Abstract
Background and Aims
The genus Salvia has traditionally included any member of the tribe Mentheae (Lamiaceae) with only two stamens and with each stamen expressing an elongate connective. The recent demonstration of the non-monophyly of the genus presents interesting implications for staminal evolution in the tribe Mentheae. In the context of a molecular phylogeny, the staminal morphology of the various lineages of Salvia and related genera is characterized and an evolutionary interpretation of staminal variation within the tribe Mentheae is presented.
Methods
Two molecular analyses are presented in order to investigate phylogenetic relationships in the tribe Mentheae and the genus Salvia. The first presents a tribal survey of the Mentheae and the second concentrates on Salvia and related genera. Schematic sketches are presented for the staminal morphology of each major lineage of Salvia and related genera.
Key Results
These analyses suggest an independent origin of the staminal elongate connective on at least three different occasions within the tribe Mentheae, each time with a distinct morphology. Each independent origin of the lever mechanism shows a similar progression of staminal change from slight elongation of the connective tissue separating two fertile thecae to abortion of the posterior thecae and fusion of adjacent posterior thecae. A monophyletic lineage within the Mentheae is characterized consisting of the genera Lepechinia, Melissa, Salvia, Dorystaechas, Meriandra, Zhumeria, Perovskia and Rosmarinus.
Conclusions
Based on these results the following are characterized: (1) the independent origin of the staminal lever mechanism on at least three different occasions in Salvia, (2) that Salvia is clearly polyphyletic, with five other genera intercalated within it, and (3) staminal evolution has proceeded in different ways in each of the three lineages of Salvia but has resulted in remarkably similar staminal morphologies.
Key words: Staminal morphology, Salvia, Mentheae, Dorystaechas, Meriandra, Perovskia, Rosmarinus, Zhumeria, Lepechinia, Melissa, key innovation, floral evolution
INTRODUCTION
The genus Salvia (Lamiaceae: tribe Mentheae) represents a cosmopolitan assemblage of nearly 1000 species displaying a remarkable diversity in growth forms, secondary compounds, floral morphology and pollination biology. Salvia has radiated extensively in three regions of the world: Central and South America (500 spp.), western Asia (200 spp.) and eastern Asia (100 spp.) (Alziar, 1988–1993). All these species display the unusual morphological character that has led to the long-standing assumption that Salvia is monophyletic: the significant elongation of the connective tissue of the two expressed anthers (Figs 1 and 2). The demonstration of the non-monophyly of the genus (Walker et al., 2004) has led to a reinvestigation of the defining character of the genus, the elongation of the connective tissue of the stamen, within Salvia and closely related genera in the Mentheae. This paper presents a molecular phylogeny of Salvia and related genera, characterizes the stamen morphology in the different clades of the genus Salvia and closely related genera, and interprets that stamen morphology in a phylogenetic context.
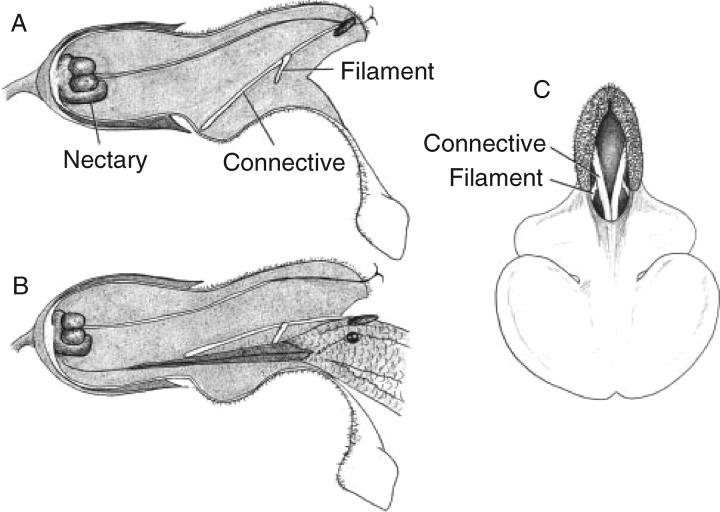
Fig. 1.
Stylized representation of the flower and lever mechanism of pollination of a hypothetical member of Salvia subgen. Calosphace (Salvia clade II). A flower prior to the activation of the lever mechanism (A). The pollinator enters the flower and activates the lever mechanism (B), depositing pollen on the head of the pollinator. (C) A ‘birds-eye’ view of the flower, with the fused posterior branches of the connective blocking access to the nectar at the base of the corolla (sketch by Cody Williams).
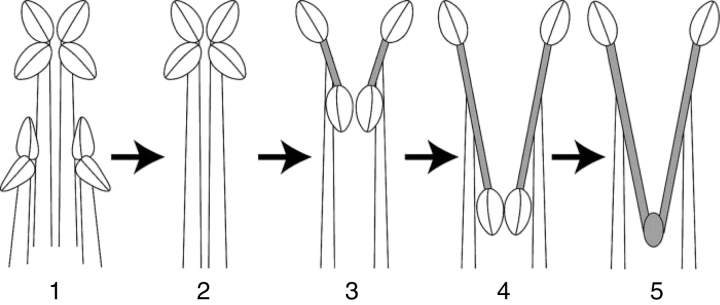
Fig. 2.
The generalized trend in stamen morphology seen within tribe Mentheae leading to that seen in Salvia. Shaded areas represent connective tissue. Step 2 (the functional loss of two of the four stamens) has apparently happened only once in the Salvia clade. The progression from step 2 to step 5 has happened on at least three independent occasions in the Salvia clade. Anterior thecae are on the top of each sketch, and the posterior thecae, which become entirely aborted and fused in step 5, are on the bottom of each sketch.
Mentheae (sensu Wagstaff et al., 1995) is a well-supported monophyletic tribe containing 73 genera within the subfamily Nepetoideae (Cantino et al., 1992; Wagstaff, 1992; Wagstaff et al., 1995; Walker et al., 2004; Bräuchler et al., 2005). Salvia is distinguished from the other 72 genera in the tribe Mentheae by having the two posterior stamens aborted, and the connective separating the thecae of the two expressed stamens significantly elongated (Fig. 2). It is the elongation of the staminal connective that allows the formation of the lever mechanism of pollination for which Salvia is best known (Fig. 1) (for thorough reviews, see Claßen-Bockhoff et al., 2003, 2004a). The significant species radiations that are correlated with the presence of the lever mechanism in Salvia (e.g. subgen. Calosphace – 500 spp.) suggest it may be the lever mechanism in a selective regime of pollination that is driving evolution in the group (Claßen-Bockhoff et al., 2004b). The significance of this lever mechanism to the reproductive biology in Salvia, first described by Sprengel (1793), has received considerable attention (Müller, 1873; Zalewska, 1928; Hruby, 1934; Werth, 1956; Baikova, 2002, 2004; Claßen-Bockhoff et al., 2003, 2004a; Reith et al., 2006; Wester and Claßen-Bockhoff, 2006). Himmelbaur and Stibal (1932–1934) directly addressed staminal evolution in Salvia, presenting a hypothesis of parallel evolution of the lever mechanism (from a common ancestor) in the New World and the Old World. This papers presents the first, robust, Salvia-wide molecular phylogeny with sampling across the tribe Mentheae directly to evaluate Himmelbaur and Stibal's (1932–1934) hypothesis of independent origins of the lever mechanism in Salvia. Additionally, the following questions are addressed and answered. How many times has an elongate connective originated in Salvia and related genera? How many times has the staminal lever mechanism originated in Mentheae? What are the most closely related genera to Salvia? What are the trends in staminal evolution within Salvia?
MATERIALS AND METHODS
Taxa sampling
Sampling within the genus Salvia attempted to include as wide a morphological and biogeographical diversity as possible. Within the New World, there is a high level of confidence that the sampling represents every major clade of Salvia. In addition to the monophyly of the 500 species in the subgenus Calosphace being supported by morphology (Bentham, 1876; Epling, 1939; Claßen-Bockhoff et al., 2004a), the monophyly is supported by molecular data collected as part of this project and by a continuing project sampling 200 species in the subgenus (our unpubl. data). Sampling included 20 of the remaining 28 non-subgenus Calosphace species of Salvia in the New World. The Old World represents a larger challenge for sampling within Salvia, as the subgeneric groups are less well established. Sampling was attempted from each of the informal subgeneric groups suggested by Hedge (1974a, b, 1982a, b) based on morphology. The 26 Old World Salvia sampled certainly do not represent every major clade of Salvia present. However, the sampling includes southern African, northern African, Mediterranean, European, west Asian, central Asian and east Asian species of Salvia.
Nomenclature for Salvia follows that suggested by Alziar (1988–1993). One hundred and forty-four trnL-F sequences, 139 nuclear rDNA internal transcribed spacer (ITS) sequences and 85 psbA-trnH sequences representing 38 genera and 144 species were obtained in this project (Table 1). Accessions, vouchers, locality and GenBank numbers are also given in Table 1. The data matrix for the ‘Mentheae-wide analysis’ combined ITS, psbA-trnH and trnL-F and consisted of 84 taxa. The data matrix for the ‘Salvia clade analysis’ combined ITS and trnL-F and comprised 93 taxa. Studies have demonstrated the monophyly of the tribe Mentheae, as well as its close relation to the tribe Ocimeae (Paton et al., 2004; Walker et al., 2004; Bräuchler et al., 2005). Outgroups chosen for the Mentheae-wide analysis were Ocimum basilicum and Hyptis alata, both from the tribe Ocimeae. Within the Mentheae, 34 genera were sampled that represented all subtribes of Mentheae. Within the ‘Salvia clade analysis’, sampling concentrated on the genus Salvia (82 species sampled) and all genera indicated by the ‘Mentheae-wide analysis’ to be closely related to Salvia. Horminum pyrenaicum was selected as the outgroup for the ‘Salvia clade analysis’ based on the results of the ‘Mentheae-wide analysis’.
Table 1.
Plant materials included in this study
| Men. | Sal. | Taxon | Locality | Voucher | psbA-trnH sequence | ITS sequence | trnL-trnF sequence |
|---|---|---|---|---|---|---|---|
| Yes | Acanthomintha lanceolata Curran | herb-MO 3133280 | Crosby&Morin 14383 | DQ667418 | DQ667333 | DQ667522 | |
| Yes | Agastache urticifolia Kunth | wild-USA (WIS) | JBW 815 | DQ667357 | DQ667247 | AY570452 | |
| Yes | Cleonia lusitanica L. | herb-F | D. Sanches & R. Garilan 20-VI-8? | DQ667395 | DQ667309 | DQ667495 | |
| Yes | Clinopodium ashei Small | wild-USA (WIS) | JBW 742 | DQ667348 | DQ667237 | DQ667437 | |
| Yes | Clinopodium coccineum Kuntze | wild-USA (WIS) | JBW 741 | DQ667344 | DQ667233 | DQ667433 | |
| Yes | Clinopodium vulgare L. | wild-USA (WIS) | JBW 3227 | DQ667409 | DQ667324 | DQ667513 | |
| Yes | Collinsonia canadensis L. | wild-USA (WIS) | JBW 958 | DQ667358 | DQ667248 | AY570453 | |
| Yes | Conradina canescens A. Gray | wild-USA (WIS) | JBW 604 | DQ667349 | DQ667238 | DQ667438 | |
| Yes | Cunila galioides Benth. | wild-Argentina (WIS) | Sytsma 7247 | DQ667391 | DQ667305 | DQ667491 | |
| Yes | Cunila incana Benth. | wild-Argentina (WIS) | Sytsma 7224 | DQ667403 | DQ667316 | DQ667504 | |
| Yes | Dicerandra oderatissima R.M. Harper | wild-USA (WIS) | JBW 1063 | DQ667345 | DQ667234 | DQ667434 | |
| Yes | Yes | Dorystaechas hastata Boiss. & Heldr. Ex Benth. | cult-RBG-Edinburgh | 1972–0177D | DQ667360 | DQ667252 | AY570454 |
| Yes | Drepanocaryum sewerzowskii (Regel) Pojark. | herb-MO 5201825 | Rinziraeva 7540 | DQ667413 | DQ667328 | DQ667517 | |
| Yes | Glechoma hederacea L. | cult-USA (WIS) | JBW 2579 | DQ667355 | DQ667245 | AY570455 | |
| Yes | Glechon marifolia Benth. | wild-Argentina (WIS) | Sytsma 7214 | DQ667390 | DQ667303 | DQ667489 | |
| Yes | Glechon thymoides Spreng. | herb-F | CA Mondin 1421 | DQ667396 | DQ667310 | DQ667496 | |
| Yes | Hedeoma costatum (Greene) Irving | wild-USA (WIS) | JBW 2143 | DQ667347 | DQ667236 | DQ667436 | |
| Yes | Hoehnea epilobioides (Epl.) Epl. | herb-F | G. Hatschbach 8/3/1984 | DQ667397 | DQ667497 | ||
| Yes | Yes | Horminum pyrenaicum L. | cult-RBG-Edinburgh | 1997–2109a | DQ667365 | DQ667257 | AY570456 |
| Yes | Hyptis alata (Raf.) Shinners | wild-USA (WIS) | JBW 1019 | DQ667346 | DQ667235 | DQ667435 | |
| Yes | Lepechinia calycina Epl. | wild-USA (WIS) | JBW 3186 | DQ667394 | DQ667308 | DQ667494 | |
| Yes | Yes | Lepechinia chamaedryoides Epl. | cult-USA (WIS) | JBW 2537 | DQ667343 | DQ667231 | AY570459 |
| Yes | Yes | Lepechinia conferta Epl. | herb-F | Alonso 8376 | DQ667393 | DQ667307 | DQ667493 |
| Yes | Yes | Lepechinia lancifolia Epl. | herb-F | Smith 444 | DQ667392 | DQ667306 | DQ667492 |
| Yes | Lycopus uniflorus Michx. | wild-USA (WIS) | JBW 2586 | DQ667389 | DQ667302 | DQ667488 | |
| Yes | Yes | Melissa officinalis L. | cult-USA (WIS) | JBW 2575 | DQ667387 | DQ667291 | DQ667477 |
| Yes | Mentha arvensis L. | wild-USA (WIS) | JBW 3228 | DQ667410 | DQ667325 | DQ667514 | |
| Yes | Mentha spicata L. | cult-USA (WIS) | JBW 2566 | DQ667354 | DQ667244 | AY570461 | |
| Yes | Yes | Meriandra bengalensis (Roxb.) Benth | herb-MO 2633828 | Lavranus & Newton 15796 | DQ667414 | DQ667329 | DQ667518 |
| Yes | Monarda fistulosa L. | wild-USA (WIS) | JBW 3223 | DQ667405 | DQ667318 | DQ667506 | |
| Yes | Nepeta cataria L. | wild-USA (WIS) | JBW 3054 | DQ667388 | DQ667301 | DQ667487 | |
| Yes | Ocimum basilium L. | cult-USA (WIS) | JBW 2557 | DQ667350 | DQ667240 | AY570462 | |
| Yes | Origanum vulgare L. | cult-USA (WIS) | JBW 2567 | DQ667353 | DQ667243 | AY570463 | |
| Yes | Perilla frutescens (L.) Britton | cult-USA (WIS) | JBW 1078 | DQ667356 | DQ667246 | DQ667439 | |
| Yes | Yes | Perovskia atriplicifolia Benth. | cult-USA (WIS) | JBW 2524 | DQ667341 | DQ667223 | AY570464 |
| Yes | Yes | Perovskia scrophulariaefolia Bunge | herb-MO 5201778 | Kinziraeva 6751 | DQ667415 | DQ667330 | DQ667519 |
| Yes | Pogogyne floribunda Jokerst | herb-MO 4282587 | Bartholemew 6021 | DQ667416 | DQ667331 | DQ667520 | |
| Yes | Poliomintha palmeri Hemsl | herb-F | Diggs Nee 2531 | DQ667398 | DQ667311 | DQ667498 | |
| Yes | Prunella vulgaris L. | wild-USA (WIS) | JBW 3225 | DQ667407 | DQ667508 | ||
| Yes | Pycnanthemum virginianum (L.) Durand & Jacks ex Rob & Fernald | wild-USA (WIS) | JBW 3224 | DQ667406 | DQ667319 | DQ667507 | |
| Yes | Rhododon ciliatus (Benth.) Epl. | herb-F | W.C. Holmes 8215 | DQ667399 | DQ667312 | DQ667499 | |
| Yes | Yes | Rosmarinus officinalis L. | cult-USA (WIS) | JBW 2558 | DQ667351 | DQ667241 | AY570465 |
| Yes | Yes | Salvia aegyptiaca L. | herb-E | McLeish 3728 | DQ667380 | DQ667285 | DQ667470 |
| Yes | Yes | Salvia aethiopis L. | wild-Armenia (MJG) | Hellwig 26/6/02 | DQ667370 | DQ667272 | AY570466 |
| Yes | Yes | Salvia apiana Jepson | wild-USA (WIS) | JBW 2509 | DQ667338 | DQ667214 | DQ667425 |
| Yes | Yes | Salvia aristata Aucher | herb-E | Wedelbo & Assadi s.n. | DQ667375 | DQ667280 | DQ667465 |
| Yes | Salvia atrocyanea Epl. | wild-Bolivia (MJG) | P. Wester 3 | DQ667270 | DQ667456 | ||
| Yes | Yes | Salvia aucheri var. canescens Benth. | herb-E | Archibald 7670 | DQ667381 | DQ667286 | DQ667471 |
| Yes | Yes | Salvia austriaca Jacq. | cult-Mainz. Bot. Gar. | Claßen-Bockhoff – 2004 | DQ667408 | DQ667323 | DQ667512 |
| Yes | Salvia axillaris Moc. et Sesse ex Benth. | Wild-Mex (WIS) | JBW 3038 | DQ667294 | DQ667480 | ||
| Yes | Yes | Salvia azurea Michx. ex Lam. | wild-USA (WIS) | JBW 3222 | DQ667404 | DQ667317 | DQ667505 |
| Yes | Salvia bangii Rusby | wild-Bolivia (MJG) | P. Wester 10 | DQ667263 | DQ667449 | ||
| Yes | Yes | Salvia cabulica Benth. | herb-E | Ghafoor & Goodman 5148 | DQ667382 | DQ667287 | DQ667472 |
| Yes | Yes | Salvia cacaliifolia Benth. | cult-RBG-Edinburgh | 1959–9358A | DQ667367 | DQ667259 | DQ667445 |
| Yes | Salvia californica Brandegee | cult-USA (WIS) | JBW 2520 | DQ667213 | DQ667424 | ||
| Yes | Yes | Salvia canariensis L. | cult-RBG-Edinburgh | 1986–0478 | DQ667364 | DQ667256 | AY570469 |
| Yes | Salvia candicans Mart. & Gal. | Wild-Mex (WIS) | JBW 3001 | DQ667299 | DQ667485 | ||
| Yes | Yes | Salvia candidissima Vahl. | cult-RBG-Edinburgh | 1999–2202A | DQ667368 | DQ667261 | DQ667447 |
| Yes | Salvia cedrosensis Greene | cult-USA (WIS) | JBW 2539 | DQ667228 | AY570470 | ||
| Yes | Salvia chionopeplica Epl. | cult-USA (WIS) | JBW 2545 | DQ667227 | AY570472 | ||
| Yes | Salvia clevelandii (Gray) Greene | wild-USA (WIS) | JBW 2508 | DQ667219 | AY570473 | ||
| Yes | Yes | Salvia cynica Dunn | herb-MO 4026698 | Boufford&Bartholemew 24763 | DQ667417 | DQ667332 | DQ667521 |
| Yes | Yes | Salvia daghestanica Sosn. | cult-RBG-Edinburgh | 1988–2283A | DQ667366 | DQ667258 | DQ667444 |
| Yes | Yes | Salvia digitaloides Diels. | cult-RBG-Edinburgh | 1999–2200A | DQ667363 | DQ667255 | AY570477 |
| Yes | Yes | Salvia disermas L. | herb-E | Goldblatt 7500 | DQ667385 | DQ667290 | DQ667475 |
| Yes | Salvia divinorum Epl. et Jativa | cult-USA (WIS) | JBW 3230 | DQ667249 | DQ667440 | ||
| Yes | Salvia dolomitica Codd | cult-USA (WIS) | JBW 3200 | DQ667322 | DQ667511 | ||
| Yes | Salvia dorrii (Kell.) Abrams | cult-USA (WIS) | JBW 2541 | DQ667229 | DQ667430 | ||
| Yes | Salvia eremostachya Jeps. | cult-USA (WIS) | JBW 2533 | DQ667232 | DQ667432 | ||
| Yes | Salvia fulgens Cav. | herb-WIS | 1967–1496A | DQ667251 | DQ667441 | ||
| Yes | Yes | Salvia garipensis E. Meyer ex Benth. | herb-E | Strohbach 149 | DQ667376 | DQ667281 | DQ667466 |
| Yes | Yes | Salvia glutinosa L. | cult-USA (WIS) | JBW 2568 | DQ667359 | DQ667250 | AY570480 |
| Yes | Yes | Salvia graciliramulosa Epl. et Jativa | wild-Bolivia (MJG) | P. Wester 14 | DQ667372 | DQ667276 | DQ667461 |
| Yes | Salvia greatai Brandegee | wild-USA (WIS) | JBW 2511 | DQ667339 | DQ667215 | AY570481 | |
| Yes | Salvia haenkei Benth. | wild-Bolivia (MJG) | P. Wester 71 | DQ667271 | DQ667457 | ||
| Yes | Salvia henryi Gray | wild-USA (WIS) | JBW 2516 | DQ667216 | AY570482 | ||
| Yes | Salvia hians Royle | cult-USA (WIS) | JBW 2577 | DQ667239 | AY570483 | ||
| Yes | Yes | Salvia hirtella Vahl. | wild-Peru (MJG) | Schmidt-Lebuhn 395 | DQ667411 | DQ667326 | DQ667515 |
| Yes | Yes | Salvia hydrangea Benth. | herb-E | Rechinger 47123 | DQ667383 | DQ667288 | DQ667473 |
| Yes | Salvia hydrangea Benth. | wild-Armenia (MJG) | Hellwig 6/18/02 | DQ667265 | DQ667451 | ||
| Yes | Salvia inconspicua Benth. | Wild-Mex (WIS) | JBW 3045 | DQ667298 | DQ667484 | ||
| Yes | Salvia lasiantha Benth. | Wild-Mex (WIS) | JBW 3009 | DQ667300 | DQ667486 | ||
| Yes | Salvia lavanduloides Kunth | Wild-Mex (WIS) | JBW 3044 | DQ667297 | DQ667483 | ||
| Yes | Salvia leucophylla Greene | Cult.-USA | JBW s.n. | DQ667210 | DQ667422 | ||
| Yes | Salvia mellifera Greene | wild-USA (WIS) | JBW 2550 | DQ667220 | DQ667427 | ||
| Yes | Salvia miltiorrhiza Bunge | herb-MO 04702028 | Wang Shilong s.n. | DQ667419 | DQ667334 | DQ667523 | |
| Yes | Salvia miltiorrhiza Bunge | herb-MO | Boufford et al. 26067 | DQ667379 | DQ667284 | DQ667469 | |
| Yes | Salvia mocinoi Benth. | wild-Mexico (MJG) | Crone 15/9/00 | DQ667274 | DQ667459 | ||
| Yes | Salvia mohavensis Greene | Cult.-USA | JBW s.n. | DQ667212 | DQ667423 | ||
| Yes | Salvia munzii Epl. | wild-USA (WIS) | JBW 2507 | DQ667224 | DQ667428 | ||
| Yes | Yes | Salvia officinalis L. | cult-USA (WIS) | JBW 2580 | DQ667342 | DQ667225 | AY570488 |
| Yes | Yes | Salvia orbignaei Benth. | wild-Bolivia (MJG) | P. Wester 43 | DQ667374 | DQ667279 | DQ667464 |
| Yes | Salvia ovalifolia St.-Hil. ex Benth | wild-Argentina (WIS) | Sytsma 7226 | DQ667315 | DQ667502 | ||
| Yes | Salvia oxyphora Briq. | wild-Bolivia (MJG) | P. Wester 16 | DQ667262 | DQ667448 | ||
| Yes | Salvia pachyphylla Epl. ex Munz | cult-USA (WIS) | JBW 2535 | DQ667230 | DQ667431 | ||
| Yes | Yes | Salvia patens Cav. | cult-RBG-Edinburgh | 1973–9197 | DQ667361 | DQ667253 | DQ667442 |
| Yes | Yes | Salvia penstemonoides Kunth et Bouche | cult-USA (WIS) | JBW 2578 | DQ667340 | DQ667221 | AY570489 |
| Yes | Salvia personata Epl. | wild-Bolivia (MJG) | P. Wester 17 | DQ667269 | DQ667455 | ||
| Yes | Yes | Salvia platystoma Epl. | wild-Bolivia (MJG) | P. Wester 18 | DQ667373 | DQ667277 | DQ667462 |
| Yes | Salvia polystachya Epl. | Wild-Mex (WIS) | JBW 3035 | DQ667292 | DQ667478 | ||
| Yes | Salvia procurrens Benth. | wild-Argentina (WIS) | Bonif 941 | DQ667304 | DQ667490 | ||
| Yes | Yes | Salvia prunelloides Kunth | wild-Mexico (MJG) | Crone 15/9/00 | DQ667371 | DQ667275 | DQ667460 |
| Yes | Yes | Salvia przewalskii Maxim. | cult-RBG-Edinburgh | 1993–2067A | DQ667362 | DQ667254 | DQ667443 |
| Yes | Salvia pubescens Benth. | Wild-Mex (WIS) | JBW 3043 | DQ667296 | DQ667482 | ||
| Yes | Salvia regla Cav. | Wild-Mex (WIS) | JBW 3019 | DQ667402 | DQ667503 | ||
| Yes | Yes | Salvia roborowskii Max. | herb-E | SBQ 852 | DQ667384 | DQ667289 | DQ667474 |
| Yes | Salvia roemeriana Scheele | wild-USA (WIS) | JBW 2515 | DQ667211 | AY570491 | ||
| Yes | Salvia rusbyi Britton ex Rusby | wild-Bolivia (MJG) | P. Wester 31 | DQ667278 | DQ667463 | ||
| Yes | Salvia rypara Briq. | wild-Bolivia (MJG) | P. Wester 32 | DQ667266 | DQ667452 | ||
| Yes | Salvia sagittata Ruiz et Pav. | herb-WIS | Weigend & Dostert 97/s.n. | DQ667260 | DQ667446 | ||
| Yes | Salvia santolinifolia Boiss. | herb-E | Runemark et al. 22255 | DQ667386 | DQ667476 | ||
| Yes | Salvia sclarea L. | cult-USA (WIS) | JBW 2527 | DQ667222 | AY570492 | ||
| Yes | Yes | Salvia scutellarioides Kunth. | wild-Peru (MJG) | Schmidt-Lebuhn 469 | DQ667412 | DQ667327 | DQ667516 |
| Yes | Salvia semiatrata Zucc. | herb-WIS | JBW 3041 | DQ667295 | DQ667481 | ||
| Yes | Yes | Salvia sessilifolia Baker | herb-E | Jongkind & Rapanarivo 929 | DQ667377 | DQ667282 | DQ667467 |
| Yes | Salvia sonomensis Greene | wild-USA (WIS) | JBW 2519 | DQ667218 | DQ667426 | ||
| Yes | Salvia sophrona Briq. | wild-Bolivia (MJG) | P. Wester 34 | DQ667268 | DQ667454 | ||
| Yes | Salvia stachydifolia Benth. | wild-Bolivia (MJG) | P. Wester 35 | DQ667267 | DQ667453 | ||
| Yes | Salvia summa A. Nelson | wild-USA (WIS) | JBW 1972 | DQ667217 | AY570496 | ||
| Yes | Yes | Salvia taraxacifolia Hook. fil. | cult-USA (WIS) | JBW 2521 | DQ667337 | DQ667209 | AY570497 |
| Yes | Yes | Salvia tetrodonta Hedge | herb-V 12403 | Podlech 18906 | DQ667421 | DQ667526 | |
| Yes | Salvia texana (Scheele) Torrey | wild-USA | P. Wester 362 | DQ667321 | DQ667510 | ||
| Yes | Salvia thymoides Benth. | wild-Mexico (MJG) | Crone 10/8/00 | DQ667273 | DQ667458 | ||
| Yes | Yes | Salvia trichocalycina Benth. | herb-E | Breckle 4963 | DQ667378 | DQ667283 | DQ667468 |
| Yes | Salvia tricuspidata Mart. & Gal. | wild-Mexico (WIS) | JBW 3037 | DQ667293 | DQ667479 | ||
| Yes | Salvia vaseyi (Porter) Parish | wild-USA (WIS) | JBW 2530 | DQ667226 | DQ667429 | ||
| Yes | Yes | Salvia verbascifolia M. Bieb. | wild-Armenia (MJG) | Hellwig 6/13/02 | DQ667369 | DQ667264 | DQ667450 |
| Yes | Salvia whitehousei Alziar | wild-USA (MJG) | P. Wester 352 | DQ667320 | DQ667509 | ||
| Yes | Schizonepeta multifida Huang, Feng & Wang | herb-F | Boyd 4805 | DQ667400 | DQ667313 | DQ667500 | |
| Yes | Thymus serpyllum L. | cult-USA (WIS) | JBW 2564 | DQ667352 | DQ667242 | AY570502 | |
| Yes | Zhumeria majudae Rech. F. & Wendelbo | herb-V 01176 | Ghazi s.n. | DQ667335 | DQ667524 | ||
| Yes | Zhumeria majudae Rech. F. & Wendelbo | herb-V 21730 | Wendelbo 15793 | DQ667420 | DQ667336 | DQ667525 | |
| Yes | Ziziphora taurica M. Bieb. | herb-F | I. Kapetariidis s.n. | DQ667401 | DQ667314 | DQ667501 |
Men., included in the Mentheae-wide analysis; Sal., included in the Salvia clade analysis. In the locality column: herb., herbarium material — herbarium code; wild, wild collected; cult., cultivated material.
Extractions, amplification and sequencing
Total genomic DNA was extracted using DNeasy Plant Mini kits (Qiagen, Valencia, CA, USA). Leaves used for DNA extractions were fresh, frozen, silica dried or obtained from herbarium specimens (see Table 1). Polymerase chain amplification (PCR) and cycle sequencing followed the methods described elsewhere (Conti et al., 1996; Givnish et al., 2000). PCR product was purified either with the QIAquick PCR purification kit (Qiagen) or with the AmPure PCR purification kit (Agencourt, Beverly, MA, USA). Sequenced products were precipitated in ethanol and sodium acetate to remove excess dye terminators or cleaned with the CleanSEQ Sequencing Reaction Clean-up system (Agencourt). Contiguous alignments were edited using Sequencher v. 3·0 (Gene Codes, Ann Arbor, MI, USA).
Sequences were aligned visually in SeAl v. 2·0a7 (Rambaut, 2001). Indels in the trnL-F data set were coded using the guidelines of Baum et al. (1994). Regions of ambiguous alignment were excluded from the analyses.
Phylogenetic analysis
Phylogenetic relationships within Salvia and Mentheae were evaluated in a two-step analysis. The first involved an 84-taxon data set (37 species of Salvia) using sequences from the chloroplast regions psbA-trnH, and trnL-F, and the nuclear ITS region (‘Mentheae-wide analysis’). The combined data sets were analysed using maximum parsimony (MP). The heuristic MP analysis (Fitch, 1971) in PAUP* 4·0b10 (Swofford, 2002) used 100 random addition sequences, with ten trees held at each step during stepwise addition, and tree bisection and reconnection (TBR) branch swapping to explore the possibility of multiple islands of most-parsimonious trees (Maddison, 1991). To assess congruence between the three data sets, 100 replicates of the partition homogeneity test (Farris et al., 1995) were conducted using a full heuristic search, simple taxon addition, TBR branch swapping and saving all most-parsimonious trees. Although the partition homogeneity test has been criticized (Yoder et al., 2001), the test has merit as a first assessment for congruence of data sets (Hipp et al., 2004). Bootstrap (Felsenstein, 1985) support values were used to evaluate support for relationships within the resulting trees. Bootstrap values were obtained through a heuristic search on all characters, with 1000 replicates and ten random addition sequences with TBR replicates with no more than 5000 trees saved per replicate.
The second analysis (the ‘Salvia clade analysis’) involved an expanded sampling within the genus Salvia (83 species of Salvia) and 11 other species representing all closely related genera. This analysis used the chloroplast trnL-F and the nuclear rDNA ITS regions and with the same methodologies used in the ‘Mentheae-wide analysis’ except for the inclusion of a maximum-likelihood (ML) analysis in addition to MP. Maximum-likelihood analyses were conducted on the ‘Salvia clade’ data set as implemented in PAUP*. Optimality criteria were explored using Modeltest v. 3·06 (Posada and Crandall, 1998). Heuristic ML searches with TBR branch-swapping were conducted.
Staminal morphological investigations
Staminal features investigated by this project are difficult to observe in herbarium specimens. Where fresh material was not available, literature that included detailed information regarding staminal morphology was used to determine the staminal form in each species (see Table 2). General stamen types were characterized for each major clade suggested by the molecular results and mapped onto the terminals in the cladograms (see Figs 4 and 5).
Table 2.
Stamen types of Salvia included in study. Types were determined by direct observation or through literature references that describe stamen form in detail
| Taxon | Stamen type | Reference* | Taxon | Stamen type | Reference |
|---|---|---|---|---|---|
| Salvia aegyptiaca L. | M | 6, 9 | Salvia mellifera Greene | H | 1, 2, 3 |
| Salvia aethiopis L. | B | 1, 8, 9 | Salvia miltiorrhiza Bunge | N | 11 |
| Salvia apiana Jepson | H | 1, 2, 3 | Salvia mocinoi Benth. | E | 13 |
| Salvia aristata Aucher | M | 9 | Salvia mohavensis Greene | H | 1, 2, 3 |
| Salvia atrocyanea Epl. | E | 13 | Salvia munzii Epl. | H | 1, 2, 3 |
| Salvia aucheri var. canescens Benth. | A | 14 | Salvia officinalis L. | A | 1 |
| Salvia austriaca Jacq. | B | 10 | Salvia orbignaei Benth. | E | 13 |
| Salvia axillaris Moc. et Sesse ex Benth. | G | 1, 13 | Salvia ovalifolia St.-Hil. ex Benth | E | 13 |
| Salvia azurea Michx. ex Lam. | E | 1, 13 | Salvia oxyphora Briq. | E | 13 |
| Salvia bangii Rusby | E | 13 | Salvia pachyphylla Epl. ex Munz | H | 1, 2, 3 |
| Salvia cabulica Benth. | A | 9 | Salvia patens Cav. | E | 1, 13 |
| Salvia cacaliifolia Benth. | E | 1, 13 | Salvia penstemonoides Kunth et Bouche | A | 1 |
| Salvia californica Brandegee | I | 1, 2, 3 | Salvia personata Epl. | E | 13 |
| Salvia canariensis L. | B | 6 | Salvia platystoma Epl. | E | 13 |
| Salvia candicans Mart. & Gal. | E | 1, 13 | Salvia polystachya Epl. | E | 1, 13 |
| Salvia candidissima Vahl. | B | 9 | Salvia procurrens Benth. | E | 13 |
| Salvia cedrosensis Greene | E | 1, 13 | Salvia prunelloides Kunth | E | 1, 13 |
| Salvia chionopeplica Epl. | H | 1, 2, 3 | Salvia przewalskii Maxim. | N | 7, 11 |
| Salvia clevelandii (Gray) Greene | H | 1, 2, 3 | Salvia pubescens Benth. | E | 1, 13 |
| Salvia cynica Dunn | N | 11 | Salvia regla Cav. | E | 1, 13 |
| Salvia daghestanica Sosn. | B | 10 | Salvia roborowskii Max. | N | 11 |
| Salvia digitaloides Diels. | N | 11 | Salvia roemeriana Scheele | A | 1, 4 |
| Salvia disermas L. | A (?) | 6 | Salvia rusbyi Britton ex Rusby | E | 13 |
| Salvia divinorum Epl. et Jativa | E | 1, 13 | Salvia rypara Briq. | E | 13 |
| Salvia dolomitica Codd | A | 1, 6 | Salvia sagittata Ruiz et Pav. | E | 1, 13 |
| Salvia dorrii (Kell.) Abrams | H | 1, 2, 3 | Salvia santolinifolia Boiss. | M | 9 |
| Salvia eremostachya Jeps. | H | 1, 2, 3 | Salvia sclarea L. | B | 1 |
| Salvia fulgens Cav. | E | 1, 13 | Salvia scutellarioides Kunth. | E | 13 |
| Salvia garipensis E. Meyer ex Benth. | B | 6 | Salvia semiatrata Zucc. | E | 1, 13 |
| Salvia glutinosa L. | N | 1, 7 | Salvia sessilifolia Baker | A | 6 |
| Salvia graciliramulosa Epl. et Jativa | E | 13 | Salvia sonomensis Greene | H | 1, 2, 3 |
| Salvia greatai Brandegee | I | 1, 2, 3 | Salvia sophrona Briq. | E | 13 |
| Salvia haenkei Benth. | E | 13 | Salvia stachydifolia Benth. | E | 13 |
| Salvia henryi Gray | A | 1, 4 | Salvia summa A. Nelson | A | 1, 4 |
| Salvia hians Royle | N | 1, 11 | Salvia taraxacifolia Hook. fil. | A | 1,6 |
| Salvia hirtella Vahl. | E | 13 | Salvia tetrodonta Hedge | M | 9, 12 |
| Salvia hydrangea Benth. | A | 9, 10 | Salvia texana (Scheele) Torrey | A | 5 |
| Salvia hydrangea Benth. | A | 9, 10 | Salvia thymoides Benth. | E | 1, 13 |
| Salvia inconspicua Benth. | E | 1, 13 | Salvia trichocalycina Benth. | M | 9 |
| Salvia lasiantha Benth. | E | 1, 13 | Salvia tricuspidata Mart. & Gal. | E | 1, 13 |
| Salvia lavanduloides Kunth | E | 1, 13 | Salvia vaseyi (Porter) Parish | H | 1, 2, 3 |
| Salvia leucophylla Greene | H | 1, 2, 3 | Salvia verbascifolia M. Bieb. | B | 10 |
| Salvia whitehousei Alziar | A | 5 |
*Reference: 1, personal observation by the first author; 2, Epling (1938); 3, Neissess (1983); 4, Walker and Elisens (2001); 5, Whitehouse (1949); 6, Hedge (1974a); 7, Claßen-Bockhoff et al. (2004b); 8, Hedge (1985); 9, Hedge (1982b); 10, Pobedimova (1954); 11, Xi-wen and Hedge (1994); 12, Hedge (1974b); 13, Epling (1939); 14, Hedge (1982a).
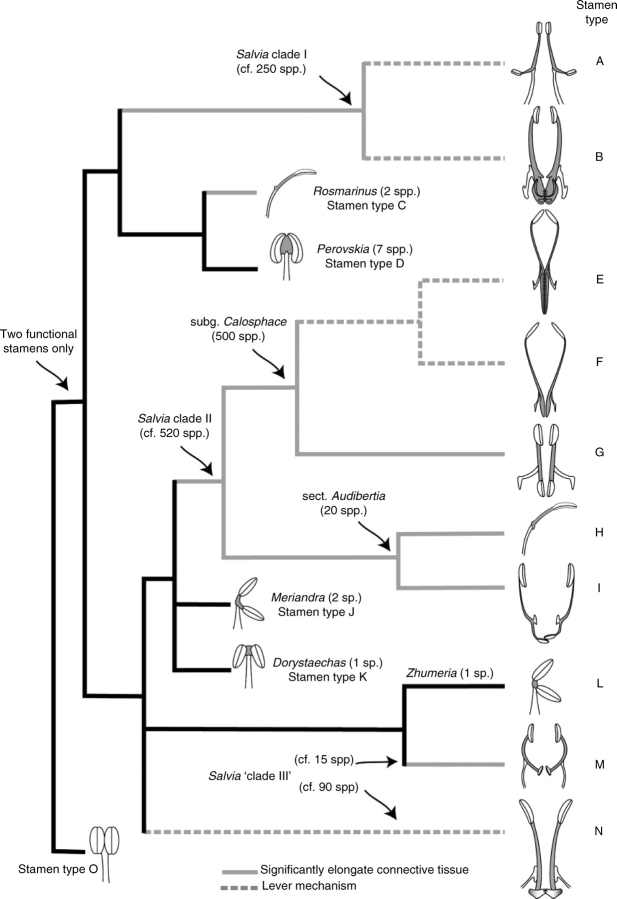
Fig. 4.
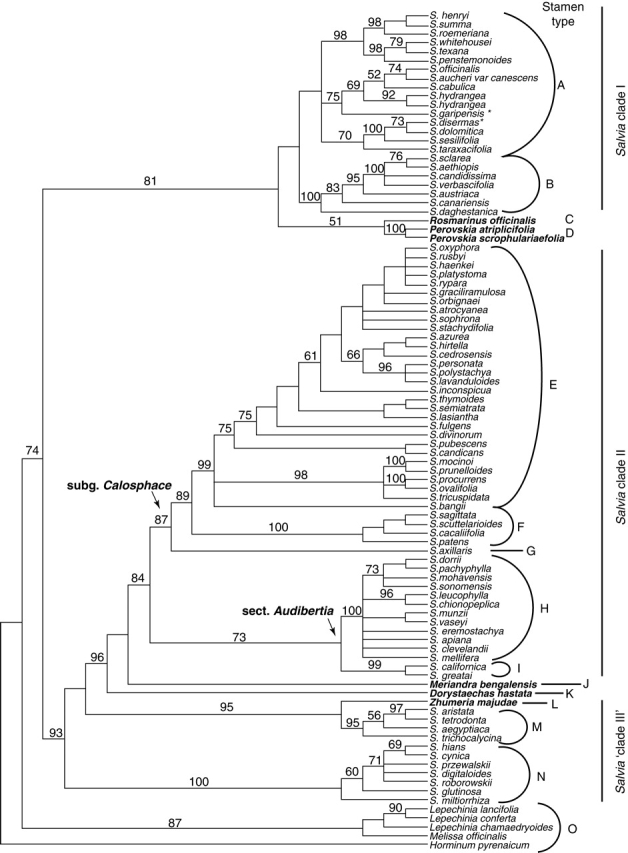
The ‘Salvia clade’ analysis. A two-region DNA, combined parsimony analysis of the chloroplast region trnL-F and the nuclear rDNA ITS. Strict consensus of over 100 000 equally parsimonious trees of 1489 steps. Bootstrap values above 50 % are shown above the branches. Stamen types corresponding to those in Fig. 5 and Table 2 are shown. Non-Salvia genera are highlighted in bold.
Fig. 5.
A summary of the cladogram shown in Fig. 4, with representations of the stamen types found in each clade. Shaded areas of the sketches represent connective tissue. Grey lines in the cladogram represent branches in which significantly elongate connectives are seen. Dashed lines in the cladogram represent lineages in which a lever mechanism is found. Total abortion of the posterior thecae and total fusion of the posterior thecae occurs only in stamen types B, E and N. Species numbers were hypothesized based on subgeneric groups suggested in the literature (Epling, 1938, 1939; Hedge 1974, 1982a, b). The two taxa with asterisks represent taxa not possessing the ‘typical’ stamen type A, and both possessing stamens with no expressed posterior thecae.
RESULTS
Analysis of Mentheae-wide data set
The aligned length of the trnL-F data set was 1137 base pairs (bp). With regions of ambiguous alignment or ambiguous sequences excluded, the total length of included characters was 1062 bp. Twenty indel events were scored for the trnL-F data set, of which 18 were parsimony- informative and were included in the analysis. Of the 1082 characters in the analysis 793 were constant, 117 variable characters were uninformative and 172 were parsimony-informative (15·9 %). Fitch parsimony analysis of the trnL-F region (uninformative characters excluded) found 4399 equally parsimonious trees of 332 steps (CI = 0·645, RI = 0·913, RC = 0·588).
The aligned length of the psbA-trnH data set was 624 bp. With regions of ambiguous alignment or ambiguous sequences excluded, the total length of included characters was 382 bp. Of the 382 characters in the analysis, 252 were constant, 58 variable characters were uninformative, and 72 were parsimony-informative (18·8 %). Fitch parsimony analysis of the psbA-trnH region (uninformative characters excluded) found 9470 equally parsimonious trees of 191 steps (CI = 0·586, RI = 0·864, RC = 0·507).
Nuclear rDNA ITS sequences were not obtained from Salvia santolinifolia, S. tetrodonta, S. regla, Hoehnea epilobioides or Prunella vulgaris. The aligned length of the nuclear ITS data set was 811 bp. With regions of ambiguous alignment or ambiguous sequences excluded, the total length of included characters was 659 bp. Of the 659 characters in the analysis, 364 were constant, 98 variable characters were uninformative and 197 were parsimony-informative (29·9 %). Fitch parsimony analysis of the ITS region found 5035 equally parsimonious trees of 1167 steps (CI = 0·336, RI = 0·652, RC = 0·219).
The combined trnL-F, psbA-trnH and nuclear ITS analysis generated 2123 characters, of which 1409 were constant, 273 were variable but uninformative and 441 were parsimony-informative (20·8 %). Fitch parsimony analysis of the three regions found 2094 equally parsimonious trees of 1737 steps (CI = 0·413, RI = 0·755, RC = 0·312).
The partition homogeneity test of the three data sets suggests significant incongruity between all three data sets (trnL-F, psbA-trnH and nuclear ITS) compared with random partitions of the same size (P < 0·01). Further analyses of the specific topological differences found between individual data sets indicate that none of the incongruent clades has bootstrap support above 50 % in the individual region analyses. The partition homogeneity test has been demonstrated to be overly sensitive in large data sets such as this (Hipp et al., 2004). Thus, the incongruence suggested by the partition homogeneity test may in fact not reflect genealogical discordance, but artefacts of the overly sensitive nature of the incongruence length difference (ILD) test in large datasets. Despite the incongruence of the data sets, all three data sets independently support the integrity of the ‘Salvia clade’ as discussed below, and the three specific clades of Salvia discussed in this paper. That is to say, each of the three data sets independently support Rosmarinus and Perovskia sister to Salvia clade I, Meriandra and Dorystaechas sister to Salvia clade II, and Zhumeria embedded in Salvia clade III (i.e. the source of the incongruence between the data sets lies elsewhere than the clades discussed herein). These facts combined with the high bootstrap support associated with each of the clades discussed in this paper in the combined analysis suggests that a ‘total evidence’, combined data set approach is justified.
The tribe Mentheae is supported at 100 % bootstrap in the strict consensus tree (Fig. 3). Within the Mentheae, a ‘Salvia clade’ is moderately supported (64 %) with the genera Lepechinia and Melissa appearing as likely sister genera (Fig. 3). For the purposes of this discussion, the term ‘Salvia clade’ is used to refer to the least inclusive clade which contains all members of Salvia. In addition to all Salvia, the ‘Salvia clade’ includes the genera Dorystaechas, Meriandra, Perovskia, Rosmarinus and Zhumeria (see Fig. 3). Three clades of Salvia are identified more closely related to one or more of these other genera than to the other major clades of Salvia; thus, Salvia is not monophyletic. Salvia clade I is strongly supported as monophyletic and together with the genera Rosmarinus and Perovskia form a monophyletic lineage (bootstrap = 94 %). Salvia clade II, likewise, forms a well-supported monophyletic lineage including two other genera, Meriandra and Dorystaechas (bootstrap = 100 %). Two remaining, well-supported lineages of Salvia, one of which includes the genus Zhumeria, occupy one of the few unresolved areas within the ‘backbone’ of the Salvia clade. These two are referred to as Salvia ‘clade III’ and could be either monophyletic or form a paraphyletic grade leading to Salvia clade II (Fig. 3).
Fig. 3.
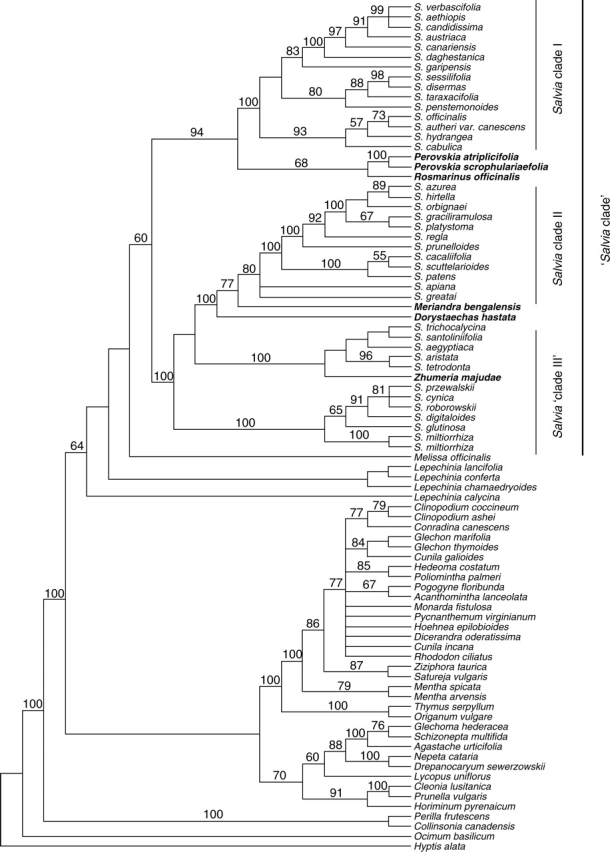
The ‘Mentheae-wide’ analysis. A three-region DNA, combined parsimony analysis of the chloroplast regions trnL-F, psbA-trnH and the nuclear rDNA ITS. Strict consensus of 2094 equally parsimonious trees of length 1737 steps. Bootstrap values above 50 % are shown above the branches. In addition to all Salvia, the ‘Salvia clade’ includes the genera highlighted in bold.
Analysis of the ‘Salvia clade’ data set
The aligned length of the trnL-F data set was 1019 bp. With regions of ambiguous alignment or ambiguous sequences excluded, the total length of included characters was 923 bp. Of the 1019 characters in the analysis, 755 were constant, 75 variable characters were uninformative and 93 were parsimony-informative (9·1 %). Fitch parsimony analysis of the trnL-F region found 26 007 equally parsimonious trees of 163 steps (CI = 0·748, RI = 0·971, RC = 0·727).
The aligned length of the nuclear ITS data set (for the 93 included taxa) was 807 bp. With regions of ambiguous alignment or ambiguous sequences excluded, the total length of included characters was 762 bp. Of the 762 characters in the analysis, 428 were constant, 101 variable characters were uninformative and 233 were parsimony-informative (30·6 %). Fitch parsimony analysis of the ITS region found over 230 000 equally parsimonious trees of 1286 steps (CI = 0·341, RI = 0·762, RC = 0·260).
The combined trnL-F and nuclear ITS analysis generated 1698 characters, of which 1183 were constant, 176 were variable but uninformative and 339 were parsimony-informative (20·0 %). Fitch parsimony analysis of the trnL-F region (uninformative characters excluded) found over 100 000 equally parsimonious trees of 1489 steps (CI = 0·376, RI = 0·814, RC = 0·306).
The partition homogeneity test of the two data sets suggests significant incongruity between the trnL-F and ITS data sets compared with random partitions of the same size (P < 0·01). Despite the incongruence of the data sets, both data sets independently support the integrity of the three clades of Salvia discussed in this project. With regard to these main clades, the topology generated from the strict consensus of the trnL-F data set does not differ from the topology of the combined analysis (although polytomies found in the trnL-F strict consensus tree are resolved in the combined analysis). None of the examples of incongruence of the data sets that would affect the interpretations included in this paper found in the ITS strict consensus tree has bootstrap support above 50 % in the ITS analysis.
ML produced a single tree with a log likelihood score of −11 859·60033. The ML analyses were performed under the K80(K2P) + G + I model of evolution: ti/tv ratio = 1·683386; proportion of invariable sites = 0·518164; nucleotide frequencies = 0·25; gamma shape parameter = 0·513370; substitution types = 2; rate categories = 4. All clades discussed in this paper were present in both the MP and ML trees, and relationships among those clades were identical under both assumptions. The only topological differences between the MP and ML trees were species relationships within the major lineages defined in this paper.
The strict consensus of all MP trees for the Salvia clade analysis (Fig. 4) exhibits the same, well-supported clades seen in the Mentheae-wide analysis. Salvia, likewise, is not monophyletic. Lepechinia together with Melissa form the sister group to the Salvia clade. Salvia ‘clade III’ still appears as a paraphyletic grade, although the branch support for paraphyly (or monophyly) is weak. Within Salvia clade II, two moderately to well-supported subclades emerge with the increased taxa sampling: sect. Audibertia from western North American sister to the large neotropical subgen. Calosphace.
Staminal morphology
Two distinct stamen types were identified in the species sampled from Salvia clade I (stamen types A and B, Fig. 5; Table 2). The two posterior thecae are expressed and not fused in stamen type A. In stamen type B, the two posterior thecae are not expressed, and the distal posterior ends of the adjacent connectives are fused into a complex structure blocking access to nectar. Five distinct stamen types were identified in Salvia clade II. In Salvia axillaris (stamen type G, Fig. 5), both posterior thecae are expressed, and not fused to one another. In sections Standleyana, Blakea and Hastatae (stamen type F, Figs 4 and 5), both posterior thecae are aborted, and the adjacent posterior thecae are not or only little fused. The remaining members of S. subgen. Calosphace (stamen type E, Fig. 5) have both posterior thecae aborted and adjacent posterior connective branches fused. Two stamen types are described for Salvia sect. Audibertia (Figs 4 and 5): those that exhibit a reduced posterior theca (stamen type I), and those with an entirely aborted posterior theca and connective arm (stamen type H). Two stamen types were recognized in Salvia ‘clade III’. The first of these (stamen type M, Figs 4 and 5) has both posterior thecae expressed and not fused to one another. The second type of stamen found in Salvia ‘clade III’ (stamen type N, Figs 4 and 5) has both posterior thecae aborted, or expressed and producing little or no pollen. The posterior thecae are flattened by growth on the abaxial side of the theca, resulting in a fan-shaped theca projected forward from the corolla throat. The two adjacent aborted thecae may be entirely fused, simply connivent, or even separated. Whereas access to the nectar is not necessarily blocked, a lever mechanism has been observed in this stamen type in at least some of these species (S. glutinosa, S. hians).
DISCUSSION
The molecular results presented here resolve a number of systematic questions within the tribe Mentheae, particularly the manner in which the lever mechanism has evolved within the Salvia clade. First, the genera Lepechinia and Melissa are closely related, and together with the ‘Salvia clade’ form a monophyletic group within the Mentheae (Fig. 3). Second, as originally demonstrated by Walker et al. (2004), there exist three distinct lineages of Salvia, each lineage more closely related to other genera in the Mentheae than to the two other major lineages of Salvia (Figs 3 and 4). And third, the staminal lever mechanism has evolved three times independently, each time with a distinct morphology (Figs 5 and 6).
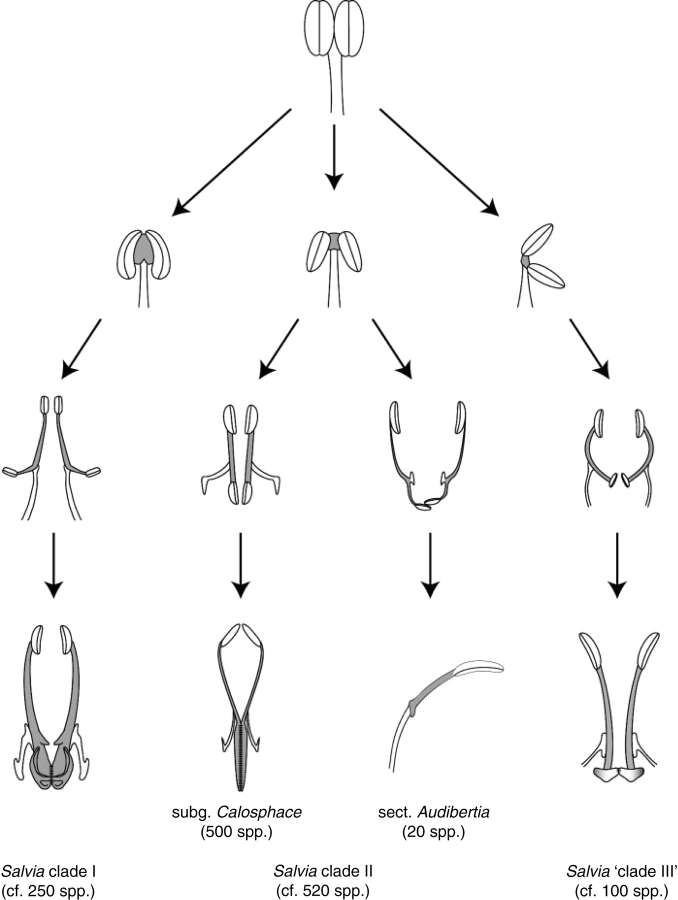
Fig. 6.
Hypothesis of evolutionary progression in the independent origin of the three different staminal lever mechanisms found in the tribe Mentheae. This figure represents a modification and revision of Himmelbaur and Stibal's (1934) original interpretation of staminal evolution in Salvia. The three lever mechanisms (Salvia clade I, clade II and ‘clade III’) are homologous in that they are derived from the connective tissue of the stamen (shaded in this figure), but have been independently derived and are morphologically distinct from one another.
Relationships within the Mentheae
This project has sampled all putative Salvia relatives, as well as representatives of all other major lineages within the tribe Mentheae. The purpose here is not to describe relationships between all genera of the Mentheae, but rather to describe the clade to which Salvia belongs. A thorough investigation into relationships within the tribe Mentheae, comprehensively sampling all genera within the tribe, is being addressed by Bräuchler et al. (2005). For the purposes of this paper, it suffices to say that our sampling within the Mentheae is thorough enough to feel confident in identifying a monophyletic lineage consisting of the genera Melissa, Lepechinia (including Chaunostoma), Salvia, Dorystaechas, Meriandra, Zhumeria, Perovskia and Rosmarinus (Fig. 3), a result also supported by Bräuchler et al. (2005). This finding is in agreement with the results of Wagstaff (1992) based on cpDNA restriction site analysis, although he did not sample Meriandra or Zhumeria, and the placement of Melissa was unresolved. Within this clade, our data support a monophyletic lineage consisting of Salvia, Dorystaechas, Meriandra, Zhumeria, Perovskia and Rosmarinus (the ‘Salvia clade’), a clade characterized morphologically by the abortion of the two adaxial stamens. Our sampling is insufficient in the genus Lepechinia to address the relationship between Lepechinia and Melissa; however, in all analyses, ‘Salvia clade’, Lepechinia and Melissa form a monophyletic group (Fig. 3). Melissa includes three species native to Iran and central Asia. Lepechinia is a New World group of approximately 40 species, historically presenting numerous taxonomic difficulties (Epling, 1944, 1948; Hart, 1983). Both Lepechinia and Melissa have four expressed stamens, each with two parallel thecae and a connective that is not elongated.
In short, we informally recognize within the larger tribe Mentheae a lineage that would correspond to a subtribe consisting of the genera Salvia, Dorystaechas, Meriandra, Zhumeria, Perovskia, Rosmarinus, Lepechinia and Melissa. This assemblage of genera warrants novel subtribal status as significant changes would have to be invoked to either Bentham's (1876) or Wunderlich's (1967) tribal and subtribal arrangements to accommodate all these genera. However, we choose to wait until relationships within the remainder of Mentheae are more completely known (e.g., Bräuchler et al., 2005) before formally naming this lineage. It is within this subtribe that we concentrate on staminal evolution within the three lineages of Salvia as suggested by the molecular phylogenetic data.
Staminal evolution in Salvia clade I
Perovskia and Rosmarinus together are well supported as sister to Salvia clade I (Figs 3 and 4). Both analyses also place Perovskia + Rosmarinus + Salvia clade I sister to the remainder of the ‘Salvia clade’. Perovskia has a slightly elongate connective in its two expressed stamens (Bentham, 1876; Bokhari and Hedge, 1971; Wagstaff, 1992; stamen type D, Fig. 5). Rosmarinus has a significantly elongated connective in its two stamens, and a total abortion of the posterior branch of the connective and the posterior theca (stamen type C, Fig. 5). The resulting appearance results in the stamen appearing essentially ‘normal’ (i.e. with no elongate connective), albeit with only one theca at the end, and a notch half way up the ‘filament’ representing where the filament ends and the connective begins (Trapp, 1956). Thus, unlike the other four genera intercalated in the genus Salvia, Rosmarinus exhibits the defining character of Salvia, a significantly elongate connective. Furthermore, this is the same staminal morphology found in Salvia sect. Audibertia from western North America, and thus, independent of phylogeny, there is no morphological basis for why Rosmarinus should not be included in the genus Salvia.
Within Salvia clade I, two lineages are identified here, each with a distinct stamen morphology. The first well-supported clade within Salvia clade I consists of S. daghestanica, S. canariensis, S. candidissima, S. verbascifolia, S. aethiopsis, S. austriaca and S. sclarea in our sampling. These species all display the staminal character of total fusion of the posterior thecae into what Bentham (1876) termed a glutinatorium, and what Claßen-Bockhoff et al. (2004a) and Himmelbaur and Stibal (1932–1934) described as ‘stamen type V’ (stamen type B, Fig. 5; Fig. 1). This morphology creates the classic Salvia lever mechanism, where the pollinator is forced to push against the fused posterior thecal tissue and activate the lever in order to access the nectar. Using the species groups established by Hedge (1974a, b, 1982a, b) and the alliances suggested by Pobedimova (1954), it can be assumed that this clade probably contains an additional 50 European and western Asian species.
The other taxa sampled from Salvia clade I produce a wide diversity of stamen types, generally including rudimentary posterior thecae, sometimes with pollen produced, and not entirely fused to the adjacent posterior theca or connective arm. Exceptions to this generality can be noted in S. disermas and S. garipensis, both of which have aborted posterior theca which are fused (Hedge, 1974a). The variations in staminal morphology present in this group is best appreciated by noting the diversity of stamens in the sketches included in Hedge's (1974a) treatment of the Salvia of Africa. Field observations by the first author suggest that a lever mechanism is employed in some of these taxa (e.g. S. taraxacifolia, S. texana) but not in others (e.g. S. summa, S. roemeriana). Using the species groups established by Hedge (1974a, b, 1982a, b) based on morphological characters and the alliances suggested by Pobedimova (1954), it can be hypothesized that essentially all central and southern African Salvia belong to this group, plus an additional at least 50 species from western Asia and the Mediterranean, and eight species in the New World (Walker and Elisens, 2001; Walker et al., 2004). These numbers would place the size of this group at over 100 species.
Staminal evolution in Salvia clade II
In both analyses, Dorystaechas and Meriandra are either sister to Salvia clade II or represent a grade toward a monophyletic Salvia clade II — a large lineage of Salvia including the New World sect. Audibertia and subgen. Calosphace. Dorystaechas and Meriandra have long been seen as somewhat anomalous genera in the Mentheae with no obvious affinities (Bokhari and Hedge, 1976). The two genera have been placed in the subtribe Meriandreae with Perovskia (Bentham, 1876), based on two expressed stamens and parallel thecae, in what Bokhari and Hedge (1976) describe as ‘… essentially an artificial assemblage of isolated relict genera united essentially only by the 2-staminate corollas’. Each of the genera also have slightly elongate connectives [in the case of Perovskia and Dorystaechas (stamen type K, Fig. 5), the connectives would probably be better described as swollen]. Dorystaechas is a monotypic genus restricted to south-west Anatolia. Meriandra has slightly elongate connectives (stamen type J, Fig. 5) and consists of two species, one native to Ethiopia and one to India (ironically, Meriandra bengalensis is the Ethiopian species).
Within the larger picture of the genus Salvia, sect. Audibertia represents an anomalous group restricted to the California Floristic Province and adjacent deserts. The separation of this group from other Salvia has been based on chemical compounds, shrubby habit with strongly lignified stems (although not present in all species), and, most importantly, on the structure of its stamens (Neissess, 1983). Sect. Audibertia is unusual within Salvia in having the posterior branch of the connective entirely aborted (although the genus Rosmarinus shows a similar phenomenon, as do some individuals of the Old World S. verticillata). Whereas the anterior branch of the connective is still elongate, functionally it acts in the same manner as would a simple filament, albeit with only a single theca at its end (Bentham, 1876; Epling, 1938; Neissess, 1983) (stamen type H, Fig. 5). Worthy of note is a difference in staminal morphology seen between Salvia sect. Audibertia and the genus Rosmarinus. Whereas the ‘joint’ between the filament and connective is indicated by a notch on the top of the stamen in Rosmarinus, an articulation circling the entire filament is found at that same ‘joint’ in sect. Audibertia. Occasionally the posterior theca and connective branch is re-expressed in members of sect. Audibertia.
Contrary to the most recent treatment of the section (Neissess, 1983), our preliminary data suggest that sect. Audibertia (sensu Bentham) is a monophyletic lineage (Figs 3 and 4), and the species included in Neissess' (1983) sect. Echinosphace probably represent a grade toward a monophyletic sect. Audibertia (sensu Neissess, 1983). The staminal morphology of sect. Echinosphace (four spp.) is distinct from sect. Audibertia (sensu Neissess) in that sect. Echinosphace displays the plesiomorphic character of the posterior branch of the connective and the posterior theca always being expressed, albeit reduced (stamen type I, Fig. 5). Section Audibertia (sensu Neissess) displays the derived character of no expressed posterior theca, and thus it is possible to define a progression from both thecae being expressed to the entire abortion of the posterior theca in this clade as well.
Salvia subgen. Calosphace consists of nearly 500 species and occurs throughout the New World, with centres of diversity in Mexico, the Andean region, and southern Brazil and Argentina. Epling (1939) created the only comprehensive treatment of the subgenus, organizing 468 species into 91 sections (and in supplementary notes, an additional 71 species and 13 sections). Stumbling blocks to past and future work in subgen. Calosphace are (1) the lack of knowledge of relationships between sections (an issue Epling did not address) and (2) the lack of faith in the monophyly of some of his larger sections. For these reasons, the only works to have been completed at the sectional level since Epling's time have generally been limited to sections of five or fewer species (Peterson, 1978; Ahlenslager, 1984; Turner, 1996). In those revisions dealing with larger sections [Serna and Ramamoorthy, 1993 (11 species); Torke, 2000 (eight species)], the monophyly of those sections was not addressed. The sampling included with this paper is part of a larger project investigating large-scale relationships within the subgenus Calosphace.
The typical staminal morphology for subgen. Calosphace consists of an elongation of the posterior connective branch, fusion of the two adjacent connective arms and no differentiation of tissue at the distal end of the connective branch (stamen type E, Fig. 5). As is well documented by, among others, Claßen-Bockhoff et al. (2004a), Baikova (2002, 2004), Epling (1939), a tooth is often present on the lower side of the posterior connective branch. Claßen-Bockhoff et al. (2004a) clearly demonstrated ontogenetically that the aborted posterior theca may be either located at the distal end of the connective arm, or in some cases represented by a dorsal outgrowth of the connective. Their finding suggests that the formations of the connective arm found within subgen. Calosphace that form the basis of the lever mechanism may not all be homologous. Despite that important difference, staminal morphology within the subgenus is uniform with respect to no posterior thecae being expressed and the two posterior connective arms, or dorsal outgrowths of the connective being fused. This uniformity is true across the entirety of subgen. Calosphace except for four of Epling's sections (sections Hastatae, Blakea, Standleyana and Axillares). Sections Hastatae (seven spp.), Blakea (four spp.) and Standleyana (one sp.) all have a total abortion of the posterior thecae; however, the connective arms do not entirely fuse. These three sections are all included within the clade represented by stamen type F (Figs 4 and 5), and form a monophyletic group. Salvia axillaris, of monotypic section Axillares, is the only member of Salvia subgen. Calosphace to have expressed posterior thecae (stamen type G, Fig. 5). The molecular phylogeny suggests that S. axillaris is sister to the remainder of subgen. Calosphace. In turn, Hastatae, Blakea and Standleyana represent a monophyletic lineage sister to remaining members of the subgenus. These four sections thus depict an evolutionary ‘trail’ of staminal morphology, showing a progression from both thecae expressed and no fusion of posterior connective branches, to abortion of posterior thecae and no fusion of posterior connective branches, and ultimately to the typical staminal morphology in subgen. Calosphace of abortion of posterior thecae and fusion of connective branches (see Figs 5 and 6).
Staminal evolution in Salvia ‘clade III’
In addition to the clearly delineated Salvia clade I and Salvia clade II, there exists a group of Salvia that fit into neither of the above groups. The molecular and morphological evidence clearly supports Salvia ‘clade III’ as having an independent origin of the lever mechanism (Fig. 5). However, this group of Salvia may represent a paraphyletic grade consisting of two monophyletic lineages rather than a single monophyletic clade (Figs 3 and 4).
One of the two lineages consists of a group of western Asian and northern African species including S. aristata, S. aegyptiaca, S. tetrodonta, S. trichocalycina and Zhumeria majudae (Fig. 4). The Salvia in this first lineage all have somewhat elongate connectives, both thecae producing pollen, and the posterior thecae never fused (stamen type M, Fig. 5). Zhumeria majudae is a shrub native to Iran with historically uncertain affinities (Bokhari and Hedge, 1976), but placed in our analyses as sister to this clade of Salvia (Fig. 4). Zhumeria is unusual within the broader ‘Salvia clade’ in that, in addition to the two fertile stamens, two large staminodes are easily identified in the corolla (Bokhari and Hedge, 1976). The thecae of the two fertile stamens are somewhat separated, though without a distinct connective (stamen type L, Fig. 5). Using the species groups established by Hedge (1974a, b, 1982a, b), based on morphological characters in addition to the species sampled here, this first lineage of Salvia ‘clade III’ probably also includes Salvia bazmanica, S. santolinifolia, S. macilenta, S. tebesana, S. eremophila, S. deserti, S. chudaei, S. pterocalyx and S. rechingeri.
The second lineage belonging to Salvia ‘clade III’ consists of a group of Asian and Mediterranean species. In our sampling, this clade consists of S. glutinosa, S. miltiorrhiza, S. hians, S. cynica, S. przewalskii, S. digitaloides and S. roborowskii. Salvia glutinosa and S. miltiorrhiza are probably the best known members of this group, and each expresses the staminal morphology typical of all members of this group. The posterior thecae are rudimentary, and produce no or very little pollen. Often (although not always) in this group, the two adjacent posterior thecae post-genitally fuse (e.g. S. glutinosa, S. przewalskii; Claßen-Bockhoff et al., 2004a). These two posterior thecae are somewhat fan-shaped and are projected forward from the corolla throat (stamen type N, Fig. 5) and a lever mechanism can be employed whether or not the posterior thecae fuse. Although this group of species probably includes nearly 100 species with a likely centre of diversity in China, it is currently impossible to define the exact extent of this clade owing to lack of familiarity with Salvia of China and the fact that the particulars of staminal morphology are rarely included in species descriptions.
Summary of staminal evolution in Salvia
The inferred progression in staminal evolution within the Salvia clade is depicted in Fig. 6 based on the tree-mapping of the stamen types defined in this project from Salvia and intercalated genera (Fig. 5). From the ancestral Mentheae stamen type without elongate connectives (stamen type O, Fig. 5), slightly elongate connectives evolved at least three times in the Salvia clade in lineages recognized as other genera (stamen types D, J, K and L, Fig. 5). The genera with these intermediate stamen types are either basal or sister to the three (or more depending on resolution within Salvia ‘clade III’) major clades of Salvia possessing the variety of stamen types described above. The staminal lever has thus independently originated three times, each time following the progression described above, and each time resulting in the functionally convergent feature of a staminal lever (Figs 5 and 6).
In hindsight, Himmelbaur and Stibal (1932–1934) presented a remarkably accurate assessment of staminal evolution in the genus Salvia. Working with limited material, and lacking the molecular evidence to suggest phylogenetic relatedness of Dorystaechas, Meriandra, Zhumeria, Perovskia and Rosmarinus to Salvia, the general progression in staminal evolution they suggested for the genus Salvia is similar in some fundamental points to that presented here. These points include their recognition of (1) the plesiomorphic staminal state as having two expressed thecae and no lever mechanism in each stamen and (2) parallel origins of the lever mechanism in the New World and the Old World. Some of the specific examples they suggest, such as Salvia sections Hastatae, Blakea and Standleyana being intermediate between the plesiomorphic state and derived state seen in core S. subgen. Calosphace, are exactly the relationships suggested by the molecular data. The molecular approach employed here clarifies the phylogenetic relationships and thus the relationships of different stamen types.
The molecular data presented in this paper strongly support at least three independent origins of the lever mechanism in Salvia. However, Claßen-Bockhoff et al. (2004a) clearly demonstrated through developmental studies the homology of the staminal lever mechanism across all major lineages of Salvia — that is, each type is derived from the elongation of the connective tissue. Do the findings of Claßen-Bockhoff et al. (2004a) concerning homology of the staminal lever contradict the findings here of three separate origins of the staminal lever mechanism? Three lines of evidence strongly support that these staminal levers, although homologous at some level, represent the evolutionary products of three separate events. First, our findings suggest that whereas the lever mechanisms in Salvia are all derived from connective tissue, the precise staminal morphology of the lever mechanism in each of the three major lineages of Salvia supports three independent origins of the lever mechanism in different ways. The ‘gubernaculum’ (Bentham, 1876; Claßen-Bockhoff et al., 2004a, stamen type III; stamen type B, Fig.5) seen in Salvia clade II is never found in Salvia clade I or III. The ‘glutinatorium’ (Bentham, 1876; Claßen-Bockhoff et al., 2004a, stamen type V; stamen type E, Fig.5) seen in Salvia clade I is never found in Salvia clade II or III. The fan-shaped, connivent posterior thecae (stamen type N, Fig.5) seen in Salvia ‘clade III’ are never found in Salvia clade I or II. Within each of the major lineages of Salvia described herein, Zalewska (1928), Himmelbaur and Stibal (1932–1934), Hedge (1974a, b, 1982a, b) and Claßen-Bockhoff et al. (2004a) have noted the uniformity of staminal morphology. Second, further support for three independent origins of the staminal lever mechanism comes from the molecular phylogeny, which strongly places each of the three clades with a lever mechanism as sister to a group of Salvia with elongate connectives, but no lever mechanism. Third, and more significantly, each of these three more inclusive lineages of Salvia is in turn sister to genera without significantly elongate connectives (in the case of Salvia ‘clade III’, the genus Zhumeria is sister to one of the two groups in ‘clade III’).
It is not only trends in staminal evolution that are consistent across the various lineages in the ‘Salvia clade’, but some of the specific stamen types are surprising in their parallel recurrence. For example, stamen type A in Salvia clade I is scarcely distinguishable from stamen types G or M in Salvia clades II and III. Another striking example of parallel recurrence of similar stamen types is the multiple origins of a stamen type exhibiting total abortion of the posterior theca and posterior connective branch. This stamen type has independently derived in Salvia sect. Audibertia (stamen type H), Rosmarinus (stamen type C) and in Salvia verticillata (not shown). Salvia verticillata belongs to the subclade of Salvia clade I expressing stamen type A (Figs 4 and 5), but itself often has the posterior branch of the connective aborted (Himmelbaur and Stibal, 1932–1934; Claßen-Bockhoff et al., 2004a, b; Walker et al., 2004). In each of these three examples, the stamens have gone through a complicated evolutionary progression only to end up with a stamen that in superficial appearance is scarcely distinguishable from the plesiomorphic state for the Salvia lineage, except in the fact that it has one theca instead of two.
This work demonstrates that the story of staminal evolution within the ‘Salvia clade’ is remarkable in its recurrent nature. On three different occasions (Salvia clade I, clade II and ‘clade III’) there is a four-step progression from slight elongation of the connective to significant elongation of the connective, to loss of fertility of the posterior thecae, and ultimately to the fusion of the posterior branches of the connectives (Figs 5 and 6).
Issues in cases of parallel evolution
That all Salvia belong to a single, well-defined lineage within the tribe Mentheae begs the question of whether Salvia is truly polyphyletic or simply paraphyletic. To make the nearly 1000 species of Salvia monophyletic would require only the inclusion of 13 species from the genera Perovskia (seven spp.), Rosmarinus (two spp.), Meriandra (two spp.), Dorystaechas (one sp.) and Zhumeria (one spp.). However, this paper demonstrates that the character that defines Salvia within the Mentheae (the significantly elongate connective) has independently originated in each of the three major Salvia lineages. The independent origin of the defining character for Salvia is supported by the molecular phylogeny, that each of the major clades of Salvia is associated with a genus that does not express the significantly elongate connective, and by the distinct staminal morphology in each of the major lineages of Salvia. Thus, this is not the case where 13 species not included in the genus Salvia represent anomalous members of the genus Salvia that have undergone character reversals (i.e. Salvia is paraphyletic). Rather, the significantly more parsimonious explanation is that the genera associated with Salvia never developed the character that defines the ‘genus’ Salvia. That is, Salvia is polyphyletic in that it is defined by a convergent character. If the genera intercalating themselves within Salvia were larger in size, or if more genera were present in the Salvia lineage, it would not be difficult to accept the polyphyly of Salvia. If the other five genera had become extinct, one could engage in a philosophical discussion as to the monophyly of a clade whose defining character evolved multiple times. However, the Salvia clade represents a wonderful example of evolution leaving a ‘trail’ as it progressed. Gould (1989) suggested that evolutionary novelties are chance occurrences, unlikely to be repeated in different times and places. This general philosophy no doubt played a role in the long-held assumption of the monophyly of Salvia based on the ‘unlikely’ origin of something as complex as the lever mechanism multiple times. However, the story of staminal evolution in Salvia presented here suggests that in the context of a selective regime, Gould's evolutionary ‘tape’ can in fact repeat itself despite long odds — perhaps in response to similar genetic canalizations, phylogenetic constraint, similar pollination-selective regimes and/or convergent tendencies.
It is certainly worth noting that the large species radiations seen in each of the three clades of Salvia are associated with the formation of a lever mechanism. Functional analyses of the lever mechanism evolved in the various lineages of Salvia, currently being addressed by Claßen-Bockhoff et al. (2004a), Thimm et al. (2005), Wester and Claßen-Bockhoff (2006) and Reith et al. (2006), will shed light on the similarity of the functional aspects of the progression in staminal evolution seen in Salvia. These functional analyses, in concert with the phylogenetic data, will, it is hoped, ultimately afford the opportunity to address the suggestion of Claßen-Bockhoff et al. (2004b) that the lever mechanism is a key innovation driving species radiations within the genus Salvia (sensu Hodges and Arnold, 1995; Hodges, 1997; Barraclough et al., 1998; Pellmyr and Krenn, 2002).
ACKNOWLEDGEMENTS
The authors thank Mike Powell, Bart O'Brien, Janet Latham, Petra Wester, Regine Claßen-Bockhoff, Ian Hedge and Richard Walker for help in obtaining plant material; Wisconsin State Herbarium, Royal Botanical Garden-Edinburgh, Field Museum Herbarium and Missouri Botanical Gardens for access to herbarium material; Kandis Elliot for help with the stamen sketches; Petra Wester for insight into staminal morphology, critical discussions and company in the field; Jocelyn Hall and Naomi Delventhal for help with laboratory work; Cody Williams for artwork; and Regine Claßen-Bockhoff and Maximilian Weigend for valuable comments and review of the manuscript. We gratefully acknowledge the support of Davis Grant funds, the California Native Plant Society, a National Science Foundation Dissertation Improvement Grant and the Botanical Society of America Karling Award for funds essential to this project.
LITERATURE CITED
- Ahlenslager K. Erythrostachys. USA: University of Montana; 1984. Systematic studies of Salvia subg. Calosphace sect. Masters thesis. [Google Scholar]
- Alziar G. Catalogue synonymique des Salvia L. du monde (Lamiaceae) Biocosme Mesogéen. 1988–1993;5(3–4):87–136. I.–VI. 6 (1–2, 4): 79–115, 163–204; 7 (1–2): 59–109; 9 (2–3): 413–497; 10 (3–4): 33–117. [Google Scholar]
- Baikova E. Two ways of stamen development in the subgenus Calosphace (Salvia, Lamiaceae) Botanicheskii-Zhurnal-(St.-Petersburg) 2002;87:71–78. [Google Scholar]
- Baikova E. Structural types and morphogenesis of stamens in the genus Salvia (Lamiaceae) Botanicheskii-Zhurnal-(St.-Petersburg) 2004;89:881–895. [Google Scholar]
- Barraclough TG, Vogler AP, Harvey PH. Revealing the factors that promote speciation. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 1998;353:241–249. [Google Scholar]
- Baum DA, Sytsma KJ, Hoch PC. The phylogeny of Epilobium L. (Onagraceae) based on nuclear ribosomal DNA sequences. Systematic Botany. 1994;19:363–388. [Google Scholar]
- Bentham G. Labiatae. in Bentham, G. and J. D. Hooker. Genera Plantarum. 1876;2:1160–1196. [Google Scholar]
- Bokhari MH, Hedge IC. Observations on the tribe Meriandreae of the Labiatae. Notes Royal Botanical Garden, Edinburgh. 1971;31:53–67. [Google Scholar]
- Bokhari MH, Hedge IC. Zhumeria (Labiatae): anatomy, taxonomy and affinities. Iran Journal of Botany. 1976;1:1–10. [Google Scholar]
- Bräuchler C, Meimberg H, Abele T, Heubl G. XVII International Botanical Congress, Vienna, Austria. 2005. A molecular perspective for tribal concepts and generic boundaries in subfamily Nepetoideae. [Google Scholar]
- Cantino PD, Harley AM, Wagstaff SJ. Genera of Labiatae: status and classification. In: Harley RM, Reynolds T, editors. Advances in Labiate science. Kew: Royal Botanic Gardens; 1992. pp. 511–522. [Google Scholar]
- Claßen-Bockhoff R, Wester P, Tweraser E. The staminal lever arm mechanism in Salvia — a review. Plant Biology. 2003;5:33–41. [Google Scholar]
- Claßen-Bockhoff R, Crone M, Baikova E. Stamen development in Salvia: homology reinvestigated. International Journal of Plant Science. 2004a;165:475–498. [Google Scholar]
- Claßen-Bockhoff R, Speck T, Tweraser E, Wester P, Thimm S, Reith M. The staminal lever mechanism in Salvia: a key innovation for adaptive radiation? Organisms Diversity & Evolution. 2004b;4:189–205. [Google Scholar]
- Conti E, Litt A, Sytsma KJ. Circumscription of Myrtales and their relationships to other rosids: evidence from rbcL sequence data. American Journal of Botany. 1996;83:221–233. [Google Scholar]
- Croizat L. Space, time, form; the biological synthesis. Caracas: published by the author; 1962. (Venezuela). [Google Scholar]
- Epling C. The Californian salvias. Annals of the Missouri Botanical Garden. 1938;25:95–188. [Google Scholar]
- Epling C. A revision of Salvia, subgenus Calosphace. Beihefte Feddes Repertorium specierum novarum regni vegetabilis. 1939;110:1–383. [Google Scholar]
- Epling C. The living mosaic. Berkeley: University of California Press; 1944. (CA). [Google Scholar]
- Epling C. A synopsis of the tribe Lepechinae (Labiatae) Brittonia. 1948;6:352–364. [Google Scholar]
- Farris JS, Kallersjo M, Kluge AG, Bult C. Testing significance and incongruence. Cladistics. 1995;10:315–319. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Zoology. 1971;20:406–416. [Google Scholar]
- Givnish TJ, Evans TM, Zjhra ML, Berry PE, Sytsma KJ. Molecular evolution, adaptive radiation and geographic diversification in the amphiatlantic family Rapateaceae: evidence from ndhF sequence data. Evolution. 2000;54:1915–1937. doi: 10.1111/j.0014-3820.2000.tb01237.x. [DOI] [PubMed] [Google Scholar]
- Gould SJ. Wonderful life: The Burgess Shale and the nature of history. New York: W.W. Norton & Co; 1989. [Google Scholar]
- Hart JA. Systematics and evolution in the genus. Lepechinia: Harvard University; 1983. PhD dissertation, Cambridge, MA, USA. [Google Scholar]
- Hedge IC. A revision of Salvia in Africa and the Canary Islands. Notes Royal Botanical Garden, Edinburgh. 1974a;33:1–121. [Google Scholar]
- Hedge IC. A further note on Salvia tetrodonta. Notes Royal Botanical Garden, Edinburgh. 1974b;33:295–299. [Google Scholar]
- Hedge IC. Labiatae. In: Davis PH, editor. Flora of Turkey and the eastern Aegaean Islands, vol. 7, Labiatae. Edinburgh: Edinburgh University Press; 1982a. pp. 000–000. [Google Scholar]
- Hedge IC. In: Salvia. Flora Iranica. vol. 150, Labiatae. Rechinger KH, editor. Graz: Akademische Druck und Verlags-Anstalt; 1982b. pp. 000–000. [Google Scholar]
- Hedge IC. In: Flora of Cyprus. Meikle RD, editor. Kew: Royal Botanic Gardens; 1985. pp. 000–000. Salvia. [Google Scholar]
- Himmelbaur W, Stibal E. Entwicklungsrichtungen in der Blutenregion der Gattung Salvia L. Biologia generalis. 1932–1934;8:449–474. I–III. 9: 129–150; 10: 17–48. [Google Scholar]
- Hipp AL, Hall JC, Sytsma KJ. Phylogenetic accuracy, congruence between data partitions, and performance of the ILD. Systematic Biology. 2004;53:81–89. doi: 10.1080/10635150490264752. [DOI] [PubMed] [Google Scholar]
- Hodges SA. Floral nectar spurs and diversification. International Journal of Plant Science. 1997;158:S81–S88. [Google Scholar]
- Hodges SA, Arnold ML. Spurring plant diversification: are floral nectar spurs a key innovation? Proceedings of the Royal Society of London Series B-Biological Sciences. 1995;262:343–348. [Google Scholar]
- Hruby K. Zytologie und Anatomie der mitteleuropäischen Salbei-Arten. Beihefte Botanisches Centralblatt. 1934;52:298–380. [Google Scholar]
- Huelsenbeck JP, Ronquist FR. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Maddison DR. The discovery and importance of multiple islands of most-parsimonious trees. Systematic Zoology. 1991;40:315–328. [Google Scholar]
- Müller H. Die Befruchtung der Blumen durch Insekten und die gegenseitige Anpassung beider. Leipzig: W. Engelmann; 1873. [Google Scholar]
- Neissess KR. Evolutions, systematics and terpene relationships of Salvia section Audibertia. Riverside, CA, USA: University of California; 1983. PhD dissertation. [Google Scholar]
- Paton AJ, Springate D, Suddee S, Otieno D, Grayer RJ, Harley MM, et al. Phylogeny and evolution of basils and allies (Ocimeae, Labiatae) based on three plastid DNA regions. Molecular Phylogenetics and Evolution. 2004;31:277–299. doi: 10.1016/j.ympev.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Pellmyr O, Krenn HW. Origin of a complex key innovation in an obligate insect-plant mutualism. Proceedings of the National Academy of Sciences of the USA. 2002;99:5498–5502. doi: 10.1073/pnas.072588699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. Farinaceae. USA: University of Maryland; 1978. Systematic studies of Salvia subgenus Calosphace in section. PhD thesis. [Google Scholar]
- Pobedimova EG. Labiatae. In: Shishkin BK, editor. Flora of the U.S.S.R., vol. 21, Labiatae. Moscow: 1954. pp. 178–260. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rambaut A. Sequence alignment editor, version 2.0a6. 2001. Available online at http://evolve.zoo.ox.ac.uk/
- Reith M, Baumann G, Claßen-Bockhoff R, Speck T. Sharing without mixing? Quantitative analyses of pollen placement on Apis mellifera as a pollinator of Salvia pratensis and Salvia nemorosa. Annals of Botany. 2006;96:000–000. [Google Scholar]
- Serna AS, Ramamoorthy TP. Revision taxonomica de Salvia seccion Sigmoideae. Acta Botanica Mexicana. 1993;23:65–102. [Google Scholar]
- Sprengel CK. Das entdeckte Geheimnis der Natur im Bau und in der Befruchtung der Pflanzen. Berlin: Friedrich Vieweg dem aeltern; 1793. [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Thimm S, Reith M, Speck T, Claßen-Bockhoff R. XVII International Botanical Congress, Vienna, Austria. 2005. Force measurements in Salvia flowers. [Google Scholar]
- Trapp A. Zur morphologie und entwicklungsgeschichte der staubblatter sympetaler bluten. Botanische Studien. 1956;5:3–93. [Google Scholar]
- Turner B. Synopsis of section Axillaris of Salvia. Phytologia. 1996;81:16–21. [Google Scholar]
- Wagstaff SJ. Phylogeny and character evolution in subfamily Nepetoideae (Labiatae) Athens, USA: Ohio University; 1992. PhD thesis. [Google Scholar]
- Wagstaff SJ, Olmstead RG, Cantino PD. Parsimony analysis of cpDNA restriction site variation in subfamily Nepetoideae (Labiatae) American Journal of Botany. 1995;82:886–892. [Google Scholar]
- Walker JB, Elisens WJ. A revision of Salvia section Heterosphace in western North America. Sida. 2001;19:571–589. [Google Scholar]
- Walker JB, Sytsma KJ, Treutlein J, Wink M. Salvia is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. American Journal of Botany. 2004;91:1115–1125. doi: 10.3732/ajb.91.7.1115. [DOI] [PubMed] [Google Scholar]
- Werth E. Zur Kenntnis des Androeceums der Gattung Salvia und seiner stammesgeschichtichen Wandlung. Berichte der Deutschen Botanischen Gesellschaft. 1956;69:381–386. [Google Scholar]
- Wester P, Claßen-Bockhoff R. Floral diversity and pollen transfer in bird-pollinated Salvia species. Annals of Botany. 2006;96:000–000. doi: 10.1093/aob/mcm036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse E. Revision of Salvia section Salviastrum. Field and Laboratory. 1949;17:151–165. [Google Scholar]
- Wunderlich R. Ein Vorschlag zu einer naturlichen Gliederung der Labiaten auf Grund der Pollenkorner, der Samenentwicklung und des reifen Samens. Öesterriechisce Botanische Zeitschrift. 1967;114:383–483. [Google Scholar]
- Xi-wen L, Hedge IC. Lamiaceae. In: Zheng-yi W, Raven PH, editors. Flora of China. St. Louis: Missouri Botanical Garden Press; 1994. (tMO). [Google Scholar]
- Yoder AD, Irwin JA, Payseur BA. Failure of the ILD to determine data combinability for slow loris phylogeny. Systematic Biology. 2001;50:408–424. [PubMed] [Google Scholar]
- Zalewska Z. Recherches sur l'évolution des étamines, considérée du point de vue de leur adaptation à la pollinisation des fleurs de la Sauge (Salvia) Bulletin internacional de l'academie polonaise des sciences et de lettres. 1928;3:133–160. [Google Scholar]