Abstract
Background and Aims
The spatial and statistical distribution of genome sizes and the adaptivity of genome size to some types of habitat, vegetation or microclimatic conditions were investigated in a tetraploid population of Festuca pallens. The population was previously documented to vary highly in genome size and is assumed as a model for the study of the initial stages of genome size differentiation.
Methods
Using DAPI flow cytometry, samples were measured repeatedly with diploid Festuca pallens as the internal standard. Altogether 172 plants from 57 plots (2·25 m2), distributed in contrasting habitats over the whole locality in South Moravia, Czech Republic, were sampled. The differences in DNA content were confirmed by the double peaks of simultaneously measured samples.
Key Results
At maximum, a 1·115-fold difference in genome size was observed. The statistical distribution of genome sizes was found to be continuous and best fits the extreme (Gumbel) distribution with rare occurrences of extremely large genomes (positive-skewed), as it is similar for the log-normal distribution of the whole Angiosperms. Even plants from the same plot frequently varied considerably in genome size and the spatial distribution of genome sizes was generally random and unautocorrelated (P > 0·05). The observed spatial pattern and the overall lack of correlations of genome size with recognized vegetation types or microclimatic conditions indicate the absence of ecological adaptivity of genome size in the studied population.
Conclusions
These experimental data on intraspecific genome size variability in Festuca pallens argue for the absence of natural selection and the selective non-significance of genome size in the initial stages of genome size differentiation, and corroborate the current hypothetical model of genome size evolution in Angiosperms (Bennetzen et al., 2005, Annals of Botany 95: 127–132).
Key words: Genome size, Poaceae; Gramineae; grasses; fescue; Festuca pallens; intrapopulation; intraspecific genome size variability; flow cytometry; transposons; evolution; ecological adaptivity; population structure; natural selection
INTRODUCTION
Changes in genome size are generally accepted as an important evolution mechanism in plants, and numerous recent works have reported evolutionary significant differences in genome size at family, generic and specific levels (Bennett et al., 2000; Voglmayr, 2000; Obermayer et al., 2002; Bureš et al., 2004b; Jakob et al., 2004; Bennett and Leitch, 2005; Kapraun, 2005; Leitch et al., 2005; Price et al., 2005; Ricroch et al., 2005; Bancheva and Greilhuber, 2006; Weiss-Schneeweiss et al., 2006). Considerable differences in genome size are also documented among species groups of different life forms and ecology (Bennett, 1972; Grotkopp et al., 2004; Hiremath and Nagasampige, 2004; Chase et al., 2005; Knight et al., 2005). An important part of genome size studies is the question of the existence of intraspecific variability, which can play a key role in the understanding of the process of genome size diversification in closely related species. Numerous works have reported intraspecific genome size variability over the last 40 years (cf. Ohri, 1998; Suda, 2004; Greilhuber, 2005). However, doubt has been cast over some of these studies due to potential instrument and methodical errors, as argued by Greilhuber (1998, 2005). Intraspecific genome size differences were found especially among geographically distant populations or among populations growing in contrasting macroclimatic conditions (Ohri, 1998; Knight et al., 2005, and citations therein). Significant relationships for intraspecific genome size have been found with altitude, latitude, temperature, precipitation, seed mass, moisture and leaf anatomical traits; however, the trends in different taxa may differ (cf. Knight et al., 2005). Available knowledge of the initial stages of genome size differentiation, on a small geographical scale and intrapopulation level, is still very poor and does not enable us to answer effectively questions regarding the origin of such differences, their microevolutionary importance and the role of natural selection in their stabilization.
Important intraspecific and also intrapopulation variability was documented in Dasypyrum villosum by Caceres et al. (1998), who found higher (11·5 % at maximum) intrapopulation genome size variability particularly in populations from higher altitudes; however, it is necessary to note that the methods and the accuracy of the results of this work were later questioned by Greilhuber (2005). Zoldoš et al. (1998) reported variation of DNA content within one population of Quercus petraea to be related to the presence of B chromosomes. Intrapopulation differences in genome size of up to 23 % were detected in Dactylis glomerata from Slovenia (Vilhar et al., 2002) and some minute microgeographic differences were also documented in Ceratonia siliqua from Israel (Bureš et al., 2004a). Considerable intraspecific and even intrapopulation genome size variability has been recently observed in several Festuca species (Šmarda, 2006; Šmarda and Bureš, 2006). The largest differences in genome size of Festuca pallens were observed between distant populations across the whole natural range (1·17-fold in diploids and 1·164-fold in tetraploids), and significant variability was also shown within separate canyon or hill systems and frequently also within populations (Šmarda and Bureš, 2006). In the most variable tetraploid population from Mikulov–Sv. Kopeček (Czech Republic, Pálava Hills), the maximum 1·121-fold difference in DNA content was almost in the same range as within all the other tetraploid populations (1·148-fold difference; Šmarda and Bureš, 2006, Fig. 3). Due to the high variability detected and the contrasting habitat conditions, this locality was chosen as a model for the analysis of microgeographic genome size differentiation, and the role of ecological adaptivity of genome size at the intrapopulation level.
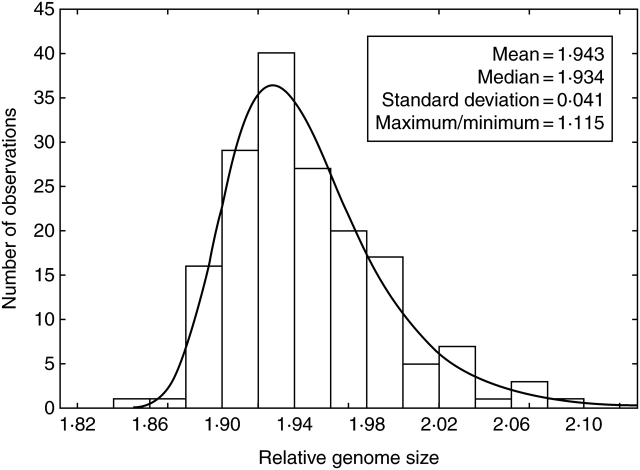
Fig. 3.
Histogram of the distribution of genome size of tetraploid F. pallens at the studied locality. The histogram is fitted by the probability function (solid line) of extreme (Gumbel) distribution. The hexaploid sample has been omitted.
The present work focused on a detailed intrapopulation study of genome size, completing the previous survey of variation in Festuca pallens, investigated on global, landscape and local spatial scales (Šmarda and Bureš, 2006). The following questions were addressed. (1) What is the range and the statistical distribution of genome size in this population? Is there any prevalent genome size category? (2) What is the spatial distribution pattern of genome size in this microclimatically and ecologically contrasting locality? Is it random or do plants with higher/lower genome size tend to concentrate in some specific parts? (3) Is the genome size adaptive to some types of habitat, vegetation or microclimatic conditions?
MATERIAL AND METHODS
Model species and the locality
Festuca pallens (Poaceae, Gramineae) is a perennial, allogamic, wind-pollinated (Auquier, 1977), tussock-forming species, growing on relict rocky outcrops, rocky slopes and rock cliffs. Its distribution is restricted to Central Europe (from northern France to southern Romania) where typically it can be found on the slopes of river canyons, valleys and gorges in rocky highlands and steppe hills in lowlands, at altitudes from 100 to 1500 m a.s.l. The studied population belongs to the Pannonian tetraploid type (cf. Tracey, 1980; Šmarda and Kočí, 2003; Šmarda and Bureš, 2006); in a narrow taxonomic concept treated recently as Festuca csikhegyensis Simonk. [syn. Festuca glaucina (Stohr) Stohr; Šmarda et al., 2007].
The Svatý Kopeček is an isolated limestone hill situated above the town of Mikulov on the south edge of the Pavlov Hills (South Moravia, Czech Republic). The altitude ranges from 250 to 360 m a.s.l. The microclimate and edaphic conditions differ sharply according to the orientation of the slopes (shady, humid on those facing north vs. sunny and xeric on those facing south-west) and the inclination of the land (flat terrain to vertical slopes). About 35 % of the hill is extremely rocky and these parts are covered with various types of dry grasslands: (1) dry rocky outcrops with Festuca pallens, Poa badensis, Fumana procumbens and Helianthemum ovatum (scattered on south and south-west exposed slopes); (2) dry rocky F. pallens steppe (south and south-west slopes); (3) Stipa capillata and S. pennata steppes (south-east slopes); (4) Festuca valesiaca steppe (central, flat part); (5) mesophilous rocky steppe (south-east exposed slopes and in the central part); (6) mesophilous rocky outcrops with Koeleria macrantha and frequent ephemerals (south-east exposed slopes and in the central part); (7) forest fringe grasslands (central flat part); and (8) Sesleria albicans grasslands (mostly west and north exposed slopes). Festuca pallens occurs frequently in all rocky grassland communities except for the forest fringes and the Stipa and Festuca valesiaca steppes.
Vegetation and habitat data sampling
Plants were collected from 57 plots, which were distributed regularly over all parts of the locality, and from all types of the plant communities occupied by F. pallens (Fig. 1). The sampling plots had an area of 2·25 m2 (usually 1·5 × 1·5 m). Three of the most morphologically contrasting tufts of F. pallens were collected from each sampling plot. A part of the sampled tuft was prepared as a herbarium specimen and stored at the Herbarium at the Department of Botany, Masaryk University in Brno (BRNU). The remaining part was stored in a plastic bag for subsequent genome size measurements. All samples were collected over 4 days.
Fig. 1.
Distribution of sample plots (stars) over the sampled locality. The three arms symbolize the three samples for each plot (four in plot 19). The lengths of arms of the stars reflect the relative genome size of the samples, expressed in a range from the minimum (0 %) to the maximum (100 %): the sample with the smallest genome size is projected on the left arm, with the median on the top arm, and that with the highest on the right arm. The only hexaploid sample is marked by a triangle (adjacent to population 2). Plot numbers are indicated.
The main gradients in the ecologic conditions at the locality seem to be most comprehensively expressed by the actual vegetation cover, and this complex character was chosen as the main descriptive character of the ecologic conditions for the sampling plots. The vegetation cover of all sampling plots/areas was documented by phytosociological relevés, recorded using the Braun–Branquet approach (Westhoff and van der Maarel, 1978). Species cover was estimated on the nine-degree Braun–Blanquet scale (Westhoff and van der Maarel, 1978; see Supplementary Information 3, available online). For all sampling plots, the total vegetation cover, cover of mosses, cover of bare rock, and the slope exposition were also recorded (see Supplementary Information 2, available online). Co-ordinates of the plots (WGS-84 system) and altitudinal data were obtained using a Garmin Ltd ‘Etrex’ GPS instrument. The treatment of taxa follows Kubát (2002).
Genome size estimations
Genome size was estimated by flow cytometry with DAPI staining. Measurements were carried out at the Institute of Botany and Zoology, Masaryk University in Brno using the same methods and the same instrument (PA-I Partec ploidy analyzer) as have been previously described for measurements of intraspecific genome size variability of Festuca pallens (Šmarda and Bureš, 2006). The only modification was the use of a single internal standard, a single tuft of diploid Festuca pallens (sample F1229; 2n = 14; Šmarda and Kočí, 2003), instead of two standards in the previous study. The earlier study of Festuca pallens (Šmarda and Bureš, 2006) indicated high resolution of flow cytometry measurements with AT-selective DAPI dye for precise determination of intraspecific genome size variability. The two extreme samples originating from the studied population in Mikulov–Sv. Kopeček were demonstrated to differ in genome size by simultaneous measurements resulting in clear double peaks, with a similar peak distance as in the separate sample measurements (Šmarda and Bureš, 2006, Fig. 4B). This study also demonstrated a high correlation of the results of base-specific DAPI staining and intercalary propidium iodide staining, as indicated by the results of several other studies (Bureš et al., 2004b; Vizintin et al., 2006), and the repeatability of the results obtained in Festuca pallens in different seasons. Use of the same method in the same taxon enables us to avoid potential biases caused by instrument errors and the concentration of cytosolic compounds (cf. Noirot et al., 2000; Loureiro et al., 2006). Samples were measured in random order, each two or three times, on different days. The final standard/sample ratio was calculated as an average (see Supplementary Information 1, avaialable online). A total of 5000 cells were analysed in each measurement. The average coefficient of variance (CV) of all sample and standard peaks of all measurements was 1·82%. The results were verified by the simultaneous measurement of some of the most contrasting samples and the differences in DNA content were demonstrated as a clear double peak (Fig. 2).
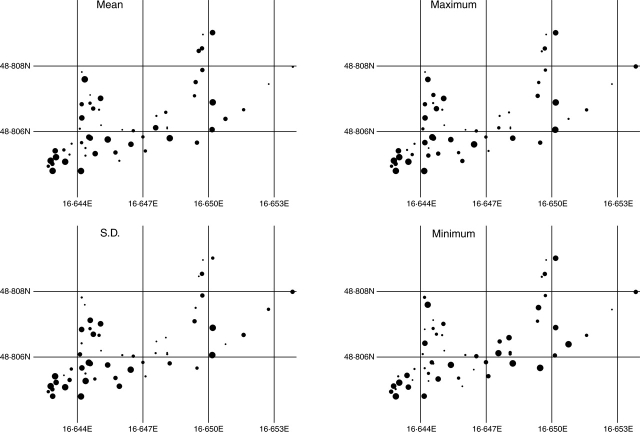
Fig. 4.
Spatial pattern of genome size parameters of the 57 sample plots. The diagrams show the mean genome size per population (Mean), the standard deviation of genome size among the samples from the same plot (S.D.), the genome size of the sample with the highest genome size in the plot (Maximum), and the genome size of the sample with the smallest genome size in the plot (Minimum). The plots were sorted in increasing order according to the genome size parameter shown and divided into six categories (9 + 9 + 10 + 10 + 10 + 9 samples), to which the size of the circles correspond.
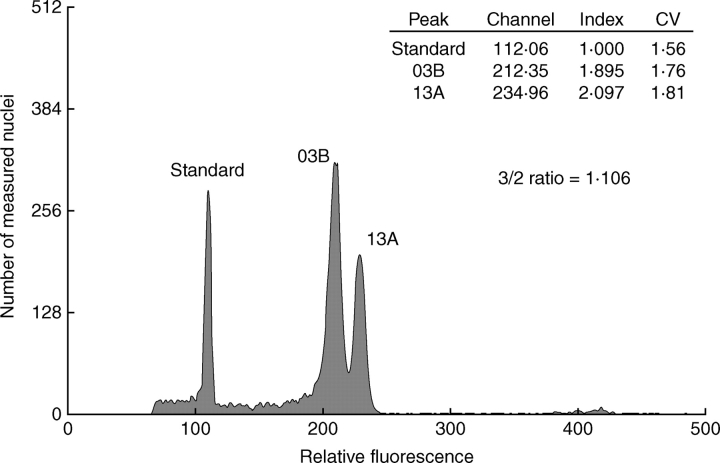
Fig. 2.
Difference in genome size in two of the most contrasting samples, 03B and 13A, proved as a clear double peak in simultaneous measurement. The diploid Festuca pallens was used as a standard.
Statistical treatment
The normality of the distribution of genome sizes of all samples investigated, and data of all Angiosperms in the Plant DNA C-value database (4427 entries; Bennett and Leitch, 2005) was tested using the Shapiro–Wilk W-test. Kolmogorov–Smirnov D-statistics (K–S D; the largest absolute difference between the observed and expected distributions) was further used to find the best distribution model among the following distribution functions: extreme value, non-normal, log-normal, normal, Weibull, Rayleigh and exponential.
To test the existence of a spatial gradient in the distribution of genome size among sampling plots, the matrix of paired-interplot geographical distances and the matrix of paired-interplot differences in a parameter of genome size were compared using the Mantel test in the PopTools 2·6·9 program (Hood, 2005). Four genome size parameters of plots were analysed: the average genome size of samples, genome size variability among samples, and maximum and minimum observed genome size. Significance of the test was calculated as one-tailed, based on 10 000 permutations. The spherical geographic distance of plots was calculated from the co-ordinates and altitudinal data using the ArcMap program (ESRI Inc., 2004).
The existence of a spatial autocorrelation of genome size was further inspected using the weighted Moran's I autocorrelation statistics and Moran correlogram, both calculated using the CrimStat III program (Levine, 2004). Analysis was conducted with all 171 tetraploid samples and with the four genome size characters of the 57 plots sampled. As only the co-ordinates of the plots themselves were available, individual co-ordinates for samples from the same plot were generated randomly ± 1·5 m around these to obtain non-zero distances in any sample pair. Distances between samples were calculated from the co-ordinates using the CrimStat III program as the ‘great circle’ distances. Ten (all samples) or six (population characters) distance classes of equal-distance ranges were compared using a Moran correlogram. One thousand Monte Carlo simulations were used to estimate the approximate confidence intervals for the actual value of Moran's I in the correlogram. The significance of global Moran's I for the whole data matrix was based on a randomization assumption.
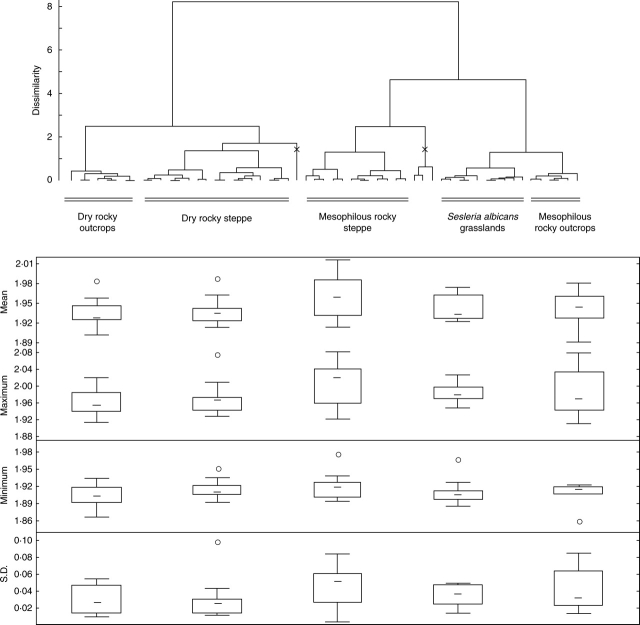
In order to test the existence of adaptivity of genome size to selected ecological conditions, as a first step, we looked for a correlation between genome size parameters and the main gradients in the actual vegetation. For this purpose, the matrix of paired interplot differences in each parameter of genome size (four parameters, as mentioned above) and the matrix of paired interplot dissimilarities in vegetation cover (phytosociologic relevés) were compared using a Mantel test. The Bray–Curtis index (percentage difference) was chosen to measure the dissimilarity of phytosociologic relevés. For an alternative hypothesis, we tested the difference of the four selected genome size parameters in several different types of vegetation. Phytosociologic relevés representing the vegetation of the plots were classified into the main vegetation types using hierarchical clustering, following the approach of Botta-Dukát et al. (2005). Principal co-ordinates analysis (PCoA; Legendre and Legendre, 1998) was performed on the original dissimilarity matrix used above. Because only the first few ordination axes in such an analysis contain interpretable ecological information (Botta-Dukát et al., 2005), the number of ordination axes was reduced. To establish the number of interpretable axes, the eigenvalues were compared with random expectations based on the broken-stick model (Jackson, 1993; Legendre and Legendre, 1998: p. 410). In this case the first 12 axes proved to be significant. They explained 74·73 % of the total variation in the data set; hence, nearly a quarter of the total variation proved to be noise. The co-ordinates along the 12 significant axes of PCoA were used instead of the raw data as input for the classification. Ward's algorithm of minimum increment of sum-of-squares was used for dendrogram construction (Ward, 1963; Legendre and Legendre, 1998) and group classification. Five vegetation groups and one group of outliers were established: (1) dry, rocky outcrops with Poa badensis; (2) dry, rocky steppe; (3) mesophilous rocky steppe; (4) Sesleria albicans grasslands; (5) mesophilous rocky outcrops; and (6) outliers. The differences of the four genome size parameters within these groups (both including and excluding the group of outliers) were tested using non-parametric Kruskal–Wallis ANOVA.
As a second step, we attempted to find relationships between genome size and several additional habitat data: altitude; deviation of slope expositions from north and south-west, corresponding to the incident solar radiation intensity; cover of shrubs, herbs, mosses and bare rock; total number of species recorded in the plot; and cover of F. pallens, corresponding to its relative fitness. Because of the non-normality of most of these characters, we used Spearman's rho non-parametric correlation for non-normally distributed and semi-quantitative data, and Kendall's tau b corrected for ties for semi-quantitative characters with many tied observations.
Common statistics and statistical tests were calculated using the Statistica software system (Statsoft Inc., 2005) and SPSS 8·0 program (SPSS Inc., 1998). Ordination and clustering methods were calculated using the SYN-TAX 2000 program (Podani, 2000).
RESULTS
Variability range and statistical distribution
Of the 172 plants investigated, 171 were found to be regular tetraploids, as previously documented from this locality by Šmarda and Kočí (2003) and Šmarda and Bureš (2006). One hexaploid plant, without any evident morphological differences from the tetraploids, was documented (Fig. 1). Because of the uniqueness of this sample and the lack of any other material to which it can be compared, it was omitted from the analyses. A maximum 1·115-fold difference in genome size was observed in the 171 tetraploid samples from the studied population (Fig. 1; see Supplementary Information 1, online); further statistical parameters are indicated in Fig. 3. The difference of some of the most contrasting samples was demonstrated as a clear double peak in the simultaneous measurement of both samples (Fig. 2), which was in a similar range to the difference between samples obtained by individual measurements (e.g. 1·106-fold difference shown in Fig. 2 compared with a 1·098-fold difference for the same samples in individual measurements, Supplementary Information 1, online).
The statistical distribution was found to be continuous and fits best to the extreme (Gumbel) distribution (K–S D = 0·039; Fig. 3). The histogram of genome sizes has a conspicuous central tendency and is slightly positively skewed (Fig. 3), in which manner it differs from the normal distribution (P < 0·001). In comparison, the distribution of genome sizes in the whole Angiosperms is much more extremely positively skewed and fits best to the log-normal distribution (K–S D = 0·058).
Spatial distribution pattern
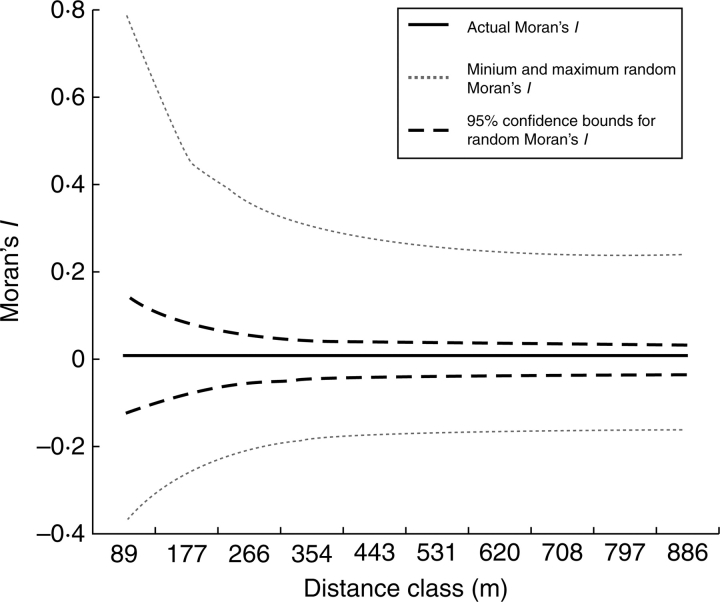
Even samples from the within the same plot were frequently found to vary considerably in genome size, and the total variability between plots was nearly the same as within plots (ANOVA, F = 1·028; P = 0·441). This result itself argues for the absence of a global, small-scale spatial correlation of the data. In addition no global spatial relation was found among the sampling plots in the distribution of: (1) average genome size in a plot; (2) standard deviation of genome sizes within a plot; (3) plants with the highest genome size within the plot; or (4) plants with the smallest genome size within the plot (Figs 1 and 4). The global Moran's spatial autocorrelation index (Moran's I) for the genome size of all the 171 tetraploid samples and for all four genome-size characters of all 57 plots was clearly non-significant for the overall locality (P > 0·05). Similarly, the spatial autocorrelation in distribution of genome size from any of the distance classes in the Moran correlograms did not differ from the random one (e.g. Fig. 5).
Fig. 5.
Moran correlogram of genome size of 171 tetraploid samples. No spatial autocorrelation was found in the distribution of genome size in any of the ten distance classes (P > 0·05). Similar spatially unautocorrelated patterns in the Moran correlograms are produced by all the four genome size parameters of the plots shown in Fig. 4.
Ecologic adaptivity
No significant correlation was found between the gradient in ecologic conditions (vegetation cover) and any of the four selected genome size parameters of F. pallens (P > 0·1). In addition, no significant global differences were found among the five vegetation types recognized (both including and excluding outliers), and the four investigated genome size parameters (Fig. 6; P > 0·05). The only notable negative correlations appear between the altitude and maximum genome size observed in plots; however, this was non-significant when the Bonferoni adjustment for multiple comparison experiments was applied (Table 1). This correlation is caused mainly by three populations with rather high genome sizes situated in the south-westernmost, bottom part of the Sv. Kopeček Hill (populations 02, 10 and 13). Although the extreme climatical conditions of south-western rocky sites gives reasons for speculation about the existence of a local optimum for survival of F. pallens here, we rather assume this situation to be a sampling artefact. From the global point of view, genome size of F. pallens at this locality does not seem to be adaptive to any habitat parameter or vegetation type.
Fig. 6.
Differences in the four investigated population genome size parameters among the five vegetation types recognized by hierarchical clustering. No significant difference among the five types was detected (P > 0·05). Outlier clusters, omitted from the analysis, are marked × on the dendrogram. Box-plots show the median (—), the 25–75 % quantile range (box), non-outlier range (whiskers), and outliers (circles).
Table 1.
Correlation of genome size parameters of plots with some habitat characters
| Genome size per plot |
||||
|---|---|---|---|---|
| Mean | Maximum | Minimum | Standard deviation | |
| Spearman's rho correlation | ||||
| Altitude | −0·173 | −0·316 | 0·006 | −0·261 |
| Cover of shrubs | 0·195 | 0·179 | 0·203 | 0·122 |
| Cover of herbs | −0·029 | −0·038 | 0·088 | −0·145 |
| Cover of mosses | −0·119 | −0·154 | −0·039 | −0·169 |
| Cover of bare rock | 0·086 | 0·029 | −0·007 | 0·126 |
| Number of species | −0·180 | −0·253 | −0·121 | −0·179 |
| Kendall's tau b correlation | ||||
| Deviation of orientation from north | −0·028 | −0·079 | 0·003 | −0·008 |
| Deviation of orientation from south-west | 0·045 | 0·063 | 0·064 | 0·031 |
| Cover of Festuca pallens | −0·107 | −0·088 | −0·036 | −0·029 |
None of the correlations are significant, even at P < 0·4, after applying Bonferroni adjustment (separately for each correlation coefficient used).
DISCUSSION
The maximum 1·115-fold difference in the range of genome size observed in this study was very similar to the 1·121-fold difference observed previously in this population by Šmarda and Bureš (2006). The double peaks from the simultaneous measurements of the most-contrasting samples at this locality (Šmarda and Bureš, 2006, Fig. 4B; this study, Fig. 2) clearly verify the high intrapopulation differences in genome size. The repeated measurements and low coefficients of variance (1·82 % on average for all measurements) guarantee that the sample genome size estimations are reasonable and that the observed pattern of genome size distribution is credible.
Potential causes of the variability: transposable elements
The reason for the observed genome size variability may be because of different mechanisms of retrotransposon proliferation and deletion, which have caused the majority of recent genome-size variability in Angiosperms (Bennetzen et al., 2005). The transposons represent an important part of the grass genome, which is otherwise relatively constant (Devos and Gale, 2000; Gaut et al., 2000; Gaut, 2002), being composed of about the same 30 chromosome blocks rearranged in the genomes of all grasses (Devos and Gale, 2000; Caetano-Anolles, 2005). Most of the differences in genome size found in grasses are caused by the different frequency and activity of transposable elements (Gaut, 2002; Bennetzen et al., 2005; Jones and Pasakinskiene, 2005). The genome-size differentiation by transposons is caused by small cumulative changes of DNA content, including almost equal amplifications and deletions (Bennetzen et al., 2005). In Hordeum spontaneum, the activity of retrotransposons may differ in contrasting microhabitat conditions (Kalendar et al., 2000), which may be the case in such a contrasting locality as that examined in the present study.
Potential causes of the variability: B chromosomes
The influence of the presence of B chromosomes on the observed pattern would be expected to be minor. B chromosomes are frequently reported in Festuca (cf. Jones and Díez, 2004); however, their frequency in Festuca pallens seems to be low. Only a single B chromosome was reported as occurring in several plants by Šmarda and Kočí (2003), whilst the 1·115-fold difference observed here would need to be caused by the addition and presence of four B chromosomes (calculating the same DNA content for B chromosome as for the normal A chromosome). In addition, the considerable intraspecific genome size variability found within Festuca pallens plants with no Bs indicates that the majority of genome size variability in Festuca pallens is probably generated by another mechanism (Šmarda and Bureš, 2006). Moreover, recent studies have indicated that B chromosomes may be a regular complement of genomes derived from A chromosomes (Jones and Houben, 2003 and citations therein), even including transcribable genes (Leach et al., 2005). Both A and B chromosomes may interchange (Nokkala and Nokkala, 2004; Ribeiro et al., 2004) and variability in B chromosomes can probably be included within regular genome-size variability.
Ecological adaptivity
The statistical distribution of genome sizes found in the study population (Gumbel extreme distribution) is in some aspects similar to the log-normal distribution of genome sizes typical of the whole Angiosperms. Both distributions are positively skewed, which in practical terms means that large genomes occur more rarely. This situation may be either the result of processes of genome-size expansion being rare, or the result of a negative selection for plants with large genomes. The potential role of negative selection of large genomes and several ecological, evolutionary and phenotype constraints of large genomes have recently been documented by Knight et al. (2005). Many correlations of genome size with ecological and physiological characters generally argue for ecological adaptivity of genome size as a whole (cf. Knight et al., 2005). However, present assumptions are based on data from interpopulation and interspecies levels and thus need not necessarily be applicable at the intrapopulation level. In a previous study on Festuca pallens, the influence of several ecological microhabitat characters on genome size was shown to be minor (Šmarda and Bureš, 2006); this fact is corroborated by the present intrapopulation study. Although some correlations appear between maximum genome size and genome-size variability with the altitude of plots, it would not be expected that the altitudinal differnece of 90 m in the present study would significantly affect the distribution of genome sizes. As indicated by detailed analysis, the existence of a gradient in the distribution of genome size in the south-western part of the locality and the overall correlation of genome size with altitude were caused by three plots with large genome sizes situated on the bottom part of the slopes. Although this may not be excluded as a case of local adaptivity or local natural selection, we have no further support for this and instead assume it to be a mathematical or sampling artefact. Based on the current observed random spatial distribution of genome sizes, and in view of the overall lack of correlations of genome sizes with habitat conditions, genome size seems to be non-adaptive in the studied population.
Selective significance
Current theory presumes that genome size cannot be of selective significance in the initial stages of genome-size diversification (Gregory, 2001; Bennetzen et al., 2005). Bennetzen et al. (2005) considered that genome size within a population could be accumulated exclusively by small and chance differences of transposon activity and similar variability in mechanisms for DNA removal, and that these differences could lead to a large degree of genome-size variation in an environment where genome size is not of selective significance. Major differences in genome size appear, and the role of selection becomes important, only when the environment of such populations changes dramatically (Bennetzen et al., 2005). In a long-term, environmentally stable population, this process may result in a continuous distribution of the genome size in which the frequency of both deviations from the common genome size, i.e. larger or smaller genomes, will occur with decreasing frequency. This expected model is very similar to the real statistical distribution of genome sizes in the Mikulov–Sv. Kopeček population studied here.
Another consequence resulting from the hypothesis of Bennetzen et al. (2005) is that relict populations, growing in long-term, stable environmental conditions, may preserve higher genome-size variability. With the exception of the highly variable population of Festuca pallens from Mikulov, Šmarda and Bureš (2006: Table 1) found the variability of the other four populations in the Pálava Hills to be very low. Of the localities in the Pálava Hills, Mikulov is the largest rocky steppe with extreme xeric conditions (especially on south-western exposed slopes), providing relatively stable conditions, which also allow the habitat's survival during cold climate periods. Compared with the other important rocky steppes and localities studied by Šmarda and Bureš (2006), the steppe in Mikulov preserves several relicts of of this habitat (e.g. Stipa eriocaulis, Iris humilis subsp. arenaria, Fumana procumbens, Globularia bisnagarica, Orobanche artemisiae-campestris), which are lacking or are only rarely distributed in the rest of the Pálava Hills. This fact may indicate that the locality is really more relict and has experienced more long-term stable environmental conditions than the remainder of the Pálava Hills, and that the variability in genome size may reflect the history of the locality. Šmarda and Bureš (2006) found that plants of Festuca pallens with larger genomes prevail in relict habitats and in refugial areas in the south-western parts of its natural range (covered by periglacial steppe at the end of last Ice Age), these being areas with relatively long-term conserved environments. In light of the previous discussion and the hypothesis of Bennetzen et al. (2005), negative selection for larger genomes during the large environmental changes of the last Ice Age is one of the possible reasons for this situation.
Although recent experimental data on intraspecific genome size variability in Festuca pallens (Šmarda and Bureš, 2006; this study) corroborate the current hypothesis of genome-size evolution in Angiosperms (Bennetzen et al., 2005), this is practically the only example avaialable, and further studies are needed to better confirm these conclusions.
SUPPLEMENTARY INFORMATION
Supplementary information is available online at http://aob.oxfordjournals.org/, and shows (1) the results of the flow cytometry measurements; (2) the characters of the plots – relative genome size data, geographical coordinates, ecological and vegetation characters; and (3) the phytosociologic relevés of the sampling plots.
ACKNOWLEDGEMENTS
This study was supported by the Ministry of Education of the Czech Republic; grants MSM0021622416 and LC06073.
LITERATURE CITED
- Auquier P. Biologie de la reproduction dans le genre Festuca L. (Poaceae): 1. Systemes de pollinisation. Bulletin de la Société Royale de Botanique de Belgique. 1977;110:129–150. [Google Scholar]
- Bancheva S, Greilhuber J. Genome size in Bulgarian Centaurea s.l. (Asteraceae) Plant Systematics and Evolution. 2006;257:95–117. [Google Scholar]
- Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society of London, Series B. 1972;181:109–135. doi: 10.1098/rspb.1972.0042. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Leitch I. Plant DNA C-values Database. 2005 doi: 10.1007/978-1-0716-3389-2_9. http://www.rbgkew.org.uk/cval/homepage.html. (release 4·0, October 2005) [DOI] [PubMed] [Google Scholar]
- Bennett MD, Bhandol P, Leitch IJ. Nuclear DNA amounts in angiosperms and their modern uses – 807 new estimates. Annals of Botany. 2000;86:859–909. [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta-Dukát Z, Chytrý M, Hájková P, Havlová M. Vegetation of lowland wet meadows along a climatic continentality gradient in Central Europe. Preslia. 2005;77:89–111. [Google Scholar]
- Bureš P, Pavlíček T, Horová L, Nevo E. Microgeographic genome size differentiation of the carob tree, Ceratonia siliqua, at ‘Evolution Canyon’, Israel. Annals of Botany. 2004a;93:529–535. doi: 10.1093/aob/mch074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureš P, Wang Y-F, Horová L, Suda J. Genome size variation in Central European species of Cirsium (Compositae) and their natural hybrids. Annals of Botany. 2004b;94:353–363. doi: 10.1093/aob/mch151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres ME, De Pace C, Scarascia Mugnozza GT, Kotsonis P, Ceccarelli M, Cionini PG. Genome size variations within Dasypyrum villosum: correlations with chromosomal traits, environmental factors and plant phenotypic characteristics and behaviour in reproduction. Theoretical and Applied Genetics. 1998;96:559–567. [Google Scholar]
- Caetano-Anolles G. Grass evolution inferred from chromosomal rearrangements and geometrical and statistical features in RNA structure. Journal of Molecular Evolution. 2005;60:635–652. doi: 10.1007/s00239-004-0244-z. [DOI] [PubMed] [Google Scholar]
- Chase MW, Hanson L, Albert VA, Whitten WM, Williams NH. Life history evolution and genome size in subtribe Oncidiinae (Orchidaceae) Annals of Botany. 2005;95:191–199. doi: 10.1093/aob/mci012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Gale MD. Genome relationships: the grass model in current research. Plant Cell. 2000;12:637–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESRI Inc. 2004 ArcMap, version 8·3. License type: ArcInfo.www.esri.com . [Google Scholar]
- Gaut BS. Evolutionary dynamics of grass genome. New Phytologist. 2002;154:15–28. [Google Scholar]
- Gaut BS, d'Ennequin ML, Peek AS, Sawkins MC. Maize as a model for the evolution of plant nuclear genomes. Proceedings of the National Academy of Sciences of the USA. 2000;97:7008–7015. doi: 10.1073/pnas.97.13.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Reviews. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size: a critical reassessment. Annals of Botany. 1998;82(Suppl. A):27–35. [Google Scholar]
- Greilhuber J. Intraspecific variation in genome size in Angiosperms: identifying its existence. Annals of Botany. 2005;95:91–98. doi: 10.1093/aob/mci004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotkopp E, Rejmanek M, Sanderson MJ, Rost TL. Evolution of genome size in pines (Pinus) and its life-history correlates: Supertree analyses. Evolution. 2004;58:1705–1729. doi: 10.1111/j.0014-3820.2004.tb00456.x. [DOI] [PubMed] [Google Scholar]
- Hiremath SC, Nagasampige MH. Genome size variation and evolution in some species of Dalbergia Linn.f. (Fabaceae) Caryologia. 2004;57:367–372. [Google Scholar]
- Hood GM. 2005 PopTools, version 2·6·9. Available on the internet. URL http://www.cse.csiro.au/poptools . [Google Scholar]
- Jackson DA. Stopping rules in principal components analysis: a comparison of heuristical and statistical approaches. Ecology. 1993;74:2204–2214. [Google Scholar]
- Jakob SS, Meister A, Blattner FR. The considerable genome size variation of Hordeum species (Poaceae) is linked to phylogeny, life form, ecology and speciation rates. Molecular Biology and Evolution. 2004;21:860–869. doi: 10.1093/molbev/msh092. [DOI] [PubMed] [Google Scholar]
- Jones N, Houben A. B chromosomes in plants: escapees from the A chromosome genome? Trends in Plant Sciences. 2003;8:417–423. doi: 10.1016/S1360-1385(03)00187-0. [DOI] [PubMed] [Google Scholar]
- Jones N, Pasakinskiene I. Genome conflict in the Gramineae. New Phytologist. 2005;165:391–409. doi: 10.1111/j.1469-8137.2004.01225.x. [DOI] [PubMed] [Google Scholar]
- Jones RN, Díez M. The B chromosome database. Cytogenetic and Genome Research. 2004;106:149–150. doi: 10.1159/000079280. (http://www.bchromosomes.org/bdb/ ) [DOI] [PubMed] [Google Scholar]
- Kalendar R, Tanskanen J, Immonen S, Nevo E, Schulman AH. Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proceedings of the National Academy of Sciences of the USA. 2000;97:6603–6607. doi: 10.1073/pnas.110587497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapraun DF. Nuclear DNA content estimates in multicellular eukaryotic green, red and brown algae: phylogenetic considerations. Annals of Botany. 2005;95:7–44. doi: 10.1093/aob/mci002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany. 2005;95:177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubát K, editor. Praha: Academia; 2002. Klíč ke květeně České republiky. [Google Scholar]
- Leach CR, Houben A, Field B, Pistrick K, Demidov D, Timmis JN. Molecular evidence for transcription of genes on a B chromosome in Crepis capillaris. Genetics. 2005;171:269–278. doi: 10.1534/genetics.105.043273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Legendre L. 2nd edition. Amsterdam: Elsevier; 1998. Numerical ecology. [Google Scholar]
- Leitch IJ, Soltis DE, Soltis PS, Bennett MD. Evolution of DNA amounts across land plants (Embryophyta) Annals of Botany. 2005;95:207–217. doi: 10.1093/aob/mci014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine N. CrimeStat III: a spatial statistics program for the analysis of crime incident locations. Houston/Washington, DC: Ned Levine & Associates/National Institute of Justice; 2004. [Google Scholar]
- Loureiro J, Rodriguez E, Doležel J, Santos C. Flow cytometric and microscopic analysis of the effect of tannic acid on plant nuclei and estimation of DNA content. Annals of Botany. 2006;98:515–527. doi: 10.1093/aob/mcl140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot M, Barre P, Louarn J, Duperray C, Hamon S. Nucleus-cytosol interactions – a source of stoichiometric error in flow cytometric estimation of nuclear DNA content in plants. Annals of Botany. 2000;86:309–316. [Google Scholar]
- Nokkala S, Nokkala C. Interaction of B chromosomes with A or B chromosomes in segregation in insects. Cytogenetic and Genome Research. 2004;106:394–397. doi: 10.1159/000079317. [DOI] [PubMed] [Google Scholar]
- Obermayer R, Leitch IJ, Hanson L, Bennett MD. Nuclear DNA C-values in 30 species double the familial representation in pteridophytes. Annals of Botany. 2002;90:209–217. doi: 10.1093/aob/mcf167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohri D. Genome size variation and plant systematics. Annals of Botany. 1998;82(A):75–83. [Google Scholar]
- Podani J. Introduction to the exploration of multivariate biological data. Leiden: Backhuys Publishers; 2000. [Google Scholar]
- Price HJ, Dillon SL, Hodnett G, Rooney WL, Ross L, Johnston JS. Genome evolution in the genus Sorghum (Poaceae) Annals of Botany. 2005;95:219–227. doi: 10.1093/aob/mci015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro T, Pires B, Delgado M, Viegas W, Jones N, Morais-Cecilio L. Evidence for ‘cross-talk’ between A and B chromosomes of rye. Proceedings of the Royal Society of London, Series B. 2004;271(6):482–484. doi: 10.1098/rsbl.2004.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricroch A, Yockteng R, Brown SC, Nadot S. Evolution of genome size across some cultivated Allium species. Genome. 2005;48:511–520. doi: 10.1139/g05-017. [DOI] [PubMed] [Google Scholar]
- Šmarda P. DNA ploidy levels and intraspecific DNA content variability in Romanian fescues (Festuca L., Poaceae) measured in fresh and herbarium material. Folia Geoboanica. 2006;41:417–432. [Google Scholar]
- Šmarda P, Bureš P. Intraspecific DNA content variability in Festuca pallens on different geographical scales and ploidy levels. Annals of Botany. 2006;98:665–678. doi: 10.1093/aob/mcl150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Kočí K. Chromosome number variability in Central European members of the Festuca ovina and F. groups (sect. Festuca) Folia Geobotanica. 2003;38:65–95. [Google Scholar]
- Šmarda P, Šmerda J, Knoll A, Bureš P, Danihelka J. Revision of Central European taxa of Festuca ser. Psammophilae Pawlus: morphometrical, karyological and AFLP analysis. Plant Systematics and Evolution. 2007 in press. [Google Scholar]
- SPSS Inc. Chicago: SPSS Inc; 1998. SPSS® Base 8·0. [Google Scholar]
- StatSoft Inc. 2005. STATISTICA (data analysis software system), version 7·1. www.statsoft.com . [Google Scholar]
- Suda J. Czech Republic: Ph.D. Thesis, Charles University, Prague; 2004. An employment of flow cytometry into plant biosystematics. [Google Scholar]
- Tracey R. Beiträge zur Karyologie, Verbreitung und Systematik des Festuca ovina Formenkreises im Osten Österreichs. Austria: University of Vienna; 1980. Ph.D. Thesis. [Google Scholar]
- Vilhar B, Vidic T, Jogan N, Dermastia M. Genome size and the nucleolar number as estimators of ploidy level in Dactylis glomerata in the Slovenian Alps. Plant Systematics and Evolution. 2002;234:1–13. [Google Scholar]
- Vizintin L, Javornik B, Bohanec B. Genetic characterization of selected Trifolium species as revealed by nuclear DNA content and ITS rDNA region analysis. Plant Science. 2006;170:859–866. [Google Scholar]
- Voglmayr H. Nuclear DNA amounts in mosses (Musci) Annals of Botany. 2000;85:531–546. [Google Scholar]
- Ward JH. Hierarchical grouping to optimize an objective function. Journal of the American Statistical Association. 1963;58:236–244. [Google Scholar]
- Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM. Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. American Journal of Botany. 2006;93:148–156. doi: 10.3732/ajb.91.3.439. [DOI] [PubMed] [Google Scholar]
- Westhoff V, van der Maarel E. The Braun–Blanquet approach. In: Whittaker RH, editor. Classification of plant communities. The Hague: D. W. Junk; 1978. pp. 287–399. [Google Scholar]
- Zoldoš V, Papeš D, Brown SC, Šiljak-Yakovev S. Genome size and base composition of seven Quercus species: inter- and intrapopulation variation. Genome. 1998;41:162–168. [Google Scholar]