Abstract
Background Aims
One of the classic examples of an allopolyploid is Iris versicolor, ‘Blue Flag’ (2n = 108), first studied by Edgar Anderson and later popularized by George Ledyard Stebbins in cytogenetics and evolutionary text-books. It is revisited here using modern molecular and cytogenetic tools to investigate its putative allopolyploid origin involving progenitors of I. virginica (2n = 70) and I. setosa (2n = 38).
Methods
Genomic in situ hybridization (GISH), fluorescent in situ hybridization (FISH) and Southern hybridization with 5S and 18–26S ribosomal DNA (rDNA) probes were used to identify the parental origin of chromosomes, and to study the unit structure, relative abundance and chromosomal location of rDNA sequences.
Key Results
GISH shows that I. versicolor has inherited the sum of the chromosome complement from the two progenitor species. In I. versicolor all the 18–26S rDNA units and loci are inherited from the progenitor of I. virginica, those loci from the I. setosa progenitor are absent. In contrast 5S rDNA loci and units from both progenitors are found, although one of the two 5S loci expected from the I. setosa progenitor is absent.
Conclusions
These data confirm Anderson's hypothesis that I. versicolor is an allopolyploid involving progenitors of I. virginica and I. setosa. The number of 18–26S rDNA loci in I. versicolor is similar to that of progenitor I. virginica, suggestive of a first stage in genome diploidization. The locus loss is targeted at the I. setosa-origin subgenome, and this is discussed in relation to other polyploidy systems.
Key words: Allopolyploid, diploidization, genome evolution, rDNA, Laevigatae irises, Iris versicolor
INTRODUCTION
There is growing interest in allopolyploidy (interspecific hybridization and genome multiplication) because of its significant role in the divergence of many angiosperm species (Leitch, Bennett, 1997; Soltis, Soltis, 1999; Wendel, 2000). Here we test Anderson's (1936) classic hypothesis that the blue flag, Iris versicolor L. (2n = 108, series Laevigatae, with the highest chromosome number in the genus) is an allopolyploid involving progenitors related to I. virginica L. southern blue flag (2n = 70, series Laevigatae) and I. setosa Pall. arctic blue flag or beach-head iris (2n = 38, series Tripetalae).
The series Laevigatae in section Limniris are a group of northern hemisphere, sub-aquatic irises with disjunct chromosome numbers and distributions. Two species are found in Japan and the Far East, Iris ensata Thunb. (2n = 24) and I. laevigata Fisch. (2n = 32), one species in Europe I. pseudacorus L. (syn. I. maackii 2n = 34), and two species in North America, I. versicolor L. (2n = 108) and I. virginica L. (2n = 70). Iris setosa has a circum-arctic distribution from coastal Alaska to the lower Lena River (Siberia) and southwards to Japan.
In nature there are reports of natural, vigorous F1 hybrids involving I. virginica and I. versicolor (I. × robusta E. Anders, 2n = 89) in areas where they grow in close proximity. But attempts to create I. virginica × I. setosa hybrids artificially proved unsuccessful. Seeds were produced but unfilled (Anderson, 1936). To date no synthetic I. versicolor has been successfully resynthesized.
In nature, several recently formed (within the last 150 years) allopolyploids have been described, these include Cardamine schultzii Urbanska (Urbanska et al., 1997), Tragopogon mirus Ownbey, and T. miscellus Ownbey (Soltis et al., 2004a) Senecio eboracensis Lowe & Abbott, S. cambrensis Rossner (Abbott and Lowe, 2004) and Spartina anglica Hubb. (Ainouche et al., 2004). The immediate genetic consequences are variable, with considerable genetic and epigenetic changes observed in some populations of Tragopogon mirus and T. miscellus (Kovarik et al., 2005; Soltis et al., 2004a, b) and limited changes observed in Spartina anglica (Ainouche et al., 2004). Over longer time periods, especially in the arctic flora, there is evidence of recurrent polyploidy involving the same progenitor species (Brochmann et al., 2004). The classic example of allopolyploidy represented by Iris versicolor is analysed here in the context of our deepening understanding of evolution involving allopolyploidy.
The 18–26S rDNA repeats can exhibit length and sequence heterogeneity both within and between species, but frequently within a species repeats show evidence of homogenization, possibly caused by processes such as unequal crossing over and gene conversion, mechanisms collectively referred to as concerted evolution (Dover, 1982). In allopolyploids the rDNA units at each of the parental rDNA loci can undergo independent evolutionary trajectories, as occurs in some Arabidopsis and Brassica allopolyploids (Hasterok et al., 2006; O' Kane et al., 1996). However, more commonly in allopolyploids, various types of genetic alterations are observed including a reduction in copy number, locus loss, intra- and intergenomic recombination (for reviews, see Alvarez and Wendel, 2003; Kovarik et al., 2004; Dadejova et al., 2007).
Here genomic in situ hybridization (GISH) is used to test the allopolyploid origin of I. versicolor and the parental origin of its chromosomes. Fluorescent in situ hybridization (FISH) and Southern hybridization with rDNA probes (5S and 18S–26S rDNA genic subunits) to I. versicolor and its putative progenitors are used to determine whether there has been divergence in the number and organization of rDNA loci since the original allopolyploidy event.
MATERIALS AND METHODS
Plant materials
All plant material used was derived from Royal Botanic Gardens (RBG), Kew (Richmond Surrey, UK): Iris virginica (RBG Kew 1994–1346-S. E. USA), I. versicolor (RBG Kew 1976–6538-New Hampshire) and I. setosa (RBG Kew 1979–4872-North Japan). Plants were grown in pots sitting in water trays at Queen Mary, University of London. All species were identified by floral morphology and chromosome counts. DNA extractions were made from young leaves using a modified Saghai-Maroof et al. (1984) protocol. For GISH probes, high quality genomic DNA was extracted using Qiagen mini kit (Qiagen Ltd) according to manufacturers instructions.
Restriction endonuclease digestion and Southern hybridization
Methods followed Sambrook et al. (1989). Genomic DNA was digested to completion with excess of restriction endonucleases and fractionated in 1% agarose by gel electrophoresis. DNA was transferred to a Hybond N + membrane (GE Healthcare, Little Chalfont, UK). Southern hybridization was carried out under high-stringency conditions (Fulnecek et al., 2002) using heat-denatured 32P-labelled 5S, 18S and 26S rDNA subunits as probes. Labelling of DNA probe was carried out by a random primed method using [32P[dCTP (DekaLabel kit; MBI, Fermentas, Vilnius, Lithuania). (a) The genic 5S rDNA probe (approx. 120 bp) containing the transcribed part of the 5S rDNA monomeric unit from Nicotiana tabacum (Fulnecek et al., 1998) was generated by excision of an insert in a cloned sequence (accession number AJ222659). (b) The 18S rDNA probe was a cloned 1·7 kb EcoRI fragment of the 18S rDNA gene subunit from Solanum lycopersicum (tomato) (Kiss et al., 1989) (accession number X51576). (3) The 26S rDNA probe was an approx. 220 bp PCR product derived from the 3′ end (region between + 2901 and + 3121) of the 26S rRNA gene of N. tabacum (Lim et al., 2000).
Chromosome preparation and fluorescent in situ hybridization
Slides with spread metaphase chromosomes were labelled by GISH and FISH according to Leitch et al. (2001) and Lim et al. (1998). Briefly, root tips of pot-grown plants were pre-treated in Lindane (Sigma-Aldrich) at room temperature for 4 h and fixed in 3 : 1 ethanol : glacial acetic acid for a few days, transferred to 70% ethanol, and stored at − 20°C. Root-tips were digested for approx. 28 min in 0·3% (w/v) cellulase R10, 0·3% (w/v) pectolyase Y23 and 0·3% (w/v) drieselase, and transferred to 1 × citrate buffer (4 mm citric acid and 6 mm sodium citrate at pH 4·8). Root meristem cells were spread onto chromic acid washed slides. Genomic and rDNA probes were labelled with digoxigenin-11-dUTP and with biotin-16-dUTP. After overnight hybridization, slides were given a stringent wash in 20% (v/v) formamide in 0·1 × SSC at 40–42°C resulting in DNA duplexes with an estimated > 85% sequence identity. Sites of probe hybridization were detected using 20 µg mL−1 fluorescein conjugated anti-digoxigenin IgG (Roche Biochemicals) and 5 µg mL−1 Cy3 conjugated avidin (GE Healthcare, Little Chalfont, UK) in 4 × SSC containing 0·2% (v/v) Tween 20 and 5% (w/v) bovine serum albumin. Chromosomes were counterstained with 2 µg mL−1 DAPI (4′, 6-diamidino-2-phenylindole), mounted in Vectashield (Vector Laboratories) medium, examined using a Leica DMRA2 epifluorescent microscope fitted with an Orca ER camera and Open Lab® software (Improvision). All images were processed using Adobe Photoshop® and treated for colour contrast and brightness uniformly.
RESULTS
Southern hybridization showing uniparental rDNA deletion
Genomic DNAs of I. virginica, I. setosa and I. versicolor were digested with restriction endonucleases Ssp1, BstN1 and EcoRV, and subjected to Southern hybridization with 5S, 26S and 18S rDNA subunit probes (Fig. 1). In I. setosa the 5S rDNA probe revealed a series of bands from 500 bp to around 6 kb representing multiples of a 500 bp 5S rDNA unit. Iris virginica generated a fewer number of bands representing multiples of a unit that is smaller than 500 bp. A few bands suggest that the Ssp site is more conserved in units of I. virginica than in those of I. setosa. The pattern of 5S rDNA bands in I. versicolor is approximately additive of that observed for I. virginica and I. setosa, but the number of rDNA signals and copies is relatively similar in all three species.
Fig. 1.
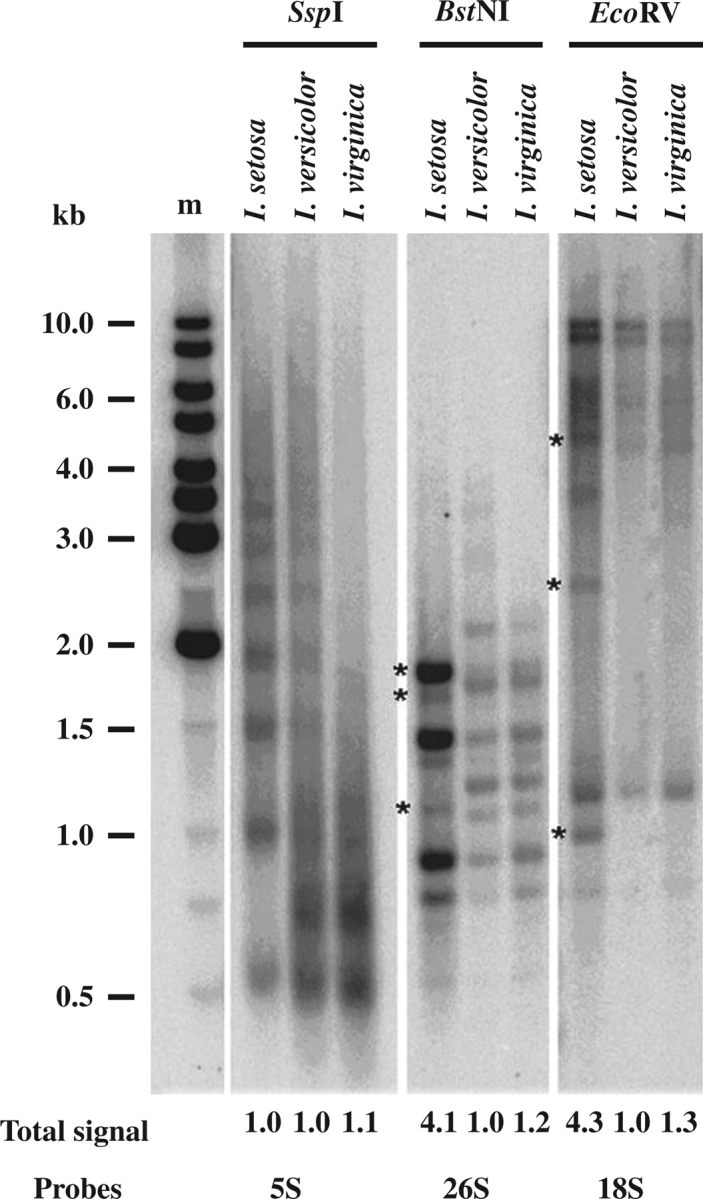
Southern restriction digests of I. setosa, I. versicolor and I. virginica with enzymes SspI, BstNI, EcoRV and probed with 5S, 26S and 18S rDNA, respectively. The total signal reflects the relative copy numbers of rDNA units, calculated by dividing the signal intensity in each lane by the intensity from the species with the lowest signal. Asterisks indicate bands of I. setosa-origin that are lost or reduced in the allopolyploid.
The pattern of bands generated for I. virginica and I. setosa when probing restricted DNA with either 26S or 18S probes is substantially different (Fig. 1). Interestingly the pattern of bands observed in I. versicolor is not additive of the pattern of bands found in the progenitor species with these probes; instead the pattern only most closely resembles that of I. virginica parent. The bands specific for an I. setosa intergenic spacer (indicated by asterisks) were completely absent in I. viriginica. The quantity of genomic DNA loaded into each lane was approximately the same for each species (data not shown). Therefore, the stronger hybridization signal generated in the I. setosa lanes with 18S and 26S rDNA probes most probably represents elevated copy numbers (approx. 3- to 4-fold) of this rDNA unit (assuming that genome sizes are approximately similar).
GISH and FISH
GISH to I. versicolor root tip metaphases reveals 38 chromosomes that label predominantly red with I. setosa genomic DNA and 70 chromosomes that label predominantly green with I. virginica total genomic DNA (Fig. 2D, E). These numbers are expected if all the chromosomes from the progenitors are inherited in I. versicolor. Two pairs of chromosomes of I. setosa-origin also carry terminal translocations from I. virginica-origin chromosomes.
Fig. 2.
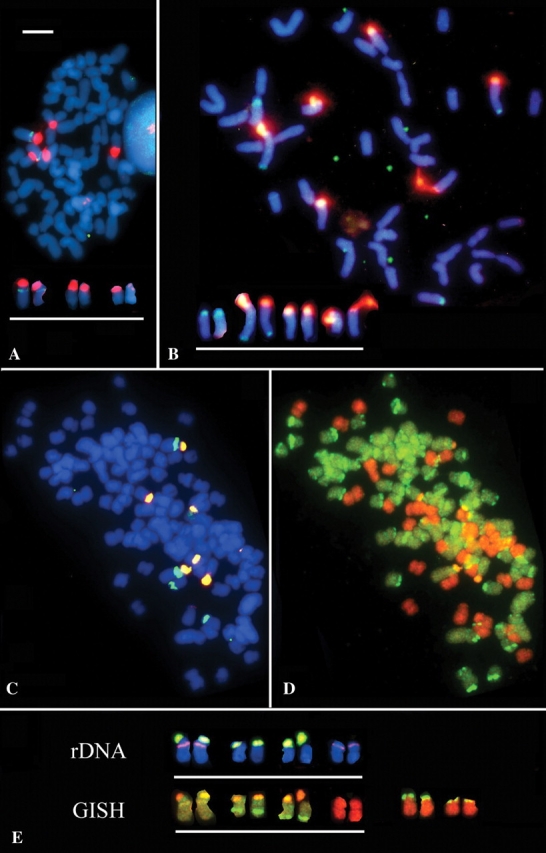
(A–C) FISH probed with 18–26S rDNA (red or orange) and 5S rDNA (green) and chromosomes counterstained with DAPI (blue) in (A) metaphase cell of I. virginica showing three loci (six signals at diploid) of 18–26S rDNA signals and one locus of 5S rDNA on one of the 18–26S carrying chromosomes (labelled chromosomes are shown arranged as a karyotype below metaphase); (B) metaphase of I. setosa with three 18–26S rDNA loci and two pairs of 5S rDNA loci of different sizes; the smallest 5S locus occurs on the same chromosome as a 18–26S rDNA locus. (C) metaphase cell of I. versicolor showing six signals of 18–26S rDNA and four signals of 5S rDNA. (D) The same cell reprobed by GISH using total genomic DNA from I. virginica (green) and from I. setosa (orange). (E) Chromosomes underlined are isolated rDNA-carrying chromosomes from an I. versicolor metaphase labelled with 18S–26S (yellow–green) and 5S (pink) rDNA, and the same chromosomes after GISH labelled in (D). From the same metaphase the two pairs of chromosomes to the right are predominantly labeled with I. setosa genomic DNA but show translocations from I. virginica-origin chromosomes. Scale bar of complete metaphases represents 10 µm.
FISH to I. virginica root-tip metaphases revealed three pairs of 18–26S rDNA carrying chromosomes, one of which also carries a proximal 5S rDNA locus (Fig. 2A). Probe labelling to I. setosa metaphases reveals two pairs of 5S rDNA loci, and three pairs of 18–26S rDNA loci; although the size of each 18–26S rDNA locus was materially larger than in I. virginica (Fig. 2B). The large sizes of I. setosa 18–26S rDNA loci are consistent with the elevated rDNA gene copy numbers observed in this species (Fig. 1). The allopolyploid I. versicolor, which has the sum of the chromosomes found in the putative progenitors; has only three 18–26S rDNA loci (expect six) and two 5S rDNA loci (expect three) (Fig. 2C, E). One pair of chromosomes carries both 5S and 18–26S rDNA loci, which is similar to I. virginica. GISH labels all I. versicolor chromosomes carrying 18–26S rDNA with I. virginica total genomic DNA (yellow) indicating that they were derived from this progenitor species. I. versicolor also carries the larger 5S rDNA locus derived from the I. setosa progenitor and it labels with I. setosa genomic DNA (orange). All those 18–26S rDNA sites derived from the I. setosa progenitor, together with the minor 5S rDNA locus in this species are absent in I. versicolor.
DISCUSSION
Genomic in situ hybridization with total genomic DNA of I. setosa and I. virginica supports Anderson's hypothesis (Anderson, 1936) that I. versicolor is an allopolyploid involving progenitors of these species. The pattern of GISH labeling suggests that I. versicolor carries the sum of the number of chromosomes found in the progenitors, i.e. 70 chromosomes from I. virginica and 38 from I. setosa. Only two pairs of chromosomes carry intergenomic translocations, a feature hypothesized to establish fertility in some newly formed allopolyploids (Gill, 1991).
FISH and GISH to I. versicolor revealed three 18–26S rDNA loci, all inherited from the progenitor related to I. virginica, and two 5S rDNA loci one from each of the progenitors of I. virginica and I. setosa. This indicates the loss of all three 18–26S rDNA loci and one 5S rDNA locus from the subgenome inherited from the I. setosa progenitor. Alternatively I. setosa may have gained a 5S rDNA site subsequent to polyploidy. These data are consistent with the results of Southern hybridization showing that I. versicolor has 5S rDNA units of both putative progenitors, and 18S-26S rDNA units that are more similar in structure and unit copy number to the I. virginica progenitor. There is no evidence for 18–26S rDNA interlocus homogenization in I. versicolor, a feature which probably has altered the length and sequence of rDNA units in other allopolyploids, changing their unit structure relative to those of both parents (e.g. in Nicotiana tabacum (Lim et al., 2000)). Thus I. versicolor 18–26S rDNA is likely to have undergone 18–26S rDNA locus loss rather than locus co-evolution, as is seen in allopolyploids of Brassica (Hasterok et al., 2006) and Arabidopsis (O' Kane et al., 1996), or concerted evolution as is seen in e.g. Nicotiana (Volkov et al., 1999), Gossypium (Wendel et al., 1995), and Glycine (Doyle et al., 2004), see Ma & Gustafson (2005) for a more comprehensive range of examples.
It may be significant that the 18–26S rDNA loci lost in I. versicolor are all of I. setosa parental origin, indicating that elimination is non-stochastic and targeted to the I. setosa-origin subgenome. Similarly, various genetic and epigenetic changes targeted to one of the subgenomes have been reported in allopolyploids of Nicotiana (Skalicka et al., 2005), Brassica (Hasterok et al., 2006) and wheat (Han et al., 2005). In I. versicolor the elimination of rDNA from the I. setosa-origin subgenome was not accompanied by any observable amplification of rDNA from the I. virginica-origin subgenome.
Based on the relative copy numbers of 18–26S rDNA units in I. setosa and I. virginica, we would have expected the units of I. setosa-origin to be in higher copy numbers, yet it is these units that have been deleted. Possibly high copy arrays of rDNA undergo more heterochromatization and transcriptional silencing than lower copy arrays. We have previously observed this phenomenon in allotetraploids of Tragopogon that formed recently (within last 100 years) in nature (Kovarik et al., 2004; Matyasek et al., 2007). Perhaps during the divergence of I. versicolor the rDNA units of I. setosa-origin were transcriptionally inactivated, heterochromatinized, and gradually eliminated.
A loss in 18–26S and perhaps 5S rDNA loci, can be considered as an early indication of genome diploidization in polyploid. In Nicotiana we have previously observed loss of rDNA loci in allopolyploids of around 1 million years of age (species of Nicotiana section Polydicleae (Lim et al., unpubl. res.). Ongoing diploidization results in the GISH-method to fail in discriminating parental origin of polyploids of 4–5 million years old (species of Nicotiana section Repandae) (Clarkson et al., 2005; Lim et al., 2007). If the rate and mode of I. versicolor allopolyploid genome divergence is similar to that observed in allopolyploids of Nicotiana, then we would expect genome divergence times of less than 1 million years. Anderson (1936) hypothesized that I. virginica and I. setosa hybridized during the Pleistocene (1·8 million-10 000 years ago) when species ranges overlapped at the advancing edge of the Wisconsin ice-sheet.
The question arises as to why rDNA loci are lost in I. versicolor and in many other polyploids, e.g. in natural polyploids of Hepatica (Weiss-Schneeweiss et al., 2007), Zingeria (Kotseruba et al., 2003), and Aegilops (Badaeva et al., 2004; Bardsley et al., 1999). Frequently polyploidy is associated with epigenetic silencing of rDNA loci (Lacadena et al., 1984; Pikaard, 2001). Perhaps those units and loci that are inactive are most vulnerable to deletion since their loss would have no selective consequence. However, there are polyploids that do not show such losses, even gains of loci have been reported e.g. in newly synthesized allopolyploids of Nicotiaina tabacum (Skalicka et al., 2003) and Arabidopsis (Pontes et al., 2004), and in natural polyploids of Helianthus (Wilson et al., 2005) and Dasypyrum (Galasso et al., 1997). Perhaps additional loci can arise through polyploidy-induced genome restructuring, while a tendency towards locus loss is a longer-term, genome wide diploidization process. An analysis of 45 species of Brassicaceae from many genera reveal considerable differences in rDNA locus number (Ali et al., 2005), and it is likely that many factors associated with species divergence can influence the number of rDNA units and their chromosomal location.
ACKNOWLEDGEMENTS
The authors thank NERC and the Grant Agency of the Czech Republic (grants 521/07/0116 and 204/05/0687) and MSMT (LC06004) for funding. We also thank Mr Tony Hall of Royal Botanic Gardens, Kew for the plant materials. We thank Mr Michael Chester and Dr Felipe Sanchez-Teyer for assistance. KYL would like to thank Dr Jack Ellis (former Ph.D. supervisor) for initiating this study.
LITERATURE CITED
- Abbott RJ, Lowe AJ. Origins, establishment and evolution of two new polyploid species: Senecio cambrensis and S. eboracensis in the British Isles. Biological Journal of the Linnean Society. 2004;82:467–474. [Google Scholar]
- Ainouche ML, Baumel A, Salmon A. Spartina anglica C. E. Hubbard: a natural model system for analysing early evolutionary changes that affect allopolyploid genomes. Biological Journal of the Linnean Society. 2004;82:475–484. [Google Scholar]
- Ali HBM, Lysak MA, Schubert I. Chromosomal localization of rDNA in the Brassicaceae. Genome. 2005;48:341–346. doi: 10.1139/g04-116. [DOI] [PubMed] [Google Scholar]
- Alvarez I, Wendel J. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29:417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Anderson E. The species problem in Iris Annals of the Missouri Botanical Gardens. 1936;23:457–509. [Google Scholar]
- Badaeva ED, Amosova AV, Samatadze TE, Zoshchuk SA, Shostak NG, Chikida NN, et al. Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Systematics and Evolution. 2004;246:45–76. [Google Scholar]
- Bardsley D, Cuadrado A, Jack P, Harrison G, Castilho A, Heslop-Harrison JS. Chromosome markers in the tetraploid wheat Aegilops ventricosa analysed by in situ hybridization. Theoretical and Applied Genetics. 1999;99:300–304. [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, Scheen A-C, Elven R. Polyploidy in arctic plants. Biological Journal of the Linnean Society. 2004;82:521–536. [Google Scholar]
- Clarkson JJ, Lim KY, Kovarik A, Chase MW, Knapp S, Leitch AR. Long-term genome diploidization in allopolyploid Nicotiana section Repandae (Solanaceae) New Phytologist. 2005;168:241–252. doi: 10.1111/j.1469-8137.2005.01480.x. [DOI] [PubMed] [Google Scholar]
- Dadejova M, Lim KY, Soucková-Skalická K, Matyášek R, Grandbastien MA, Leitch AR, Kovařík A. Transciption activity of rRNA genes correlates with a tendency towards intergenomic homogenisation in Nicotiana allotetraploids. New Phytologist. 2007;174:658–668. doi: 10.1111/j.1469-8137.2007.02034.x. [DOI] [PubMed] [Google Scholar]
- Dover G. Molecular drive: cohesive mode of species evolution. Nature. 1982;9:111–116. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Rauscher JT, Brown AHD. Evolution of the perennial soybean polyploid complex (Glycine subgenus Glycine): a study of contrasts. Biological Journal of the Linnean Society. 2004;82:583–597. [Google Scholar]
- Fulnecek J, Matyasek R, Kovarik A, Bezdek M. Mapping of 5-methylcytosine residues in Nicotiana tabacum 5S rRNA genes by genomic sequencing. Molecular and General Genetics. 1998;259:133–141. doi: 10.1007/s004380050798. [DOI] [PubMed] [Google Scholar]
- Fulnecek J, Lim KY, Leitch AR, Kovarik A, Matyasek R. Evolution and structure of 5S rDNA loci in allotetraploid Nicotiana tabacum and its putative parental species. Heredity. 2002;88:19–25. doi: 10.1038/sj.hdy.6800001. [DOI] [PubMed] [Google Scholar]
- Galasso I, Blanco A, Katsiotis A, Pignone D, HeslopHarrison JS. Genomic organization and phylogenetic relationships in the genus Dasypyrum analysed by Southern and in situ hybridization of total genomic and cloned DNA probes. Chromosoma. 1997;106:53–61. doi: 10.1007/s004120050224. [DOI] [PubMed] [Google Scholar]
- Gill BS. Nucleocytoplasmic interaction (NCI) hypothesis of genome evolution and speciation in polyploid plants. In: Sasakuma T, Kinoshita T, editors. Proceedings of the Kihara Memorial International Symposium on Cytoplasmic Engineering in Wheat. Japan: Yokohama; 1991. [Google Scholar]
- Han F, Fedak G, Guo W, Liu B. Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics. 2005;170:1239–1245. doi: 10.1534/genetics.104.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, et al. Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Annals of Botany. 2006;97:205–216. doi: 10.1093/aob/mcj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T, Szkulek A, Solomosy F. Nucleotide sequence of a 17S (18S) rDNA gene from tomato. Nucleic Acids Research. 1989;17:21–27. doi: 10.1093/nar/17.5.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotseruba V, Gernand D, Meister A, Houben A. Uniparental loss of ribosomal DNA in the allotetraploid grass Zingeria trichopoda (2n = 8) Genome. 2003;46:156–163. doi: 10.1139/g02-104. [DOI] [PubMed] [Google Scholar]
- Kovarik A, Matyasek R, Lim KY, Skalicka K, Koukalova B, Knapp S, et al. Concerted evolution of 18–5·8–26S rDNA repeats in Nicotiana allotetraploids. Biological Journal of the Linnean Society. 2004;82:615–625. [Google Scholar]
- Kovarik A, Pires JC, Leitch AR, Lim K, Sherwood A, Matyasek R, et al. Rapid concerted evolution of nuclear ribosomal DNA in two allopolyploids of recent and recurrent origin. Genetics. 2005;169:931–944. doi: 10.1534/genetics.104.032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacadena JR, Cermeno MC, Orellana J, Santos JL. Evidence for wheat-rye nucleolar competition (amphyplasty) in Triticale by silver-staining procedure. Theoretical and Applied Genetics. 1984;67:207–213. doi: 10.1007/BF00317037. [DOI] [PubMed] [Google Scholar]
- Leitch AR, Lim KY, Webb DR, McFadden GI. In situ hybridisation. In: Hawes C, Satiat-Jeunemaitre B, editors. Plant cell biology: a practical approach. Oxford: Oxford University Press; 2001. [Google Scholar]
- Leitch IJ, Bennett MD. Polyploidy in angiosperms. Trends in Plant Science. 1997;2:470–476. [Google Scholar]
- Lim KY, Leitch IJ, Leitch AR. Genomic characterisation and the detection of raspberry chromatin in polyploid Rubus. Theoretical and Applied Genetics. 1998;97:1027–1033. [Google Scholar]
- Lim KY, Kovarik A, Matyasek R, Bezdek M, Lichtenstein CP, Leitch AR. Gene conversion of ribosomal DNA in Nicotiana tabacum is associated with undermethylated, decondensed and probably active gene units. Chromosoma. 2000;109:161–172. doi: 10.1007/s004120050424. [DOI] [PubMed] [Google Scholar]
- Lim KY, Kovařík A, Matyášek R, Chase MW, Clarkson JJ, Grandbastien MA, Leitch AR. Determining the sequence of events leading to plant genome turnover in five million years. New Phytologist. 2007 doi: 10.1111/j.1469-8137.2007.02121.x. (in press) [DOI] [PubMed] [Google Scholar]
- Ma XF, Gustafson JP. Genome evolution of allopolyploids: a process of cytological and genetic diploidization. Cytogenetic and Genome Research. 2005;109:236–249. doi: 10.1159/000082406. [DOI] [PubMed] [Google Scholar]
- Matyášek R, Tate JA, Lim KY, Srubarova H, Koh J, Leitch AR, Soltis DE, Soltis PS, Kovařík A. Concerted evolution of rDNA in recently formed Tragopogon allotetraploids is typically associated with an inverse correlation between gene copy number and expression. Genetics in press. 2007 doi: 10.1534/genetics.107.072751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O' Kane SL, Schaal BA, AlShehbaz IA. The origins of Arabidopsis suecica (Brassicaceae) as indicated by nuclear rDNA sequences. Systematic Botany. 1996;21:559–566. [Google Scholar]
- Pikaard CS. Genomic change and gene silencing in polyploids. Trends in Genetics. 2001;17:675–677. doi: 10.1016/s0168-9525(01)02545-8. [DOI] [PubMed] [Google Scholar]
- Pontes O, Neves N, Silva M, Lewis MS, Madlung A, Comai L, et al. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proceedings of the National Academy of Sciences of the USA. 2004;101:18240–18245. doi: 10.1073/pnas.0407258102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population-dynamics. Proceedings of the National Academy of Sciences of the USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor laboratory Press; 1989. [Google Scholar]
- Skalicka K, Lim KY, Matyasek R, Koukalova B, Leitch AR, Kovarik A. Rapid evolution of parental rDNA in a synthetic tobacco allotetraploid line. American Journal of Botany. 2003;90:988–996. doi: 10.3732/ajb.90.7.988. [DOI] [PubMed] [Google Scholar]
- Skalicka K, Lim Y, Matyasek R, Matzke M, Leitch AR, Kovarik A. Preferential elimination of repeated DNA sequences from the paternal, N. tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytologist. 2005;166:291–303. doi: 10.1111/j.1469-8137.2004.01297.x. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends in Ecology & Evolution. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Pires JC, Kovarik A, Tate JA, Mavrodiev E. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic, genomic and genetic comparisons. Biological Journal of the Linnean Society. 2004a;82:485–501. [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since plant speciation. New Phytologist. 2004b;161:173–191. [Google Scholar]
- Urbanska KM, Hurka H, Landolt E, Neuffer B, Mummenhoff K. Hybridization and evolution in Cardamine (Brassicaceae) at Urnerboden, central Switzerland: biosystematic and molecular evidence. Plant Systematics and Evolution. 1997;204:233–256. [Google Scholar]
- Volkov RA, Borisjuk NV, Panchuk II, Schweizer D, Hembeleben V. Elimination and rearrangement of parental rDNA in the allotetraploid Nicotiana tabacum. Molecular Biology and Evolution. 1999;16:311–320. doi: 10.1093/oxfordjournals.molbev.a026112. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Schneeweiss GM, Stuessy TF, Mabuchi T, Park JM, Jang CG, Sun BY. Chromosomal stasis in diploids contrasts with restructuring in auto- and allopolyploid taxa of Hepatica (Ranunculaceae) New Phytologist. 2007;174:669–682. doi: 10.1111/j.1469-8137.2007.02019.x. [DOI] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Molecular Biology. 2000;42:225–249. [PubMed] [Google Scholar]
- Wendel JF, Schnabel A, Seelanan T. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium) Proceedings of the National Academy of Sciences of the USA. 1995;92:280–284. doi: 10.1073/pnas.92.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson BMD, San-Martin JAB, Boneventi P, Torezan JMD, Vanzela ALL. Functionality of major and minor 45S rDNA sites in different diploid wild species and varieties of sunflowers. Caryologia. 2005;58:374–379. [Google Scholar]


