marker development. A reference molecular genetic linkage map of cacao is available (Pugh et al., 2004) allowing the mapping and possible co-localization of candidate genes with QTLs associated with witches' broom resistance. This will also provide important information about their role in the defence mechanisms and provide a new source of markers for marker-assisted selection in the development of new cacao varieties with durable resistance to witches' broom disease.
ACKNOWLEDGEMENTS
The work of ASG and ACS was supported by the Research Support Foundation of the State of Bahia, the work of NC was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico, and the work of JNM was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior. We thank Frederico Evangelista dos Santos and Robson José Costa Dias (Laboratório de Genética, UESC) for technical assistance and Dr Claudia Fortes Ferreira (Embrapa, Brazil) and Dr Martin Brendel (UESC, Brazil) for helpful advice and critical reading of the manuscript. This research was supported by FAPESB, Banco do Nordeste and Conselho Nacional de Desenvolvimento Científico e Tecnológico.
LITERATURE CITED
- Aime MC, Phillips-Mora W. The causal agents of witches' broom and frosty pod rot of cacao (chocolate, Theobroma cacao) form a new lineage of Marasmiaceae. Mycologia. 2005;97:1012–1022. doi: 10.3852/mycologia.97.5.1012. [DOI] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S. Characteristics of the Lotus japonicus gene repertoire deduced from large-scale expressed sequence tag (EST) analysis? Plant Molecular Biology. 2004;54:405–414. doi: 10.1023/B:PLAN.0000036372.46942.b8. [DOI] [PubMed] [Google Scholar]
- Austin R, Provart NJ, Sacadura NT, Nugent KG, Babu M, Saville BJ. A comparative genomic analysis of ESTs from Ustilago maydis. Functional and Integrative Genomics. 2004;4:207–218. doi: 10.1007/s10142-004-0118-x. [DOI] [PubMed] [Google Scholar]
- Bantignies B, Séguin J, Muzac I, Dédaldéchamp F, Gulick P, Ibrahim R. Direct evidence for ribonucleolytic activity of a PR-10-like protein from white lupin roots. Plant Molecular Biology. 2000;42:871–881. doi: 10.1023/a:1006475303115. [DOI] [PubMed] [Google Scholar]
- Bortoluzzi S, Danieli GA. Towards an in silico analysis of transcription patterns. Trends in Genetics. 1999;15:118–119. doi: 10.1016/s0168-9525(98)01682-5. [DOI] [PubMed] [Google Scholar]
- Bräutigam M, Lindlöf A, Zakhrabekova S, Gharti-Chhetri G, Olsson B, Olsson O. Generation and analysis of 9792 EST sequences from cold acclimated oat, Avena sativa. BMC Plant Biology. 2005;5 doi: 10.1186/1471-2229-5-18. 18 (doi:10·1186/1471-2229-5-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporale C, Di Berardino I, Leonardi L, Bertini L, Cascone A, Buonocore V, Caruso C. Wheat pathogenesis-related proteins of class 4 have ribonuclease activity. FEBS Letters. 2004;575:71–76. doi: 10.1016/j.febslet.2004.07.091. [DOI] [PubMed] [Google Scholar]
- Ceita GO. Brazil: Universidade Estadual de Santa Cruz; 2004. Analise do processo de morte celular em Theobroma cacao L. induzido por Crinipellis perniciosa (Stahel) Singer. Masters Dissertation. [Google Scholar]
- Ceita GO, Macêdo JNA, Santos TB, Alemanno L, Gesteira AS, Micheli F, et al. Involvement of calcium oxalate degradation during programmed cell death in Theobroma cacao tissues triggered by the hemibiotrophic fungus Moniliophthora perniciosa. Plant Science. 2007 (doi:10·1016/j.plantsci.2007·04·06) [Google Scholar]
- Chen Z-Y, Brown RL, Lax AR, Cleveland TE, Russin JS. Inhibition of plant-pathogenic fungi by a corn trypsin inhibitor overexpressed in Escherichia coli. Applied and Environmental Microbiology. 1999;65:1320–1324. doi: 10.1128/aem.65.3.1320-1324.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilosi G, Caruso C, Caporale C, Leonardi L, Bertini L, Buzi A, et al. Antifungal activity of a bowman-birk-type trypsin inhibitor from wheat kernel. Journal of Phytopathology. 2000;148:477–481. [Google Scholar]
- Clement D, Lanaud C, Sabau X, Fouet O, Le Cunff L, Ruiz E, et al. Creation of BAC genomic resources for cocoa (Theobroma cacao L.) for physical mapping of RGA containing BAC clones. Theoretical and Applied Genetics. 2004;108:1627–1630. doi: 10.1007/s00122-004-1593-0. [DOI] [PubMed] [Google Scholar]
- Corpet F, Servant F, Gouzy J, Kahn D. ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Research. 2000;28:267–269. doi: 10.1093/nar/28.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. The role of membrane-bound ankyrin-repeat protein acd6 in programmed cell death and plant defense. Science's STKE. 2004 doi: 10.1126/stke.2212004pe6. pe6. [DOI] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl M, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Forment J, Gadea J, Huerta L, Abizanda L, Agusti J, Alamar S, et al. Development of a citrus genome-wide EST collection and cDNA microarray as resources for genomic studies. Plant Molecular Biology. 2005;57:375–391. doi: 10.1007/s11103-004-7926-1. [DOI] [PubMed] [Google Scholar]
- Frias GA, Purdy LH, Schmidt RA. An inoculation method for evaluating resistance of cacao to Crinipellis perniciosa. Plant Disease. 1995;79:787. [Google Scholar]
- Garcia O, Macedo JN, Tibúrcio R, Zaparoli G, Rincones J, Bittencourt LMC, et al. Characterization of necrosis and ethylene-inducing proteins (NEP) in the basidiomycete Moniliophthora perniciosa, the causal agent of witches' broom in Theobroma cacao. Mycological Research. 2007 doi: 10.1016/j.mycres.2007.01.017. (doi:10·1016/j.mycres.2007·01·017) [DOI] [PubMed] [Google Scholar]
- Gebhardt C, Valkonen JPT. Organization of genes controlling disease resistance in the potato genome. Annual Review of Phytopathology. 2001;39:79–102. doi: 10.1146/annurev.phyto.39.1.79. [DOI] [PubMed] [Google Scholar]
- Gesteira AS, Micheli F, Ferreira CF, Cascardo JCM. Isolation and purification of functional total RNA from different organs of cocoa tree during its interaction with the pathogen Crinipellis perniciosa. BioTechniques. 2003;35:494–500. doi: 10.2144/03353st02. [DOI] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, et al. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science. 2004;305:855–858. doi: 10.1126/science.1099859. [DOI] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Research. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PG, Allaway D, Gilmour DM, Harris C, Rankin D, Retzel ER, Jones CA. Gene discovery and microarray analysis of cacao (Theobroma cacao L.) varieties. Planta. 2002;216:255–264. doi: 10.1007/s00425-002-0882-6. [DOI] [PubMed] [Google Scholar]
- Journet E-P, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer M-J, et al. Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Research. 2002;30:5579–5592. doi: 10.1093/nar/gkf685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragh KM, Hendriks T, de Jong AJ, Lo Schiavo F, Bucherna N, Hojrup P, et al. Characterization of chitinases able to rescue somatic embryos of the temperature-sensitive carrot variant t s l l. Plant Molecular Biology. 1996;31:631–645. doi: 10.1007/BF00042235. [DOI] [PubMed] [Google Scholar]
- Kris-Etherton P, Keen C. Evidence that the antioxidant flavonoids in tea and cocoa are beneficial for cardiovascular health. Current Opinion in Lipidology. 2002;13:41–49. doi: 10.1097/00041433-200202000-00007. [DOI] [PubMed] [Google Scholar]
- Kuhlmann M, Horvay K, Strathmann A, Heinekamp T, Fischer U, Böttner S, et al. The-helical d1 domain of the tobacco bzip transcription factor bzi-1 interacts with the ankyrin-repeat protein ank1 and is important for bzi-1 function, both in auxin signaling and pathogen response. Journal of Biological Chemistry. 2003;278:8786–8794. doi: 10.1074/jbc.M210292200. [DOI] [PubMed] [Google Scholar]
- van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annual Review of Phytopathology. 2006;44:1–28. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- Lopez C, Jorge V, Piégu B, Mba C, Cortes D, Restrepo S, et al. A unigene catalogue of 5700 expressed genes in cassava. Plant Molecular Biology. 2004;56:541–554. doi: 10.1007/s11103-004-0123-4. [DOI] [PubMed] [Google Scholar]
- Lu H, Liu Y, Greenberg JT. Structure–function analysis of the plasma membrane localized Arabidopsis defense component ACD6. The Plant Journal. 2005;44:798–809. doi: 10.1111/j.1365-313X.2005.02567.x. [DOI] [PubMed] [Google Scholar]
- Meinhardt LW, Bellato M, Rincones J, Azevedo RA, Cascardo JC, Pereira GA. Current Microbiology. 2006;52:191–196. doi: 10.1007/s00284-005-0182-z. In vitro production of biotrophic-like cultures of Crinipellis perniciosa, the causal agent of witches' broom disease of Theobroma cacao. [DOI] [PubMed] [Google Scholar]
- Mir G, Domenech J, Huguet G, Guo WJ, Goldsbrough P, Atrian S, et al. A plant type 2 metallothionein (MT) from cork tissue responds to oxidative stress. Journal of Experimental Botany. 2004;55:2483–2493. doi: 10.1093/jxb/erh254. [DOI] [PubMed] [Google Scholar]
- Park CJ, Kim KJ, Shin R, Park JM, Shin YC, Paek KH. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. The Plant Journal. 2004;37:186–198. doi: 10.1046/j.1365-313x.2003.01951.x. [DOI] [PubMed] [Google Scholar]
- Penman D, Britton G, Hardwick K, Collin HA, Isaac S. Chitin as a measure of biomass of Crinipellis perniciosa, causal agent of witches' broom disease of Theobroma cacao. Mycological Research. 2000;104:671–675. [Google Scholar]
- Pugh T, Fouet O, Risterucci AM, Brottier P, Abouladze M, Deletrez C, et al. A new cacao linkage map based on codominant markers: development and integration of 201 new microsatellite markers. Theoretical and Applied Genetics. 2004;108:1151–1161. doi: 10.1007/s00122-003-1533-4. [DOI] [PubMed] [Google Scholar]
- Purdy LH, Schmidt RA. Status of cocoa witches broom: biology, epidemiology, and management. Annual Review of Phytopathology. 1996;34:573–594. doi: 10.1146/annurev.phyto.34.1.573. [DOI] [PubMed] [Google Scholar]
- Rincones J, Mazotti GD, Griffith GW, Pomela A, Figueira A, Leal GA, Jr, et al. Genetic variability and chromosome-length polymorphisms of the witches' broom pathogen Crinipellis perniciosa from various plant hosts in South America. Mycological Research. 2006;110:821–32. doi: 10.1016/j.mycres.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Rios-Ruiz RA. Melhoramento para resistência a doenças. In: Dias LAS,, editor. Melhoramento genético do cacaueiro. Viçosa: Funape-UFG, Editora Folha de Viçosa Ltda; 2001. pp. 289–324. [Google Scholar]
- Rocha HM, Miranda RAC, Sgrillo RB, Setubal RA. Witches' broom in Bahia, Brazil. In: Rudgard SA, Madison AC, Andebrhan T, editors. Disease and management in cocoa: comparative epidemiology of witches' broom. London: Chapman and Hall; 1993. pp. 189–200. [Google Scholar]
- Ronning CM, Stegalkina SS, Ascenzi RA, Bougri O, Hart AL, Utterbach TR, et al. Comparative analyses of potato expressed sequence tag libraries. Plant Physiology. 2003;131:419–429. doi: 10.1104/pp.013581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouze P, Pavy N, Rombauts S. Genome annotation: which tools do we have for it? Current Opinion in Plant Biology. 1999;2:90–95. doi: 10.1016/S1369-5266(99)80019-3. [DOI] [PubMed] [Google Scholar]
- Rudd S. Expressed sequence tags: alternative or complement to whole genome sequences? Trends in Plant Science. 2003;8:321–329. doi: 10.1016/S1360-1385(03)00131-6. [DOI] [PubMed] [Google Scholar]
- Rudgard SA, Andebrhan T, Maddison AC, Schmidt RA. Disease management: recommendations. In: Rudgard SA, Madison AC, Andebrhan T, editors. Disease and management in cocoa: comparative epidemiology of witches' broom. London: Chapman and Hall; 1993. pp. 201–212. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis EFT. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular cloning – a laboratory manual. [Google Scholar]
- Scarpari LM, Meinhardt LW, Mazzafera P, Pomella AWV, Schiavinato MA, Cascardo JMC, et al. Biochemical changes during the development of witches' broom: the most important disease of cocoa in Brazil caused by Crinipellis perniciosa. Journal of Experimental Botany. 2005;56:865–877. doi: 10.1093/jxb/eri079. [DOI] [PubMed] [Google Scholar]
- Silva SD, Luz EDMN, Almeida OC, Gramacho K, Bezerra JL. Redescrição da sintomatologia causada por Crinipellis perniciosa em cacaueiro. Agrotropica. 2002;1:1–23. [Google Scholar]
- Stekel DJ, Git Y, Falciani F. The comparison of gene expression from multiple cDNA libraries. Genome Research. 2000;10:2055–2061. doi: 10.1101/gr.gr-1325rr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica JA, Maximova SN, Strem MD, Carlson JE, Bailey BA, Guiltinan MJ. Isolation of ESTs from cacao (Theobroma cacao L.) leaves treated with inducers of the defense response. Plant Cell Reporter. 2004;23:404–413. doi: 10.1007/s00299-004-0852-5. [DOI] [PubMed] [Google Scholar]
- Wheeler BEJ, Mepsted R. Pathogenic variability amongst isolates of Crinipellis perniciosa from cocoa (Theobroma cacao) Plant Pathology. 1988;37:475–488. [Google Scholar]